ABSTRACT
We find that clusters of β4 integrin, organized into distinct puncta, localize along vimentin filaments within lamellipodia at the cell edge of A549 cells, as assessed by interferometric photoactivated localization microscopy. Moreover, puncta and vimentin filaments exhibit a dynamic interplay in live cells, as viewed by structured-illumination microscopy, with β4 integrin puncta that associate with vimentin persisting for longer than those that do not. Interestingly, in A549 cells β4 integrin regulates vimentin cytoskeleton organization. When β4 integrin is knocked down there is a loss of vimentin filaments from lamellipodia. However, in these conditions, vimentin filaments instead concentrate around the nucleus. Although β4 integrin organization is unaffected in vimentin-deficient A549 cells, such cells move in a less-directed fashion and exhibit reduced Rac1 activity, mimicking the phenotype of β4 integrin-deficient A549 cells. Moreover, in vimentin-deficient cells, Rac1 fails to cluster at sites enriched in α6β4 integrin heterodimers. The aberrant motility of both β4 integrin and vimentin-deficient cells is rescued by expression of active Rac1, leading us to propose that complexes of β4 integrin and vimentin act as signaling hubs, regulating cell motility behavior.
KEY WORDS: Cytoskeleton, Directed motility, Matrix receptor, Vimentin, Integrin
Summary: β4 integrin associates with vimentin at the periphery of migrating epithelial cells, and such β4 integrin–vimentin complexes support signaling through Rac1, leading to directed cell migration.
INTRODUCTION
β4 integrin is involved in mediating cell–matrix adhesion as a component of the hemidesmosome (de Pereda et al., 2009; Hopkinson et al., 2014). The hemidesmosome is a protein complex that anchors epithelial cells to the basement membrane in many epithelial tissues and indirectly links keratin intermediate filaments to laminin in the extracellular matrix (Sawamura et al., 2010; Hopkinson et al., 2014). β4 integrin has only one α-integrin binding partner, α6 integrin (Humphries et al., 2006). Interestingly, a number of studies have noted that α6β4 integrin is found in cells, particularly cancer cells, lacking hemidesmosomes (Hieda et al., 1992; Langhofer et al., 1993). Indeed, α6β4 integrin expression has been reported to be upregulated in many aggressive cancers and is associated with reduced cancer survival (Stewart and O'Connor, 2015; Stewart et al., 2016; Leng et al., 2016). The function of α6β4 integrin in such cells remains controversial (Geuijen and Sonnenberg, 2002; Sehgal et al., 2006; Hiroyasu et al., 2016; Seltmann et al., 2013; Kawano et al., 2010; Hsu et al., 2013). This controversy stems from contradictory data regarding α6β4 integrin function, especially with regards to cell motility. For example, we, and others, have provided evidence that α6β4 integrin enhances both single- and collective-cell migration (Sehgal et al., 2006; Colburn and Jones, 2017; Mercurio et al., 2001; Mainiero et al., 1996). In contrast, others have reported that the loss of β4 integrin or its destabilization promotes migration (Raymond et al., 2005; Dmello et al., 2015).
Following expression of α6β4 integrin in endothelial cells, it has been demonstrated that α6β4 integrin associates with the type III intermediate filament vimentin, which forms a filamentous network that extends from a dense perinuclear mesh toward the cell edge (Homan et al., 1998, 2002). In such cells, α6β4 integrin localizes in arrays at the basal surface, where it colocalizes with vimentin filaments, as assayed by immunocytochemistry (Homan et al., 1998). Moreover, Homan and co-workers also hypothesized that β4 integrin associates with vimentin filaments via the cytolinker plectin. To test this hypothesis, they expressed β4 integrin mutants with deletions at two sites in the cytoplasmic tail known to bind plectin. They found that deletion of the N-terminal plectin-binding site reduced the association of β4 integrin with vimentin (Homan et al., 2002).
Vimentin is expressed in a number of cell types, including microvascular endothelial cells and fibroblasts (Homan et al., 2002; Helfand et al., 2011). Its expression is also a hallmark of epithelial-to-mesenchymal transition and is characteristic of many cancers (Chaw et al., 2012; Cy et al., 2015). As with β4 integrin, vimentin expression is correlated with reduced cancer survival (Al-Saad et al., 2008). Moreover, vimentin has been identified as a positive regulator of motility and wound healing (Eckes et al., 2000; Hyder et al., 2015; Cy et al., 2015; Lanier et al., 2015; Hong et al., 2009; Mendez et al., 2010). Indeed, a recent publication has provided a possible mechanism via which vimentin acts, since the authors presented data revealing that vimentin is necessary for the proper organization of the microtubule network, which, in turn, is necessary for cell polarization and directed migration (Gan et al., 2016).
No study to date has tested the hypothesis that the interaction of β4 integrin with vimentin influences cell motility. Thus, in the present study, we used super-resolution microscopy to characterize the association of β4 integrin and vimentin in the lung cancer cell line A549. Since we have previously shown that β4 integrin positively regulates the directed migration of A549 cells, we also assessed how the interaction of β4 integrin with vimentin controls motile behavior (Colburn and Jones, 2017). Our results reveal not only that they each modulate the dynamics of the other but also that β4 integrin–vimentin complexes dictate directed cell migration through maintaining the activity of Rac1.
RESULTS
β4 integrin associates with vimentin
β4 integrin and vimentin have been reported to associate in a number of cell types (Homan et al., 1998, 2002; Dmello et al., 2015). In A549 cells β4 integrin is found in distinct puncta in lamellipodia (Colburn and Jones, 2017). Since A549 cells co-express vimentin and keratin, we hypothesized that they both could associate with β4 integrin puncta in these cells. As expected, keratin associates with β4 integrin puncta (Fig. S1A). Moreover, confocal analyses revealed that β4 integrin puncta are frequently distributed along vimentin filaments (Fig. 1A–C). To determine whether this occurs more often than would be expected by chance, which is indicative that the association is specific, we examined regions near the cell edge. We focused on these regions because of their substantial role in cell motility (Hopkinson et al., 2014) and because the relative sparseness of filaments in these regions enables accurate segmentation. We segmented both vimentin filaments and puncta in such regions and determined how often puncta colocalized with vimentin. We then compared these frequencies to the frequencies observed when the analysis was performed on images in which the puncta were randomly assigned to new positions (Fig. 1A–D). In only 87 of 30,000 such simulations did β4 integrin localize with vimentin more often in the simulation than in the real image (Fig. 1D). In contrast to what was seen with vimentin, β4 integrin puncta failed to exhibit robust association with actin filaments in A549 cells (Fig. 1E). We confirmed this by performing another simulation. In 12,000 of 30,000 simulations, β4 integrin associated with actin more often in the simulated data than in the real data (Fig. 1F). Since integrins typically associate with actin at the tips of filaments rather than along their lengths, this method could potentially overestimate the rate of colocalization in the real data compared with the simulated data. Therefore, we performed a follow-up analysis in which we assessed the frequency at which β4 integrin localized to within 240 nm of actin filament tips (Fig. 1G). β4 integrin localized to actin filament tips more frequently in the real data than in the simulated data in only 1% more simulations than would be expected by chance. Thus, it is extremely unlikely that β4 integrin puncta exhibit a real association with actin. This is in sharp contrast to our observation of specific β4 integrin association with vimentin, which we emphasize is also observed when the cells are plated on collagen I- or laminin-332-rich matrices (Fig. S2).
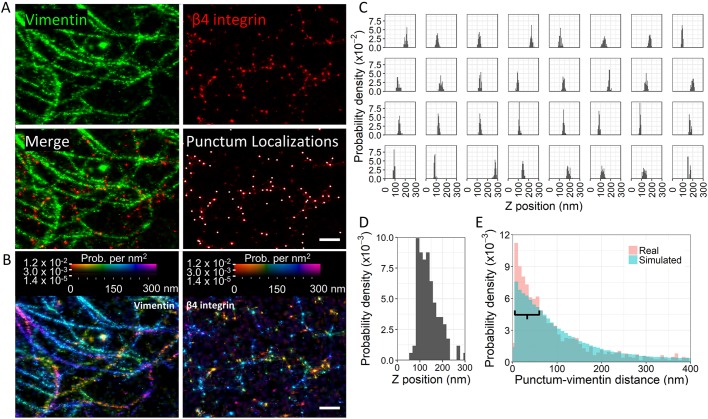
Fig. 1.
