Abstract
The entrainment of circadian rhythms, physiological cycles with a period of about 24 h, is regulated by a variety of mechanisms, including nonvisual photoreception. While circadian rhythms have been shown to be integral to many processes in multicellular organisms, including immune regulation, the effect of circadian rhythms on symbiosis, or host–microbe interactions, has only recently begun to be studied. This review summarizes recent work in the interactions of both pathogenic and mutualistic associations with host and symbiont circadian rhythms, focusing specifically on three mutualistic systems in which this phenomenon has been best studied. One important theme taken from these studies is the fact that mutualisms are profoundly affected by the circadian rhythms of the host, but that the microbial symbionts in these associations can, in turn, manipulate host rhythms. The interplay between circadian rhythms and symbiosis is a promising new field with effects that should be kept in mind when designing future studies across biology.
Non-visual photoreception is integral to many processes throughout all domains of life. One such function is to provide input into the circadian clock, often through the action of blue light on photosensitive proteins such as cryptochromes or melanopsins. By the activation of blue-light receptors and other cues that vary over the day/night cycle, the circadian clock regulates a large number of physiological processes, including the immune system, the means by which an organism interacts with symbiotic (both mutualistic and pathogenic) microbes. This review will offer a view into the study of the interplay of circadian rhythms and host–symbiont interactions, with a focus on the effects of circadian rhythms on mutualisms and vice versa.
Circadian rhythms and the immune system
Circadian rhythms are cyclic changes in a given activity that exhibit a period of about 24 h and are normally entrained to this period by exposure to daily cues, such as the presence of sunlight or intake of food (Johnson 1992). In organisms with robust circadian rhythms, which are found in all domains of life (Edgar et al. 2012), these cycles are usually controlled and maintained by transcriptional oscillators that can remain free running, i.e., the periodicity degrades only gradually in the absence of external cues. However, recent studies have shown that perhaps the most phylogenetically widespread type of oscillator is one based on the oxidation state of peroxiredoxin proteins (Edgar et al. 2012), so there may be more types of circadian oscillators than was previously thought.
Circadian rhythms can provide input to many physiological processes, such as sleep/wake cycles, food intake, and body temperature; in fact, it has been shown that about 10% of an animal’s transcriptome is controlled in a circadian manner (Storch et al. 2002). The evolutionarily convergent set of functions that is commonly known as the immune system is no exception, as connections between circadian circuitry and immunity have been found in many organisms, including the plant Arabidopsis thaliana (Wang et al. 2011) and mammals such as mice and humans (reviewed in Arjona et al. 2012; Scheiermann et al. 2013). In mammals, where this connection has been most studied, the influence of time of day on immunity has been apparent since 1927, when Shaw observed that “the leucocytes of man exhibit twice daily a tidal rhythm of about twelve hours’ duration which is independent of certain recognized physiological stimuli” (Shaw 1927). Almost a century later, this connection between circulating immune cells and circadian rhythms was supported when it was shown that splenic macrophages in mammals contain free-running circadian clocks that control inflammatory processes (Keller et al. 2009). This link is not restricted to mammals: It was also shown that the phagocytic activity of immune cells is circadian in the insect Drosophila melanogaster (Stone et al. 2012).
Animal organ systems function in concert with their bacterial partners
Immune system functions have, until recently, been studied primarily in their role as host defense mechanisms during pathogenic insult. However, the mammalian immune system likely expends more energy managing and communicating with the microbial communities that colonize various tissues in the body than defending against dedicated pathogens. In fact, it is possible that the evolution of the vertebrate adaptive immune system was driven by the evolutionary advantages of allowing vertebrates to interact with complex beneficial microbial consortia instead of with only one or a few species of microorganisms, as is common for invertebrates, which lack an adaptive immune system (McFall-Ngai 2007). As such, these consortia have profound effects on the physiology of organs and organ systems, cincluding stimulating proper development of the immune system (Bouskra et al. 2008).
The gut consortium, which is by far the best studied, also assists in the digestion of food and can deliver signals to sites as distant as the mammalian brain (Cryan and Dinan 2012). Since many aspects of gut physiology, such as processivity and the transcription of many genes in gut epithelial and immune cells, is highly circadian (Froy and Chapnik 2007; Hoogerwerf et al. 2008; Keller et al. 2009), there is likely an interface between the circadian rhythmicity of the gut and the physiological effects of symbionts therein.
