Abstract
BACKGROUND: COP9 signalosome subunit 2 (CSN2) is believed to be involved in human cancer, but its prognostic significance in colorectal cancer (CRC) has not been elucidated. PATIENTS AND METHODS: We retrospectively analyzed the expression of CSN2 andCD8+ tumor-infiltrating lymphocytes (TILs), and mismatch repair (MMR) status in 267 paraffin-embedded specimens using immunohistochemistry in a training cohort. A number of risk factors were used to form nomograms to evaluate survival, and Harrell's concordance index (C-index) was used to evaluate the predictive accuracy. Further validation was performed in an independent cohort of 238cases. RESULTS: Low CSN2 expression and a low number of CD8 + TILs were significantly associated with diminished disease-free survival (DFS) and overall survival (OS) in CRC patients, and patients with MMR-deficient CRC had enhanced DFS and OS. Moreover, the multivariate Cox analysis identified CSN2, CD8 + TILs, and MMR status as independent prognostic factors for DFS and OS. Using these three markers and four clinicopathological risk variables, two nomograms were constructed and validated for predicting DFS and OS (C-index: training cohort, 0.836 (95% CI:0.804–0.868) and 0.841 (0.808–0.874), respectively; validation cohort, 0.801 (0.760–843) and 0.843 (0.806–0.881), respectively). CONCLUSIONS: CSN2, CD8+ TILs, and MMR status were independent prognostic factors. The nomograms could be used to generate individualized predictions for DFS and OS.
Introduction
Colorectal cancer (CRC) continues to be one of the most common causes of cancer death worldwide [1]. Although prognosis and the need for postoperative treatment are presently determined by pathological staging, obvious heterogeneity in outcomes exists among patients with a similar disease stage [2], [3], [4]. Indeed, other tumor-associated characteristics, intrinsic to the tumor cell and pertaining to both the tumor microenvironment and the patient, may similarly influence outcome and be used to determine the need for further treatment [5], [6].
COP9 signalosome (CSN), composed of eight subunits (CSN1 to CSN8), is a regulator of the Skp1-cullin-F-box protein ubiquitin ligases in the ubiquitin-proteasome system [7]. Many tumor suppressor and oncogene products are regulated by ubiquitination- and proteasome-mediated protein degradation. Therefore, it is conceivable that CSN plays a significant role in cancer, regulating processes relevant to carcinogenesis and cancer progression (e.g., cell cycle control, signal transduction and apoptosis) [8], [9]. The role of CSN subunits in cancer biology is emerging [9]. Recent evidence implicates CSN5 and CSN6 in the ubiquitin-mediated proteolysis of important mediators in carcinogenesis and cancer progression [8], [10]. CSN2 and its variant, Alien, can be transcriptional corepressors, which could facilitate pluripotency maintenance [11], [12]. Serum CSN2 may serve as a noninvasive diagnostic biomarker for gastric cancer, and low serum CSN2 is associated with unfavorable survival [13]. CSN2 may be a tumor suppressor gene and has reduced expression in tumor tissues [13], [14]. However, the molecular and prognostic functions of CSN2 in CRC are not yet known.
Extensive studies in CRC have suggested that the density of tumor-infiltrating lymphocytes (TILs) strongly influences clinical outcome [15], [16], [17]. A high intratumoral density of CD3+ and cytotoxic CD8+ lymphocytes has been associated with a reduced incidence of tumor metastasis and favorable prognosis [17]. Among TILs, CD8+ T cells, also known as cytotoxic T cells, play a critical role in antitumor immunity by killing cancer cells. An immunoscore of colon cancer, derived from a measure of CD3+ and CD8+ T cell densities in the invasive margin and center of tumor, had a larger relative prognostic value than pT stage, pN stage, lympho-vascular invasion, tumor differentiation, and microsatellite instability (MSI) status [18]. Moreover, the density of CD8+ TILs is associated with chemotherapy efficacy and prognosis of CRC, particularly if combined with microsatellite status [19]. The extent of T lymphocyte infiltration in human CRC differs based on mismatch repair (MMR) status [20].
In this study, we investigated CSN2 expression in patients with CRC by IHC and explored its associations with clinicopathological factors and prognosis. We also investigated the prognostic value of CD8+ TILs and MMR status and the interrelationship of CSN2, CD8+ TILs and MMR status. Moreover, we generated two predictive nomograms integrating CSN2 expression, CD8+ TILs, MMR status, the level of CEA, tumor depth, lymph node metastasis and distant metastasis to assess the risk score for disease-free survival (DFS) and overall survival (OS) of CRC patients.
Methods and Materials
Patients and Tissue Specimens
We used 505 formalin-fixed paraffin-embedded (FFPE) specimens from 505CRC patients in this study. For the training cohort, data were obtained from 267 patients with incident, primary, biopsy-confirmed CRC diagnosed between June 2006 and September 2007 at the first affiliated Hospital of Bengbu Medical University, Bengbu, China. Inclusion criteria were the availability of hematoxylin and eosin slides with invasive tumor components, the availability of follow-up data and clinicopathological characteristics, no history of treated cancer, and appropriate patient informed consent. We excluded patients if they had received preoperative treatment with any anticancer therapy. We also included an additional 238 patients as the internal validation cohort, with the same criteria as above, between October 2008 and December 2009. And all these patients with metastatic CRC included in this study had undergone radical resection of both the primary and metastatic sites. TNM staging was reclassified according to the AJCC staging manual (eighth edition). All participants were Han Chinese (self-reported). Two independent pathologists reassessed all samples. The institutional review board of the first affiliated Hospital of Bengbu Medical University approved the retrospective analysis of the anonymous data.
