Abstract
RNA-binding proteins are central to posttranscriptional gene regulation and play an important role in a number of major human diseases. Cloning such proteins is a crucial but often difficult step in elucidating the biological function of RNA regulatory elements. To make it easier to clone proteins that specifically bind RNA elements of interest, we have developed a rapid and broadly applicable in vitro genetic selection method based on T7 phage display. Using hairpin II of U1 small nuclear RNA (U1hpII) or the 3′ stem loop of histone mRNA as bait, we could selectively amplify T7 phage that display either the spliceosomal protein U1A or the histone stem loop-binding protein from a lung cDNA phage library containing more than 107 independent clones. The use of U1hpII mutants with various affinities for U1A revealed that this method allows the selection even of proteins that bind their cognate RNA targets with relatively weak affinities (Kd as high as the micromolar range). Experiments with a mixture of recombinant phage displaying U1A or the closely related protein U2B" demonstrated that addition of a competitor RNA can suppress selection of a protein with a higher affinity for a given RNA target, thereby allowing the preferential amplification of a lower affinity protein. Together, these findings suggest that T7 phage display can be used to rapidly and selectively clone virtually any protein that binds a known RNA regulatory element, including those that bind with low affinity or that must compete for binding with other proteins.
RNA-binding proteins (RNA-BPs) play a key role in a variety of posttranscriptional regulatory processes, including RNA processing, nucleocytoplasmic transport, translation, and mRNA decay (1–3). Moreover, a burgeoning body of evidence has implicated a number of RNA-BPs in the genetic etiology of human diseases such as fragile X mental retardation (4), paraneoplastic neurologic disorders (5), and spinal muscular atrophy (6), as well as in many microbial infections, including AIDS (7) and influenza (8).
An essential step in elucidating the regulatory mechanism of a given RNA element is to identify and characterize the protein(s) that binds to this element and mediates its effect. Characterization of such an RNA-BP is most readily accomplished by cloning its cDNA. Traditionally, this has been achieved by protein purification, peptide microsequencing, and cDNA amplification with PCR primers designed on the basis of the amino acid sequence information. This labor-intensive strategy requires an enormous effort and can be particularly difficult in the case of RNA-BPs present at a low cellular concentration.
Some genetic screening methods have been developed to facilitate the cloning of RNA-BP cDNAs, most notably the yeast three-hybrid system (9, 10) and plaque-lift analysis (11, 12). Both techniques have allowed the cloning of previously unidentified RNA-BPs (13–16). However, several limitations may interfere with the general applicability of these procedures. For example, when using the three-hybrid system, the RNA–protein interaction takes place in vivo, where it can be influenced by a multitude of cellular parameters. False-positive clones frequently predominate, and these must be eliminated by additional time-consuming screening steps. For their part, plaque-lift assays are often severely hampered by inefficient or nonspecific binding of the radiolabeled RNA probe to the filter-bound plaques.
Another potential, but as yet untested method for rapidly cloning RNA-BP cDNAs is phage display (17, 18), which should combine high selectivity with the rapid amplification of a particular clone. As the RNA–protein interaction would occur in vitro, the binding parameters could be completely controlled. Previous mutational studies of RNA-BPs using M13 phage display (19, 20) have demonstrated the utility of this method for identifying which protein residues of a known RNA-BP are critical for its RNA-binding affinity.
A pivotal drawback of using M13 phage display for cloning RNA-BPs from a cDNA library is that the nonlytic propagation mechanism of this phage requires that all components of the phage particle be exported through the bacterial inner membrane before phage assembly. Consequently, only proteins that are capable of such export can be displayed on the M13 phage capsid. This property depends not only on the presence of a signal peptide, which can be provided by the cloning vector, but also on the length, sequence, and folding characteristics of each protein to be displayed. Therefore, only a subset of proteins encoded by a given cDNA library will be capable of display on M13 phage.
In principle, this limitation of protein display on M13 could be obviated by using a lytic phage vector, as phage assembly would take place entirely in the cytoplasm. Recently, phage display vectors based on the lytic phage T7 have been developed (21), which would eliminate the need for protein export. However, unlike display on M13 phage, which replicate without lysing the host cell, RNA-BP display on lytic phage could pose a serious new problem, as the bacterial ribonucleases released on cell lysis may contaminate T7 phage preparations and degrade the RNA used as bait for selection. Here, we show that this difficulty can be overcome by constructing a new ribonuclease-deficient host strain to minimize contamination by cellular ribonucleases. With this important modification, it was possible to demonstrate the effectiveness of T7 phage display as a strategy for the selective isolation of RNA-BPs from a complex cDNA library. This procedure is faster and more specific than previous methods for cloning RNA-BP cDNAs.
Materials and Methods
Preparation of Recombinant T7 Phage.