While rhythms of the gut have been implicated in diseases such as obesity and diabetes (Konturek et al. 2011; Lamia et al. 2011; Paschos et al. 2012), gastroenterologists are now beginning to integrate this information with the known roles of the microbiota in these same disorders (Turnbaugh et al. 2006; Wen et al. 2008). In addition, recent studies of gut–brain interactions have demonstrated that behavioral disorders such as depression, which have been known to affect and be affected by circadian rhythms (McCarthy and Welsh 2012), are also influenced by the microbiota (e.g., Holzer et al. 2012). Taken together, the current data strongly implicate host–microbiota interactions in the maintenance of healthy circadian behaviors.
The effect of circadian rhythms on host–symbiont interactions pathogenesis
The ability of an organism to defend against pathogens is dependent upon its immune state, so day/night variations in immune state can lead to circadian variation in the outcome of pathogenesis. This was first shown in 1969, when it was reported that mice challenged with pneumococci in the early morning survived longer than animals inoculated at any other time (Feigin et al. 1969); this phenomenon was later also shown in mouse Coxsackie virus infection (Feigin et al. 1972). More recently, it was shown that the inflammatory response to Salmonella typhimurium is strongly affected by circadian circuitry in mice (Bellet et al. 2013). Once again, this variation is not restricted to mammals, since D. melanogaster also exhibits diel variation in its ability to survive infection with Pseudomonas aeruginosa or Staphylococcus aureus (Lee and Edery 2008) and plants such as A. thaliana require intact circadian circuitry to fight off infections such as those caused by Pseudomonas syringae (Bhardwaj et al. 2011; reviewed in Roden and Ingle 2009). These data suggest that the influence of circadian rhythms on pathogenesis is a trait that is found throughout multicellular eukaryotic life. In addition, these findings emphasize the need to consider the time of day and the day/night light cycle experienced by experimental subjects in research concerning host–symbiont interactions.
While it is tempting to assume that circadian rhythms in symbiosis are only experienced by the host, it should be appreciated that circadian circuitries of both the host and microbe can affect symbiont behavior. This is perhaps best exemplified by parasitic infections that require transmission by a vector, as the infectious stage of the parasite must be presented to the vector during the animal’s active period. In malaria, the emergence of merozoites from primate red blood cells en masse occurs soon after midday, which allows infective gametocytes to develop in time for peak mosquito feeding time in the evening (Hawking et al. 1968; reviewed in Mideo et al. 2013). Evidence suggests that this synchronization of parasite development is driven by the host melatonin cycle (Hotta et al. 2000), which represents a co-option of host signaling cascades for the adaptive benefit of the parasite, since de-coupling the parasite from the circadian rhythm of the host results in a net fitness cost to the malaria cells (O'Donnell et al. 2011). Probing the exact mechanism of the integration of the parasite and host circadian rhythms has so far been hampered by the fact that erythrocytes do not contain DNA and so cannot produce transcriptional oscillators. However, erythrocytes maintain a robust circadian rhythm through the oxidative state of a peroxiredoxin protein (O'Neill and Reddy 2011), and so erythrocyte rhythms may prove to be a fruitful avenue of research for malaria biology. Other eukaryotic parasites also exhibit circadian behavior, as shown by the coordination of daily parasite emergence by Schistosoma japonicum with the activity cycles of its likely reservoir, the snail Oncomelania hupensis (Lu et al. 2009) and by the late afternoon shedding of Isospora turdi oocysts from its bird hosts, which likely allows for increased oocyst survival in the environment (Martinaud et al. 2009). Moreover, the use of circadian cues by parasites is not restricted to these unicellular eukaryotes. A recent study showed that the fungus Cercospora zeae-maydis requires both a cycle of exogenous light and its own circadian machinery to time its infection to when the host plant stomata, through which the pathogen infects the plant, are open (Kim et al. 2011).
Mutualisms
Though the bulk of research on the effect of circadian rhythms on symbiosis has been performed in pathogenic associations, day/night cycles can and do have a profound effect on mutualisms (Fig. 1). While several mutualisms have shown single instances of ties to circadian rhythms, such as the nocturnal spore formation of parrotfish gut symbionts (Flint et al. 2005) and the diurnal variation of bacterial number in the cow rumen (Leedle et al. 1982), the bulk of research concerning the integration of symbiosis has been performed in three systems: The Symbiodinium–Cnidarian association, the mutualism between the Hawaiian bobtail squid and Vibrio fischeri, and the mammalian gut microbiome.
