Malformations of cortical development are associated with significant morbidity and mortality. Alcantara et al. report that mosaic and constitutional mutations of the AKT3 gene are associated with a wide spectrum of brain disorders including focal and bilateral cortical malformations, diffuse megalencephaly and autism spectrum disorders.
Keywords: AKT3, megalencephaly, polymicrogyria, hemimegalencephaly, epilepsy
Abstract
Mutations of genes within the phosphatidylinositol-3-kinase (PI3K)-AKT-MTOR pathway are well known causes of brain overgrowth (megalencephaly) as well as segmental cortical dysplasia (such as hemimegalencephaly, focal cortical dysplasia and polymicrogyria). Mutations of the AKT3 gene have been reported in a few individuals with brain malformations, to date. Therefore, our understanding regarding the clinical and molecular spectrum associated with mutations of this critical gene is limited, with no clear genotype–phenotype correlations. We sought to further delineate this spectrum, study levels of mosaicism and identify genotype–phenotype correlations of AKT3-related disorders. We performed targeted sequencing of AKT3 on individuals with these phenotypes by molecular inversion probes and/or Sanger sequencing to determine the type and level of mosaicism of mutations. We analysed all clinical and brain imaging data of mutation-positive individuals including neuropathological analysis in one instance. We performed ex vivo kinase assays on AKT3 engineered with the patient mutations and examined the phospholipid binding profile of pleckstrin homology domain localizing mutations. We identified 14 new individuals with AKT3 mutations with several phenotypes dependent on the type of mutation and level of mosaicism. Our comprehensive clinical characterization, and review of all previously published patients, broadly segregates individuals with AKT3 mutations into two groups: patients with highly asymmetric cortical dysplasia caused by the common p.E17K mutation, and patients with constitutional AKT3 mutations exhibiting more variable phenotypes including bilateral cortical malformations, polymicrogyria, periventricular nodular heterotopia and diffuse megalencephaly without cortical dysplasia. All mutations increased kinase activity, and pleckstrin homology domain mutants exhibited enhanced phospholipid binding. Overall, our study shows that activating mutations of the critical AKT3 gene are associated with a wide spectrum of brain involvement ranging from focal or segmental brain malformations (such as hemimegalencephaly and polymicrogyria) predominantly due to mosaic AKT3 mutations, to diffuse bilateral cortical malformations, megalencephaly and heterotopia due to constitutional AKT3 mutations. We also provide the first detailed neuropathological examination of a child with extreme megalencephaly due to a constitutional AKT3 mutation. This child has one of the largest documented paediatric brain sizes, to our knowledge. Finally, our data show that constitutional AKT3 mutations are associated with megalencephaly, with or without autism, similar to PTEN-related disorders. Recognition of this broad clinical and molecular spectrum of AKT3 mutations is important for providing early diagnosis and appropriate management of affected individuals, and will facilitate targeted design of future human clinical trials using PI3K-AKT pathway inhibitors.
Introduction
Mutations of multiple genes within the phosphatidylinositol-3-kinase (PI3K)-AKT-MTOR pathway are well known causes of brain overgrowth (megalencephaly) as well as segmental cortical dysplasia (such as hemimegalencephaly, focal cortical dysplasia and polymicrogyria) (Mirzaa et al., 2013). The AKT complex is one of the most central molecules of this pathway that mediates many of its functions by regulating several downstream pathways. Mutations of the AKT3 gene have been reported in several patients to date, including seven with constitutional mutations causing megalencephaly with polymicrogyria (Riviere et al., 2012; Jamuar et al., 2014; Nakamura et al., 2014; Harada et al., 2015; Nellist et al., 2015; Negishi et al., 2017), and four with the mosaic p.E17K mutation causing hemimegalencephaly (Lee et al., 2012; Poduri et al., 2012; Riviere et al., 2012; Jansen et al., 2015). However, our understanding regarding the clinical and molecular spectrum associated with mutations of this critical gene is limited, with no clear genotype–phenotype correlations. We therefore sought to further delineate this spectrum, study levels of mosaicism and identify genotype–phenotype correlations of AKT3-related disorders. Here, we report 14 additional patients with AKT3 mutations, including five with novel mutations, who have more diverse phenotypes including bilateral perisylvian polymicrogyria, bilateral periventricular nodular heterotopia, and megalencephaly with autism but without any cortical dysplasia. We further provide the first detailed neuropathological characterization of extreme megalencephaly caused by a constitutional AKT3 mutation in a previously reported child (Riviere et al., 2012). This child has one of the largest documented brain sizes in the paediatric population, to our knowledge. We also report on the first child with bilateral multifocal cortical dysplasia caused by the mosaic p.E17K mutation that was detectable in skin-derived DNA. Our report substantially expands the clinical, molecular and biochemical spectrum of AKT3-related disorders and shows that activating mutations of this critical gene are associated with a broader spectrum of developmental brain disorders. Knowledge of this spectrum has important implications for the clinical and molecular diagnosis of affected individuals, recurrence risk counselling, and design of future human clinical trials using PI3K-AKT-MTOR pathway inhibitors.
Materials and methods
Human subjects and samples
The Institutional Review Board at Seattle Children’s Hospital approved this study. Individuals with megalencephaly and focal malformations of cortical development were enrolled as part of the developmental brain disorders research project. Informed consent was obtained from subjects prior to enrolment in the study. Genomic DNA was extracted from various tissues (blood, saliva, skin fibroblasts, brain) using standard protocols. Brain tissue was obtained during clinically indicated epilepsy surgery and appropriate samples were analysed by our molecular methods.
Brain MRI
All subjects underwent brain MRI as part of their routine clinical care. The investigators reviewed all images and relevant clinical and phenotypic data.
Molecular methods
Multiplex targeted sequencing using single molecule molecular inversion probes
We designed a pool of 26 single molecule molecular inversion oligonucleotide probes (smMIPs) targeting the coding sequences of AKT3. SmMIPs tiled across a total of 2937 bp of genomic sequence, including 100% of the 1498 coding base pairs of AKT3. Capture reactions (100 ng) were performed in parallel. Massively parallel sequencing was performed on an Illumina HiSeq system. Variant analysis was performed using our previously published pipeline (Mirzaa et al., 2016a). All missense, nonsense and splice site variants seen in two or more capture events that had a frequency <1% in public databases were retained for analysis.
Sanger sequencing
We performed confirmation of constitutional mutations by direct Sanger sequencing. PCR amplification was done with 50 ng of genomic DNA using Taq DNA polymerase (Applied Biosystems). Primers used to amplify the coding and flanking non-coding regions of AKT3 were designed using Primer 3. Double-stranded DNA sequence analysis was done with the BigDye® Terminator chemistry (Applied Biosystems), and reactions were run on the ABI 3730_l Genetic Analyzer (Applied Biosystems). Sequence chromatograms were analysed with Mutation Surveyor software version 3.30. Sequences were compared with normal control samples and the reference sequences for AKT3.
Overgrowth next generation sequencing
The multiplex polymerase chain reaction (PCR) panel was followed by next generation sequencing (NGS) performed on an Ion Torrent PGM platform. Allele detection limit was 1% at 1000× and 10% at 200× coverage. The threshold for mutation detection was set at 10× without strand bias.