A U1A-displaying phage was constructed in several steps. First, an EcoRI site (5′-GGAATTCC-3′) was inserted into the NcoI site (filled in) of the expression vector pU1A101.ex (19), which encodes amino acids 1–101 of human U1A preceded by a tether of 10 amino acids (PMAQVQLQVD). A FLAG epitope-tag (DYKDDDDK) was inserted into the NotI site downstream of the U1A cDNA. The resulting EcoRI–NotI restriction fragment was cloned between the corresponding sites of the T7 phage vector T7Select1–2c (Novagen). DNA (1 μg) was packaged by using a T7 packaging extract (Novagen). Phage titers were determined by plaque assays, as described (21). The inserts of recombinant clones were examined by heating portions of randomly chosen plaques at 65°C for 5 min in 10 mM EDTA (50 μl) and analyzing the DNA by PCR using a pair of primers that flank the site of cDNA insertion: 5′-GGAGCTGTCGTATTCCAGTC-3′ (forward) and 5′-GGCTGATACCACCCTTCAAG-3′ (reverse). Recombinant phage were amplified in liquid culture (5 ml) according to ref. 21, and their identity was confirmed by DNA sequencing. Display of U1A on the phage surface was verified by immunoblot analysis using the M2 anti-FLAG antibody (Sigma) or a monoclonal anti-U1A antibody (MAb 12E12) (22).
The U2B"-displaying phage was identical to the U1A phage except that (i) the U1A cDNA was replaced with a DNA segment encoding amino acids 1–98 of a previously described human U2B" variant bearing eight U1A-derived amino acid substitutions near its carboxyl terminus (23), and (ii) an untranslated 100-bp spacer was inserted between the NotI and XhoI sites to allow the U2B" phage to be distinguished from U1A phage by PCR.
Construction of a T7 Human Lung cDNA Library.
A T7 phage display library of human lung cDNAs was constructed from an existing human lung cDNA plasmid library (displayGREEN cDNA library, Display Systems Biotech, Vista, CA) generated from WI-38 lung fibroblasts. The cDNA inserts of the plasmid library were excised by digestion with EcoRI and NotI and inserted between the corresponding sites of an equimolar mixture of T7Select1–2a, -2b, and -2c DNA (Novagen) to allow translation of each cDNA in all three reading frames. The resulting phage library contained 1.2 × 107 independent clones, as determined by plaque assays. The library was amplified once by infecting a mid-log-phase Escherichia coli culture (250 ml, OD600 ≈0.6) with the phage library at a multiplicity of infection of 0.001. After cell lysis, the phage lysate was made 0.5 M in NaCl, clarified by centrifugation, and stored at −80°C. The insert sizes of 64 individual clones as well as of the complete library were analyzed by PCR with the forward primer 5′-GGAGCTGTCGTATTCCAGTC-3′ (see above) and the reverse primer 5′-AACCCCTCAAGACCCGTTTA-3′. The same PCR primers were used to analyze the phage pools isolated after each round of selection from the library.
RNA Synthesis.
The in vitro transcription templates pEP-U1hpII and pEP-U2hpIV have been described previously (19), as have the two pEP-U1hpII variants mut1 and mut2 (23). The DNA template for in vitro synthesis of the histone 3′ stem loop was prepared by inserting a synthetic HindIII–PstI fragment (5′-AAGCTTAAAAGGCCCTTTTCAGGGCCACCCTGCAG-3′) into the polylinker of pT7/T3α-19 (GIBCO/BRL). RNA was synthesized with a T7-MEGAshortscript kit (Ambion, Austin, TX), using DNA templates linearized with AvaI and a trace amount of [α-32P]CTP to aid in visualization of RNA bands. The radiolabeled transcripts were separated on denaturing 8% polyacrylamide gels, and the RNA bands were excised and eluted overnight in 0.3 mM sodium acetate, pH 5.0/1 mM EDTA/0.1% SDS at 37°C. The sequences of the RNAs prepared in this manner and used for in vitro selection were as follows: U1hpII, 5′-GGGAGACCCAAGCUUAUCCAUUGCACUCCGGAUGAGCUGCAGGUCGACUCUAGAGGAUCCCCGG-3′; U2hpIV, 5′-GGGAGACCCAAGCUUCCUGGUAUUGCAGUACCUCCAGGAAGX-3′; and 3′ histone RNA stem loop, 5′-GGGAAAGCUUAAAAGGCCCUUUUCAGGGCCACCX-3′ (where X denotes the 3′ tail underlined in the U1hpII sequence). The U1hpIImut1 and U1hpIImut2 RNAs were each identical to U1hpII except for the nucleotide substitutions shown in Figs. 3 and 5.