Fig. 1.
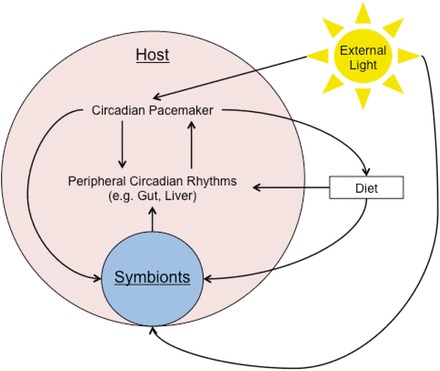
Interactions between host, symbionts, and external day/night cycles in mutualisms. Shown is a schematic of interactions between host circadian rhythm regulators, microbial symbionts, and external day/night cycles. Interactions shown have experimental support in one or more mutualisms.
Symbiodinium–Cnidarian symbioses
Many cnidarians form associations with unicellular algae called dinoflagellates of the genus Symbiodinium. These unicellular algae can be acquired vertically (through maternal inheritance) or horizontally (anew each generation) by acquisition of symbionts from the water column. Once acquired, the dinoflagellates reside within specialized, membrane-bound compartments in the gastrodermis of the animal and the association continues throughout the lifetime of the host. The algal symbionts are photosynthetic and provide sugars for the host in exchange for nutrients, a stable location for access to sunlight, and protection from predators (Davy et al. 2012). While many species of cnidarians form this type of association, it is perhaps best studied in reef building corals. Coral–algal symbioses are of critical ecological importance as the loss of these symbionts (so-called coral bleaching) can lead to the death of the coral and therefore loss of coral reefs, habitats that are crucial to the survival of many species.
As the dinoflagellates in these associations are photosynthetic, it is not surprising that the activities of the two partners are regulated on the day/night cycle (reviewed in Sorek et al. 2014). The photosynthetic capabilities of Symbiodinium as measured by physiological and transcriptional metrics are regulated in a circadian manner both in association with its coral host and in its free-living niche (Sorek et al. 2013). In addition, the daily rhythm of photosynthesis in these algae exhibits temperature compensation, a hallmark of circadian oscillators (Sorek and Levy 2012a). While in a 2015 study the Symbiodinium genome was not found to contain any homologs of known transcriptional oscillators, it does contain genes for photoreceptors such as cryptochromes (blue-light receptors), phytochromes (red-light receptors), and rhodopsin, suggesting that it has the capability to perceive light in a manner independent from its photosynthetic capabilities (Noordally and Millar 2015). Indeed, the cryptochrome and phytochrome genes are transcriptionally responsive to blue and red light, respectively (Sorek and Levy 2012b).
The host corals also undergo circadian regulation of symbiosis-related genes such as those encoding stress-related chaperones and antioxidants (Levy et al. 2011). As these genes are transcribed before peak oxygen production on the part of the symbiont, it is thought that the host uses its circadian circuitry to anticipate oxygen stress and thereby ameliorate the potentially harmful effects of the symbiosis. While it is likely that the coral uses the output of the dinoflagellates as one set of circadian cues, it has been shown that corals themselves encode cryptochromes, which may also be used in the setting of their circadian clocks (Levy et al. 2007). Indeed, Reitzel et al. showed that Nematostella vectensis, another anthozoan, undergoes regulation of a large number of circadian clock-related genes in response to external light cycles, suggesting that anthozoans have a robust circadian gene network (Reitzel et al. 2010). The presence of a known circadian gene repertoire in sea anemones is important for future work since reef-building corals are at times difficult to manipulate in laboratory; thus cell biological studies of Dinoflagellate–Cnidarian interactions are often performed in anemones such as Aptasia sp. as a proxy for corals (Oakley et al. 2015). Using these systems, the community will be able to begin determining the pathways by which the host and symbiont affect each other’s circadian activities. Understanding how circadian rhythms in corals are intertwined with symbiosis may be of broad significance, because the hallmarks of global warming that lead to coral bleaching, such as increased temperature and decreased pH, have been shown to alter expression of circadian genes in the coral Acropora millepora (Kaniewska et al. 2015).
Euprymna scolopes—Vibrio fischeri symbiosis
Another symbiosis that is tied to the presentation of light is the association between Euprymna scolopes, the Hawaiian bobtail squid, and its luminescent gram-negative bacterial symbiont Vibrio fischeri (reviewed in McFall-Ngai 2014). E. scolopes first encounters its symbiont upon hatching, when the animal’s first ventilatory movements bring planktonic V. fischeri cells along with water into its mantle cavity and thus into contact with host tissue. Through a process requiring both host and symbiont chemical communication, the symbiont colonizes a ventral, epithelium-lined organ called the “light organ” within hours of the animal’s hatching. Every day at dawn, the squid expels 90% of its bacterial symbionts into the water column in a process known as “venting” (Nyholm and McFall-Ngai 1998). The remaining V. fischeri cells then regrow throughout the day so that by sunset the light organ contains the full complement of symbionts, at a density high enough for the bacteria to produce light through quorum sensing pathways (reviewed in Miyashiro and Ruby 2012). This leads to the squid being exposed to two different light cycles: that of the sun and that of the bacterial symbionts inside its body. One study performed a transcriptomic analysis of both the host light organ and the resident symbiont cells in the adult animal at four different time points throughout the day (Wier et al. 2010). This study found that the transcriptomes of both the host and symbiont are regulated over the day/night cycle and showed reciprocal patterns. As one example, the upregulation of genes related to chitin synthesis in the host was followed by those involved in chitin catabolism on the part of the symbiont. It was shown later that this is due to the presence of chitin in the host hemocytes (Heath-Heckman and McFall-Ngai 2011), which migrate into the crypts at dusk. The chitin is then released from the hemocytes and catabolized by the bacterial symbionts, leading to acidification of the crypt spaces and modulation of the light-sensitive bacterial luminescence apparatus (Schwartzman et al. 2015). The finding that the migration of blood cells into the light organ varies over the day/night cycle suggests that circadian regulation of immune cell abundance in different body sites may be a shared trait of many animals, as humans exhibit daily variation in immune cell migration as well.
Although the cyclical regulation suggested that these changes in transcription were circadian, the mechanisms by which transcription was controlled were not shown. A subsequent study found that blue-light receptors called cryptochromes were not only transcribed and produced in the light organ, but that the transcription of one cryptochrome gene was regulated by the presentation of bacterial light to the host on a rhythm different from that found in the head, suggesting that the light produced by bacterial symbionts was being used as a circadian cue (Heath-Heckman et al. 2013). This was the first instance of symbionts directly controlling regulators of circadian rhythms in a host. Subsequently, other types of mutualisms, such as those found in the mammalian gut, have been shown to involve similar regulatory interactions, suggesting that the influence of symbionts on host circadian rhythms may be evident in many mutualisms. As the details of the circadian circuitry in cephalopods have not yet been elucidated, it will be of great interest to determine the core components of the squid’s circadian clock and how the output of that clock affects the light organ and its activities.
Mammalian gut symbioses
In mammals, most major organ systems, including, but not limited to, the gut, skin, mouth, and reproductive tracts, maintain their own consortia of archaeal, bacterial, fungal, and viral species (Dethlefsen et al. 2007). These consortia have profound effects on the physiology of their respective host organs and organ systems, such as stimulating proper development of the immune system (Bouskra et al. 2008). The gut consortium, which is by far the most well-studied, also assists in the digestion of food and can deliver signals to sites as distant as the mammalian brain (Cryan and Dinan 2012). Many aspects of gut physiology, such as processivity and transcription of many genes in gut epithelial and immune cells, are highly circadian (Froy and Chapnik 2007; Hoogerwerf et al. 2008; Keller et al. 2009). Thus there seems likely to be an interface between the circadian rhythmicity of the gut and the physiological effects of symbionts therein. Indeed, a 2013 study showed that when mice were treated with antibiotics the cycle of clock gene transcription in the gut was all but abolished due to a lack of local Toll-like receptor signaling (Mukherji et al. 2013). While this study suggested that the rhythmicity in signaling stemmed from rhythmic Toll-like receptor transcription and arrhythmic presentation of bacterial cues, later studies showed that the composition and activity of the gut community is regulated over the day/night cycle as well. In mice, for example, the composition of the microbiome changes over the day/night cycle in response to feeding. Genetic perturbations