Cell culture
HEK293 cells were grown at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal calf serum, L-GLN and antibiotics (penicillin-streptomycin).
Expression vector, site-directed mutagenesis, transfection and immunoprecipitation
AKT3 expression vector was obtained from OriGene (RC221051) as pCMV6-FLAG-MYC tagged Human cDNA ORF Clone containing AKT3 (NM_005465). Patient mutations were introduced using the QuikChange® Site-Directed Mutagenesis Kit (200518) from Agilent Technologies (Stratagene) using custom-designed primer pairs (Supplementary Table 1). AKT3-containing plasmids were expressed and transfected into HEK293 cells using calcium phosphate. Briefly, 5 µg of DNA was added to 61 µl of 2 M CaCl2 in 500 µl ddH2O. This was added dropwise to 500 µl of 2× HBS (NaCl, Na2HPO4, HEPES pH to 7.0) before adding to adherent cells, which were harvested 48 h later. Protein extracts were prepared by incubating the cell pellet on ice (1 h) in detergent extraction buffer [50 mM Tris HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 50 mM NaF, 25 mM β-glycerolphosphate, 0.1 mM Na-orthovanadate, 0.2% Triton™ X-100, 0.3% IGEPAL with protease inhibitor cocktail (Roche)]. Insoluble material was precipitated by centrifugation at 4°C and the supernatant used for immunoprecipitation. Ectopically expressed FLAG-tagged AKT3 was then immunoprecipitated using ANTI-FLAG® M2 Affinity Gel (A2220, Sigma-Aldrich) according to manufacturers’ instructions.
Kinase assay
AKT3-specific kinase assay using FLAG-captured ectopically expressed AKT3 was assessed using the Non-radioactive AKT Kinase Assay Kit (9840) from Cell Signaling Technology according to the manufacturers’ instructions, using phospho-GSK-3α/β (Ser21/9) (37F11) rabbit monoclonal antibodyt (9327) and mouse monoclonal ANTI-FLAG® M2 antibody (F3165) from Sigma-Aldrich.
Phosphoinositide dot blot binding analysis
FLAG immunoprecipitated AKT3 was eluted from the FLAG beads using FLAG peptide (3× FLAG® Peptide, F4799 Sigma-Aldrich). PIP strip membranes were incubated with purified AKT3 protein in 5 ml of PBS-Tween 3% bovine serum albumin according to manufacturer’s instructions (P-6001 PIP STRIPS and P-6100 PIP Array, Echelon Biosciences). Binding was detected by incubation with mouse monoclonal ANTI-FLAG® M2 antibody with chemiluminescence detection.
Statistics
P-values were calculated by use of Fisher’s exact test. A P-value < 0.05 was considered statistically significant.
Results
Clinical results
We identified AKT3 mutations in 14 new and four previously reported subjects, who collectively demonstrate a wide spectrum of features (Riviere et al., 2012; Jansen et al., 2015; Nellist et al., 2015). The clinical and molecular data of our 14 new mutation-positive children, as well as the previously published 11 patients, are summarized in Table 1. The neuroimaging features of these children are shown in Fig. 1. More comprehensive clinical, neuroimaging and molecular data are provided in Supplementary Tables 2–5.
Table 1.
Summary of the clinical and molecular findings of AKT3 mutation-positive patients (n = 24) [AKT3: NM_005465.4]
| Subject ID | Diagnosis | cDNA change | Amino acid change | Functional domain | Inheritance | Mutation type, AAP (tissue) |
|---|---|---|---|---|---|---|
| Mosaic AKT3 mutations (n = 5) | ||||||
| LR15-262 | DMEG/HMEG | c.49G>A | p.Glu17Lys | PH | NA | Mosaic, ∼8–14% (brain, skin) |
| HME-1565 | HMEG | c.49G>A | p.Glu17Lys | PH | De novo | Mosaic, ∼16–30% (brain) |
| (Lee et al., 2012) | ||||||
| Patient 3 | DMEG/HMEG | c.49G>A | p.Glu17Lys | PH | De novo | Mosaic, 35% (brain) |
| (Poduri et al., 2012) | ||||||
| LR11-443 | DMEG/HMEG | c.49G>A | p.Glu17Lys | PH | De novo | Mosaic, 1–18% (brain, skin) |
| (Jansen et al., 2015) | ||||||
| LR16-251 | DMEG/multifocal | c.49G>A | p.Glu17Lys | PH | NA | Mosaic, 1.8% (skin) |
| Constitutional AKT3 mutations (by functional domain; n = 20) | ||||||
| Patient | MEG | c.118G>A | p.Glu40Lys | PH | De novo | Constitutional |
| (Takagi et al., 2017) | ||||||
| LR16-372 | MEG | c.159C>A | p.Asn53Lys | PH | De novo | Constitutional |
| LR16-301 | MEG-PMG-PNH | c.161T>A | p.Phe54Tyr | PH | De novo | Constitutional |
| LR17-245 | MEG | c.237G>T | p.Trp79Cys | PH | De novo | Constitutional |
| LP96-103 | MEG-PMG-PNH | c.548T>A | p.Val183Asp | Kinase | De novo | Constitutional |
| LR12-314 | MEG-PMG-PNH | c.548T>A | p.Val183Asp | Kinase | De novo | Constitutional |
| (Nellist et al., 2015) | ||||||
| LR11-354 | MEG-PMG | c.686A>G | p.Asn229Ser | Kinase | De novo | Constitutional |
| (Riviere et al., 2012) | ||||||
| Patient | MEG-PMG | c.686A>G | p.Asn229Ser | Kinase | De novo | Presumed constitutionala |
| (Harada et al., 2015) | ||||||
| Patient 2 | MEG-PMG (MCAP) | c.686A>G | p.Asn229Ser | Kinase | De novo | Constitutional |
| (Nakamura et al., 2014) | ||||||
| Patient 1 | MEG-PMG | c.686A>G | p.Asn229Ser | Kinase | De novo | Constitutional |
| (Negishi et al., 2014) | ||||||
| LR13-041 | MEG-PMG | c.803T>C | p.Val268Ala | Kinase | De novo | Presumed constitutionala |
| LR14-271 | MEG-PMG | c.964G>A | p.Asp322Asn | Kinase | De novo | Constitutional |
| LR14-254 | MEG-PMG | c.964G>A | p.Asp322Asn | Kinase | De novo | Constitutional |
| LR12-412 | MEG-PMG | c.1393C>T | p.Arg465Trp | C-terminal | NA | Constitutional |
| LR14-025 | MEG-PMG | c.1393C>T | p.Arg465Trp | C-terminal | De novo | Constitutional |
| LR12-470 | MEG | c.1393C>T | p.Arg465Trp | C-terminal | De novo | Presumed constitutionala |
| LR13-008 | MEG-autism | c.1393C>T | p.Arg465Trp | C-terminal | De novo | Presumed constitutionala |
| LR14-112 | MEG-PMG-PNH | c.1393C>T | p.Arg465Trp | C-terminal | De novo | Constitutional |
| LR08-018 | MEG-PMG | c.1393C>T | p.Arg465Trp | C-terminal | De novo | Constitutional |
| (Riviere et al., 2012) | ||||||
| PMG-3801 | MEG-PMG | c.1393C>T | p.Arg465Trp | C-terminal | De novo | Constitutional |
| (Jamuar et al., 2014) | ||||||
AAP = alternate allele percentage (i.e. mutation level); DMEG = dysplastic megalencephaly; HMEG = hemimegalencephaly; MEG = megalencephaly; NA = not available; PH = pleckstrin homology; PMG = polymicrogyria; PNH = periventricular nodular heterotopia.
aMutations are presumed to be constitutional or germline due to one tissue only being molecularly analysed, with no evidence of mosaicism in the analysed tissue.