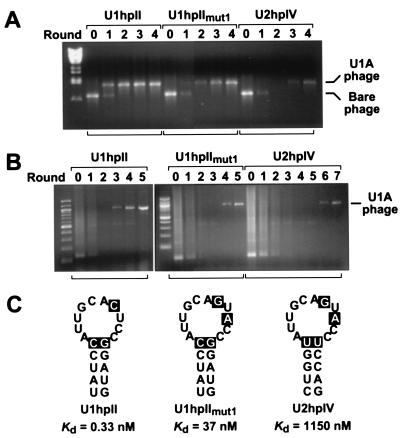
Figure 3.
Selection of U1A-displaying phage from phage pools of various complexities. (A) Selection of recombinant U1A-displaying phage after dilution with a 106-fold excess of bare phage. The phage mixture (2 × 109 pfu) was incubated with each of three bait RNAs (100 nM): U1hpII, a U1hpII variant (U1hpIImut1), or U2hpIV. Multiple rounds of in vitro selection were performed as outlined in Fig. 1. The relative abundance of U1A-displaying phage in the original phage mixture (denoted round 0) and after each round of selection was monitored by PCR analysis with primers flanking the site of cDNA insertion. Signals corresponding to the U1A-displaying phage and the bare phage are indicated. (B) Isolation of U1A-displaying phage from a human lung cDNA library comprising 1.2 × 107 independent clones. In vitro selection from the phage library (2.5 × 109 pfu) was performed as in A, using the same three bait RNAs (100 nM) but a different reverse primer for PCR analysis. The cDNA insert of the phage clone isolated in this manner was shown by sequence analysis to encode full-length U1A. (C) Schematic representation of the three RNA stem loop structures used in these experiments. The previously determined affinity of each for U1A (Kd) is indicated (23). The nucleotide changes in U1hpIImut1 and U2hpIV that are responsible for the diminished affinity of U1A for these stem loops are highlighted. In addition to the stem loop structures shown, each RNA contained four or six additional base pairs at the bottom of the stem, a short 5′ leader, and a 3′ tail that had been annealed to a complementary 5′-biotinylated DNA oligonucleotide before selection.
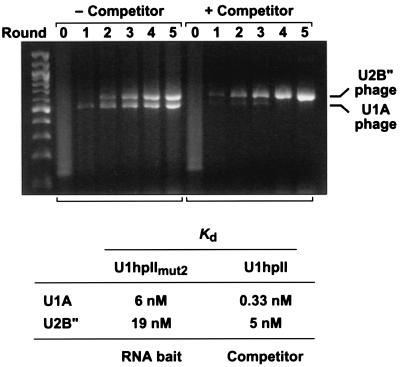
Figure 5.
Suppression of a high affinity phage clone by the use of a competitor RNA. An equimolar mixture of recombinant phage displaying U1A or U2B" was diluted with a 106-fold excess of bare phage. U1hpIImut2 (50 nM) bearing a 3′ tail annealed to a biotinylated DNA oligonucleotide was used as the RNA bait. Wild-type U1hpII (0 or 25 nM) that had not been annealed to biotinylated DNA served as a competitor. All other conditions of in vitro selection were identical to those described for Fig. 3A. The Kd values previously determined for dissociation of U1hpIImut2 and U1hpII from U1A and U2B" are indicated (23).
Construction of an RNase I-Deficient E. coli Host Strain.
A derivative of E. coli BLT5615 (Novagen) lacking the RNase I gene (rna) was generated by P1 transduction from E. coli DK533 (24) and selection for the kanamycin resistance phenotype of its Δrna∷kan allele. The resulting E. coli strain RNA5615 has the genotype F− rna ompT gal hsdSB (rB− mB−) dcm lac pAR5615 (ampR). Synthesis of the wild-type phage capsid protein encoded by pAR5615 was induced by adding isopropyl β-D-thiogalactoside (1 mM) to the medium about 30 min before phage infection.
For the RNA degradation experiments, purified, radiolabeled RNA (5 pmol) was mixed with a T7 phage lysate [5 × 109 plaque-forming units (pfu)] grown on BLT5615 or RNA5615 and incubated at room temperature. The samples were analyzed by electrophoresis on nondenaturing 8% polyacrylamide gels, and the radioactive bands were visualized with a Molecular Dynamics PhosphorImager.
Phage Affinity Selection.