Antibodies
This study used the following commercially available monoclonal antibodies: an anti-human COPS2 antibody (clone NBP1–90190 Novus Biologicals, Canada) and a CD8 antibody (clone SP16; NeoMarker, USA). Staining for MMR proteins was performed using the following primary antibodies: mouse anti-human MLH-1 (cloneG168–728; Cell Marque Corporation, Rocklin, CA);mouse anti-human MSH-2 (clone G219–1129; Cell Marque Corporation, Rocklin, CA); mouse anti-humanMSH-6 (clone BC/44; Biocare, Concord, CA); and rabbit anti-human PMS2 (clone EPR3947; Cell Marque Corporation, Rocklin, CA).
IHC Staining
FFPE samples were processed for IHC as previously described [21], [22], [23]. Sections were cut at a thickness of 4 μm, de-waxed in xylene, and rehydrated in decreasing concentrations of ethanol. Prior to staining, the sections were subjected to endogenous peroxidase blocking in a 1% H2O2 solution in methanol for 10 min and then heated in a microwave for 30 min in 10 mM citrate buffer, pH 6.0. Serum blocking was performed using 10% normal rabbit serum for 30 min. The slides were incubated with the primary antibody (anti-CSN2 was used at a 1:100 dilution; anti-CD8 at 1:200; anti-MLH1 at 1:200, anti-MSH2 at 1:200, anti-PMS2 at 1:200 and anti-MSH6 at 1:200) overnight at 4°C and then incubated with a labeled polymer/HRP amplification system (EnVision™, DakoCytomation, Denmark) for 30 min. To visualize the sites of bound peroxidase, 0.05% 3,3′-diaminobenzidine tetrahydrochloride (DAB) was used prior to counterstaining with modified Harris hematoxylin.
Evaluation of IHC Staining
Two pathologists who were unaware of the clinical parameters or outcomes for each patient independently reviewed the IHC-stained sections. For the scoring of all molecules, 10 fields in the tumor region were randomly selected and examined with high-power magnification. All discrepancies were resolved by a joint review of the slides in question.
For CSN2 expression, the staining of each specimen was evaluated using a semiquantitative H score, which was calculated by multiplying the result of a 4-stepscale (0, negative; 0.5, weak staining; 1, moderate staining; 1.5, strong staining) and the fraction of positive-stained cells from 0 to 100%, and ranged from 0 to 150 [24]. For CSN2, the H score was dichotomized at the median and categorized as high vs. low.
To evaluate the density of stained CD8-positive cells, the nucleated stained cells in each area were quantified and expressed as the number of cells per field at 200× magnification, as previously described [21], [22]. The CD8-positive cell density was dichotomized at the median and categorized as high vs. low.
The loss of MMR protein was defined as the absence of nuclear staining of tumor cells in the presence of positive nuclear staining in normal epithelial cells and lymphocytes. Tumors showing a complete loss of expression of at least one MMR protein (MLH1, MSH2, MSH6, or PMS2) were classified as dMMR, and tumors without a loss of MMR protein expression were classified as MMR-proficient (pMMR).
Construction of the Nomograms
In the training cohort, survival curves for different variable values were generated using the Kaplan–Meier estimates and compared using the log-rank test. Variables that achieved significance at P < .05 were entered into the multivariable analyses via the Cox regression model. Statistical analyses to identify independent prognostic factors were conducted in SPSS 19.0 for Windows (SPSS, Chicago, IL). On the basis of the results of the multivariable analysis, two nomograms were formulated by R 3.2.3 (http://www.r-project.org) with the survival and rms package. Backward step-wise selection was applied using the likelihood ratio test with Akaike's information criterion as the stopping rule [25].
Validation and Calibration of the Nomograms
The performance of the developed nomograms was tested in the validation cohort. The model performance for predicting outcome was evaluated by calculating the concordance index (C-index) [26]. The value of the C-index ranges from 0.5 to 1.0, with 0.5 indicating a random chance, and 1.0 indicating a perfect ability to correctly discriminate the outcome with the model. The calibration of the nomogram for1-, 3-, and 5-year DFS and OS was performed by comparing the predicted survival with the observed survival after bias correction.
Clinical Use
Decision curve analysis was conducted to determine the clinical usefulness of the nomograms by quantifying the net benefits at different threshold probabilities [27], [28].
Risk Group Stratification Based on the Nomogram
Using X-tile [29], the composite scoring of the nomograms was divided into three risk groups that accurately discriminated patients with good, intermediate, and poor prognosis.
Statistical Analyses
We compared two groups using the t test for continuous variables and χ2 test for categorical variables. The DFS and OS were calculated in months from the date of surgery to the date of regional recurrence or distant metastasis (for DFS) and death or final clinical follow-up (for OS). The Kaplan–Meier method and log-rank test were used to estimate DFS and OS. A multivariate Cox proportional hazards regression analysis was performed for all variables found to be significant in a univariate analysis. All the other statistical tests were performed with R software (version 3.2.3) and SPSS software (version 19.0). Statistical significance was set at 0.05.