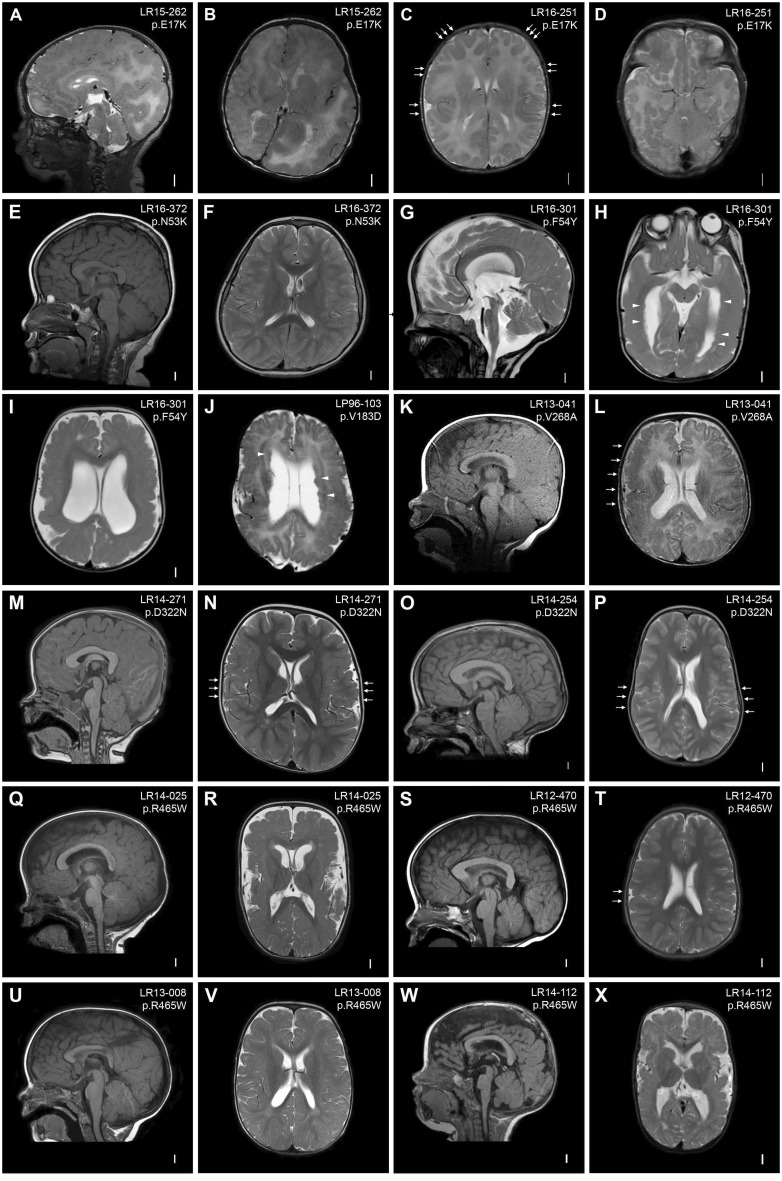
Figure 1.
Brain MRIs of AKT3 mutation-positive children. (A and B) Brain MRI of Patient LR15-262 showing markedly enlarged and dysplastic right cerebral hemisphere with diffuse cortical dysplasia and dysmyelination consistent with hemimegalencephaly. The contralateral hemisphere is markedly decreased in size with areas of cortical dysplasia (hemimicroencephaly). (C and D) Images of Patient LR16-251 showing multifocal areas of dysplastic cortex in the perisylvian, frontal, temporal and occipital regions (arrows). (E and F) Images of Patient LR16-372 showing a thick and dysplastic corpus callosum and deeply infolded perisylvian regions. (G–I) Images of Patient LR16-301 showing striking megalencephaly, ventriculomegaly, stretched but thick corpus callosum, diffuse polymicrogyria with deep infolding in the right occipital lobe, and bilateral periventricular nodular heterotopia (arrowheads). (J) Image of Patient LP96-103 showing diffuse bilateral perisylvian polymicrogyria, ventriculomegaly, cavum septum pellucidum et vergae and diffuse periventricular nodular heterotopia (arrowheads). (K and L) Images of Patient LR13-041 showing a large cerebellum with cerebellar tonsillar ectopia, bilateral polymicrogyria predominantly in the perisylvian region (more severe on the right, arrows) with dysmyelination. (M and N) Images of Patient LR14-271 showing diffuse megalencephaly with a thick corpus callosum and deep infolding in the perisylvian region suggestive of polymicrogyria (arrows). (O and P) Images of Patient LR14-254 showing diffuse megalencephaly, thick corpus callosum and bilaterally diffuse infolding of the perisylvian region suggestive of polymicrogyria (arrows). (Q and R) Images of Patient LR14-025 showing megalencephaly, thick corpus callosum and bilateral diffuse polymicrogyria with increased extra-axial space. (S and T) Images of Patient LR12-470 showing megalencephaly and thick corpus callosum. This patient also had deep infolding in the right perisylvian region suspicious for polymicrogyria, with very limited involvement (arrows). (U and V) Images of Patient LR13-008 showing diffuse megalencephaly and possible area of cortical dysplasia in the right perisylvian region. (W and X) Images of Patient LR14-112 showing diffuse megalencephaly, bilateral perisylvian polymicrogyria and bilateral ventriculomegaly.
Neuroimaging features
First, we identified the mosaic p.E17K AKT3 mutation in three children. The first child (Patient LR15-262) had hemimegalencephaly with contralateral hemimicroencephaly (Fig. 1A and B), and several cutaneous capillary malformations. This child was born in status epilepticus, had early onset intractable epilepsy, and underwent hemispherectomy at age 2 weeks. The second patient (Patient LR16-251) harbouring the p.E17K mutation had a distinctive phenotype characterized by megalencephaly with multifocal but bilateral cortical dysplasia (Fig. 1C and D). This child had intractable epilepsy and passed away at the age of 10 months due to his deteriorating neurologic status. He also had three capillary-lymphatic malformations. The third child (Patient LR11-443) had a massively enlarged and dysplastic cerebral hemisphere with dysplasia identified in the contralateral hemisphere as well. She also had a distinctive vascular malformation on the lower leg characterized by cutis marmorata telangiectatica congenita (CMTC) (Jansen et al., 2015).