Purified RNA was annealed via its 3′ tail to a complementary 5′-biotinylated DNA oligonucleotide (5′-ACCGGGGATCCTCTAGAGTC-3′). Phage-binding experiments were performed in a volume of 60 μl containing RNA (10–120 nM), T7 phage (1–5 × 109 pfu), E. coli tRNA (50 μg, Sigma), and SUPERase⋅IN (20 units, Ambion) in TENT buffer (10 mM Tris-HCl, pH 8.0/1 mM EDTA/250 mM NaCl/0.5% Triton X-100). After a 20-min incubation period on a tube rotator at room temperature, the binding reaction was mixed with prewashed streptavidin-coated paramagnetic beads (Dynabeads M-280 Streptavidin, Dynal, Lake Success, NY) and incubated for another 30 min at room temperature to allow the capture of RNA-bound phage on the beads. The beads were separated from the supernatant with a magnet (Dynal), washed twice, and resuspended in TENT buffer. The bound phage were then used without release from the beads to infect a log-phase E. coli culture (5 ml). The infected cells lysed within about 2 h, whereupon the phage lysates were titered via plaque assays, and an aliquot was used for the next round of selection. In vitro selection of phage was monitored by PCR using the procedure and primers described above.
Results
Display of the Amino-Terminal RNA-Binding Domain of U1A on T7 Phage.
As a model system for the development of a method for rapidly cloning RNA-BPs from T7 phage display libraries, we chose the well studied interaction of the spliceosomal protein U1A with its RNA target, the U1 snRNA stem loop U1hpII (25–27). U1A comprises two tandem RNA-binding domains belonging to the RRM (RNA recognition motif) family (2, 28). Because the amino-terminal RRM domain, comprising U1A residues 1–101, alone is responsible for binding U1hpII (29), the initial experiments were performed with this single domain. A human cDNA fragment encoding this U1A domain was inserted together with a FLAG epitope tag into the phage cloning vector T7Select1–2c (21) to generate a gene fusion encoding the amino-terminal half of U1A fused to the carboxyl terminus of the phage capsid protein 10B (Fig. 1) via a peptide tether 10 residues in length. The presence of U1A on the surface of the recombinant phage was confirmed by Western analysis of phage lysates, using anti-FLAG or anti-U1A antibodies (data not shown).
Figure 1.
In vitro selection of RNA-BP cDNAs by using T7 phage display. An RNA-BP cDNA is inserted into a T7 cloning vector and packaged in a phage capsid to generate a recombinant phage in which the RNA-BP is displayed on the surface as a carboxyl-terminal fusion to the T7 capsid protein 10B. The resulting phage is allowed to bind to the RNA bait, which itself is annealed to a biotinylated DNA oligonucleotide. RNA-bound T7 phage are captured on streptavidin-coated paramagnetic beads, separated from other members of the phage mixture with a magnet, and used to infect E. coli without prior release from the beads. After replication, the phage progeny can be subjected to additional rounds of selection.
Stability of RNA in Phage Lysates.
For use as bait, U1hpII RNA was synthesized by in vitro transcription and hybridized via its 3′ tail to a complementary 5′-biotinylated DNA oligonucleotide. The biotin modification made it possible to couple the RNA/DNA duplex to streptavidin-coated paramagnetic beads, thereby providing a selective means for isolating from a phage pool any recombinant T7 phage that can bind the bait RNA (Fig. 1).
A parameter critical for the success of this cloning method was the stability of the bait RNA in the phage lysate. Unlike M13 phage, which grow nonlytically and are exported from intact E. coli cells, the lytic release of T7 phage from E. coli would be expected to contaminate such phage preparations with host-cell ribonucleases released at the same time. Consistent with this prediction, incubating U1hpII RNA with T7 phage grown in E. coli BLT5615 cells resulted in the degradation of more than 90% of the RNA within 60 min at room temperature (Fig. 2 Left). Such RNA degradation would seriously complicate efforts to select T7 phage that display a protein with RNA-binding properties of interest. Prior purification of the phage particles via polyethylene glycol precipitation and CsCl step-gradient centrifugation resulted in only a minimal improvement in RNA stability (data not shown).
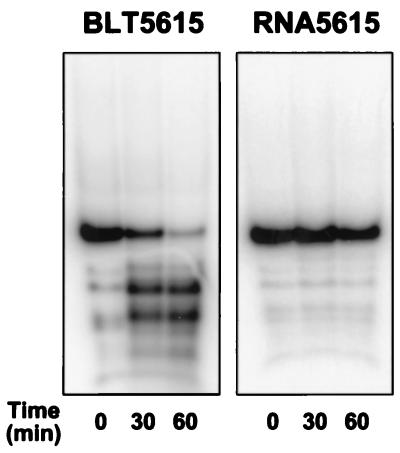
Figure 2.
RNA stability in phage lysates generated from different E. coli host strains. Radiolabeled U1hpII RNA (5 pmol) was mixed with either of two T7 phage lysates (5 × 109 pfu) prepared from E. coli BLT5615 or RNA5615 (an isogenic strain deficient in RNase I). After incubation at room temperature for the times indicated, RNA samples were analyzed by electrophoresis on a nondenaturing polyacrylamide gel.