Results
Clinicopathologic Correlations
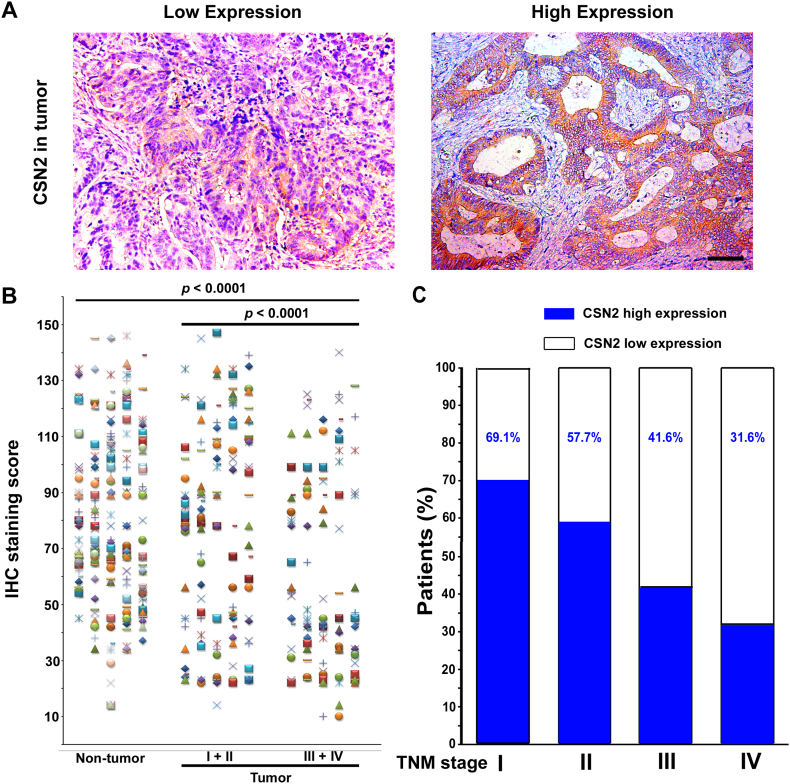
Tables 1 and S1–S2 list the clinical characteristics of the patients and the clinicopathologic correlations with CSN2, CD8, and MMR status in the training and validation cohorts. Specimens from 505 patients with CRC were obtained for this study. The specific expression of cytoplasmic CSN2 was observed, and the staining intensity was variable (Figure 1A). Compared with the non-tumoral CSN2 density in epithelial cells, intratumoral CSN2 expression in cancer cells was lower (P < .0001; Figure 1B). CSN2 expression was substantially lower in advanced stages [stages I–II (n = 150) vs. stages III–IV (n = 117), P < .0001]. Furthermore, the percentage of patients with high CSN2 expression reduced moderately with accompanying disease progression from TNM stage I to IV (Figure 1C, Table 1). Low CSN2 expression was associated with fewer CD8 + TILs, a higher N stage, and a higher TNM stage (Figure S1, Table 1). CD8+ TILs were associated with CSN2 expression, MMR status, depth of invasion, lymph node metastasis, distant metastasis, and TNM stage (Table S1). pMMR was associated with less CD8+ TIL infiltration, a higher T stage, a higher N stage, a higher M stage, and a higher TNM stage (Figure S1, Table S2).
Table 1.
Clinical Characteristics of Patients According to CSN2 in the Training and Validation Cohorts
| Variables | Training Cohort (n = 267) |
Validation Cohort (n =238) |
||||||
|---|---|---|---|---|---|---|---|---|
| N | Low CSN2 (%) | High CSN2(%) | P Value | N | Low CSN2 (%) | High CSN2 (%) | P Value | |
| Gender | 0.497 | 0.354 | ||||||
| Male | 166 | 80(48.2%) | 86(51.8%) | 143 | 75(52.4%) | 68(47.6%) | ||
| Female | 101 | 53(52.5%) | 48(47.5%) | 95 | 44(46.3%) | 51(53.7%) | ||
| Age(years) | 0.425 | 0.697 | ||||||
| <60 | 138 | 72(52.2%) | 66(47.8%) | 117 | 60(51.3%) | 57(48.7%) | ||
| ≧60 | 129 | 61(47.3%) | 68(52.7%) | 121 | 59(48.8%) | 62(51.2%) | ||
| Tumor location | 0.299 | 0.357 | ||||||
| Colon | 120 | 64(53.3%) | 56(46.7%) | 99 | 53(53.5%) | 46(46.5%) | ||
| Rectum | 147 | 69(46.9%) | 78(53.1%) | 139 | 66(47.5%) | 73(52.5%) | ||
| Differentiation status | 0.99 | 0.164 | ||||||
| Well | 66 | 33(50%) | 33(50%) | 53 | 22(41.5%) | 31(58.5%) | ||
| Moderate | 160 | 80(50%) | 80(50%) | 137 | 68(49.6%) | 69(50.4%) | ||
| Poor and undifferentiated | 41 | 20(48.8%) | 21(51.2%) | 48 | 29(60.4%) | 19(39.6%) | ||
| CEA | 0.08 | 0.167 | ||||||
| Elevated | 85 | 49(57.6%) | 36(42.4%) | 78 | 44(56.4%) | 34(43.6%) | ||
| Normal | 182 | 84(46.2%) | 98(53.8%) | 160 | 75(46.9%) | 85(53.1%) | ||
| CA199 | 0.094 | 0.325 | ||||||
| Elevated | 44 | 27(61.4%) | 17(38.6%) | 46 | 26(56.5%) | 20(43.5%) | ||
| Nomal | 223 | 106(47.5%) | 117(52.5%) | 192 | 93(48.4%) | 99(51.6%) | ||
| Depth of invasion | 0.009 | 0.323 | ||||||
| T1 | 10 | 2(20%) | 8(80%) | 14 | 4(28.6%) | 10(71.4%) | ||
| T2 | 37 | 14(37.8%) | 23(62.2%) | 41 | 18(43.9%) | 23(56.1%) | ||
| T3 | 166 | 80(48.2%) | 86(51.8%) | 108 | 57(52.8%) | 51(47.2%) | ||
| T4a | 18 | 12(66.7%) | 6(33.3%) | 26 | 12(46.2%) | 14(53.8%) | ||
| T4b | 36 | 25(69.4%) | 11(30.6%) | 49 | 28(57.1%) | 21(42.9%) | ||