Second, we identified constitutional AKT3 mutations in the remaining patients who can be clinically segregated into three groups. The first group includes children with megalencephaly and polymicrogyria (n = 6). These children had bilateral perisylvian polymicrogyria with variable ventriculomegaly (Fig. 1K–R). Among this group, one child had hydrocephalus requiring neurosurgical shunting and Chiari malformation requiring posterior fossa decompression (Patient LR14-254; Fig. 1O and P). This child showed global psychomotor delay at 2 years of age (due to mainly language and motor delays). However, on preschool evaluation at age 6 years, he showed normal cognitive skills (Wechsler Preschool and Primary Scale of Intelligence, WPPSI, score = 92). The second group consists of children with megalencephaly and diffuse cortical dysplasia (also termed dysplastic megalencephaly) with diffuse and bilateral periventricular nodular heterotopia, a rare subgroup not previously associated with any genes (n = 3) (Fig. 1G–J, W and X). Patients LR16-301 and LP96-103 had extensive heterotopia all along the ventricular surface (Fig. 1G–J), whereas Patient LR14-112 had fewer and more discrete heterotopia (Fig. 1W and X). Interestingly, all three patients within this group had moderate-to-severe ventriculomegaly. The third group of children with constitutional mutations had megalencephaly with no or subtle (often unilateral) cortical malformations and variable ventriculomegaly (n = 4) (Fig. 1E, F and S–V). One child within this group had megalencephaly, mildly thick corpus callosum and very subtle cortical dysplasia with unilateral prominent in cortical infolding into the perisylvian region (Patient LR12-470, Fig. 1S and T). This child had mild learning issues and communication problems. Another child within this group (Patient LR13-008) was formally diagnosed with autism spectrum disorders (ASD). Finally, one child (Patient LR17-245), also within this group, had megalencephaly only, with normal tone, and no developmental or neurological issues at age 3 years (Supplementary material).
Neuropathological abnormalities
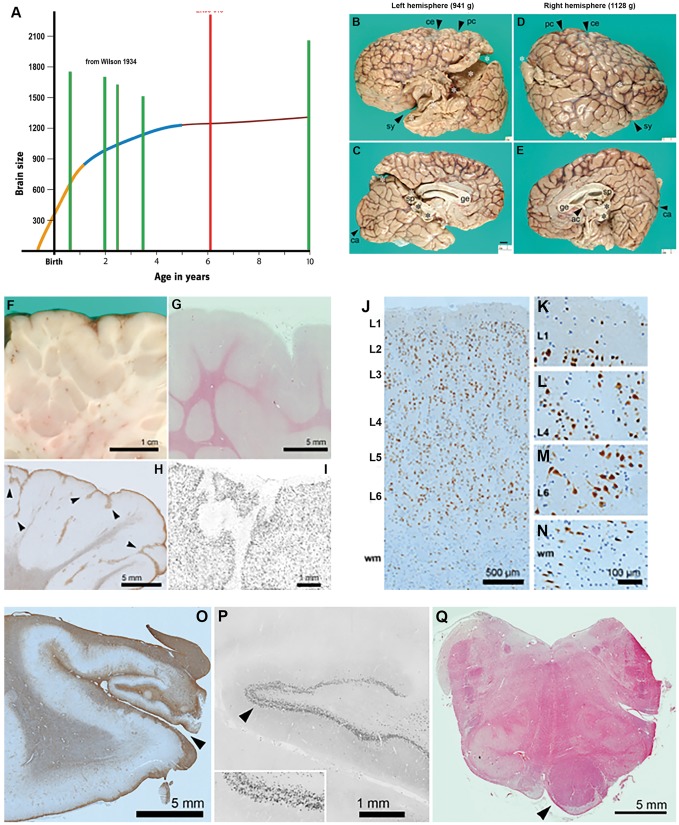
We examined the neuropathological features of megalencephaly-associated polymicrogyria in a previously reported AKT3 mutation-positive child (Patient LR08-018) who died unexpectedly at age 6 years. This boy had congenital megalencephaly, bilateral perisylvian polymicrogyria, cerebellar tonsillar ectopia, somatic asymmetry and connective tissue dysplasia (Mirzaa et al., 2012). He was identified to have a de novo p.R465W mutation in AKT3 (Riviere et al., 2012). This child passed away during sleep presumably due to sudden unexpected death in epilepsy (SUDEP) (further details provided in the Supplementary material). Neuropathological analysis on post-mortem brain tissue revealed that his brain weighed twice the normal weight of adult brains (total weight = 2313 g) and was asymmetrically enlarged (Fig. 2A and B). There was evidence of diffuse cortical dysplasia with irregular hyperconvoluted gyri suggestive of diffuse polymicrogyria, including anomalous branching and fusion of gliotic layer 1. There were also increased numbers of neurons in layer 6 and within the white matter that appeared disorganized and maloriented. However, neurons were not strikingly enlarged or dysplastic, and no balloon cells were identified. The hippocampus was grossly small and gliotic, and the dentate gyrus exhibited focal ‘tram-track’ splitting of the granule cell layer, typically associated with chronic epilepsy (Fig. 2B–Q).
Figure 2.
Pathologic examination of the brain of Patient LR08-018 (p.R465W). (A) Brain size. Graph depicting the largest paediatric brain sizes (in grams) previously reported in the literature (green bars) relative to the brain size of Patient LR08-018 (with the p.R465W mutation, red bar) demonstrating that brain size for this patient is markedly larger than these patients (Wilson, 1934). The graph is an adaptation of the human brain growth diagram from the Smithsonian Institute (http://humanorigins.si.edu/human-characteristics/brains). The graph shows the periods of rapid brain growth (in orange) plus the period of decreased brain growth (in blue) followed by the plateau in brain growth. (B–E) Cerebral hemispheres. The massive brain (2313 g; approximately twice normal weight for age) was asymmetrically enlarged. The left hemisphere (B and C) weighed 941 g, and the right (D and E) weighed 1128 g. Primary fissures such as the Sylvian (sy), central (ce), postcentral (pc), and calcarine (ca) sulci were recognizable, but secondary and tertiary sulci were abnormal. Gyri appeared irregular and overall hyperconvoluted. The corpus callosum was present including genu (ge), body, and splenium (sp). The anterior commissure (ac) was present but small. White asterisks: artefactual disruption of hemispheres due to brain removal and transport. Black asterisks: torn junction of hemispheres and midbrain. All panels are at the same magnification. (F–I) Hyperconvolution and polymicrogyria in cerebral cortex. (F) Brain slice through parietal cortex showed redundant folds of cortex extending deep into white matter. (G) Histological section (haematoxylin and eosin) through the same region showed relatively sharp grey-white junctions. (H) Inferior temporal cortex exhibited features of polymicrogyria, including anomalous branching and fusion of gliotic layer 1 (GFAP immunohistochemistry). (I) Layer 1 fusion and branching were confirmed by NeuN immunohistochemistry. (J–N) Abnormal layering of cerebral cortex, and excessive white matter neurons. In foci not involved with polymicrogyria, such as right posterior perisylvian cortex, cortical layering was moderately disorganized. (J) Cortical layers were identified based on cell size and density. NeuN immunohistochemistry. (K) Layer 1 was cell-sparse and contained only small neurons. (L) Layer 4 contained typical small (granular) neurons. (M) Layer 6 neurons were particularly disorganized and maloriented. (N) Increased neurons in white matter (wm). Interestingly, neurons in this case were not strikingly enlarged or dysplastic, nor were any balloon cells present. (O–Q) Hippocampal and brainstem abnormalities of Patient LR08-018. (O) The left hippocampus was very small and gliotic, and the hippocampal sulcus (arrowhead) was open, suggesting a deficiency of perforant pathway fibres, which would normally cross the fused sulcus. GFAP immunohistochemistry. (P) Histologically, the dentate gyrus exhibited focal ‘tram-track’ splitting of the granule cell layer (arrowhead; enlarged 2× in inset), a finding usually associated with chronic epilepsy. (Q) The upper medulla showed marked asymmetry of the pyramidal tract, essentially limited to one side (arrowhead). The adjacent inferior olives were moderately hypoconvoluted (haematoxylin and eosin).