β4 integrin localizes along vimentin filaments in lamellipodia. (A) Confocal images of an A549 cell immunostained for vimentin and β4 integrin. Scale bar: 10 µm. Note that the brightness of the vimentin channel was increased to visualize filaments near the cell edge. The cell outline is marked in blue. The region where individual filaments are discernable is outlined in white. (B) Vimentin and β4 integrin images from A were segmented. Simulated puncta were generated by randomly assigning puncta in the region of interest to new positions in the same region. The result of one simulation is shown in the right panel. (C) Enlargement of the segmented images in B and the region bounded by the white box in A. Scale bar: 3 µm. (D) Computation of the ratio of the frequency of vimentin filament–punctum colocalization for images relative to that in their corresponding simulations. Points to the right of the dashed red line indicate more frequent colocalization in the real image compared with that in the simulation. Results are for 30 images with 1000 simulations each. (E) Confocal image of the edge of an A549 cell dual-labeled for actin and β4 integrin. Scale bar: 5 µm. (F) Histogram depicting the degree of actin filament–β4 integrin punctum colocalization in real images relative to that in their corresponding simulations, computed as in B. Results are for 30 images with 1000 simulations each. (G) Since β4 integrin could associate with the tips of actin filaments rather than along their length, the analysis in F was modified to evaluate the localization of β4 integrin within 240 nm of actin filament tips. Results are for 30 images with 1000 simulations each.
β4 integrin cannot bind vimentin filaments directly (Wiche, 1998). Rather, their association requires a cytolinker such as plectin (Geerts et al., 1999; Nievers et al., 2000; Wiche, 1998). We have confirmed that plectin colocalizes with β4 integrin puncta in A549 cells (Fig. S3). Plectin is a very large protein, over 500 kDa, with two 9 nm globular domains separated by a 200 nm rod domain (Wiche, 1998; Foisner and Wiche, 1987). The N-terminal globular domain has an actin-binding domain, while the C-terminal domain has an intermediate-filament-binding domain (Wiche, 1998). Both globular domains also have β4 integrin-binding sites (Wiche, 1998). Thus, β4 integrin and vimentin could be indirectly associating with each other through binding globular domains at the same or opposite ends of the plectin molecule. Consequently, β4 integrin and vimentin could be separated by ∼9 nm or 200 nm. Since plectin forms dimers, however, the separation distance could be even larger (Wiche et al., 2015; Walko et al., 2011). Therefore, we assayed β4 integrin puncta and vimentin association via super resolution microscopy. We performed interferometric photoactivated localization microscopy (iPALM) on cells expressing mEos3.2-tagged vimentin and immunostained for β4 integrin. We then grouped localizations into puncta by undertaking single-linkage clustering (Fig. 2A).
Fig. 2.
Super-resolution imaging of vimentin and β4 integrin in A549 cells. A549 cells transfected with mEos3.2–vimentin and immunostained for β4 integrin (Alexa Fluor 647-conjugated secondary antibody) were subjected to iPALM. (A) Rendering of iPALM localizations of vimentin and β4 integrin near the edge of a cell are shown. In the image marked ‘Punctum Localizations’, white spots indicate calculated punctum centroids. Scale bar: 500 nm. (B) Heatmaps indicating the variability in z-axis position and localization density for vimentin and β4 integrin localizations in the same field of view as in A. Scale bar: 500 nm. (C) Panel of histograms indicating the z-axis distribution of localizations for randomly selected puncta depicted in A. (D) Histogram of the z-axis position of punctum centroids. (E) Overlay of normalized histograms depicting the separation distance between each punctum and vimentin for both real and simulated data. The real data was generated from 16 regions of interest in eight cells, involving 1537 puncta; 160,000 simulated puncta were evaluated. The distributions are significantly different as determined with a Kolmogorov–Smirnov test (P=4×10−11). More puncta lie within ∼70 nm of vimentin in the real than in the simulated data (peak at 8 nm). This region is indicated by the black bar.
Interestingly, even puncta in the same field of view displayed substantial variability in their z positions (Fig. 2B–D). The reason for this is not clear. These puncta could represent molecules being inserted into the membrane or undergoing internalization. Alternatively, the variation may be due to buckling of the membrane. In addition, since the β4 integrin tail is over 1000 amino acids long, we cannot rule out the possibility that we have detected differences in the β4 integrin conformational state. However, a recent publication suggests that the cytoplasmic tail of β4 integrin does not undergo conformational changes in the same way that other integrins do (Miyazaki et al., 2018). Since the majority of puncta localized in an ∼100 nm range between 100 and 200 nm above the fiducial markers, we suspect that puncta less than 200 nm from the fiducial markers are incorporated into the membrane (Fig. 2D).
Through iPALM, we determined the distance between the localizations of each β4 integrin punctum and the nearest vimentin filament (Fig. 2E). The distribution was strongly right skewed with a prominent peak at 8 nm. Approximately 50% of β4 integrin puncta were less than 70 nm from a vimentin filament. To determine whether the peak represents a real phenomenon as opposed to being the result of a chance association of puncta and filaments in a limited space, we performed a simulation (Fig. 2E). The substantial peak observed between 8 and 50 nm in the real data was absent in the simulated data, suggesting that β4 integrin–vimentin association is a real phenomenon. In support of this notion, the real and simulated distributions were significantly different as determined by a Kolmogorov–Smirnov test (P=4×10−11). Since the peak occurred at a distance much closer to 9 nm than 200 nm and the globular domain at the C-terminal end of a plectin molecule is ∼9 nm in size, these data indicate that β4 integrin and vimentin likely bind to plectin via the same globular domain or juxtaposed globular domains in a plectin dimer. This is consistent with the ∼9 nm size of a plectin globular domain (Foisner et al., 1988).
Vimentin regulates β4 integrin dynamics
To determine whether the association of β4 integrin and vimentin detailed above has functional consequences, we evaluated the dynamics of β4 integrin puncta and vimentin filaments in live cells. We did this by performing time-lapse total internal reflection-structured illumination microscopy (TIRF-SIM) on cells expressing green fluorescent protein-tagged β4 integrin (GFPβ4) and mCherry-tagged vimentin (Fig. 3A). The resolution of this technique is on the order of 40 nm.
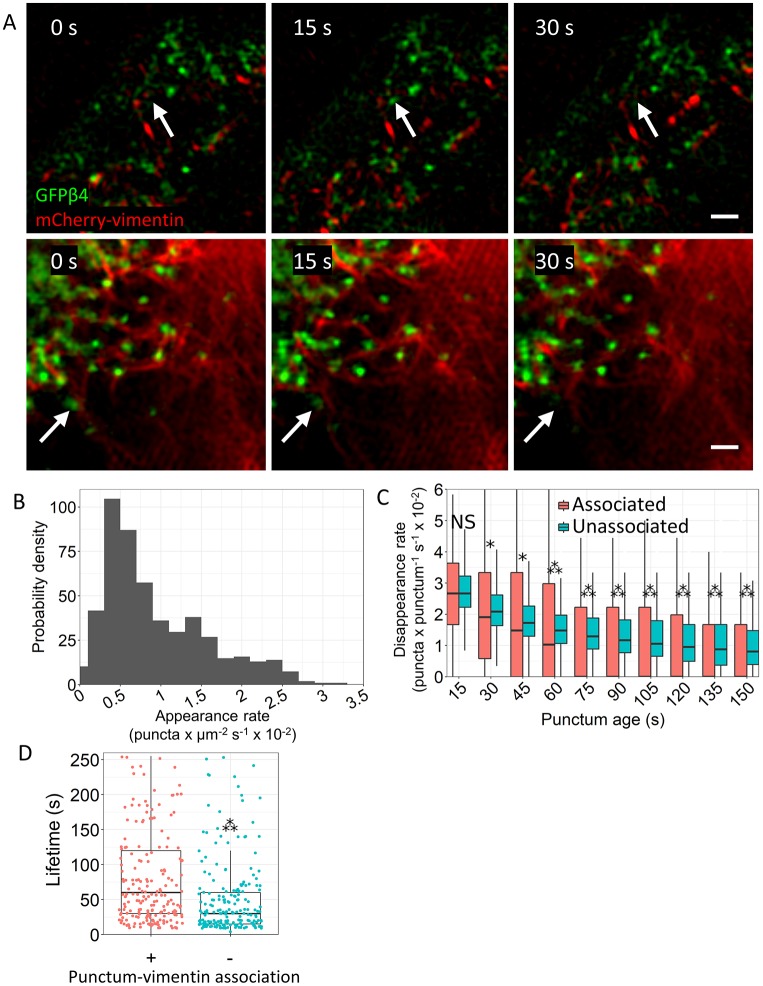
Fig. 3.