We therefore investigated whether RNA degradation in T7 phage lysates could be prevented by genetically inactivating one or more ribonucleases of the host strain. RNase I was chosen as an initial target because it is a major, yet nonessential, ribonuclease activity in E. coli (30). The RNase I gene (rna) of strain BLT5615 was disrupted by P1 transduction to generate a new E. coli host strain (RNA5615) lacking this ribonuclease. In contrast to the extensive RNA degradation observed in phage lysates prepared from the original host strain, RNA was almost completely stable in phage lysates of the RNase I-deficient strain (Fig. 2 Right). Even in the presence of magnesium ions, which can activate many ribonucleases, RNA remained intact in the phage lysates prepared from RNA5615 (data not shown). These findings demonstrate the potential usefulness of this RNase I-deficient host strain for T7 phage display studies of both magnesium-dependent and magnesium-independent RNA–protein interactions.
Affinity Selection of U1A-Displaying Phage.
As a first test of the feasibility of using T7 phage display to clone RNA-BP cDNAs, we performed a pilot experiment in which we set out to isolate recombinant U1A-displaying phage from a 106-fold excess of T7 phage lacking an insert (bare phage) (Fig. 3A). The phage mixture (≈2 × 109 pfu) was incubated with U1hpII (Fig. 3C) at an RNA concentration (100 nM) well above the Kd of the U1A–U1hpII complex (0.33 nM) (19). The RNA-bound phage were recovered by addition of streptavidin-coated paramagnetic beads, washed twice, and amplified by growth in E. coli. The new phage lysate was then titered and subjected to another round of selection. In all, four selection cycles were performed. After each round, the phage population was analyzed by PCR using phage-specific primers flanking the U1A cDNA insert. U1A-displaying phage were detectable after just one round of selection, and only these phage remained by the end of the second round, demonstrating the specificity of the selection method. These results suggest a selective enrichment of the U1A-phage of 104- to 105-fold per round.
Many RNA-BPs of potential interest would bind their RNA targets with an affinity significantly less than that of U1A for U1hpII. To assess the usefulness of T7 phage display for isolating phage that display proteins with a low RNA-binding affinity, we simulated such weak interactions by using low affinity RNA ligands to select for U1A phage diluted into a 106-fold excess of bare phage. Two such ligands were used. One was a U1hpII variant (U1hpIImut1; Fig. 3C) bearing two point mutations that reduce its affinity for U1A by two orders of magnitude (Kd = 37 nM). The other was a U2 snRNA stem loop (U2hpIV; Fig. 3C) that resembles U1hpII but has a markedly lower affinity for U1A (Kd = 1150 nM). Three rounds of selection with U1hpIImut1 were sufficient to produce a U1A phage signal comparable in strength to that observed after one round with wild-type U1hpII (Fig. 3A Center). With U2hpIV, a U1A phage signal was detected after four rounds of selection (Fig. 3A Right). These findings demonstrate not only that T7 phage display allows the selection of low affinity RNA-BPs but also that an inverse correlation exists between the affinity of an RNA-BP for the RNA bait and the number of selection cycles needed for phage isolation.
Selection of RNA-BP-Displaying Phage from a Lung cDNA Library.
We next investigated whether U1A phage could be isolated from a complex phage pool displaying an entire cDNA library. A T7 phage display library of human lung cDNAs was constructed by subcloning the cDNA inserts of a commercial oligo(dT)-primed plasmid library into an equimolar mixture of three T7 phage vectors, T7Select1–2a, -2b, and -2c, which should allow translation of each cDNA in all three reading frames. The resulting phage library contained 1.2 × 107 independent recombinants (about twice the number of independent clones in the original plasmid library), with insert sizes of 0.3–3 kb (0.6–0.8 kb on average).
Initial selection from the cDNA library was performed by using U1hpII as bait (Fig. 3B Left). After three rounds of selection, a unique clone bearing a 1.1-kb cDNA insert could be detected. Sequence analysis of this insert showed it to comprise the complete coding sequence and 3′-untranslated region of human U1A cDNA. As the U1A RNA-binding domain that recognizes U1hpII is located at the extreme amino terminus of the protein, very few truncated forms of U1A encoded by shorter cDNAs would have been able to bind the RNA bait. Subsequent selection experiments with the low affinity ligands U1hpIImut1 and U2hpIV led to isolation of the same U1A-displaying phage clone, albeit after a greater number of selection cycles (Fig. 3B Center and Right).
Surprisingly, when using U2hpIV as bait, we failed to isolate phage displaying U2B", the natural protein ligand of this snRNA stem loop (31–33), even though U2B" binds to U2hpIV >200-fold more tightly than does U1A (23). PCR analysis with U2B"-specific primers revealed that the T7 cDNA library used in these experiments did not contain a U2B" cDNA clone of sufficient length to be capable of binding U2hpIV (data not shown).