Somatic features
Several children in this series had vascular malformations including CMTC (Patient LR11-443), capillary malformations and/or prominent veins (n = 5), and connective tissue abnormalities including aplasia cutis congenita (n = 2). One child (Patient LR13-008) had a prenatal stroke due to occlusion of the right cerebral artery, with no evidence of thrombophilia. Several patients had endocrine issues including recurrent hypoglycaemia (n = 2) and hypothyroidism (n = 1). Other notable features include immunological issues including recurrent infections (n = 2). In one of these patients (Patient LR14-112), recurrent infections were due to combined IgA and IgE deficiency. Patient LR13-008 also had severe vitamin A malabsorption (Supplementary Table 4).
Molecular results
All 14 AKT3 mutations reported in this series, and our previously reported four mutations, were identified or confirmed by targeted next generation and/or Sanger sequencing. Levels of mosaicism, tissues tested and methods of detection are provided in Supplementary Table 5. Mutations in two children with megalencephaly and megalencephaly with heterotopia (Patients LR16-372 and LR16-301) were identified by clinical whole exome sequencing (WES) on the proband, followed by targeted parental mutation analysis by Sanger sequencing.
The p.E17K mutation detected in our three patients was low-level mosaic, as mentioned previously. In Patient LR15-262, the alternate allele percentage (AAP) was 12.6–13.9% in affected brain tissue from the more dysplastic hemisphere resected during epilepsy surgery. Interestingly, the mutation was detectable in skin fibroblasts from normal-appearing skin (at 8.6–9.5% AAP) but was undetectable in peripheral blood-derived DNA. The mutation in Patient LR11-443 was present in 20–36% of cells from several affected brain regions from the more severely affected hemisphere. It was also detectable at a very low level (AAP 1.3%) from skin fibroblast-derived DNA (also from healthy-appearing skin). In Patient LR16-251 who had bilateral multifocal cortical dysplasia, the p.E17K mutation was present at a very low level in skin fibroblasts (AAP 1.8%). This child did not undergo epilepsy surgery due to his bilateral malformations, and no post-mortem brain tissue was available for molecular analysis. The other AKT3 mutations identified in our series were all constitutional, and were confirmed to be de novo when parental DNA was available. Five novel constitutional AKT3 mutations—p.N53K, p.F54Y, p.V268A, p.D322N, and p.W79C—were detected in six children in this series. Mutations in two patients (Patients LR12-470 and LR13-008) are presumed to be constitutional as DNA derived from saliva was used for molecular analysis, and peripheral blood-derived DNA was unavailable.
Functional analysis of AKT3 mutants
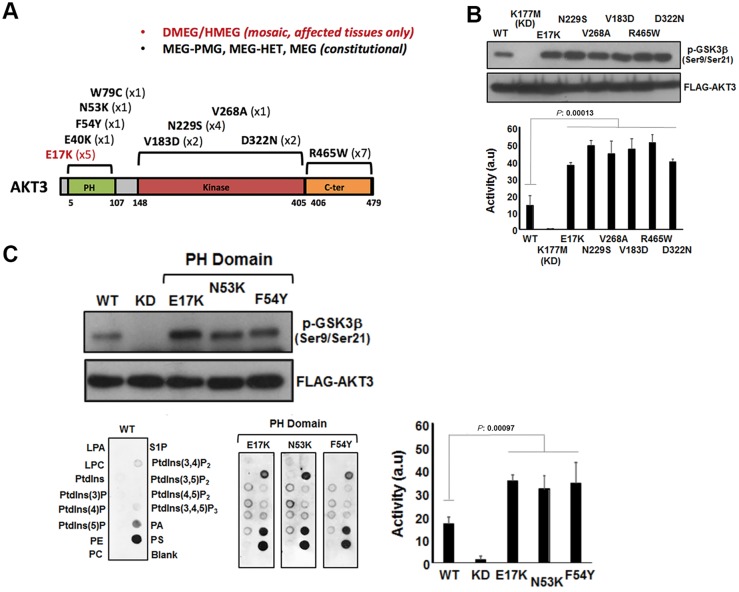
With the exception of p.R465W, which localizes to the C-terminal region of AKT3, the remaining mutations are within the pleckstrin homology domain or the catalytic kinase domain (Fig. 3A). Kinase activity analysis following ectopic overexpression clearly showed that all identified patient mutations caused increased activity compared to wild-type AKT3 (Fig. 3B and C). The p.E17K pleckstrin homology mutant domain has previously been shown to result in elevated kinase activity and to be oncogenic via enhanced pathological localization to the plasma membrane (Carpten et al., 2007; Parikh et al., 2012). As the pleckstrin homology domain is critical for phospholipid binding and consequent kinase activation, we assessed whether the novel mutations localized in the pleckstrin homology domain described here (p.N53K and p.F54Y) have a similar impact upon phospholipid binding as p.E17K (Park et al., 2008; Parikh et al., 2012). We used dot-blot analysis using an array of different phospholipids immobilized on wild-type AKT3. Indeed, these pleckstrin homology mutant domains exhibited markedly elevated binding to phosphatidylinositol-(3,4)-biphosphate [PtdIns(3,4)P2] in particular, which is a key plasma membrane constituent and substrate of phosphatidylinostitol-3-kinase.
Figure 3.
Analysis of AKT3 activity in vitro. (A) The primary structure of AKT3 showing the relative positions of the pleckstrin homology (PH) domain for lipid binding the catalytic kinase domain and C-terminal (C-ter) region. Mutations identified to date are shown along with the numbers of patients with these mutations in brackets. (B) Catalytic kinase domain and C-terminal localizing patient-derived AKT3 mutations are associated with elevated kinase activity. Ectopically expressed wild-type (WT) AKT, a kinase dead variant K177M, the E17K activating pleckstrin homology domain mutant and various patient mutants were assessed for kinase activity using a GSK3β peptide as a substrate in an ex vivo kinase assay. The upper panel shows immune detection of phosphorylated GSK3β peptide following western blotting with anti-phospho-GSK3β (Ser9/Ser21) antibody. The patient mutants all exhibit elevated phospho-activity compared to wild-type. The graph depicts quantitation of phospho-GSK3β (Ser9/Ser21) signal (a.u. = arbitrary units). Error bars represent mean ± SD (n = 4), P-values were determined using Student’s t-test. (C) Pleckstrin homology domain localizing patient mutations are associated with elevated kinase activity and altered phospholipid-binding profile. Left panels show western blot analysis of phospho-GSK3β (Ser9/Ser21) of ectopically expressed wild-type, K177M kinase dead and three pleckstrin homology domain patient mutants; E17K, N53K and F54Y. The graph depicts quantitation of phospho-GSK3β (Ser9/Ser21) signal. Error bars represent mean ± SD (n = 4), P-values were determined using Student’s t-test. The bottom panels depict PIP-membranes seeded with various lipids and phospholipids for dot blot binding analysis. Ectopically expressed FLAG-tagged wild-type and AKT3 pleckstrin homology domain mutants were incubated with the PIP Strips and bound protein detected by western blotting using anti-FLAG. All three pleckstrin homology domain mutants exhibit altered and elevated binding to specific phospholipids compared to wild-type. DMEG = dysplastic megalencephaly; HMEG = hemimegalencephaly; LPA = lysophophatidic acid; LPC = lysophosphocholine; MEG = megalencephaly; P = phosphate; PA = phosphatidic acid; PC = phosphatidylcholine; PE = phosphatidylethanolamine; PMG = polymicrogryria; PS = phosphatidylserine; PtdIns = phosphatidylinositol; S1P = sphingosine-1-phosphate.
Discussion
Malformations of cortical development comprise a wide range of disorders characterized by aberrant neuronal migration, proliferation and organization, and result in significant childhood morbidity and mortality (Barkovich et al., 2012). A growing spectrum of these malformations is now known to be caused by germline or mosaic mutations of genes within the PI3K-AKT-MTOR signalling network (Mirzaa et al., 2013; Jamuar et al., 2014; Mirzaa and Poduri, 2014). The post-zygotic (mosaic) mutations are most readily identified in affected (surgically removed) brain tissues (Mirzaa et al., 2016b) (Supplementary Fig. 1).
AKT3 is one of three AKT homologues (AKT1–3), the central effector of the PI3K-AKT-MTOR pathway (Yang et al., 2004). Mutations of AKT1 and AKT2 have been identified in somatic overgrowth disorders such as Proteus syndrome and in somatic overgrowth with hypoglycaemia, respectively (Hussain et al., 2011; Lindhurst et al., 2011). To date, reported children with these phenotypes all harboured the p.E17K mutation in these respective genes. The paralogous mutation in the brain-enriched isoform, AKT3, has been identified in mosaic form in children with hemimegalencephaly (Lee et al., 2012; Poduri et al., 2012; Jansen et al., 2015). Constitutional mutations of AKT3 have been identified in children with diffuse megalencephaly in syndromic forms such as megalencephaly polymicrogyria polydactyly hydrocephalus (MPPH) syndrome, and somatic duplications of the AKT3 locus have been identified in a few children with hemimegalencephaly and diffuse megalencephaly without cortical dysplasia (Wang et al., 2013; Chung et al., 2014; Conti et al., 2015; Hemming et al., 2016). To date, 11 children have been reported with AKT3 mutations in case reports of one or a few individuals. Here, we report on the clinical and neuroimaging spectrum of 14 children identified to have constitutional or mosaic AKT3 mutations, including six children with five novel AKT3 mutations, adding to our previously published data on four AKT3 mutation-positive children (Riviere et al., 2012; Jansen et al., 2015; Nellist et al., 2015). Using in vitro kinase assay, we show that these mutations constitutively activate AKT3.
Our series provides several important insights into the clinical and molecular spectrum of mutations associated with this critical gene. First, our series shows that there are important and emerging genotype–phenotype correlations of AKT3 mutations whereby the highly mosaic p.E17K mutation is associated with very segmental brain malformations (i.e. hemimegalencephaly), whereas constitutional mutations are associated with more widespread cortical malformations with bilateral (but often asymmetric) findings (P < 0.05; Supplementary Table 6). We describe two new patients with the recurrent, mosaic p.E17K mutation. Importantly, one of these children had a novel phenotype characterized by multifocal and bilateral focal cortical dysplasia lacking a severity gradient between right and left hemispheres, unlike all previously reported children with the p.E17K AKT3 mutation who had highly asymmetric cortical dysplasia (regarded as ‘classic hemimegalencephaly’). Second, our series substantially broadens the clinical spectrum of constitutional AKT3 mutations to include diffuse, most often perisylvian, polymicrogyria and periventricular nodular heterotopia (PNH), with hydrocephalus and cerebellar tonsillar ectopia occurring in a subset of individuals. Therefore, this report establishes AKT3 mutations as the first genetic cause of megalencephaly with PNH and polymicrogyria; a clinical phenotype previously reported as a distinct entity (Wieck et al., 2005). Importantly, our series further shows that constitutional AKT3 mutations, which can also occur in the pleckstrin homology domain where p.E17K mutation localizes, are associated with diffuse megalencephaly without cortical malformations, with normal cognitive development in two patients, one of whom had autistic features—resembling the PTEN-hamartoma tumour syndrome—and suggesting that mutations of the AKT3 gene are associated with autism spectrum disorders (ASD).
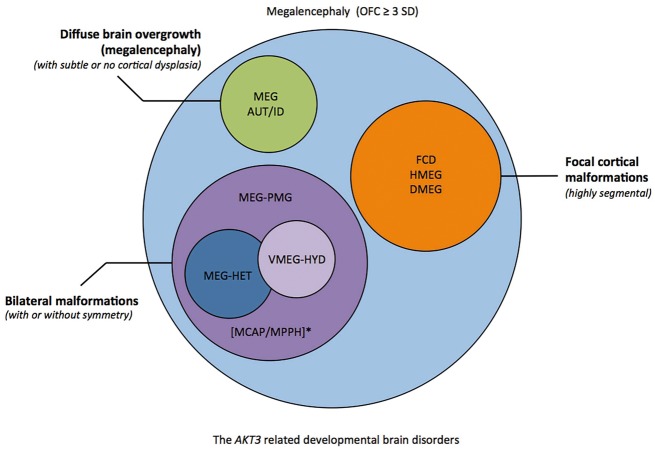
Collectively our data suggest that there are several distinct brain malformation syndromes caused by AKT3 mutations that can be broadly categorized into: (i) highly segmental cortical dysplasia (including hemimegalencephaly) and vascular malformations associated with the mosaic p.E17K AKT3 mutation. We believe that this comprises a clinically recognizable subgroup with megalencephaly, extensive focal cortical dysplasia (either hemimegalencephaly or bilateral, multifocal FCD), and cutaneous vascular malformations; (ii) megalencephaly-polymicrogyria with frequent asymmetry and occasionally patchy somatic findings [as occurs in MPPH and megalencephaly-capillary malformation syndrome (MCAP)]; (iii) megalencephaly-polymicrogyria with periventricular nodular heterotopia (PNH); and (iv) megalencephaly with normal or minimal cortical dysplasia, and ASD or autistic features (Fig. 4). We believe this clinical stratification is diagnostically important as resection of epileptic brain tissue is likely to be clinically warranted for children in Group 1, but less likely in Groups 2 and 3. Hydrocephalus and Chiari malformations, on the other hand, are complications likely to occur in children with diffuse megalencephaly occurring in Groups 2 and 3, depending on the severity of ventriculomegaly and cerebellar tonsillar enlargement, respectively.
Figure 4.
Diagram showing the spectrum of AKT3-associated phenotypes. Several groups of partially overlapping developmental brain disorders are associated with AKT3 mutations that include the following phenotypes (i) focal malformations of cortical development that are highly segmental [e.g. focal cortical dysplasia (FCD), hemimegalencephaly (HMEG), dysplastic megalencephaly DMEG; orange]; (ii) bilateral polymicrogyria (PMG) (purple) with or without ventriculomegaly or hydrocephalus (VMEG-HYD; light purple), and heterotopia (HET; blue); (iii) diffuse megalencephaly with intellectual disability (ID) and/or autistic features (AUT) with subtle or no cortical dysplasia (green). *Of note, MCAP and MPPH fit within the second group of AKT3-related disorders, from the brain phenotype perspective. MCAP can be further clinically distinguished by somatic findings (somatic overgrowth, vascular or lymphatic abnormalities), and MPPH by the occurrence of polydactyly in a subset of affected individuals. A general molecular diagnostic workflow for megalencephaly has been proposed (Supplementary Fig. 1). OFC = orbitofrontal cortex.
Seizures are a common feature of our cohort and have been reported in Akt mouse models with activating Akt3 mutations (Tokuda et al., 2011; Baek et al., 2015). Seizures are especially severe in children with the p.E17K mutation, likely due to the severity of the underlying cortical malformation. However, one of our patients (Patient LR08-018) with the constitutional p.R465W AKT3 mutation had intractable epilepsy as well, and may have had SUDEP; an interesting association that suggests that aggressive anti-epilepsy treatment and management may be necessary with this spectrum. This association will require further investigation in future children identified to have AKT3 mutations.
With regards to non-neurological findings, our series shows that vascular malformations are common with AKT3-related disorders and may provide useful diagnostic clues. Vascular malformations in all AKT3 mutation patients reported to date were patchy, few, and best characterized as capillary malformations, although one patient (Patient LR11-443) had CMTC, which is clinically distinct from capillary malformations. Unlike PIK3CA-related disorders, these vascular malformations were not associated with marked somatic overgrowth, lipomatosis, or soft tissue hypertrophy (Keppler-Noreuil et al., 2015; Mirzaa et al., 2016a). The occurrence of a prenatal stroke in one child is clinically noteworthy, as prenatal strokes and/or thrombophilia have been reported in some children with MCAP caused by PIK3CA mutations, as well as in Proteus syndrome caused by AKT1 mutations (Slavotinek et al., 2000; Mirzaa et al., 2012).
Two children in our series also had hypoglycaemia. These data add to a previous report of hypoglycaemia with AKT3-related disorders and further support this gene’s roles in glucose regulation (Nellist et al., 2015). Mutations of AKT2 are known to cause asymmetric overgrowth with hypoglycaemia, exemplifying the critical role of this gene in insulin-dependent glucose regulation (Cho et al., 2001; Hussain et al., 2011; Arya et al., 2014; Garg et al., 2015). While the cause of hypoglycaemia in our patients remains undetermined, this potential association warrants further studies as this may have important clinical implications for AKT3 mutation-positive children identified in the future as well.
Our functional analysis of the patient mutations showed a robust elevation of catalytic kinase activity. This is consistent with the elevated PI3K-AKT-MTOR signalling underlying these types of brain overgrowth disorders. Furthermore, pleckstrin homology domain-localizing mutations all exhibited elevated phospholipid binding compared to wild-type. This feature of the p.E17K mutation has been proposed to explain why this mutation is oncogenic and recurrent in a wide range of cancers (Carpten et al., 2007). Considering the similar phospholipid binding profile, we have uncovered here for p.N53K and p.F54Y compared to p.E17K, our findings suggest that careful monitoring of such individuals for cancers is warranted, although formal cancer surveillance guidelines cannot be proposed at this time due to lack of clinical evidence for these complications.
Among the limitations of our study was the ability to determine whether mutations of other genes (i.e. a two-hit phenomenon) are present in any of the patients in our series, and that may also explain the clinical heterogeneity of the brain phenotype. Indeed, this phenomenon has long been suspected for focal malformations of cortical development, especially PI3K-AKT-MTOR related disorders, and has been shown for specific ones such as the cortical tubers of tuberous sclerosis, for example (Crino et al., 2010). Therefore, this phenomenon is clearly of relevance to this spectrum of developmental brain disorders. However, there are several challenges towards identifying second hit mosaicism in this patient population. The first is the accessibility of affected tissues (i.e. brain) for molecular analysis, and the yield of DNA from these tissues. Only one of our two patients with focal malformations of cortical development due to AKT3 mutations underwent epilepsy surgery allowing molecular analysis on affected brain tissues. Second, the depth of coverage of the molecular method used. The ability to detect low-level mutations requires ultra-deep sequencing. Therefore, methods such as the ones we have used (deep next generation sequencing) would be necessary. To achieve the necessary depth of coverage, these methods often must be targeted (by testing a panel or subset of genes), and therefore, performing more global genomic assays to look for second hit mosaicism broadly (such as by whole exome or whole genome sequencing) may either not be feasible or technically possible (due to insufficient DNA yield). Finally, the most ideal approach to delineate the occurrence of second hit mosaicism would be to isolate specific cell populations by single cell sequencing. Therefore, while we were unable to detect the presence of second hit mosaicism in the individuals reported in our study due to some of challenges highlighted above, we anticipate that future advances in our molecular methods will undoubtedly improve our ability to detect second hit mosaicism in this spectrum of disorders.
In summary, we show that activating mutations of AKT3 are associated with a much broader spectrum of developmental brain disorders in children, with several clinical phenotypes determined partially by the type of mutation and level of mosaicism. Our series suggests that the mosaic p.E17K AKT3 mutation is associated with highly segmental brain malformations that may warrant surgical resection to achieve seizure control, whereas constitutional mutations of AKT3 are associated with bilateral brain malformations including bilateral perisylvian polymicrogyria, periventricular nodular heterotopia and megalencephaly without cortical dysplasia. In this series, we also report on the neuropathological abnormalities caused by a constitutional AKT3 mutation in one of the largest documented brain sizes in the paediatric population, to date. Our data also suggest that monitoring for hypoglycaemia in affected individuals may also be warranted.
Supplementary Material
Acknowledgements
We thank the patients, their families and referring providers for their contribution and support of our research. We thank Dr Louanne Hudgins from the Department of Pediatrics at Stanford University School of Medicine for her collaboration and for providing clinical data.
Glossary
Abbreviations
- MCAP
megalencephaly-capillary malformation syndrome
- MPPH
megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome
Funding
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke (NINDS) and the National Heart, Lung and Blood Institute (NHLBI) and the National Institutes of Health (NIH) under award numbers K08NS092898 (to G.M.), R01NS092772 and R01HL130996 (to W.B.D.), a Cancer Research UK Programme award C24110/A15394 (to M.O’D.) and grant N602531 from the European Union Seventh Framework Program under project DESIRE (to R.G.); RF-2013-02355240 (to R.G.), and The Stanford Department of Pediatrics (to G. B.).
The content is solely the responsibility of the authors, and does not necessarily represent the official views of the National Institutes of Health. The funding sources had no role in the design and conduct of the study, collection, management, analysis and interpretation of the data, preparation, review or approval of the manuscript, or decision to submit the manuscript for publication.
Supplementary material
Supplementary material is available at Brain online.
References
- Arya VB, Flanagan SE, Schober E, Rami-Merhar B, Ellard S, Hussain K. Activating AKT2 mutation: hypoinsulinemic hypoketotic hypoglycemia. J Clin Endocrinol Metab 2014; 99: 391–4. [DOI] [PubMed] [Google Scholar]
- Baek ST, Copeland B, Yun EJ, Kwon SK, Guemez-Gamboa A, Schaffer AE, et al. An AKT3-FOXG1-reelin network underlies defective migration in human focal malformations of cortical development. Nat Med 2015; 21: 1445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. A developmental and genetic classification for malformations of cortical development: update 2012. Brain 2012; 135: 1348–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007; 448: 439–44. [DOI] [PubMed] [Google Scholar]
- Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB 3rd, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 2001; 292: 1728–31. [DOI] [PubMed] [Google Scholar]
- Chung BK, Eydoux P, Van Karnebeek CD, Gibson WT. Duplication of AKT3 is associated with macrocephaly and speech delay. Am J Med Genet A 2014; 164A: 1868–9. [DOI] [PubMed] [Google Scholar]
- Conti V, Pantaleo M, Barba C, Baroni G, Mei D, Buccoliero AM, et al. Focal dysplasia of the cerebral cortex and infantile spasms associated with somatic 1q21.1-q44 duplication including the AKT3 gene. Clin Genet 2015; 88: 241–7. [DOI] [PubMed] [Google Scholar]
- Crino PB, Aronica E, Baltuch G, Nathanson KL. Biallelic TSC gene inactivation in tuberous sclerosis complex. Neurology 2010; 74: 1716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, Bademci G, Foster J 2nd, Sıklar Z, Berberoglu M, Tekin M. MORFAN Syndrome: an infantile hypoinsulinemic hypoketotic hypoglycemia due to an AKT2 mutation. J Pediatr 2015; 167: 489–91. [DOI] [PubMed] [Google Scholar]
- Harada A, Miya F, Utsunomiya H, Kato M, Yamanaka T, Tsunoda T, et al. Sudden death in a case of megalencephaly capillary malformation associated with a de novo mutation in AKT3. Childs Nerv Syst 2015; 31: 465–71. [DOI] [PubMed] [Google Scholar]
- Hemming IA, Forrest AR, Shipman P, Woodward KJ, Walsh P, Ravine DG, et al. Reinforcing the association between distal 1q CNVs and structural brain disorder: a case of a complex 1q43-q44 CNV and a review of the literature. Am J Med Genet B Neuropsychiatr Genet 2016; 171B: 458–67. [DOI] [PubMed] [Google Scholar]
- Hussain K, Challis B, Rocha N, Payne F, Minic M, Thompson A, et al. An activating mutation of AKT2 and human hypoglycemia. Science 2011; 334: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamuar SS, Lam ATN, Kircher M, D'Gama AM, Wang J, Barry BJ, et al. Somatic mutations in cerebral cortical malformations. N Engl J Med 2014; 371: 733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LA, Mirzaa GM, Ishak GE, O'Roak BJ, Hiatt JB, Roden WH, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 2015; 138: 1613–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Noreuil KM, Rios JJ, Parker VE, Semple RK, Lindhurst MJ, Sapp JC, et al. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am J Med Genet A 2015; 167A: 287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 2012; 44: 941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, Peters K, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med 2011; 365: 611–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa G, Timms AE, Conti V, Boyle EA, Girisha KM, Martin B, et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight 2016a; 1: e87623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa GM, Conway RL, Gripp KW, Lerman-Sagie T, Siegel DH, deVries LS, et al. Megalencephaly-capillary malformation (MCAP) and megalencephaly-polydactyly-polymicrogyria-hydrocephalus (MPPH) syndromes: two closely related disorders of brain overgrowth and abnormal brain and body morphogenesis. Am J Med Genet A 2012; 158A: 269–91. [DOI] [PubMed] [Google Scholar]
- Mirzaa GM, Riviere JB, Dobyns WB. Megalencephaly syndromes and activating mutations in the PI3K-AKT pathway: MPPH and MCAP. Am J Med Genet C Semin Med Genet 2013; 163C: 122–30. [DOI] [PubMed] [Google Scholar]
- Mirzaa GM, Poduri A. Megalencephaly and hemimegalencephaly: breakthroughs in molecular etiology. Am J Med Genet C Semin Med Genet 2014; 166C: 156–72. [DOI] [PubMed] [Google Scholar]
- Mirzaa GM, Campbell CD, Solovieff N, Goold C, Jansen LA, Menon S, et al. Association of MTOR mutations with developmental brain disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA Neurol 2016b; 73: 836–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kato M, Tohyama J, Shiohama T, Hayasaka K, Nishiyama K, et al. AKT3 and PIK3R2 mutations in two patients with megalencephaly-related syndromes: MCAP and MPPH. Clin Genet 2014; 85: 396–8. [DOI] [PubMed] [Google Scholar]
- Negishi Y, Miya F, Hattori A, Johmura Y, Nakagawa M, Ando N, et al. A combination of genetic and biochemical analyses for the diagnosis of PI3K-AKT-mTOR pathway-associated megalencephaly. BMC Med Genet 2017; 18: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellist M, Schot R, Hoogeveen-Westerveld M, Neuteboom RF, van der Louw EJ, Lequin MH, et al. Germline activating AKT3 mutation associated with megalencephaly, polymicrogyria, epilepsy and hypoglycemia. Mol Genet Metab 2015; 114: 467–73. [DOI] [PubMed] [Google Scholar]
- Parikh C, Janakiraman V, Wu WI, Foo CK, Kljavin NM, Chaudhuri S, et al. Disruption of PH–kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc Natl Acad Sci USA 2012; 109: 19368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WS, Heo WD, Whalen JH, O'Rourke NA, Bryan HM, Meyer T, et al. Comprehensive Identification of PIP3-Regulated PH Domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell 2008; 30: 381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 2012; 74: 41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere JB, Mirzaa GM, O'Roak BJ, Beddaoui M, Alcantara D, Conway RL, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet 2012; 44: 934–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM, Vacha SJ, Peters KF, Biesecker LG. Sudden death caused by pulmonary thromboembolism in Proteus syndrome. Clin Genet 2000; 58: 386–9. [DOI] [PubMed] [Google Scholar]
- Takagi M, Dobashi K, Nagahara K, Kato M, Nishimura G, Fukuzawa R, et al. A novel de novo germline mutation Glu40Lys in AKT3 causes megalencephaly with growth hormone deficiency. Am J Med Genet A 2017; 173: 1071–6. [DOI] [PubMed] [Google Scholar]
- Tokuda S, Mahaffey CL, Monks B, Faulkner CR, Birnbaum MJ, Danzer SC, et al. A novel Akt3 mutation associated with enhanced kinase activity and seizure susceptibility in mice. Hum Mol Genet 2011; 20: 988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zeesman S, Tarnopolsky MA, Nowaczyk MJ. Duplication of AKT3 as a cause of macrocephaly in duplication 1q43q44. Am J Med Genet A 2013; 161A: 2016–9. [DOI] [PubMed] [Google Scholar]
- Wieck G, Leventer RJ, Squier WM, Jansen A, Andermann E, Dubeau F, et al. Periventricular nodular heterotopia with overlying polymicrogyria. Brain 2005; 128: 2811–21. [DOI] [PubMed] [Google Scholar]
- Wilson SA. Megalencephaly. J Neurol Psychopathol 1934; 14: 193–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZZ, Tschopp O, Baudry A, Dümmler B, Hynx D, Hemmings BA. Physiological functions of protein kinase B/Akt. Biochem Soc Trans 2004; 32: 350–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.