of the core clock circuitry lead to aberrant feeding schedules which then cause dysbiosis (altered bacterial consortia) in the host that can lead to disease states such as obesity and glucose intolerance (Thaiss et al. 2014). It was later shown that, in addition to the diel regulation of the composition of the microbiome, the total number of bacteria in the gut fluctuates over the day/night cycle as well, a phenomenon that requires the mammalian core clock component Bmal1 and is more pronounced in females (Liang et al. 2015). Another study concluded that altered light regimes did not cause dysbiosis in the host without a high-fat diet; this interpretation may result from the fact that the animals were not sampled over the day/night cycle (Voigt et al. 2014). However, the association between diet and circadian rhythms in gut symbiosis was robust, as it was shown in a different study that in conventionally raised mice, the microbiome responds both to the external light cycle and to dietary composition: Mice that fed a high-fat diet exhibit a lack of proper rhythmic changes in the composition of the microbiome and become obese due to the presentation of aberrant microbial signals, such as short-chain fatty acids, due to this dysbiosis (Leone et al. 2015). However, if the mice are raised in the absence of a microbiome, they undergo disruptions in the transcription of core clock genes in the liver and brain but do not become obese. It was later shown that the microbiome is necessary for both the presence and amplitude of normal transcriptional rhythms in the liver as well as its metabolomic output (Montagner et al. 2016). Taken together, the current research suggests that the presence and proper circadian behavior of the gut microbiome is critical for normal diel transcriptional activity of the liver and gut, and that perturbation of microbial cues can lead to disease states such as obesity and diabetes.
While the study of circadian rhythms in gut symbiosis is still young, it has already brought to light the many connections between symbiosis, circadian rhythms, and disease. While much of the present work focuses on the signaling pathways by which the host and bacterial symbionts communicate with each other to reinforce each other’s rhythms, it is important to remember that the gut microbiome is also composed of fungi and viruses that also affect host physiology (Dethlefsen et al. 2007), and so it will be of great interest to determine whether these lesser-studied mammalian partners also exhibit variation over the day/night cycle.
Implications for research and future directions
Understanding the integration of symbiosis and circadian rhythms has profound implications for how scientific research should be conducted. Because the time of day can profoundly influence the outcome of both pathogenic and beneficial host–symbiont associations, researchers should be mindful of the time of day at which experiments are performed and be sure to accurately record and report the timing of experiments. In this vein, animal facilities may also benefit from being maintained under day/night conditions as close to natural as possible in order to minimize the disruption of subject circadian behaviors. It seems likely that certain discrepancies in the literature may be due to unappreciated differences among animal facilities in day/night cycle and irradiance regimes and/or that researchers perturb the animals at different times of day to perform experiments.
One final important aspect of our increasing knowledge of the integration of circadian and immune functions is its potential to improve our approach to human health. For example, transcription of the murine Toll-like receptor 9 gene exhibits circadian variation; in a recent study, this was shown to result in a diurnal fluctuation in vaccine efficacy (Silver et al. 2012). This leads to the idea that tuning vaccine delivery to a certain time of day may improve immunization outcomes in humans. TH17 cell differentiation is also under the control of the circadian clock, suggesting that inflammatory diseases mediated by these cells are also affected by circadian dysfunction (Yu et al. 2013). Likewise, a study has shown that placing circadian mutants on a strict feeding regimen can stop the animals from developing dysbiosis that can in turn cause obesity and glucose intolerance (Thaiss et al. 2014). With the development of new treatments and technologies that can impact our microbial communities, such as fecal transplants and probiotics, it would behoove us as a community to understand and respect the interplay between symbiosis and our body’s internal clock to ensure that these treatments are implemented as effectively as possible.
Acknowledgments
I would like to thank James Cronin and Sonke Johnsen for organizing this symposium and for the opportunity to present my work. In addition, I am grateful to David Weisblat and Nathan Whitehorn for their assistance in the preparation of this manuscript and to Margaret McFall-Ngai and Edward Ruby for many fruitful conversations on the topic of circadian rhythms and host–microbe interactions.
Funding
This work was supported by the National Institutes of Health [1 F32 NS095665-01].