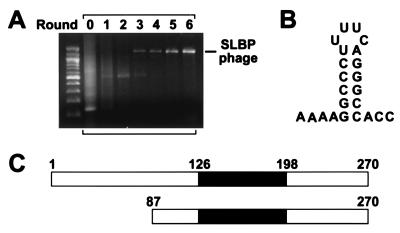
As a further test of the general utility of this method, we used the same cDNA library to select for phage displaying an RNA-BP of a different type: the histone stem loop-binding protein (SLBP). This protein, which is essential for the 3′ processing and stability of histone mRNAs, uses an RNA-binding domain entirely unlike that of U1A to bind a stem loop near the 3′ terminus of these transcripts (Fig. 4B) (34). Three rounds of selection with this stem loop led to the emergence of a unique phage clone containing a cDNA insert 1.3 kb in length (Fig. 4A). Sequence analysis revealed that this cDNA encoded amino acid residues 87–270 of human SLBP (Fig. 4C) together with the 3′-untranslated region of SLBP mRNA. This portion of SLBP contains the complete RNA-binding domain (residues 126–198), which alone is sufficient to bind the histone mRNA stem loop with high affinity (Kd = 4 nM) (35, 36). PCR and sequence analyses of the cDNA library used for these experiments showed that it contained no other in-frame SLBP clone encoding the RNA-binding domain.
Figure 4.
Isolation of an SLBP-displaying phage from a lung cDNA phage library. (A) In vitro selection from a human lung cDNA library was performed as in Fig. 3, except that the 3′-terminal stem loop of histone mRNA (120 nM) was used as the RNA bait. Sequence analysis confirmed that the cDNA insert in the amplified phage clone encoded SLBP. (B) Structure of the 3′ histone RNA stem loop used in this experiment. In addition, this RNA bore a 5′ leader and a 3′ tail annealed to a complementary biotinylated DNA oligonucleotide (the same as in Fig. 3C). (C) Schematic representation of full-length human SLBP (13, 14) and of the amino-terminally truncated form of SLBP (amino acid residues 87–270) displayed on the surface of the isolated phage clone. The region of SLBP comprising its minimal RNA-binding domain (residues 126–198) is shown in black (13).
Together, these findings demonstrate that T7 phage display allows the efficient cloning of a variety of RNA-BPs from a complex cDNA library, including those that bind their RNA targets with modest affinities corresponding to Kd values as high as the micromolar range.
Use of a High Bait Concentration or a Competitor RNA to Aid Selection of a Lower Affinity RNA-BP.
A potential impediment to cloning some RNA-BPs on the basis of their RNA-binding specificity is that the RNA ligand of interest may have a greater affinity for a protein other than the one being sought, making it difficult to isolate proteins besides that with the highest affinity. The control over selection conditions afforded by phage display offers an opportunity to circumvent such complications by using a high concentration of the bait RNA to neutralize the affinity difference or a competitor RNA to sequester the interfering high affinity protein.
To investigate the feasibility of these approaches, we used U1A and the closely related protein U2B" as a model system. U2B" is a homologous spliceosomal protein that uses its amino-terminal RRM domain to bind a U2 snRNA stem loop (U2hpIV) closely related to U1hpII (23, 31–33). A U1hpII variant (U1hpIImut2) bearing a single C → G substitution in the loop (AUUGCACUCC→AUUGCAGUCC) binds U1A three times more tightly than U2B" (Kd = 6 nM for U1A versus 19 nM for U2B") (23). Nevertheless, when an equimolar mixture of U1A- and U2B"-displaying phage was diluted into a 106-fold excess of bare T7 phage and subjected to in vitro selection with U1hpIImut2 at a concentration (50 nM) in excess of either of these Kd values, the two displaying phage were amplified with approximately equal efficiency, even after multiple rounds of selection (Fig. 5 Left).
We next tested the effect of including an unbiotinylated competitor RNA in the selection mixture. Wild-type U1hpII binds 15 times more tightly to U1A than to U2B" (Kd = 0.33 nM for U1A versus 5 nM for U2B") (23). When selection of the U1A- and U2B"-displaying phage was repeated using a combination of biotinylated U1hpIImut2 RNA (bait, 50 nM) and unbiotinylated U1hpII RNA (competitor, 25 nM), the U2B" phage exhibited a marked selective advantage over the U1A phage (Fig. 5 Right). This finding demonstrates the possibility of using an RNA competitor to suppress selection of an unwanted high affinity RNA-BP clone, thereby facilitating the isolation of T7 phage that display RNA-BPs of lower affinity for the RNA bait.