TIRF-SIM of live A549 cells indicates that vimentin association stabilizes β4 integrin puncta. A549 cells were transfected with plasmid encoding mCherry–vimentin and infected with adenovirus encoding GFP–β4. (A) In the upper panels, three images from Movie 1 are presented. The arrow marks a punctum that appears in a single frame of the movie. In the lower panels, three images from Movie 2 are shown. This area is enriched in vimentin filaments. A punctum, indicated by the arrow, loses association with vimentin and disappears. Other puncta persist over the same time period. Scale bars: 0.5 µm. (B) Histogram of punctum appearance rate (5 experiments, 30 cells, 540 data points). (C) Disappearance rate of puncta exhibiting association or lacking association with vimentin. The number of puncta per category from left to right are: 521, 600, 455, 569, 398, 535, 356, 500, 318, 466, 284, 434, 242, 406, 232, 370, 188, 338, 147 and 302. *P<0.05; ***P<0.001; NS, not significant (pairwise one-tailed Wilcoxon rank sum tests followed by Bonferroni correction). (D) Lifetime of puncta exhibiting or lacking association with vimentin (five experiments with 30 cells; 3563 puncta associated; 28,243 puncta unassociated). 200 randomly selected points are plotted per category. ***P<2.2×10−16 (one-tailed Wilcoxon rank sum test). In C and D, the box represents the 25–75th percentiles, and the median is indicated. The whiskers show the 10th to 90th percentiles.
Over the course of our experiments, we observed the appearance and disappearance of β4 integrin puncta (Fig. 3A; Movies 1 and 2). Puncta that associate with vimentin appeared more persistent than those that did not (Movies 1 and 2). Interestingly, some puncta existed only transiently and were visible in only a single frame (Fig. 3A). We also noted that β4 integrin puncta were remarkably immobile in A549 cells (Fig. 3A; Movies 1 and 2). This is consistent with previous reports that certain clusters of β4 integrin are quite stable in the plane of the membrane (Tsuruta et al., 2003; Geuijen and Sonnenberg, 2002).
We next quantified the appearance and disappearance of puncta over time. The distribution of appearance rates of β4 integrin puncta was right skewed in these cells (Fig. 3B). The peak of the distribution occurred at a rate of 0.005 puncta per square micron per second, which equates to ∼10 puncta per second in an average-sized cell. Puncta localizing with vimentin exhibited approximately the same disappearance rate as puncta far from vimentin for the first 60 s after their appearance (Fig. 3C). However, for longer-lived puncta, the disappearance rate when not associated with vimentin was significantly higher than for those associated with vimentin (Fig. 3C). Consequently, the lifetime of β4 integrin puncta that exhibited an association with vimentin was greater than the lifetime of β4 integrin puncta that did not (Fig. 3D).
Vimentin regulates focal adhesion formation but not β4 integrin punctum distribution
Our TIRF-SIM observations not only indicated that vimentin promoted the persistence of β4 integrin in the plane of the membrane, but also suggested that the establishment of persistent β4 integrin puncta was a highly stochastic process. Thus, we suspect, as others have suggested, that this process was dependent on conditions in the local microenvironment of deposited integrin heterodimers (Sastry and Burridge, 2000; Harburger and Calderwood, 2009). We therefore hypothesized that vimentin was one factor influencing punctum establishment. To test this hypothesis, we knocked down vimentin using two different siRNAs targeting vimentin (vimentin siRNAs 1 and 2). As assessed via western blotting, A549 cells treated with either vimentin siRNA exhibited ∼20% of the level of vimentin seen in control siRNA-treated cells (Fig. 4A,B). The cells were also 20% smaller (Fig. 4C,D). Interestingly, however, knocking down vimentin did not affect β4 integrin localization or puncta density (number per unit area) (Fig. 4C,E), which contrasts with the phenotype observed in breast cancer cells (Vuoriluoto et al., 2011). Moreover, vimentin loss had no obvious effect on keratin organization or its association with β4 integrin near the cell edge (Fig. S1B). On the other hand, paxillin-positive focal adhesions in the vimentin-knockdown cells exhibited reduced density, even though focal adhesion area was unchanged (Fig. 4F,G). This effect was likely indirect since focal adhesion–vimentin association is exceedingly rare in A549 cells, with only 3% of focal adhesions associating with vimentin filaments as assessed by confocal microscopy (Fig. S4). Moreover, the disappearance rate of β4 integrin puncta was increased in vimentin-knockdown cells (Fig. 4H).
Fig. 4.
Vimentin expression and matrix adhesions in A549 cells. A549 cells were treated with control (Ctrl) or vimentin (VIM) siRNAs. (A) A representative immunoblot for vimentin and GAPDH (of three experiments). (B) Quantification of results in A (n=3). (C) Confocal images of control and vimentin-knockdown cells immunostained for β4 integrin and the focal adhesion (FA) protein paxillin. The right column presents magnified views of the regions indicated by the white boxes. Scale bars: 10 µm (main images); 2 µm (magnified images). Quantifications of these images are presented in D–G. (D) Quantification of cell area. *P<0.05 (pairwise one-tailed Wilcoxon rank sum tests with Bonferroni correction). (E) Quantification of punctum density. NS, not significant (Kruskal–Wallis test followed by pairwise two-tailed Wilcoxon rank sum tests with Bonferroni correction). (F) Focal adhesion (FA) density. *P<0.05; ***P<0.001 (one-tailed Welch's t-tests with Bonferroni correction). (G) Quantification of FA area. NS, not significant (Kruskal–Wallis test followed by two-tailed Wilcoxon rank sum tests with Bonferroni correction). All quantifications are from three experiments with n=76 (Ctrl-siRNA), n=30 (VIM-siRNA1) and n=28 (VIM-siRNA2) cells. (H) The effect of vimentin expression on the disappearance rate of β4 integrin puncta was determined by performing TIRF microscopy on cells expressing GFP–β4 that were treated with the indicated siRNA. Results are from three experiments with n=24 (Ctrl-siRNA), n=18 (VIM-siRNA1), n=23 (VIM-siRNA2) cells. ***P<0.001 (one-tailed Welch's t-tests with Bonferroni correction). All graphs show mean±s.e.m.
β4 integrin controls vimentin cytoskeleton organization
Even though loss of vimentin did not affect β4 integrin localization, it was nevertheless possible that β4 integrin affected vimentin distribution. We tested this possibility by using three previously characterized A549 clones expressing shRNA targeted against β4 integrin [β4 integrin knockdown (β4KD) cells] (Fig. 5A) (Colburn and Jones, 2017). A line of A549 cells stably expressing control shRNA was used as a control (Fig. 5A). To assay the effect of β4 integrin expression on vimentin distribution, we immunostained for vimentin in control and β4KD cells. Vimentin filaments were mostly absent from the edge of β4KD cells, in sharp contrast to the well-spread vimentin networks of control cells (Fig. 5B). This was the case regardless of the serum content of the medium or whether the cells were plated onto uncoated coverslips or coverslips coated with either collagen I or laminin-332 (Fig. S5). Interestingly, the microtubule networks of β4KD cells were unaffected (Fig. 5B). We quantified the effect of β4 integrin expression on vimentin network organization by categorizing cells based on whether they exhibited extension of vimentin filaments into their lamellipodia (Fig. 5C). Interestingly, vimentin filaments extended into the lamellipodia in 80% of control A549 cells, but only 60% of β4KD cells (Fig. 5C). To confirm that the observed vimentin distribution differences were not the result of altered vimentin expression, we assayed vimentin expression by western blotting (Fig. 5D,E). There were no obvious differences between the expression levels of vimentin in control and β4KD cells, indicating that β4 integrin expression affects the vimentin cytoskeleton distribution, not vimentin expression. We also wondered whether attachment to β4 integrin puncta might make vimentin resistant to collapse triggered by nocodazole-induced microtubule depolymerization. However, in nocodazole-treated A549 cells, the absence of vimentin at the edge of cells was not accompanied by any obvious change in β4 integrin localization (Fig. S6).
Fig. 5.
β4 integrin regulates vimentin distribution. (A) Extracts of A549 cells (WT) or A549 cells expressing scrambled (Scr) or β4 integrin targeted shRNA (β4KD) were processed for immunoblotting with antibodies against β4 integrin and lamin A/C (a representative blot from three experiments is shown). (B) Control and β4KD cells were fixed and triple labeled for vimentin, tubulin, and actin. Scale bar: 10 µm. (C) Quantification of cells exhibiting extension of vimentin filaments into their lamellipodia. Results are from three experiments, n=303 cells. ***P<0.001; NS, not significant (one-sided Fisher's tests with Bonferroni correction). (D) Cell extracts processed for immunoblotting with antibodies against vimentin and lamin A. (E) Quantification of D (three experiments). All graphs show mean±s.e.m.
Vimentin expression regulates the motility of A549 cells
We previously reported that β4 integrin regulates the processivity of A549 cells (Colburn and Jones, 2017). Since β4 integrin closely associates with vimentin and β4 integrin loss alters vimentin cytoskeleton organization, we next tested the hypothesis that the ability of β4 integrin to regulate cell motility is mediated by vimentin or vice versa. In support of this possibility, vimentin has been reported to be a regulator of motility by numerous groups (Eckes et al., 2000; Hyder et al., 2015; Cy et al., 2015; Lanier et al., 2015; Hong et al., 2009; Helfand et al., 2011; Gan et al., 2016; Mendez et al., 2010).