| Lymph node metastasis | 0.003 | 0.001 | ||||||
| N0 | 159 | 66(41.5%) | 93(58.5%) | 116 | 44(37.9%) | 72(62.1%) | ||
| N1 | 74 | 44(59.5%) | 30(40.5%) | 77 | 46(59.7%) | 31(40.3%) | ||
| N2 | 34 | 23(67.6%) | 11(32.4%) | 45 | 29(64.4%) | 16(35.6%) | ||
| Metastasis | 0.015 | 0.011 | ||||||
| M0 | 234 | 110(47.0%) | 124(53.0%) | 195 | 90(46.2%) | 105(53.8%) | ||
| M1 | 33 | 23(69.7%) | 10(30.3%) | 43 | 29(67.4%) | 14(32.6%) | ||
| TNM stage | 0.001 | 0.001 | ||||||
| I | 38 | 12(31.6%) | 26(68.4%) | 43 | 13(30.2%) | 30(69.8%) | ||
| II | 112 | 48(42.9%) | 64(57.1%) | 63 | 26(41.3%) | 37(58.7%) | ||
| III | 84 | 50(59.5%) | 34(40.5%) | 89 | 51(57.3%) | 38(42.7%) | ||
| IV | 33 | 23(69.7%) | 10(30.3%) | 43 | 29(67.4%) | 14(32.6%) | ||
| CD8+ cells/field (mean±S.D.) | 74.7±59.3 | 95.0±58.4 | 0.005 | 75.8±46.1 | 95.1±54.1 | 0.003 | ||
| CD8 | <0.001 | 0.028 | ||||||
| low | 133 | 81(60.9%) | 52(39.1%) | 119 | 68(57.1%) | 51(42.9%) | ||
| high | 134 | 52(38.8%) | 82(61.2%) | 119 | 51(42.9%) | 68(57.1%) | ||
| MMR | <0.001 | 0.23 | ||||||
| dMMR | 45 | 9(20%) | 36(80%) | 41 | 17(41.5%) | 24(58.5%) | ||
| pMMR | 222 | 124(55.9%) | 98(44.1%) | 197 | 102(51.8%) | 95(48.2%) | ||
Figure 1.
CSN2 expression in CRC tissues. (A) Representative IHC images ofCSN2expression in tumor tissue. (B) Scatter plots for IHC staining score in unpaired non-tumor tissue (n = 267) and tumor tissue (n = 267) from the training cohort. The P value was determined using the nonparametric Mann–Whitney test. (C) Percentage of patients with high intratumoral CSN2 expression increased moderately with disease progression from TNM stage I–IV (data from the training and validation cohort). Scale bar, 100 μm.
Prognostic Value of CSN2, CD8, and MMR Status
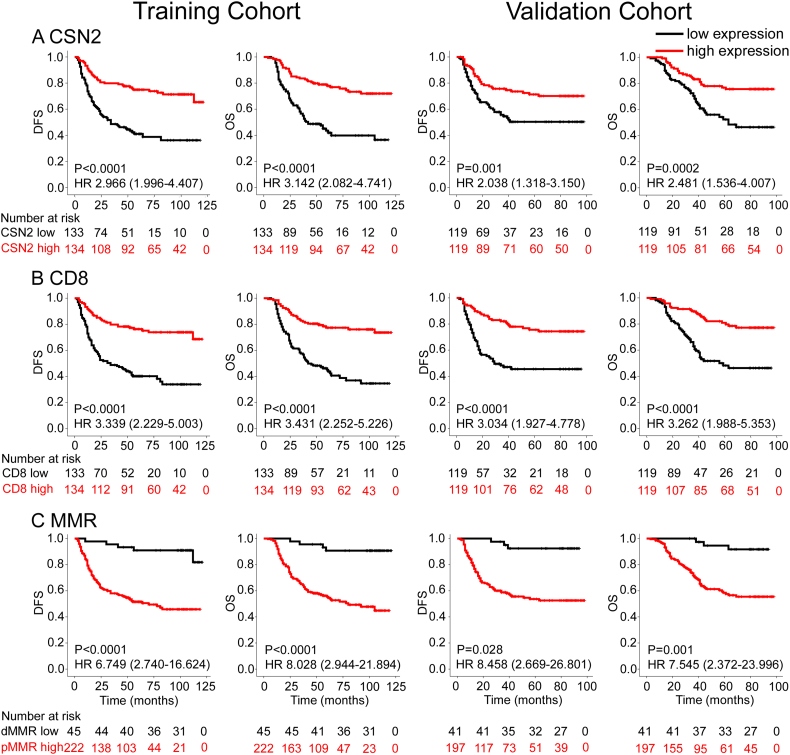
In the training cohort, patients with low expression levels of CSN2 and CD8 showed significantly unfavorable DFS and OS (Figure 2). Patients with dMMR CRC had better clinical outcomes than patients with pMMR carcinomas (Figure 2). Similar results were observed in the validation cohort. The clinicopathological parameters for the prediction of DFS and OS were further investigated by univariate analysis with the Cox regression model. In the univariate analysis, T stage, N stage, the level of CEA, the expression of CSN2, and CD8 and MMR status were significantly associated with DFS and OS (P < .05, Tables S3, S4). These significantly associated variables were used for the multivariate Cox regression model. In the DFS and OS models, CSN2, CD8 and MMR status remained powerful and independent prognostic factors for patients with CRC (Table 2). When stratified by clinicopathological risk factors, CSN2 remained a clinically and statistically significant prognostic marker (Figures S2, S3).
Figure 2.
Kaplan–Meier survival analysis of disease-free survival (DFS) and overall survival (OS) according to CSN2 expression (A), CD8 expression (B), and MMR status (C) of CRC patients in the training cohort and validation cohort. The left panel shows the results from the training cohort, and the right panel shows the results from the validation cohort.
Table 2.
Multivariable Cox Regression Analysis in the Training Cohort
| Variables | Overall Survival |
Disease-Free Survival |
||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | P Value | |
| CEA(ng/ml) (elevated vs. normal) | 2.070 (1.285-3.336) | 0.003 | 2.403 (1.531-3.771) | 0.001 |
| Depth of invasion | 0.093 | 0.031 | ||
| T4b | Reference | Reference | ||
| T1 | 0.001(0.0001-999) | 0.962 | 0.001(0.0001-999) | 0.959 |
| T2 | 0.343 (0.132-0.894) | 0.029 | 0.310 (0.125-0.769) | 0.012 |
| T3 | 0.463 (0.257-0.836) | 0.011 | 0.388 (0.214-0.702) | 0.002 |
| T4a | 0.859 (0.422-1.750) | 0.676 | 0.648 (0.325-1.290) | 0.217 |
| Lymph node metastasis | 0.012 | 0.012 | ||
| N0 | Reference | Reference | ||
| N1 | 1.736 (1.074-2.808) | 0.024 | 1.736 (1.074-2.808) | 0.024 |
| N2 | 2.287 (1.263-4.142) | 0.006 | 2.287 (1.263-4.142) | 0.006 |
| Metastasis (M1 vs. M0) | 2.739 (1.532-4.895) | 0.001 | 2.739 (1.532-4.895) | 0.001 |
| CSN2 (high vs. low) | 0.546 (0.354-0.842) | 0.006 | 0.624 (0.410-0.949) | 0.027 |
| CD8 (high vs. low) | 0.627 (0.396-0.993) | 0.047 | 0.574 (0.372-0.886) | 0.012 |
| MMR status (pMMR vs. dMMR) | 4.065 (1.415-11.677) | 0.009 | 3.580 (1.376-9.310) | 0.009 |
CEA: carcinoembryonic antigen.
Development and Validation of Nomograms for Predicting CRC Prognosis
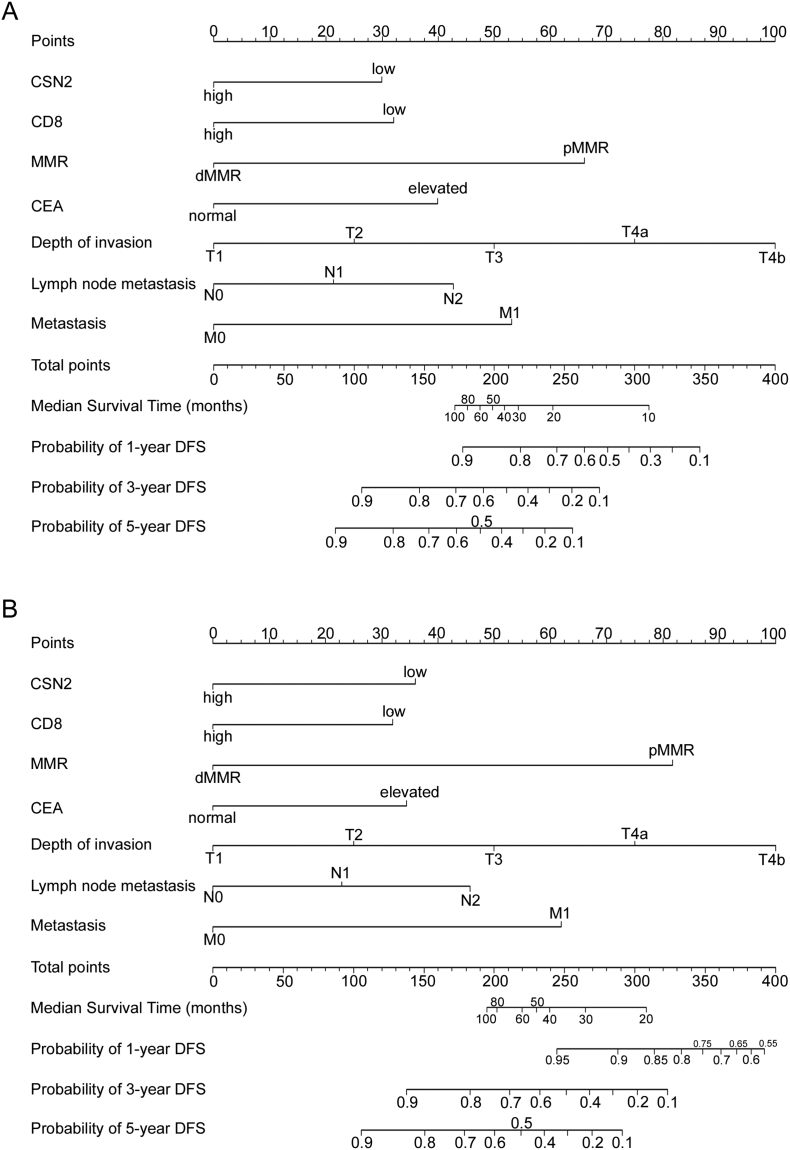
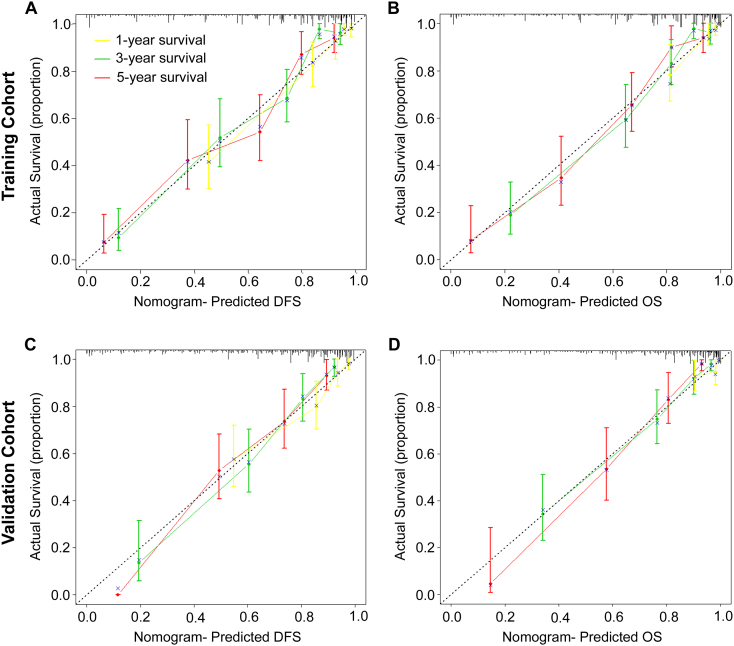
To predict DFS and OS for patients with CRC, two nomograms were established using the multivariate Cox regression model according to all significantly independent factors for DFS and OS (Figure 3, A and B). Nomograms can be interpreted by summing up the points assigned to each variable, which are indicated at the top of the scale. The total points can be converted to predicted 1-, 3-, and 5-year DFS and OS for a patient in the lowest scale [3]. In the training cohort, the C-indexes for the prediction of DFS and OS were 0.836 (95% CI: 0.804–0.868) and 0.841 (0.808–0.874), respectively. Calibration curves for the two nomograms (Figure 4, A and B) revealed no deviations from the reference line and no need for recalibration. In the validation cohort, the C-indexes for the prediction of DFS and OS were 0.801 (0.760–843) and 0.843 (0.806–0.881), respectively. The calibration curves yielded good agreement between the predicted and observed outcomes for 1-, 3-, and 5-year DFS and OS (Figure 4, C and D).
Figure 3.
Nomogram for predicting disease-free survival (DFS) and overall survival (OS): Locate the grade of the patient on the grade axis and then draw a straight line upward to the Points axis to determine how many points toward survival the patient receives for her/his grade. Repeat this process for the other axes, each time drawing a straight line upward toward the Points axis. Take the sum of the points received for each predictor and locate this sum on the Total Points axis. Draw a straight line down to the survival-probability axis to find the patient's probability of surviving colorectal cancer.
Figure 4.
Calibration curves for the nomogram. The calibration curve for predicting patient DFS (A, C) and OS (B, D) at 1-year, 3-year, and 5-year in the training and validation cohorts. Nomogram-predicted OS and DFS are plotted on the x-axis, and actual OS and DFS are plotted on the y-axis. The dotted line represents an ideal nomogram, and the solid blue line represents the current nomogram. The vertical bars are 95% CIs, and the ×s are bootstrap-corrected estimates.
Furthermore, we compared the discrimination of our nomograms with that of the AJCC TNM classification. The discrimination of our nomogram was superior to that of the AJCC TNM classification (AJCC classification C-index, training cohort: DFS 0.734 (95% CI 0.703–0.784), OS 0.748(0.705–0.791), both P < .001; validation cohort: DFS 0.730 (0.683–0.778), OS 0.785 (0.742–0.829), both P < .001; respectively).
Using X-tile, the composite scoring was divided into three risk groups that accurately discriminated between patients with good, intermediate, and poor prognosis (Figures 5 and S4). Therefore, we further analyzed subgroups of CRC patients in stages II, III, and IV. The three risk groups were able to significantly distinguish between CRC patients with different prognoses in stage II, III, or IV (Figures S5–S7).
Figure 5.
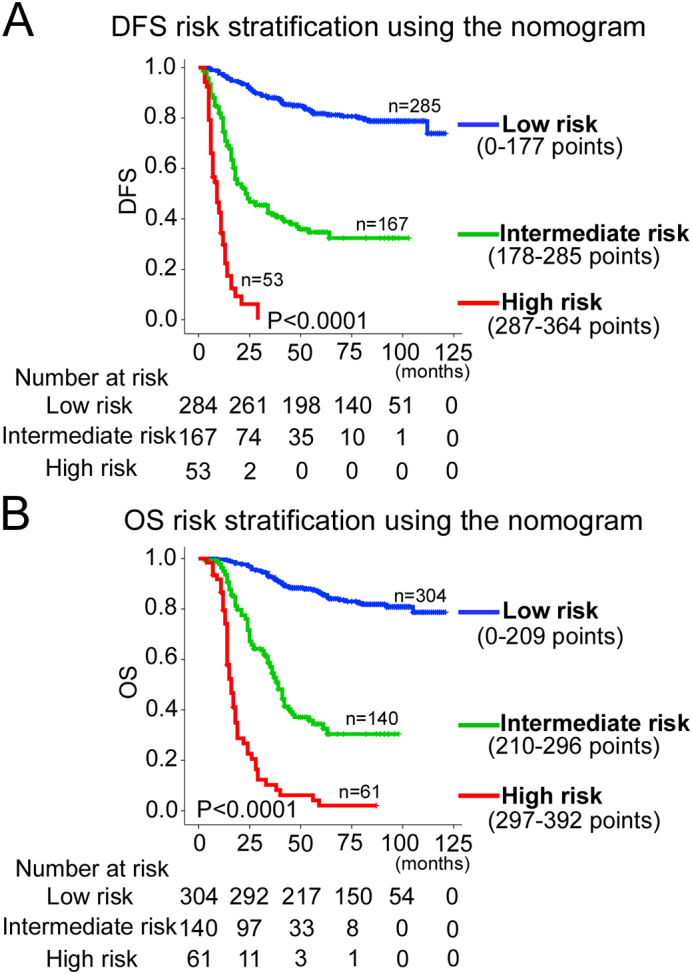
Kaplan–Meier survival analysis of OS and DFS according to three risk groups. The entire population was divided into 3 subgroups according to the total number of points given by the nomograms. (A): DFS nomogram and (B): OS nomogram.
Clinical Use
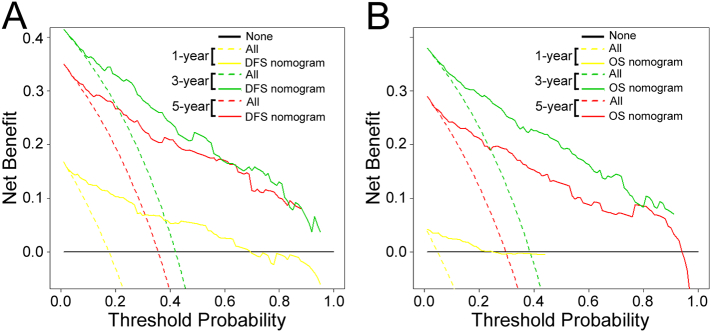
The decision curve analysis for the two nomograms is presented in Figures 6 and S8. The decision curve showed that if the threshold probability of a patient or doctor was >10%, using the two nomograms to predict 1-, 3-, 5-year DFS and OS added more benefit than either the treat-all-patients scheme or the treat-none scheme. Within this range, the net benefit was comparable to several overlaps on the basis of the nomograms.
Figure 6.
Decision curve analysis for the two nomograms in the training cohort. The y-axis measures the net benefit. The solid lines (yellow, blue and red) represent the nomogram. The dotted lines (yellow, blue and red) represent the assumption that all patients have1-, 3-, or 5-year survival, respectively. The thin black line represents the assumption that no patients have 1-, 3-, or 5-year survival. The net benefit was calculated by subtracting the proportion of all patients who are false positive from the proportion who are true positive, weighting by the relative harm of forgoing treatment compared with the negative consequences of an unnecessary treatment [27], [28].Here, the relative harm was calculated by [pt/(1–pt)]. The “pt” (threshold probability) is where the expected benefit of treatment is equal to the expected benefit of avoiding treatment; at which time a patient will opt for treatment informs us of how a patient weighs the relative harms of false-positive results and false-negative results ([a-c]/[b-d] = [1-pt]/pt); a-c is the harm from a false-negative result; b-d is the harm from a false-positive result. a, b, c and d give, respectively, the value of true positive, false positive, false negative, and true negative [27], [28]. (A): DFS nomogram and (B): OS nomogram.
Discussion
In the present study, we found that CSN2 is a prognosis-associated marker that has reduced expression in CRC. The results indicated that lowCSN2 expression and low CD8 + TILs were associated with poor prognosis, and patients with dMMR CRC had better clinical outcomes. Using these three markers and four clinicopathological risk variables, two nomograms were constructed and externally validated for predicting 1-, 3-, and 5-year DFS and OS probabilities after curative resection. The nomograms performed well with good discrimination and calibration, identifying this model as a simple and easy tool for estimating the survival of individual patients with CRC.
CSN is a multiprotein complex involved in protein degradation, transcriptional activation, signal transduction, and tumorigenesis [8], [9]. Although CSN functions as a complex, CSN subunits also have their own functions independent of the CSN complex. Fang et al. reported that ERK2-activated CSN6 regulates β-Trcp and stabilized β-catenin expression by blocking the ubiquitin-proteasome pathway, thereby promoting CRC development [8].CSN2 integrates into the CSN complex via its C-terminal region, and its N-terminal region is necessary for direct interaction with Cul1 [7]. Moreover, CSN2 inhibits p27kip1 degradation and impedes G1-S phase progression via the deneddylation of SCF Cul1. Zhang et al. found that CSN2 promotes pluripotency maintenance by stabilizing Nanog protein and repressing transcription [11]. Yang et al. showed that the level of serum CSN2 was higher in GC patients than healthy people; however, low serum CSN2 was associated with unfavorable survival [13]. In addition, CSN2 has reduced expression in GC tissues and has been identified as a tumor suppressor gene [13], [14]. Carvalho et al. showed that CSN2 is significantly decreased in colorectal adenomas and carcinomas with a 13q gain compared with those without, and the oncogenic role of miR-15a-3p in 13q amplicon-driven colorectal adenoma-to-carcinoma progression involves UCP2 and CSN2 as candidate target genes [30]. In the present study, we found that CSN2 had reduced expression in CRC tissues, and the percentage of patients with low intratumoral CSN2 expression increased as disease progression increased from TNM stage I–IV (Figure 1C). Furthermore, low intratumoral CSN2 expression was significantly associated with a poor prognosis.
Tumor MMR deficiency maybe detected as MSI or loss of MMR protein expression by IHC. Previous studies have demonstrated the prognostic value of MMR protein expression in CRC [31], [32], [33]. Most previous studies evaluating the prognostic or predictive role of MMR status in CRC have been performed using microsatellite analysis to assess tumor phenotype. IHC testing is generally preferred because of lower costs, faster turnaround time, a wider availability in routine diagnostic laboratories, reported increased sensitivity, and the ability to perform direct germline mutation testing. In our study, we evaluated the prognostic role of the IHC expression of MMR status in a large series of CRC patients. In this investigation, patients with dMMR CRC whose tumors demonstrated a loss of expression of at least one MMR protein (MLH1, MSH2, MSH6, or PMS2) had a better outcome than patients with pMMR tumors. Moreover, in the multivariate analysis, the survival advantage for dMMR patients was independent of clinicopathological factors.
A randomized retrospective study showed a survival advantage in 5-FU-treated CRC patients with MSI-L and MSS cancers but not in patients with MSI-H tumors [34]. In a pooled molecular reanalysis of randomized chemotherapy trials (n = 341), MMR deficiency was shown to be a predictive marker for a lack of benefit from 5-FU-based chemotherapy for patients with stage II or III colon cancer [35]. However, in a randomized trial of 491 CRC patients who received adjuvant chemotherapy, MMR protein expression did not have predictive value for response to 5-FU treatment with respect to OS [32]. Even if the use of MMR status to predict the outcomes of adjuvant chemotherapy was still controversial, the results from the previous trials are very promising and indicate that 5-FU is beneficial for CRC patients with MSI tumors [34], [36], [37]. A large multicenter AGEO study showed that high-risk stage II dMMR colon cancer tended to have better outcomes with oxaliplatin-based adjuvant chemotherapy than with surgery alone [38]. However, before MMR status can be applied as a prognostic and predictive biomarker in clinical practice, its value must be proven in large, high-powered prospective trials.
Extensive literature has suggested that the TILs in cancer are clinically important [15], [17], [22]. A recent meta-analysis summarized the impact of immune cells, including all subsets of T cells, NK cells, B cells, macrophages, and MDSCs on clinical outcome from more than 120 published articles [39]. Most importantly, the beneficial impact of the immune infiltration with cytotoxic and memory T cell phenotypes have been demonstrated in cancers of diverse anatomical sites, including not only CRC but also malignant melanoma, gastric, lung, esophageal, breast, and bladder cancers, et al. [39]. Based on the numeration of lymphocyte populations in both the core of tumors and the invasive margin, the immunoscore demonstrates the prevalence of immune infiltrates and has been defined as a new component for the classification of CRC, designated TNM-Immune [15], [16]. MSI-H tumors are thought to have higher TIL densities relative to MSS tumors because of their higher mutational load [40], [41]. Next-generation sequencing studies have shown that MSI-H tumors typically harbor more than 1000 coding somatic mutations per tumor cell genome, compared with the 50 to 100 somatic mutations found in MSS tumors [42]. Zlobec et al. revealed that an infiltrative tumor margin and absence of CD8+ TILs are highly predictive of local recurrence in node-negative pMMR colon cancer and may help identify high-risk patients who could benefit from adjuvant chemotherapy [43]. In our study, pMMR tumors have far fewer CD8 + TILs than dMMR tumors. We also found that high CSN2-expression tumors have more CD8+ TILs than low CSN2-expression tumors, and the mechanism remains unknown.
Division of CRCs into molecular subsets by The Cancer Genome Atlas (TCGA) project yields important consequences for prognosis and therapeutic response [40], [44], [45]. The MSI immune subgroup, accounting for 15% of all CRCs, are a result of deficient cellular DNA mismatch repair (dMMR) mechanisms [40]. Deficient MMR can result from a germline mutation in an MMR gene (MLH1, MSH2, MSH6, PMS2), ie, Lynch syndrome (LS). Membranous PD-L1 expression occurs only in patients with dMMR CRC and is prominent on TILs and tumor-associated macrophages located at the invasive fronts of the tumor [45]. Both PD-1 expression in TILs and PD-L1 expression on tumor cells differ in MSI-H from that in MSS tumors [46]; 77% of TILs from MSI-H tumors express PD-1, compared with 39% from MSS tumors. In addition, 32% of MSI-H tumors express PD-L1, compared with 13% of MSS tumors. Immune checkpoint blockades directed against PD-1 have recently shown excellent activity with a response rate to single-agent therapy of 55%in preliminary studies involving patients with stage IV dMMR disease [45]. According to a phase II study, dMMR renders different solid tumors highly sensitive to immune checkpoint blockades in patients treated with the PD-1 inhibitor pembrolizumab [40], [45]. With the ability to fix DNA replication errors compromised, dMMR tumors accumulate numerous somatic mutations, which could produce neoantigens triggering a potent antitumor immune response in the presence of the PD-1 blockade [45]. If these observations are confirmed in randomized trials of stage II and III CRC, MMR status may be applied as a predictive biomarker of all disease stages, and adjuvant treatment for dMMR patients through immuno- rather than chemotherapy may re-emerge.
The TNM stage is the most commonly used system to predict survival for patients who have undergone curative resection for CRC. However, CRC patients within the same stage have different cellular, genetic, and clinicopathological characteristics and outcomes [47]. To provide a more individualized staging system, nomograms have been developed to evaluate a large number of significant clinicopathologic predictors to better predict the prognosis of individual patients. Improved prediction of individual outcomes would be useful for counseling patients, personalizing treatment, and scheduling patient follow-ups [48]. Although there are several CRC nomograms available, no particular nomogram has been widely used in the clinic [3], [4], [48]. In this study, we developed and validated two nomograms including IHC expression of CSN2, CD8 and MMR status, T stage, N stage, M stage, and the level of CEA to improve prognosis prediction for CRC patients. The nomograms can be used to better predict an individual patient's probability of 1-, 3-, and 5-year DFS and OS. Validation of the nomograms was performed using calibration plots and the C-index. The nomograms performed well with a good calibration. Furthermore, the C-index for DFS and OS was satisfactory [0.836 (0.804–0.868) and 0.841 (0.808–0.874), respectively, in the training cohort]. Compared with previous studies, our nomograms included three prognosis biomarkers (CSN2, CD8 and MMR status) that greatly improved accuracy.
Moreover, the improved survival estimates calculated using the nomograms may help identify patients with a high risk of poor clinical outcome within known TNM stages and facilitate the choice of therapies. