Abstract
Optical coherence tomography (OCT) is able to provide high resolution volumetric data for biological tissues. However, the field of view (FOV) of OCT is sometimes smaller than the field of interest, which limits the clinical application of OCT. One way to overcome the drawback is to stitch multiple 3D volumes. In this paper, we propose a novel method to register multiple overlapped volumetric OCT data into a single volume. The relative positions of overlapped volumes were estimated on en face plane and at depth. On en face plane, scale invariant feature transform (SIFT) was implemented to extract the keypoints in each volume. Based on the invariant features, volumes were paired through keypoint matching. Then, we formulated the relationship between paired offsets and absolute positions as a linear model and estimated the centroid of each volume using least square method. Moreover, we calibrated the depth displacement in each paired volume and aligned the z coordinates of volumes globally. The algorithm was validated through stitching multiple volumetric OCT datasets of human cervix tissue and of swine heart. The experimental results demonstrated that our method is capable of visualizing biological samples over a wider FOV, which enhances the investigation of tissue structure such as fiber orientation.
I. Introduction
Optical coherence tomography (OCT) has experienced rapid advancement in recent years. In comparison with existing image modalities, OCT is able to achieve high penetration, high sensitivity, fast data acquisition, and high axial resolution simultaneously. OCT is a promising medical modality for a wide range clinical application such as imaging the myocardium [1], cervix [2], and retina[3]. However, the field of view (FOV) of current OCT systems is limited to the magnitude of millimeters. For some clinical applications, e.g. monitoring the mechanical properties of human cervix tissue [4] or assessing the ventricular septum, such FOV is not sufficient to visualize the whole tissue structure. There is thus an increasing need to extend the FOV. Without complicating the current OCT system design, stitching multiple overlapped OCT volumes into a single three dimensional dataset serves as a possibility to enlarge the FOV.
Though many image registration methods are proposed for image stitching [5, 6], general methods to stitch volumetric OCT data are still scarce. Stitching OCT volumes is challenging because the signal noise ratio (SNR) in OCT image is much weaker than most of other image modalities, such as computed tomography and magnetic resonance imaging. A weak SNR indicates relatively strong noise, which can change the texture of the overlapped area and result in poor correlation and matching between volumes. In addition, in OCT, the volumes should be aligned in three dimensions, which increase the difficulty of global optimization algorithms. An method for mosaicking OCT volumes from bladder tissue is proposed in [7]. Zawadzki et al [8] register multiple retinal OCT volumes through manual stitching, then develop a ray cast method to render the volume and use an annealing algorithm to optimize the alignment among multiple retinal imaging volumes [9]. Multiple OCT volumes are stitched in [10] based on blood vessel matching algorithm. In [3], a B-spline based free form deformation method was used to register multiple volumes for a motion-free composite image of the retinal vessels. Most of current OCT stitching methods are developed for retinal imaging, where blood vessels are a good reference for registration. Furthermore, the retinal tissues have a clear layered structure, which enables the layer information to help register B-scans in 2D.
In this paper, we propose a generic method that can be applied to register multiple volumes of biological tissues. Since the features within en face images sometimes fail to be a reliable reference for registration, we rely upon the scale invariant features from camera images that can be easily obtained through OCT system with integrated microscopes. Scale invariant transform (SIFT) was applied to extract the features that are robust to contrast, brightness, and movements. Though weak SNR undeniably exists in OCT image, the contrast at the surface of tissue is strong due to surface reflection. Therefore, the edge information of OCT is used to quantify the displacement at depth. The three dimensional offsets were considered in a linear model and globally optimized using a least square estimation. The stitched volume is used to enhance the analysis of fiber organization in heart tissue [11, 12] and cervix [13–15] over large FOV.
II. Methodology
A. Data collocation
Three dimensional volumetric data was collected from human cervix and swine heart. The cervix specimen was acquired from a nonpregnant hysterectomy patient using an Institutional Review Board (IRB) approved protocol at the Columbia University Medical Center [16]. The swine heart was obtained from Green Village Packing, Green Village, NJ. The data were acquired using a commercial spectral domain OCT system, Telesto (Thorlabs GmbH, Germany). It is an InGaAs based system centering at 1325 nm, with a bandwidth of 150 nm. The axial and lateral resolutions in water are 4.9 μm and 9.8 μm, respectively. The maximum axial line rate is 92 kHz. In our experiment, each volume consists of 800 × 800 × 512 pixels, corresponding to a tissue volume of 4 mm × 4 mm × 1.8 mm. Samples were placed in a linear transition stage underneath the objective. For each sample, we obtained multiple volumes. There was an overlap proportion of 10% to 20% between two consecutive volumes. Camera images were taken simultaneously with the same FOV. The camera images and OCT images were calibrated by the default factory setting in Thorlab pre-experiment.
B. Algorithm flow
To stitch multiple volumes, our algorithm consists of the registrations on both the en face plane and depth, as shown in Fig. 1. On the en face plane, we first paired individual volumes based on their scale invariant features. Then, the offset between each pair of volumes was measured. We then formulated a linear regression model between the measured offsets and the centroid of each volume in a global coordinate system. The least square estimation of the regression model was the global optimization registration results of en face plane. Based on the estimation of offsets on x-axis and y-axis, we calculated the overlapped area between any pair of volumes and measured displacements at depth through comparing the measured edge in each volume. Another linear regression model for depth estimation was then derived and the global offset of depth was obtained. With the global offsets in x, y, and z axis, all volumes were stitched and visualized in 3D.
Figure 1.
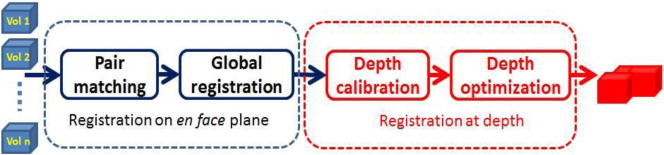
Flowchart of registration algorithm. Scale invariant transform (SIFT) is used for pair matching. Linear regression models are used and least square estimations are calculated in global registration and depth optimization. Estimation in first block and edge detection is used in depth calibration.
C. Registration on en face plane
We consider the camera image of each volume as the reference of registration on en face plane. SIFT algorithm in [5] was applied to each image. Over one hundred keypoints were extracted in each image. A normalized descriptor, with a dimension of 128, was assigned to each keypoint. The local descriptor was based on the gradient in a small area and thereby was robust to illumination and brightness changes caused by movement of transition stage. A Best-Bin-First (BBF) algorithm [6] was used to search a matched keypoint from other image. Denote DESi and DESj are the descriptors of keypoint sets in volume i and j respectively. For a keypoint des ∈ DESi, the set of distance is defined as:
| (1) |
where the ● is the dot product of two vectors. The calculated distance represents the angle between two normalized descriptors. In our experiment, a match between des and the set DESj is established only if the distance to the nearest neighbor is less than 0.45 times of the distance to the second-nearest neighbor. If at least one of match relationship can be established between keypoints in volume i and volume j, we consider the two volumes are paired. Suppose there are K keypoints matched between two volumes. The offsets, Dxij and Dyij, can be estimated as
| (2) |
| (3) |
where (xik, yik) and (xjk, yjk) are the coordinates of the kth matched keypoint. Fig. 2 (a) shows a typical match between volume i and volume j.
Figure 2.
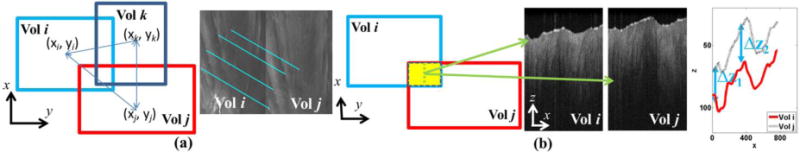
Schematics of registration on en face plane (a) and at depth (b). (a) On en face plane, the offsets between two volumes are measured through mataching its corresponding camera image. The centroids of multiple volumes are estimated globally by considering their relative offsets. (b) To register the OCT volumes at depth, the Bscans at overlaped area are compared. The displacement of detected edge shows the offset of depth among volumes.
Next, we stitched all volumes in a global space. Denoting the centroid of volume i (i = 1, 2, …, N) as (xi, yi), we wrote the centroids in all volumes as a vector c = [x1 x2 … xN y1 y2 … yN]T. Since the samples were moved along x axis and/or y axis, the offsets in each pair were maintained in global space. The geometric relationship between offsets and centroid of OCT volumes were shown in Fig. 2 (a). Suppose there are M pairs of volumes. The measured offset, d = [dx1 dx2 … dxM dy1 dy2 … dyM]T, had a linear relationship with centroid vector c as
| (4) |
W is a matrix with dimension of 2M×2N. Let the mth pair be the offsets between volume i and volume j. The mth row and (m+M)th row of W is given by the following rows:
The ith column in mth row and (i+N)th column in (m+M)th row equal to 1 while The jth column in mth row and (j+N)th column in (m+M)th row equal to −1. Each row in W represents the offset between two volumes in either x or y axis, with a relationship of xi − xj = dxij. The least square estimates of (4) is:
| (5) |
In our experiment, the matrix WTW may not be full rank. A generalized inverse can be used to solve (5). Here, we use Moor-Penrose pseudo inverse of matrix during implementation.
D. Registration at depth
Based on the estimation of ĉ, we can arrange the volumes as overlapped tiles in a single space. However, the volumes were not perfectly stitched because of variations at depth. To register multiple volumes at depth, we used the edge of B-scans as reference. We calculated the overlapped region between each volume pairs as the gray area in Fig. 2(b). To calibrate the displacement at depth, we obtained B-scans from two volumes at the same position within the overlapped region. The edge of each B-scans was determined by smooth filter followed by searching maximum gradient point. The displacements of two edges were measured along x axis. A typical scenario of overlayed edges was drawn in Fig. 2 (b). The measured offset Δzij between volume i and volume j was computed as the median of all displacements between edges in volume i and j along x axis. Similar to Section II-B, a linear relationship can be formulated between the z coordinates of centroid in all volumes, z = [z1 z2 … zN]T, and the measured offset, d2 = [dz1 dz2 … dzM]T as following:
| (6) |
where mth row of V can be written as [… 0 1 0 … 0 −1 0 … 0]. The entry of ith column is 1 while the entry of jth column is −1. The least square result of (6) is:
| (7) |
Finally, we aligned all volumetric dataset on the basis of estimation ĉ and . Specifically, we used a hard combination of all individual volumes. All non-zero voxel values are maintained when added to the stitched volume.
III. Experiments and Results
We set up an evaluation test to validate the accuracy of our method. We used an OCT volume of 4 mm × 4 mm × 1.8 mm (800 × 800 × 512 pixels) as ground truth. Sticking to the same space, we additionally imaged four overlapped volumes of 2.5 mm × 2.5 mm × 1.8 mm (500 × 500 × 512 pixels). The volumes were stitched by our methods. We set three pairs of landmarks at ground truth and then specified the same landmarks at stitched volume. The 3D distances in each pair were measured. We found that difference of measured distance between stitched volume and ground truth is 10.57±8.51 pixels. Our methods are further validated in human cervix tissue and swine heart. The two dimensional results are shown in Fig. 3. For cervix data, a total of 17 camera images are stitched in Fig. 3 (a) in a semi-circular shape. The combined OCT image was visualized in Fig. 3 (b) based on the offsets measured in Fig. 3 (a). More importantly, the inner canal of the cervix is clearly observable at the center of stitched image. The detailed collagen fiber structure can be directly viewed at depth. Similarly, the structure of ventricular septum is shown on the camera image in Fig. 3 (c) and OCT image in Fig. 3 (d), respectively. In total, 16 small volumes are aligned as a rectangle shape. The inner structure of myocardium is observable. At each B-scan, Fig 3. (e) and (f) show the stitching results. We found that the surfaces of multiple B-scans are stitched smoothly at a range of over 1 cm. Three dimensional registration results are shown in Fig. 4. The cylinder shape of cervix is shown in Fig. 4 (a), and the representative B-scans are overlayed with 3D volume in Fig. 4 (b). Square volumes of ventricular septum are given in Fig. 4 (d) and Fig. 4 (e). As an application of registration, we show the fiber estimation results in a large volume using algorithm in [11, 12]. Fig. 4 (c) and (f) show the fiber trend in cervix tissue and myocardium, respectively.
Figure 3.
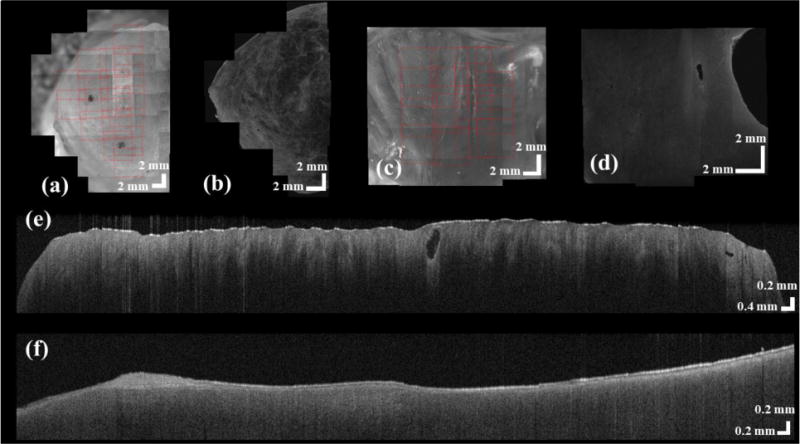
Registration results of human cervix (a, b, and e) and swine heart (c,d, and f). Based on the registration algorithm on en face plane, we register the camera images of human cervix (a) and swine heart (c). The OCT images can be registered correspondingly on human cervix (b) and swine heart (d). Using the registering algorithm at depth, multiple B-scans in human cervix (e) and swine heart (f) are aligned.
Figure 4.
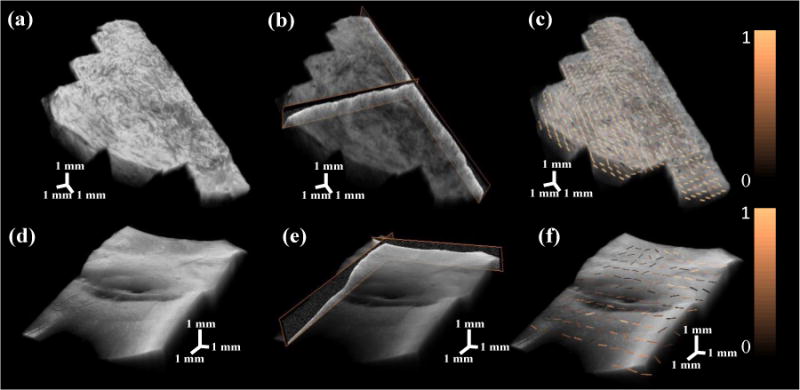
Three dimensional registration results of human cervix (a, b, and c) and swine heart (d, e, and f). The stitched OCT volumes of cervix sample is shown in (a) with a size of 21.6 mm × 12.5 mm × 1.5 mm. The stitched myocardium sample is presented in (d) with a size of 12.5 mm × 12.5 mm × 1.5 mm. Typical B-scans are overlayed with OCT volumes in (b) and (e). Fiber orientation estimation on en face plane is processed and ovelayed with original OCT volumes in (c) and (f). The fiber orientation estimation is color coded based on confidence level from high (bright) to low (dark).
IV. Conclusion
We present an automatic registration method to stitch multiple OCT volumes based on SIFT and least square estimation in three dimensions. By showing a large view of cervix and myocardium and investigating the structure property within a large area, we demonstrate our algorithms’ ability in automatic stitching.
Future work will be concentrated on the compensation of overlapped area post registration and the extension of the algorithm on catheter-based OCT system.
Acknowledgments
The authors will like to thank Joy Vink, MD, and Ronald Wapner, MD, from Columbia University Medical Center for providing the cervical tissue sample for the study.
This project is supported by the following sources: National Science Foundation EEC-1342273, BRIGE1125670, Columbia University Provost Small Grants, and National Institute of Health Loan Repayment Award.
Contributor Information
Yu Gan, Department of Electrical Engineering, Columbia University, NY, USA.
Wang Yao, Department of Mechanical Engineering, Columbia University, NY, USA.
Kristin M. Myers, Department of Mechanical Engineering, Columbia University, NY, USA
Christine P. Hendon, Department of Electrical Engineering, Columbia University, NY, USA
References
- 1.Fleming CP, Quan KJ, Rollins AM. Toward guidance of epicardial cardiac radiofrequency ablation therapy using optical coherence tomography. Journal of Biomedical Optics. 2010;15:041510–041510. doi: 10.1117/1.3449569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang W, Qi X, Tresser NJ, Kareta M, Belinson JL, Rollins AM. Diagnostic efficacy of computer extracted image features in optical coherence tomography of the precancerous cervix. Medical Physics. 2011;38:107–113. doi: 10.1118/1.3523098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendargo HC, Estrada R, Chiu SJ, Tomasi C, Farsiu S, Izatt JA. Automated non-rigid registration and mosaicing for robust imaging of distinct retinal capillary beds using speckle variance optical coherence tomography. Biomedical optics express. 2013;4:803–821. doi: 10.1364/BOE.4.000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myers KM, Paskaleva A, House M, Socrate S. Mechanical and biochemical properties of human cervical tissue. Acta Biomaterialia. 2008;4:104–116. doi: 10.1016/j.actbio.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Lowe DG. Distinctive image features from scale-invariant keypoints. International journal of computer vision. 2004;60:91–110. [Google Scholar]
- 6.Jian C, Tian J, Lee N, Jian Z, Smith RT, Laine AF. A Partial Intensity Invariant Feature Descriptor for Multimodal Retinal Image Registration. Biomedical Engineering, IEEE Transactions on. 2010;57:1707–1718. doi: 10.1109/TBME.2010.2042169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lurie KL, Angst R, Ellerbee AK. Automated mosaicing of feature-poor optical coherence tomography volumes with an integrated white light imaging system. Biomedical Engineering, IEEE Transactions. 2014:1–1. doi: 10.1109/TBME.2014.2316535. [DOI] [PubMed] [Google Scholar]
- 8.Zawadzki RJ, Choi SS, Fuller AR, Evans JW, Hamann B, Werner JS. Cellular resolution volumetric in vivo retinal imaging with adaptive optics?optical coherence tomography. Optics Express. 2009;17:4084–4094. doi: 10.1364/oe.17.004084. 2009/03/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capps AG, Zawadzki RJ, Werner JS, Hamann B. Combined volume registration and visualization. Visualization in Medicine and Life Sciences. 2013:7–11. [Google Scholar]
- 10.Li Y, Gregori G, Lam BL, Rosenfeld PJ. Automatic montage of SD-OCT data sets. Optics Express. 2011;19:26239–26248. doi: 10.1364/OE.19.026239. 2011/12/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan Y, Fleming CP. Extracting three-dimensional orientation and tractography of myofibers using optical coherence tomography. Biomedical Optics Express. 2013;4:2150–2165. doi: 10.1364/BOE.4.002150. 2013/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming CP, Ripplinger CM, Webb B, Efimov IR, Rollins AM. Quantification of cardiac fiber orientation using optical coherence tomography. J Biomed Opt. 2008;13:030505. doi: 10.1117/1.2937470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arifler D, Pavlova I, Gillenwater A, Richards-Kortum R. Light Scattering from Collagen Fiber Networks: Micro-Optical Properties of Normal and Neoplastic Stroma. Biophysical Journal. 92:3260–3274. doi: 10.1529/biophysj.106.089839. 5/1/2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers KM, Socrate S, Paskaleva A, House M. A study of the anisotropy and tension/compression behavior of human cervical tissue. Journal of biomechanical engineering. 2010;132 doi: 10.1115/1.3197847. [DOI] [PubMed] [Google Scholar]
- 15.Aspden RM. Collagen Organisation in the Cervix and its Relation to Mechanical Function. Collagen and Related Research. 1988;8 doi: 10.1016/s0174-173x(88)80022-0. [DOI] [PubMed] [Google Scholar]
- 16.Yao W, Yoshida K, Fernandez M, Vink J, Wapner RJ, Ananth CV, et al. Measuring the compressive viscoelastic mechanical properties of human cervical tissue using indentation. Journal of the Mechanical Behavior of Biomedical Materials. 2014;34 doi: 10.1016/j.jmbbm.2014.01.016. [DOI] [PubMed] [Google Scholar]