We first performed a retrospective motility assay in which time-lapse microscopy of A549 cells was immediately followed by immunostaining the same cells for vimentin. We categorized cells based on whether vimentin filaments extended into their leading lamellipodia (Fig. S7). Those cells with lamellipodia containing vimentin showed 40% greater processivity but only 7% greater speed than those cells whose lamellipodia were devoid of vimentin filaments (Fig. 6A). To provide additional experimental support for a role of vimentin in A549 cell motility, we evaluated the migration of vimentin-deficient cells (Figs 4A and 6B). Compared with cells treated with control siRNA, cells treated with two different vimentin siRNAs exhibited reduced speed and processivity. Specifically, speed was reduced 14 and 28%, while processivity was reduced 37 and 28%, for vimentin siRNAs 1 and 2, respectively (Fig. 6B). In this regard, we previously reported that knocking down β4 integrin reduced the processivity but not the speed of A549 cells (Colburn and Jones, 2017). We also assayed the motility of A549 cells treated with plectin siRNA to disrupt vimentin–β4 integrin association (Homan et al., 2002) (Fig. 6C). In such cells, plectin levels were only 20% that of controls (Fig. 6D). Like A549 cells deficient in β4 integrin, the processivity but not the speed of plectin-knockdown cells was reduced (Fig. 6B).
Fig. 6.
β4 integrin and vimentin regulate motility via Rac1. (A) In this retrospective motility assay, time-lapse microscopy was performed on cells plated on coverslips. Immediately following imaging, cells were fixed and immunostained for vimentin. Cells were then categorized based on whether they exhibited extension of their vimentin filaments into their lamellipodia (–, no extension; +, extension present) (Fig. S5). Results are from three experiments, n=259 (−), n=48 (+) cells. *P<0.05 (one-tailed Wilcoxon rank sum tests). (B) Quantification of the motility of A549 cells expressing the indicated siRNAs. Results are from three experiments, n=167 (Ctrl-siRNA), n=842 (VIM-siRNA1), n=243 (VIM-siRNA2) and n=268 (PLEC-siRNA) cells. ***P<0.001 (one-tailed Wilcoxon rank sum tests with Bonferroni correction). (C) Extracts of A549 cells expressing control or plectin siRNAs were processed for immunoblotting with antibodies against plectin and GAPDH (a representative blot from three experiments is shown). (D) Quantification of C. (E) Rac1 activity assay performed using a G-LISA on extracts of the indicated cells. Results are from two experiments with three technical replicates. *P<0.05; ***P<0.001 (pairwise one-tailed Wilcoxon rank sum tests with Benjamini–Hochberg correction). (F) Quantification of the motility of vimentin- and β4 integrin-knockdown cells expressing the indicated constructs. CA Rac, constitutively active Rac1. Results are from three experiments. Number of cells analyzed from left to right are: 108, 80, 106, 104, 86, 94 and 54. ***P<0.001 (Kruskal–Wallis test followed by pairwise one-tailed Wilcoxon rank sum tests with Bonferroni correction). All graphs show mean±s.e.m.
Vimentin or plectin loss results in reduced Rac1 activity
Our previous study presented evidence that β4 integrin deficient A549 cells exhibit a reduction in Rac1 activation and a consequent decrease in processivity (Colburn and Jones, 2017). Thus, we assayed Rac1 activity in A549 cells treated with vimentin or plectin siRNA. Rac1 activity was reduced in vimentin and plectin knockdown cells (Fig. 6E). Finally, the motility phenotypes of both β4 integrin- and vimentin-deficient A549 cells were rescued by expression of constitutively active Rac1 (Fig. 6F).
Recruitment of Rac to α6β4 integrin puncta depends on vimentin and plectin
The reduction of Rac1 activity we observed in vimentin- and plectin-knockdown cells could be the result of either reduced ability to activate Rac1 or reduced ability to recruit Rac1 to their sites of activation. To test the latter hypothesis, we immunolabeled α6 integrin in cells treated with different siRNAs and expressing GFP-tagged WT Rac1 (GFP–Rac). We then imaged these cells using TIRF microscopy (Fig. 7A). Compared with cells treated with control siRNA, cells treated with vimentin or plectin siRNA exhibited reduced localization of GFP–Rac to puncta (Fig. 7A). We quantified these images by segmenting α6 integrin puncta and determining the average fluorescence of GFP–Rac in puncta relative to the average fluorescence across the entire basal surface of each cell. The results revealed loss of Rac association with puncta in plectin- or vimentin-deficient cells (Fig. 7B).
Fig. 7.
Loss of vimentin or plectin inhibits recruitment of Rac1 to α6β4 integrin puncta. (A) A549 cells infected with an adenovirus encoding GFP–Rac and expressing the indicated siRNA were immunostained for α6 integrin and imaged by TIRF microscopy. A magnified image of the boxed areas are shown in the right-hand panels. The yellow color in the inset in the top set of panels indicates colocalization, whereas there is little, if any, yellow in the images of cells in which vimentin or plectin were knocked down. Scale bars: 10 µm (main images) and 5 µm (magnified images). (B) Quantification of the relative intensity of GFP–Rac at puncta. 3 experiments, n=106 (Ctrl-siRNA), n=59 (VIM-siRNA1), n=32 (VIM-siRNA2), n=55 (PLEC-siRNA). ***P<0.001 (pairwise one-tailed Wilcoxon rank sum tests with Bonferroni correction). Graph shows mean±s.e.m. (C) A model depicting the role of α6β4 integrin in localizing vimentin filaments to the cell edge and the effect on cell signaling and directed migration.
DISCUSSION
Previous work has demonstrated that, in keratinocytes, keratin intermediate filaments interact with hemidesmosomes via plectin and β4 integrin (de Pereda et al., 2009; Hopkinson et al., 2014). Moreover, these interactions favor stabilization of the adhesion of cells to their extracellular matrix (de Pereda et al., 2009; Hopkinson et al., 2014). There is also evidence that keratin associates with focal adhesions, which appear to act as nucleation centers for keratin filament assembly (Windoffer et al., 2006). Likewise, vimentin filaments have been demonstrated to show interaction with focal adhesions in endothelial cells, strengthening cell–matrix adhesion (Tsuruta and Jones, 2003; Bhattacharya et al., 2009). Moreover, focal adhesions may nucleate vimentin filament assembly (Tsuruta and Jones, 2003). In sharp contrast, in A549 cells, we see little association of focal adhesions and vimentin. Rather, puncta, enriched in β4 integrin, display close association with vimentin filaments in the latter cells. By using confocal and super resolution microscopy techniques, we have demonstrated that β4 integrin localizes along vimentin filaments more often than would be expected by random chance. This observation is consistent with other reports showing that β4 integrin and vimentin filaments can associate in endothelial cells (Homan et al., 1998, 2002). However, in these published reports β4 integrin expression was induced in the cells whereas in A549 cells, as in certain other tumor cells including oral squamous cell carcinoma cells, β4 integrin is endogenously expressed alongside vimentin (Dmello et al., 2015).
Our immunofluorescence results indicate that β4 integrin in A549 cells is incorporated into puncta near the cell edge, rather than into the cat paw pattern often observed in keratinocytes (Colburn and Jones, 2017; Tsuruta et al., 2003). Moreover, these puncta are distinct from focal adhesions (Colburn and Jones, 2017). Previous reports have shown that β4 integrin interacts with the cytolinker protein plectin which, in turn, has the ability to interact directly with vimentin (Homan et al., 2002). Plectin is dumbbell shaped, possessing two binding sites for β4 integrin located within the globular domains at each end of the 200 nm long molecule (Foisner and Wiche, 1987; Walko et al., 2011; Castañón et al., 2013). The C-terminal globular domain of plectin also contains a binding site for vimentin (Castañón et al., 2013). If β4 integrin and vimentin interact with plectin at different ends of the molecule, we would have predicted that β4 integrin and vimentin would be separated by a distance of ∼200 nm. However, our super resolution (iPALM) analyses indicate that β4 integrin and vimentin localize within a distance of between 8 and 50 nm from each other, leading us to suggest that both vimentin and β4 integrin interact with the C-terminal globular domain of plectin.
Our functional assays reveal that vimentin, in concert with β4 integrin, regulates the processivity of A549 cells. Several mechanisms have been described to explain how vimentin might modulate motility and/or directed cell migration. Goldman and his colleagues have presented evidence showing that retraction of vimentin from regions close to the cell edge is a prerequisite to extension of a lamellipodium in 3T3 fibroblasts (Helfand et al., 2011). However, our data, albeit in a different cell type, indicate that lamellipodia-containing vimentin filaments are found predominantly in those A549 cells undergoing processive migration. In other words, we find no evidence that an absence of vimentin from cell edges correlates with either lamellipodial development or directed motility. It has also been shown that vimentin loss in non-small cell lung cancer cells results in depletion of focal adhesions, which could be considered to be the engine of cell motility (Havel et al., 2014). Interestingly, in our studies, vimentin-knockdown but not β4 integrin-knockdown cells show both a loss of focal adhesions and a reduction in speed. We suggest this is a consequence of the effects on focal adhesions induced by vimentin depletion. Finally, a recent report has proposed that the vimentin cytoskeleton regulates the directed motility of retinal pigmented epithelial cells by functioning as a template for the microtubule cytoskeleton and vice versa (Gan et al., 2016). Our results do not contradict this model. Rather, they emphasize that directed migration is a highly complex process involving not only multiple cytoskeletal systems, but also cytoskeletal anchorage sites located at the cell surface. We propose that β4 integrin puncta, which are predominantly assembled near the cell edge, recruit vimentin filaments, a process that likely depends on microtubules as proposed by Gan and colleagues (2016). For the first time, to our knowledge, we show that the resulting complexes of vimentin and β4 integrin then participate as signaling hubs by regulating the activity of Rac1 (Fig. 7C). Moreover, remarkably, our TIRF data indicate that vimentin and plectin not only regulate the activity of Rac1 but also its recruitment to the basal surface of the cell where it colocalizes with α6β4 integrin-rich puncta. In our model, puncta that recruit vimentin filaments persist for longer than those that do not show such interaction. Moreover, by promoting Rac1 activation, these longer-lived puncta increase actin polymerization and cell polarization, thereby promoting directed migration.
Some, if not all, β4 integrin puncta in A549 cells associate with both keratin and vimentin filaments. Intriguingly, when vimentin is knocked down there does not appear to be any effect on keratin–β4 integrin association near the cell edge. However, despite β4 integrin retaining interaction with the keratin cytoskeleton, this interaction does not compensate for the loss of vimentin, at least with regards to the ability to mediate signal transduction and directed migration. Assuming β4 integrin, rather than the cytoskeleton, mediates signaling, one explanation for our results is that vimentin facilitates β4 integrin-mediated signaling, whereas keratin does not. Alternatively, the loss of vimentin may reduce the total number of intermediate filaments that interact with β4 integrin complexes. This loss could impact the ability of β4 integrin to mediate signaling, possibly by modulating conformational changes in the β4 integrin cytoplasmic domain that are necessary for Rac–β4 integrin interaction. However, we emphasize that such interaction is likely indirect and involves a guanine nucleotide exchange factor (GEF). The identification of this GEF will be an interesting focus of future studies.
MATERIALS AND METHODS
Cell culture
A549 cells were purchased from American Type Culture Collection (ATCC) and were screened negatively for Mycoplasma by using the Universal Mycoplasma Detection kit (ATCC, 30-1012K). Scrambled (SCR) and β4 integrin knockdown (β4KD) cells were generated by transduction of wild-type (WT) cells with a lentiviral vector, encoding scrambled shRNA or β4 integrin-specific shRNA, respectively (Sigma-Aldrich, St Louis, MO) (Colburn and Jones, 2017). Cells were cultured in minimum essential medium supplemented with 4 mM L-glutamine and 10% fetal bovine serum at 37°C. SCR and β4KD cells were kept under puromycin selection (10 µg/ml, P8833, Sigma-Aldrich). Control (D-001210-01) and pooled plectin (L-003945-00) siRNAs were purchased from Dharmacon (Lafayette, CO). Vimentin siRNAs (#1 SI04201890 and #2 SI00302190) were purchased from QIAGEN Sciences (Germantown, MD). siRNA transfections were performed using Lipofectamine 2000 reagent (100014469), purchased from Life Technologies (Carlsbad, CA) and used according to the manufacturer's protocol. Adenoviruses encoding green fluorescent protein (GFP), GFP-tagged β4 integrin (GFP–β4), GFP-tagged WT Rac1 (GFP–Rac), or GFP-tagged constitutively active Rac1 (GFP–CA-Rac) were as described previously (Sehgal et al., 2006; Colburn and Jones, 2017). Plasmids encoding mCherry–Vimentin-7 and mEos3.2–Vimentin-7 were Addgene plasmids #55156 and #57485, respectively (deposited by Michael Davidson; Shaner et al., 2007). Plasmids were transfected using Lipofectamine LTX reagent (94754) with PLUS reagent (10964021) according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). Assays were performed on cells maintained on either tissue culture plastic or glass. In some experiments, cells were plated onto coverslips coated with matrix ligands and immunostained 6 h later. Such coverslips were coated with rat tail collagen I (354236) purchased from BD Biosciences (San Jose, CA) as described by the manufacturer or were coated with laminin-332 rich conditioned medium derived from 804G cells as described previously (Baker et al., 1997).
Immunocytochemistry
Immunocytochemistry was performed using previously described protocols (Kligys et al., 2012). Briefly, 6 h after plating onto matrix-coated coverslips, or 12 h otherwise, cells were fixed with 3.7% formaldehyde for 5 min and then cells extracts were prepared with 0.5% Triton X-100 for 7 min. Primary antibodies, diluted 1:100, were incubated with 5% normal goat serum (005-000-001, Jackson ImmunoResearch Laboratories, West Grove, PA) in 0.05% Tween-20 in PBS for 1 h at 37°C. A mouse monoclonal antibody against keratin (4545T) was purchased from Cell Signaling Technology (Danvers, MA). Mouse monoclonal antibodies against β4 integrin (450-11A) and vimentin (V9) were purchased from BD Biosciences and Sigma-Aldrich, respectively. Rabbit monoclonal antibodies against β4 integrin (ab133682), paxillin (ab32084), and plectin (ab32528) were purchased from Abcam (Cambridge, MA). A rat antibody against α6 integrin (J1b5), described elsewhere (Damsky et al., 1992), was a kind gift of Dr Caroline Damsky (University of California San Francisco, CA). A monoclonal rat antibody against tubulin (NB600-506) was purchased from Novus Biologicals (Littleton, CO). Actin was visualized using Alexa Fluor 647-conjugated phalloidin (A22287) purchased from Thermo Fisher (Rockford, IL, USA) and Rhodamine-conjugated phalloidin (R415) purchased from Invitrogen. Fluorophore-conjugated secondary antibodies were incubated on coverslips for 1 h at 37°C. Goat anti-mouse-IgG antibodies used were conjugated to fluorescein (115-095-166, Jackson ImmunoResearch Laboratories), Rhodamine (115-025-166, Jackson ImmunoResearch Laboratories), or Alexa Fluor 647 (A21235, Molecular Probes, Eugene, OR). Goat anti-rabbit-IgG antibodies conjugated to fluorescein (111-095-144) and Dylight 649 (111-025-144) were purchased from Jackson ImmunoResearch Laboratories. A Rhodamine-conjugated goat anti-rat-IgG antibody (112-025-102) was purchased from Jackson ImmunoResearch Laboratories. Confocal images of fixed samples were acquired at room temperature with a TCS SP5 microscope, equipped with a 63×1.4 NA objective, and running LASAF 2.4.1 build 6384 software (Leica Microsystems, Buffalo Grove, IL, USA). For each experiment, the settings (laser intensity, gain etc.) were identical across samples.
Confocal colocalization analyses
To determine the propensity of vimentin to associate with β4 integrin in confocal images, we compared the association of β4 integrin with vimentin to the association observed when β4 integrin puncta near the cell edge were randomly redistributed. Similar methods have been described previously (Fletcher et al., 2010). The advantage of this method over more traditional colocalization analyses is that it accounts for the possibility that observed associations occur only as often as would be expected by chance. We first segmented β4 integrin puncta and vimentin filaments in images of dual-labeled samples by using Fiji software (Schindelin et al., 2012). Specifically, this was accomplished by a combination of background subtraction, local contrast enhancement (CLAHE; S. Saalfeld, 2009; http://rsbweb.nih.gov/ij/plugins/clahe/index.html, date of access: 30 June 2017) and thresholding, the parameters for which were adjusted on an image-by-image basis to minimize segmentation error. The regions near the edge of each cell in which filaments were sufficiently sparse so as to be distinguishable from each other were selected. All puncta inside this region were selected and randomly assigned to new positions inside the same region. This was repeated 1000 times on each of 30 real images. We then determined for each punctum whether any pixels were shared with vimentin. Finally, we determined the relative frequencies at which puncta associated with vimentin in the real and simulated images. The resulting ratios were log2-transformed for visualization purposes.