Discussion
Although several genetic methods have been developed to study RNA–protein interactions (9–12, 19, 20, 37–40), so far only two of them, the yeast three-hybrid system and a plaque-lift screening assay, have been used successfully to clone RNA-BPs from cDNA libraries (13–16). In this study, we have devised a powerful selection strategy for isolating RNA-BP cDNAs. This procedure, which is based on T7 phage display and iterative in vitro selection, combines speed with high sensitivity and seems to overcome many of the limitations of existing RNA-BP cloning strategies.
The use of a lytic phage vector circumvents a major drawback encountered with attempts to display cDNA libraries on M13, namely incomplete representation of the protein library on the phage surface. Many proteins cannot be displayed on M13 because merely fusing them to a signal peptide is insufficient to allow their export through the bacterial inner membrane, as required for assembly into phage particles. Because T7 phage assemble within the cytoplasm, there is no such impediment to protein display. Furthermore, joining cDNAs to the promoter-distal end of T7 gene 10 (one translated DNA junction) is significantly more likely to yield in-frame gene fusions than using M13 cloning vectors in which cDNAs must be sandwiched between vector sequences that encode a signal peptide and the M13 capsid protein (two translated junctions).
A potential problem of using lytic phage display vectors to clone RNA-BPs is that the phage preparations become contaminated with bacterial ribonucleases released on phage-induced cell lysis. These ribonucleases can degrade the RNA used as bait during selection. This problem was solved by constructing a new E. coli host strain that lacks the periplasmic ribonuclease RNase I. T7 phage grown in this strain are free of significant ribonuclease contamination.
A key advantage of T7 phage display for isolating specific RNA-BP cDNAs is the speed of the selection process. Two cycles of selection can be performed per day, making it possible to isolate a unique cDNA clone in as little as 2–3 days, a significantly shorter time than is required for other cloning methods. A second important benefit of phage display is that selection is performed in vitro, where conditions for the RNA–protein interaction can be controlled precisely. In contrast, the three-hybrid cloning system relies on screening in yeast cells, where the binding environment is poorly defined and selection is susceptible to interference by unknown cellular parameters. Consequently, the selection of false positive clones, which is a significant problem for the yeast three-hybrid system (9, 10, 13–15), is unlikely in the case of T7 phage display.
An important parameter that can be controlled during in vitro selection is the concentration of the RNA bait. As the binding affinity of a novel RNA-BP is not likely to be known before its cloning and characterization, it is advisable initially to use a moderately high RNA concentration (e.g., 100 nM) for selection. Our success in using the low affinity ligand U2hpIV to isolate U1A phage from a cDNA library suggests that an RNA bait concentration as low as one-tenth of the Kd is sufficient. It should be noted that the ease with which a cDNA clone can be isolated will depend not only on its equilibrium binding affinity for the RNA bait but also on its rate of dissociation from the ligand during subsequent steps in which bound phage are immobilized on beads and unbound phage are washed away.
Our data show that T7 phage display can be used to clone RNA-BPs that bind their targets with a wide variety of affinities, including those that associate weakly with a Kd value in the micromolar range. The sensitivity and specificity of this selection strategy is due in large measure to the iterative nature of the selection process, which allows the cumulative amplification of even modest selective advantages. A potential disadvantage of a cloning method with such a high degree of specificity is that it may prevent isolation of any but the clone with the greatest affinity for a particular bait RNA. Such an outcome could be problematic in cases where a lower affinity clone is of greater interest. One possible solution is to use a bait concentration that is likely to exceed the dissociation constants of all binding partners. As illustrated in Fig. 5 (Left), despite their 3-fold greater affinity for the RNA bait (U1hpIImut2), the U1A phage did not have a selective advantage over the U2B" phage when the bait concentration exceeded the Kd for both proteins. A second solution is to include a nonbiotinylated competitor RNA that preferentially binds RNA-BPs of lesser interest. By sequestering these clones, such a competitor can facilitate the isolation of additional RNA-BP clones that might otherwise be obscured because of their intrinsically lower affinity for the biotinylated RNA bait (Fig. 5 Right). Alternatively, a predominant RNA-BP clone could be removed immunochemically.
In conclusion, our findings suggest that in vitro selection from T7 phage display libraries of cellular cDNAs should allow the protein ligand of virtually any RNA regulatory element to be readily cloned, provided that the protein can bind as a monomer or homomultimer. Furthermore, the potential of lytic phage to display all types of RNA-binding domains makes T7 superior to M13 as a phage display vector for isolating novel protein variants with customized RNA-binding specificities from combinatorial libraries of RNA-BP mutants. By targeting specific mRNAs, such customized RNA-BPs could be of value as tools for manipulating gene expression.
Acknowledgments
We thank Carol Lutz for providing the anti-U1A antibody. We also thank Chaitanya Jain and Daniel Chagnovich for their helpful comments. These studies were supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (to S.D.) and by National Institutes of Health Research Grant GM55624 (to J.G.B.).
Abbreviations
- RNA-BP
RNA-binding protein
- SLBP
histone stem loop-binding protein
- U1hpII
hairpin II of U1 small nuclear RNA
- U2hpIV
hairpin IV of U2 small nuclear RNA
- pfu
plaque-forming unit
References
- 1.Mattaj I W. Cell. 1993;73:837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- 2.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 3.Siomi H, Dreyfuss G. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 4.Ashley C T, Wilkinson K D, Reines D, Warren S T. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 5.Musunuru K, Darnell R B. Annu Rev Neurosci. 2001;24:239–262. doi: 10.1146/annurev.neuro.24.1.239. [DOI] [PubMed] [Google Scholar]
- 6.Fischer U, Liu Q, Dreyfuss G. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 7.Cullen B R. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Sastre A. Virology. 2001;279:375–384. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- 9.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putz U, Skehel P, Kuhl D. Nucleic Acids Res. 1996;24:4838–4840. doi: 10.1093/nar/24.23.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Z, Wilusz J. Anal Biochem. 1993;212:547–554. doi: 10.1006/abio.1993.1367. [DOI] [PubMed] [Google Scholar]
- 12.Sägesser R, Martinez E, Tsagris M, Tabler M. Nucleic Acids Res. 1997;25:3816–3822. doi: 10.1093/nar/25.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z F, Whitfield M L, Ingledue T C, Dominski Z, Marzluff W F. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 14.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens M. Nature (London) 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 16.Denti A, Martinez de Alba A E, Sägesser R, Tsagris M, Tabler M. Nucleic Acids Res. 2000;28:1045–1052. doi: 10.1093/nar/28.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowman H B. Annu Rev Biophys Biomol Struct. 1997;26:401–424. doi: 10.1146/annurev.biophys.26.1.401. [DOI] [PubMed] [Google Scholar]
- 18.Rodi D J, Makowski L. Curr Opin Biotechnol. 1999;10:87–93. doi: 10.1016/s0958-1669(99)80016-0. [DOI] [PubMed] [Google Scholar]
- 19.Laird-Offringa I A, Belasco J G. Proc Natl Acad Sci USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laird-Offringa I A, Belasco J G. Methods Enzymol. 1996;267:149–168. doi: 10.1016/s0076-6879(96)67011-6. [DOI] [PubMed] [Google Scholar]
- 21.Novagen. T7Select System Manual TB178. Madison, WI: Novagen; 2000. [Google Scholar]
- 22.O'Connor J P, Alwine J C, Lutz C S. RNA. 1997;3:1444–1455. [PMC free article] [PubMed] [Google Scholar]
- 23.Rimmele M E, Belasco J G. RNA. 1998;4:1386–1396. doi: 10.1017/s1355838298981171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannistraro V J, Kennell D. J Bacteriol. 1991;173:4653–4659. doi: 10.1128/jb.173.15.4653-4659.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherly D, Boelens W, van Venrooij W J, Dathan N A, Hamm J, Mattaj I W. EMBO J. 1989;8:4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jessen T H, Oubridge C, Teo C H, Pritchard C, Nagai K. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allain F H T, Howe P W A, Neuhaus D, Varani G. EMBO J. 1997;16:5764–5774. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai K, Oubridge C, Ito N, Avis J, Evans P. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 29.Lutz-Freyermuth C, Query C C, Keene J D. Proc Natl Acad Sci USA. 1990;87:6393–6397. doi: 10.1073/pnas.87.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson A W. FEMS Microbiol Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 31.Scherly D, Boelens W, Dathan N A, Van Venrooij W J, Mattaj I W. Nature (London) 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 32.Bentley R C, Keene J D. Mol Cell Biol. 1991;11:1829–1839. doi: 10.1128/mcb.11.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price S R, Evans P R, Nagai K. Nature (London) 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 34.Dominski Z, Marzluff W F. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 35.Battle D J, Doudna J A. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel F, Schümperli D, Müller B. RNA. 2000;6:1539–1550. doi: 10.1017/s135583820000056x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacWilliams M P, Celander D W, Gardner J F. Nucleic Acids Res. 1993;21:5754–5760. doi: 10.1093/nar/21.24.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada K, Martin S S, Frankel A D. Nature (London) 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 39.Jain C, Belasco J G. Cell. 1996;87:115–125. doi: 10.1016/s0092-8674(00)81328-8. [DOI] [PubMed] [Google Scholar]
- 40.Paraskeva E, Atzberger A, Hentze M W. Proc Natl Acad Sci USA. 1998;95:951–956. doi: 10.1073/pnas.95.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]