References
- Arjona A, Silver AC, Walker WE, Fikrig E. 2012. Immunity’s fourth dimension: approaching the circadian-immune connection. Tren Immunol 33:607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, Edwards RA, Sahar S, Dandekar S, Baldi P. et al. 2013. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci USA 110:9897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC. 2011. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One 6:e26968.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, Eberl G. 2008. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456:507–10. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 701–12. [DOI] [PubMed] [Google Scholar]
- Davy SK, Allemand D, Weis VM. 2012. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev 76:229–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA. et al. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin RD, Middelkamp JN, Reed C. 1972. Circadian rhythmicity in susceptibility of mice to sublethal Coxsackie B3 infection. Nat New Biol 240:57–8. [DOI] [PubMed] [Google Scholar]
- Feigin RD, San Joaquin VH, Haymond MW, Wyatt RG. 1969. Daily periodicity of susceptibility of mice to pneumococcal infection. Nature 224:379–80. [DOI] [PubMed] [Google Scholar]
- Flint JF, Drzymalski D, Montgomery WL, Southam G, Angert ER. 2005. Nocturnal production of endospores in natural populations of Epulopiscium-like surgeonfish symbionts. J Bacteriol 187:7460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O, Chapnik N. 2007. Circadian oscillation of innate immunity components in mouse small intestine. Mol Immunol 44:1954–60. [DOI] [PubMed] [Google Scholar]
- Hawking F, Worms MJ, Gammage K. 1968. 24- and 48-hour cycles of malaria parasites in the blood; their purpose, production and control. Trans R Soc Trop Med Hyg 62:731–65. [DOI] [PubMed] [Google Scholar]
- Heath-Heckman EA, McFall-Ngai MJ. 2011. The occurrence of chitin in the hemocytes of invertebrates. Zoology (Jena) 114:191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath-Heckman EAC, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. 2013. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. MBio 4:e00167–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer P, Reichmann F, Farzi A. 2012. Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut–brain axis. Neuropeptides 46:261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogerwerf WA, Sinha M, Conesa A, Luxon BA, Shahinian VB, Cornelissen G, Halberg F, Bostwick J, Timm J, Cassone VM. 2008. Transcriptional Profiling of mRNA Expression in the Mouse Distal Colon. Gastroenterology 135:2019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta CT, Gazarini ML, Beraldo FH, Varotti FP, Lopes C, Markus RP, Pozzan T, Garcia CR. 2000. Calcium-dependent modulation by melatonin of the circadian rhythm in malarial parasites. Nat Cell Biol 2:466–8. [DOI] [PubMed] [Google Scholar]
- Johnson BC. 1992. Nutrient intake as a time signal for circadian rhythm. J Nutr 122:1753–9. [DOI] [PubMed] [Google Scholar]
- Kaniewska P, Chan C-KK, Kline D, Ling EYS, Rosic N, Edwards D, Hoegh-Guldberg O, Dove S. 2015. Transcriptomic changes in coral holobionts provide insights into physiological challenges of future climate and ocean change. PLoS One 10:e0139223.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk H-D, Kramer A, Maier B. 2009. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA 106:21407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ridenour JB, Dunkle LD, Bluhm BH. 2011. Regulation of stomatal tropism and infection by light in Cercospora zeae-maydis: evidence for coordinated host/pathogen responses to photoperiod? PLoS Pathog 7:e1002113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek PC, Brzozowski T, Konturek SJ. 2011. Gut clock: implication of circadian rhythms in the gastrointestinal tract. J Physiol Pharmacol 62:139–50. [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. 2011. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480:552–U183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-E, Edery I. 2008. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr Biol 18:195–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leedle JA, Bryant MP, Hespell RB. 1982. Diurnal variations in bacterial numbers and fluid parameters in ruminal contents of animals fed low- or high-forage diets. Appl Environ Microbiol 44:402–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N. et al. 2015. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17:681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Appelbaum L, Leggat W, Gothlif Y, Hayward DC, Miller DJ, Hoegh-Guldberg O. 2007. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 318:467–70. [DOI] [PubMed] [Google Scholar]
- Levy O, Kaniewska P, Alon S, Eisenberg E, Karako-Lampert S, Bay LK, Reef R, Rodriguez-Lanetty M, Miller DJ, Hoegh-Guldberg O. 2011. Complex diel cycles of gene expression in coral-algal symbiosis. Science 331:175–175.. [DOI] [PubMed] [Google Scholar]
- Liang X, Bushman FD, Fitzgerald GA. 2015. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA 112:10479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DB, Wang TP, Rudge JW, Donnelly CA. 2009. Evolution in a multi-host parasite: Chronobiological circadian rhythm and population genetics of Schistosoma japonicum cercariae indicates contrasting definitive host reservoirs by habitat. Int J Parasitol 39:1581–8. [DOI] [PubMed] [Google Scholar]
- Martinaud G, Billaudelle M, Moreau J. 2009. Circadian variation in shedding of the oocysts of Isospora turdi (Apicomplexa) in blackbirds (Turdus merula): an adaptative trait against desiccation and ultraviolet radiation. Int J Parasitol 39:735–9. [DOI] [PubMed] [Google Scholar]
- McCarthy MJ, Welsh DK. 2012. Cellular circadian clocks in mood disorders. J Biol Rhy 27:339–52. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. 2007. Adaptive immunity: care for the community. Nature 445:153. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mideo N, Reece SE, Smith AL, Metcalf CJE. 2013. The Cinderella syndrome: why do malaria-infected cells burst at midnight? Tren Parasitol 29:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro T, Ruby EG. 2012. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol 84:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner A, Korecka A, Polizzi A, Lippi Y, Blum Y, Canlet C, Tremblay-Franco M, Gautier-Stein A, Burcelin R, Yen Y-C. et al. 2016. Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci Rep 6:20127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, Ye T, Chambon P. 2013. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153:812–27. [DOI] [PubMed] [Google Scholar]
- Noordally ZB, Millar AJ. 2015. Clocks in algae. Biochemistry 54:171–83. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. 1998. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull 195:89–97. [DOI] [PubMed] [Google Scholar]
- O'Donnell AJ, Schneider P, McWatters HG, Reece SE. 2011. Fitness costs of disrupting circadian rhythms in malaria parasites. Proc Biol Sci 278:2429–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill JS, Reddy AB. 2011. Circadian clocks in human red blood cells. Nature 469:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley CA, Ameismeier MF, Peng L, Weis VM, Grossman AR, Davy SK. 2015. Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia. Cell Microbiol 18:1009–23. [DOI] [PubMed] [Google Scholar]
- Paschos GK, Ibrahim S, Song W-L, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M. et al. 2012. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med 18:1768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Behrendt L, Tarrant AM. 2010. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: The evolution of the animal circadian clock. PLoS One 5: :e12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden LC, Ingle RA. 2009. Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21:2546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann C, Kunisaki Y, Frenette PS. 2013. Circadian control of the immune system. Nat Rev Immunol 13:190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman JA, Koch E, Heath-Heckman EAC, Zhou L, Kremer N, McFall-Ngai MJ, Ruby EG. 2015. The chemistry of negotiation: rhythmic, glycan-driven acidification in a symbiotic conversation. Proc Natl Acad Sci USA 112:566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AFB. 1927. The diurnal tides of the leucocytes of man. J Pathol 30:1–19. [Google Scholar]
- Silver AC, Arjona A, Walker WE, Fikrig E. 2012. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36:251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek M, Díaz-Almeyda EM, Medina M, Levy O. 2014. Circadian clocks in symbiotic corals: the duet between Symbiodinium algae and their coral host. Mar Gen 14:47–57. [DOI] [PubMed] [Google Scholar]
- Sorek M, Levy O. 2012a. The effect of temperature compensation on the circadian rhythmicity of photosynthesis in Symbiodinium, coral-symbiotic alga. Sci Rep 2:536.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek M, Levy O. 2012b. Influence of the quantity and quality of light on photosynthetic periodicity in coral endosymbiotic algae. PLoS One 7:e43264.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek M, Yacobi YZ, Roopin M, Berman-Frank I, Levy O. 2013. Photosynthetic circadian rhythmicity patterns of Symbiodium, the coral endosymbiotic algae. Proc Biol Sci 280:20122942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, Shirasu-Hiza MM. 2012. The circadian clock protein timeless regulates phagocytosis of bacteria in Drosophila PLoS Pathog 8:e1002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch K-F, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417:78–83. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N. et al. 2014. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159:514–29. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–31. [DOI] [PubMed] [Google Scholar]
- Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. 2014. Circadian disorganization alters intestinal microbiota. PLoS One 9:e97500.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Toer M, Caldelari D, Lee D-U, Fu X-D, Dong X. 2011. Timing of plant immune responses by a central circadian regulator. Nature 470:110–U126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB. 2008. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455:1109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E. et al. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA 107:2259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, Hooper LV. 2013. TH17 cell differentiation is regulated by the circadian clock. Science 342:727–30. [DOI] [PMC free article] [PubMed] [Google Scholar]