We similarly analyzed β4 integrin–actin filament colocalization in images dual labeled for β4 integrin and actin. Actin filaments were segmented using a combination of background subtraction, local contrast enhancement (CLAHE as above) and ridge detection (Steger, 1998; T. Wagner, 2017, https://imagej.net/Ridge_Detection, date of access: 24 October 2017) on an image-by-image basis. The locations of the tips of actin filaments were identified from such images using the 2D skeleton analysis feature in Fiji (Schindelin et al., 2012). All subsequent processing was performed as described for vimentin.
iPALM
At 24 h after plating onto gold bead-coated coverslips (Shtengel et al., 2009), cells were transfected with mEos3.2–vimentin. After 48 h, cells were immunostained for β4 integrin using a mouse monoclonal antibody (1:100, 450-11A, BD Biosciences) and an Alexa Fluor 647-conjugated secondary antibody. The microscope set up is described in detail elsewhere (Shtengel et al., 2009). Briefly, two Nikon CFI Apochromat TIRF 60×1.49 NA objectives were situated in conjunction with three Andor iXon DU-897 BI EMCCD cameras (Andor Technology, Belfast, UK) in order to perform interferometry. A total of 20,000 images were captured for the β4 integrin channel and 50,000 images were captured for the vimentin channel. Three-dimensional localizations were calculated using methods described elsewhere and the computing resources available at the Janelia research campus (Shtengel et al., 2009; Kanchanawong et al., 2010). Since our interest was primarily in β4 integrin puncta in lamellipodia, we exported the x, y and z (unwrapped group z) coordinates of localizations in these regions to text files. β4 integrin localizations were grouped into puncta using a Euclidean single-linkage clustering approach with a maximum linking distance of 15 nm. This was implemented using a custom R package, Bioi v0.1.0 (github.com/zcolburn/Bioi). Clusters with very low localization frequency (those composed of fewer than 15 localizations) were excluded as noise.
We estimated the distance separating β4 integrin puncta and vimentin filaments by comparing the real distances observed with distances measured in simulated iPALM data. Simulated iPALM data was generated by randomly assigning puncta to new positions. Positions in the lateral axes were drawn from uniform distributions, whereas positions in the z-axis were drawn from the real z-axis distribution of punctum centroids. For each randomly generated centroid, localizations were generated corresponding to the localizations of a real punctum. Thus, each randomly generated punctum corresponded to a spatially translated real punctum. For both real and simulated puncta, the distance to the nearest vimentin localization was calculated.
TIRF-SIM and conventional TIRF
At 24 h after plating onto coverslips, cells were transfected with mCherry–vimentin. Control cells were also infected with adenovirus encoding GFP–β4. At 48 h post transfection, cells, maintained at 37°C and 5% CO2 (H301 stage-top incubation chamber, okolab, Naples, Italy), were imaged every 15 s for 5 min with a TIRF-SIM microscope equipped with an Olympus Plan-Apochromat 100×1.49 NA objective (Nikon Inc., Melville, NY) at the Advanced Imaging Center (HHMI Janelia, Ashburn, VA). Excitation patterns were generated with a phase-only spatial light modulator (BVO AHWP3, Bolder Vision Optik, Boulder, CO, USA) and using a mask system only the ±1 diffraction orders were focused onto the back focal plane of the objective. By using a liquid crystal variable retarder (LC, Meadowlark, SWIFT) and wave plates, the polarity of the light was rotated to match the excitation pattern angle. Emitted light was captured using interference filters and two sCMOS cameras (Hamamatsu, Orca Flash 4.0 v2 sCMOS). These images were used to reconstruct high-resolution images by taking into account their interference patterns as described elsewhere (Gustafsson et al., 2008). The Wiener filter used during reconstruction was selected on an image-by-image basis according to the guidelines outlined by Demmerle et al. (2017). β4 integrin puncta were tracked using the TrackMate plugin (http://fiji.sc/TrackMate) (Tinevez et al., 2017) for Fiji (Schindelin et al., 2012). Vimentin filaments were segmented by performing thresholding following background subtraction as described for confocal image analysis.
Cells treated with control or vimentin siRNA and expressing GFP–β4 were imaged every 15 s for 5 min with a Leica SD6000 AF spinning disc confocal microscope (Leica Microsystems) equipped with a 100×1.47 NA objective and an iXon3 Andor DU-897E-C00-#BV camera (Andor Technology). Puncta were identified and linked between consecutive frames using the TrackMate plugin for Fiji as described above.
For Rac1–integrin association studies, A549 cells plated on glass coverslips were transfected with control, vimentin or plectin siRNA. After 24 h, they were infected with an adenovirus encoding GFP-tagged WT Rac1 (GFP–Rac). Finally, following 48 h of incubation they were immunostained for α6 integrin. Samples were imaged on a Leica SD6000 AF Spinning Disc Confocal microscope using total internal reflection microscopy as described above.
To evaluate effects of vimentin and plectin knockdown on the association of GFP–Rac with puncta, α6 integrin puncta were segmented, and their GFP–Rac intensities compared to the average at the basal surface. Segmentation was performed as described for the confocal colocalization analyses. To compare the association of GFP–Rac with puncta, the average intensity of GFP–Rac at each punctum relative to the average intensity of GFP–Rac at the basal surface was calculated and normalized to the Ctrl-siRNA condition.
Western blotting
Western blotting was performed as described elsewhere (Hiroyasu et al., 2016). Briefly, sample buffer (12 M urea, 1% SDS, 10% glycerol, 1 M Tris-HCl) was supplemented with Halt protease inhibitor cocktail (87786, Thermo Scientific, Rockford, IL) and 10% β-mercaptoethanol immediately before solubilizing cells. Extracts of cells were briefly sonicated and then processed for SDS-PAGE. Separated proteins were transferred to nitrocellulose membranes and subsequently blocked with 5% non-fat milk in 0.05% Tween-20 in PBS at room temperature for 1 h. Mouse monoclonal antibodies against β4 integrin (450-11A) and vimentin (V9) were purchased from BD Biosciences and Sigma-Aldrich, respectively. A rabbit monoclonal antibody against plectin (ab32528) was purchased from Abcam. A mouse monoclonal antibody against GAPDH (MAB374) was purchased from EMD Millipore (Burlington, MA). A polyclonal rabbit antibody against lamin A and C (2032) was purchased from Cell Signaling Technology. Primary antibodies, diluted 1:1000, were incubated overnight at 4°C, while the peroxidase-conjugated secondary antibodies against mouse IgG (115-035-146, Jackson ImmunoResearch Laboratories) and rabbit IgG (7074S, Cell Signaling Technology) were incubated for 1 h at room temperature. Immnoblots were developed using Pierce ECL Western blotting substrate (32106, Thermo Scientific) with a MyECL Imager (Thermo Scientific).
Single-cell motility assays
Cells maintained at 37°C in a heated chamber (05-11-0035, Pathology Devices, Westminster, MD) on a DMi8 conventional microscope (Leica Microsystems) were viewed with a 5×0.12 NA objective and imaged every 5 min for 2 h with a DFC365 FX FCAM2 camera using version 1.0.0.12269 of the LAS X image acquisition suite (Leica Microsystems). Following acquisition, cells were tracked using image cross-correlation velocimetry implemented in MetaMorph software (version 7.8.4.0, Molecular Devices, Sunnyvale, CA). These data were used to calculate speed and processivity, a metric of directed migration. Processivity was defined as the maximum displacement from the origin of a cell divided by the path length. For retrospective motility assays, time-lapse microscopy was performed as described above on A549 cells cultured on etched grid coverslips (1916-92525, Bellco Glass, Vineland, NJ, USA). Immediately after imaging, cells were fixed and immunostained for vimentin. Cells in stained images were matched to the same cells in the videos.
Rac1 activity assay
A Rac1 G-LISA kit (Cytoskeleton Inc., Denver, CO) was used to assay Rac1 activity in extracts of cultured cells (Colburn and Jones, 2017). Extracts of cells transfected with siRNA were made 72 h after transfection.
Additional methods
In all immunostaining experiments, primary antibodies were diluted 1:100. A549 cells were imaged following immunostaining for β4 integrin, vimentin, and keratin, then imaged using confocal microscopy with a Leica TCS SP5 confocal microscope (Fig. S1A). Vimentin was labeled with a Cy3-conjugated primary antibody (C9080) purchased from Sigma-Aldrich. Similarly, 48 h after treatment with control or vimentin-targeted siRNA, A549 cells were immunostained for β4 integrin and keratin (Fig. S1B). Confocal microscopy with a Leica TCS SP5 microscope was performed on A549 cells plated onto coverslips coated with collagen I or laminin-332 rich conditioned medium and, 6 h later, triple-labeled for β4 integrin, vimentin and actin (Fig. S2A,B). Such images were quantified (Fig. S2C,D) as in Fig. 1. TIRF images of cells expressing GFP-tagged β4 integrin and immunostained for plectin were captured at room temperature with a Leica SD6000 AF Spinning disc confocal microscope (Fig. S3A). The acquisition software used was MetaMorph 7.8.4.0 (Molecular Devices). Plectin–β4 integrin colocalization was quantified using Spearman correlation for pixels comprising GFP–β4 puncta (Fig. S3B). The lack of focal adhesion–vimentin association was demonstrated by performing confocal microscopy with a Leica TCS SP5 microscope on A549 cells immunostained for paxillin and vimentin (Fig. S4). To determine whether matrix composition or serum affects the loss of vimentin filaments from the cell edge of β4KD cells, WT and β4KD A549 cells were plated onto uncoated coverslips or coverslips coated with either collagen I or laminin-332 in medium containing or lacking serum. Such samples were then imaged using a TCS-SP5 confocal microscope (Fig. S5). To determine the effect of the microtubule cytoskeleton on vimentin, A549 cells were treated with DMSO or 10 µM nocodazole (M1404, Sigma-Aldrich) for 1 h then immunostained and imaged using a TCS-SP5 confocal microscope (Fig. S6). Following time-lapse observation of single cells on etched grid coverslips, the preparations were fixed and immunostained for vimentin (Fig. S7).
Statistics
Statistical analyses were undertaken using data derived from a minimum of three independent experiments (with the exception of the Rac1 activity assays, which were performed twice). Statistical methodologies are indicated in the corresponding legends. For tests requiring normally distributed data, the assumption of normality was evaluated visually and by performing the Shapiro–Wilk test with α=0.05.
Supplementary Material
Acknowledgements
Imaging data used in this publication was produced in collaboration with the Advanced Imaging Center, a facility jointly supported by the Gordon and Betty Moore Foundation and HHMI at HHMI's Janelia Research Campus. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no competing financial interests.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.C.R.J.; Methodology: Z.T.C., J.C.R.J.; Software: Z.T.C.; Formal analysis: Z.T.C.; Investigation: Z.T.C.; Data curation: Z.T.C.; Writing - original draft: Z.T.C.; Writing - review & editing: J.C.R.J.; Visualization: Z.T.C.; Supervision: J.C.R.J.; Project administration: J.C.R.J.; Funding acquisition: J.C.R.J.
Funding
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (National Institutes of Health) under award number RO1 AR AR054184. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.214593.supplemental
References
- Al-Saad S., Al-Shibli K., Donnem T., Persson M., Bremnes R. M. and Busund L.-T. (2008). The prognostic impact of NF-κB p105, vimentin, E-cadherin and Par6 expression in epithelial and stromal compartment in non-small-cell lung cancer. Br. J. Cancer 99, 1476-1483. 10.1038/sj.bjc.6604713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. E., Skalli O., Goldman R. D. and Jones J. C. R. (1997). Laminin-5 and modulation of keratin cytoskeleton arrangement in FG pancreatic carcinoma cells: involvement of IFAP300 and evidence that laminin-5/cell interactions correlate with a dephosphorylation of α6A integrin. Cell Motil. Cytoskeleton 37, 271-286. [DOI] [PubMed] [Google Scholar]
- Bhattacharya R., Gonzalez A. M., DeBiase P. J., Trejo H. E., Goldman R. D., Flitney F. W. and Jones J. C. R. (2009). Recruitment of vimentin to the cell surface by β3 integrin and plectin mediates adhesion strength. J. Cell Sci. 122, 1390-1400. 10.1242/jcs.043042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañón M. J., Walko G., Winter L. and Wiche G. (2013). Plectin-intermediate filament partnership in skin, skeletal muscle, and peripheral nerve. Histochem. Cell Biol. 140, 33-53. 10.1007/s00418-013-1102-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw S. Y., Abdul Majeed A., Dalley A. J., Chan A., Stein S. and Farah C. S. (2012). Epithelial to mesenchymal transition (EMT) biomarkers - E-cadherin, beta-catenin, APC and vimentin - in oral squamous cell carcinogenesis and transformation. Oral Oncol. 48, 997-1006. 10.1016/j.oraloncology.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Colburn Z. T. and Jones J. C. R. (2017). α6β4 integrin regulates the collective migration of epithelial cells. Am. J. Respir. Cell Mol. Biol. 56, 443-452. 10.1165/rcmb.2016-0313OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cy L., Hh L., Mj T. and Yk W. (2015). Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget 6, 15966-15983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky C. H., Fitzgerald M. L. and Fisher S. J. (1992). Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J. Clin. Invest. 89, 210-222. 10.1172/JCI115565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pereda J. M., Ortega E., Alonso-García N., Gómez-Hernández M. and Sonnenberg A. (2009). Advances and perspectives of the architecture of hemidesmosomes. Cell Adhes. Migr. 3, 361-364. 10.4161/cam.3.4.9525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmerle J., Innocent C., North A. J., Ball G., Müller M., Miron E., Matsuda A., Dobbie I. M., Markaki Y. and Schermelleh L. (2017). Strategic and practical guidelines for successful structured illumination microscopy. Nat. Protoc. 12, 988-1010. 10.1038/nprot.2017.019 [DOI] [PubMed] [Google Scholar]
- Dmello C., Sawant S., Alam H., Gangadaran P., Tiwari R., Dongre H., Rana N., Barve S., Costea D. E., Chaukar D. et al. (2015). Vimentin-mediated regulation of cell motility through modulation of beta4 integrin protein levels in oral tumor derived cells. Int. J. Biochem. Cell Biol. 70, 161-172. 10.1016/j.biocel.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Eckes B., Colucci-Guyon E., Smola H., Nodder S., Babinet C., Krieg T. and Martin P. (2000). Impaired wound healing in embryonic and adult mice lacking vimentin. J. Cell Sci. 113, 2455-2462. [DOI] [PubMed] [Google Scholar]
- Fletcher P. A., Scriven D. R. L., Schulson M. N. and Moore E. D. W. (2010). Multi-image colocalization and its statistical significance. Biophys. J. 99, 1996-2005. 10.1016/j.bpj.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R. and Wiche G. (1987). Structure and hydrodynamic properties of plectin molecules. J. Mol. Biol. 198, 515-531. 10.1016/0022-2836(87)90297-X [DOI] [PubMed] [Google Scholar]
- Foisner R., Leichtfried F. E., Herrmann H., Small J. V., Lawson D. and Wiche G. (1988). Cytoskeleton-associated plectin: in situ localization, in vitro reconstitution, and binding to immobilized intermediate filament proteins. J. Cell Biol. 106, 723-733. 10.1083/jcb.106.3.723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Z., Ding L., Burckhardt C. J., Lowery J., Zaritsky A., Sitterley K., Mota A., Costigliola N., Starker C. G., Voytas D. F. et al. (2016). Vimentin intermediate filaments template microtubule networks to enhance persistence in cell polarity and directed migration. Cell Syst. 3, 252-263.e8. 10.1016/j.cels.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts D., Fontao L., Nievers M. G., Schaapveld R. Q. J., Purkis P. E., Wheeler G. N., Lane E. B., Leigh I. M. and Sonnenberg A. (1999). Binding of integrin α6β4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J. Cell Biol. 147, 417-434. 10.1083/jcb.147.2.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuijen C. A. W. and Sonnenberg A. (2002). Dynamics of the α6β4 integrin in keratinocytes. Mol. Biol. Cell 13, 3845-3858. 10.1091/mbc.02-01-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M. G. L., Shao L., Carlton P. M., Wang C. J. R., Golubovskaya I. N., Cande W. Z., Agard D. A. and Sedat J. W. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957-4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harburger D. S. and Calderwood D. A. (2009). Integrin signalling at a glance. J. Cell Sci. 122, 159-163. 10.1242/jcs.018093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel L. S., Kline E. R., Salgueiro A. M. and Marcus A. I. (2014). Vimentin regulates lung cancer cell adhesion through a VAV2-Rac1 pathway to control focal adhesion kinase activity. Oncogene 34, 1979-1990. 10.1038/onc.2014.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand B. T., Mendez M. G., Murthy S. N. P., Shumaker D. K., Grin B., Mahammad S., Aebi U., Wedig T., Wu Y. I., Hahn K. M. et al. (2011). Vimentin organization modulates the formation of lamellipodia. Mol. Biol. Cell 22, 1274-1289. 10.1091/mbc.e10-08-0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieda Y., Nishizawa Y., Uematsu J. and Owaribe K. (1992). Identification of a new hemidesmosomal protein, HD1: a major, high molecular mass component of isolated hemidesmosomes. J. Cell Biol. 116, 1497-1506. 10.1083/jcb.116.6.1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroyasu S., Colburn Z. T. and Jones J. C. R. (2016). A hemidesmosomal protein regulates actin dynamics and traction forces in motile keratinocytes. FASEB J. 30, 2298-2310. 10.1096/fj.201500160R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan S. M., Mercurio A. M. and LaFlamme S. E. (1998). Endothelial cells assemble two distinct α6β4-containing vimentin-associated structures: roles for ligand binding and the β4 cytoplasmic tail. J. Cell Sci. 111, 2717-2728. [DOI] [PubMed] [Google Scholar]
- Homan S. M., Martinez R., Benware A. and LaFlamme S. E. (2002). Regulation of the association of α6β4 with vimentin intermediate filaments in endothelial cells. Exp. Cell Res. 281, 107-114. 10.1006/excr.2002.5643 [DOI] [PubMed] [Google Scholar]
- Hong L., Sun H., Lv X., Yang D., Zhang J. and Shi Y. (2009). Expression of periostin in the serum of NSCLC and its function on proliferation and migration of human lung adenocarcinoma cell line (A549) in vitro. Mol. Biol. Rep. 37, 2285-2293. 10.1007/s11033-009-9721-1 [DOI] [PubMed] [Google Scholar]
- Hopkinson S. B., Hamill K. J., Wu Y., Eisenberg J. L., Hiroyasu S. and Jones J. C. R. (2014). Focal contact and hemidesmosomal proteins in keratinocyte migration and wound repair. Adv. Wound Care 3, 247-263. 10.1089/wound.2013.0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-L., Wu C.-Y., Hung J.-Y., Lin Y.-S., Huang M.-S. and Kuo P.-L. (2013). Galectin-1 promotes lung cancer tumor metastasis by potentiating integrin α6β4 and Notch1/Jagged2 signaling pathway. Carcinogenesis 34, 1370-1381. 10.1093/carcin/bgt040 [DOI] [PubMed] [Google Scholar]
- Humphries J. D., Byron A. and Humphries M. J. (2006). Integrin ligands at a glance. J. Cell Sci. 119, 3901-3903. 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyder C. L., Kemppainen K., Isoniemi K. O., Imanishi S. Y., Goto H., Inagaki M., Fazeli E., Eriksson J. E. and Törnquist K. (2015). Sphingolipids inhibit vimentin-dependent cell migration. J. Cell Sci. 128, 2057-2069. 10.1242/jcs.160341 [DOI] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F. and Waterman C. M. (2010). Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580-584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S., Mizutani K., Miyata M., Ikeda W. and Takai Y. (2010). Interaction of integrin α6β4 with ErbB3 and implication in heregulin-induced ErbB3/ErbB2-mediated DNA synthesis. Genes Cells 15, 995-1001. 10.1111/j.1365-2443.2010.01438.x [DOI] [PubMed] [Google Scholar]
- Kligys K. R., Wu Y., Hopkinson S. B., Kaur S., Platanias L. C. and Jones J. C. R. (2012). α6β4 integrin, a master regulator of expression of integrins in human keratinocytes. J. Biol. Chem. 287, 17975-17984. 10.1074/jbc.M111.310458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhofer M., Hopkinson S. B. and Jones J. C. (1993). The matrix secreted by 804G cells contains laminin-related components that participate in hemidesmosome assembly in vitro. J. Cell Sci. 105, 753-764. [DOI] [PubMed] [Google Scholar]
- Lanier M. H., Kim T. and Cooper J. A. (2015). CARMIL2 is a novel molecular connection between vimentin and actin essential for cell migration and invadopodia formation. Mol. Biol. Cell 26, 4577-4588. 10.1091/mbc.e15-08-0552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng C., Zhang Z., Chen W., Luo H., Song J., Dong W., Zhu X., Chen X., Liang H. and Zhang B. (2016). An Integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK-AKT pathway. Cancer Lett. 376, 188-196. 10.1016/j.canlet.2016.03.023 [DOI] [PubMed] [Google Scholar]
- Mainiero F., Pepe A., Yeon M., Ren Y. and Giancotti F. G. (1996). The intracellular functions of α6β4 integrin are regulated by EGF. J. Cell Biol. 134, 241-253. 10.1083/jcb.134.1.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez M. G., Kojima S.-I. and Goldman R. D. (2010). Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 24, 1838-1851. 10.1096/fj.09-151639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio A. M., Rabinovitz I. and Shaw L. M. (2001). The α6β4 integrin and epithelial cell migration. Curr. Opin. Cell Biol. 13, 541-545. 10.1016/S0955-0674(00)00249-0 [DOI] [PubMed] [Google Scholar]
- Miyazaki N., Iwasaki K. and Takagi J. (2018). A systematic survey of the conformational states in β1 and β4 integrins using negative-stain electron microscopy. J. Cell Sci. 131, jcs216754 10.1242/jcs.216754 [DOI] [PubMed] [Google Scholar]
- Nievers M. G., Kuikman I., Geerts D., Leigh I. M. and Sonnenberg A. (2000). Formation of hemidesmosome-like structures in the absence of ligand binding by the α6β4 integrin requires binding of HD1/plectin to the cytoplasmic domain of the β4 integrin subunit. J. Cell Sci. 113, 963-973. [DOI] [PubMed] [Google Scholar]
- Raymond K., Kreft M., Janssen H., Calafat J. and Sonnenberg A. (2005). Keratinocytes display normal proliferation, survival and differentiation in conditional β4-integrin knockout mice. J. Cell Sci. 118, 1045-1060. 10.1242/jcs.01689 [DOI] [PubMed] [Google Scholar]
- Sastry S. K. and Burridge K. (2000). Focal Adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp. Cell Res. 261, 25-36. 10.1006/excr.2000.5043 [DOI] [PubMed] [Google Scholar]
- Sawamura D., Nakano H. and Matsuzaki Y. (2010). Overview of epidermolysis bullosa. J. Dermatol. 37, 214-219. 10.1111/j.1346-8138.2009.00800.x [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal B. U., DeBiase P. J., Matzno S., Chew T.-L., Claiborne J. N., Hopkinson S. B., Russell A., Marinkovich M. P. and Jones J. C. R. (2006). Integrin β4 regulates migratory behavior of keratinocytes by determining laminin-332 organization. J. Biol. Chem. 281, 35487-35498. 10.1074/jbc.M606317200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann K., Roth W., Kröger C., Loschke F., Lederer M., Hüttelmaier S. and Magin T. M. (2013). Keratins mediate localization of hemidesmosomes and repress cell motility. J. Invest. Dermatol. 133, 181-190. 10.1038/jid.2012.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Patterson G. H. and Davidson M. W. (2007). Advances in fluorescent protein technology. J. Cell Sci. 120, 4247-4260. 10.1242/jcs.005801 [DOI] [PubMed] [Google Scholar]
- Shtengel G., Galbraith J. A., Galbraith C. G., Lippincott-Schwartz J., Gillette J. M., Manley S., Sougrat R., Waterman C. M., Kanchanawong P., Davidson M. W. et al. (2009). Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl. Acad. Sci. USA 106, 3125-3130. 10.1073/pnas.0813131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger C. (1998). An unbiased detector of curvilinear structures. IEEE Trans. Pattern Anal. Mach. Intell. 20, 113-125. 10.1109/34.659930 [DOI] [Google Scholar]
- Stewart R. L. and O'Connor K. L. (2015). Clinical significance of the integrin α6β4 in human malignancies. Lab. Invest. 95, 976-986. 10.1038/labinvest.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R. L., West D., Wang C., Weiss H. L., Gal T., Durbin E. B., O'Connor W., Chen M. and O'Connor K. L. (2016). Elevated integrin α6β4 expression is associated with venous invasion and decreased overall survival in non-small cell lung cancer. Hum. Pathol. 54, 174-183. 10.1016/j.humpath.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.-Y., Perry N., Schindelin J., Hoopes G. M., Reynolds G. D., Laplantine E., Bednarek S. Y., Shorte S. L. and Eliceiri K. W. (2017). TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80-90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Tsuruta D. and Jones J. C. R. (2003). The vimentin cytoskeleton regulates focal contact size and adhesion of endothelial cells subjected to shear stress. J. Cell Sci. 116, 4977-4984. 10.1242/jcs.00823 [DOI] [PubMed] [Google Scholar]
- Tsuruta D., Hopkinson S. B. and Jones J. C. R. (2003). Hemidesmosome protein dynamics in live epithelial cells. Cell Motil. Cytoskeleton 54, 122-134. 10.1002/cm.10089 [DOI] [PubMed] [Google Scholar]
- Vuoriluoto K., Haugen H., Kiviluoto S., Mpindi J.-P., Nevo J., Gjerdrum C., Tiron C., Lorens J. B. and Ivaska J. (2011). Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 30, 1436-1448. 10.1038/onc.2010.509 [DOI] [PubMed] [Google Scholar]
- Walko G., Vukasinovic N., Gross K., Fischer I., Sibitz S., Fuchs P., Reipert S., Jungwirth U., Berger W., Salzer U. et al. (2011). Targeted proteolysis of plectin isoform 1a accounts for hemidesmosome dysfunction in mice mimicking the dominant skin blistering disease EBS-Ogna. PLoS Genet. 7, e1002396 10.1371/journal.pgen.1002396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche G. (1998). Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 111, 2477-2486. [DOI] [PubMed] [Google Scholar]
- Wiche G., Osmanagic-Myers S. and Castañón M. J. (2015). Networking and anchoring through plectin: a key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 32, 21-29. 10.1016/j.ceb.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Windoffer R., Kölsch A., Wöll S. and Leube R. E. (2006). Focal adhesions are hotspots for keratin filament precursor formation. J. Cell Biol. 173, 341-348. 10.1083/jcb.200511124 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.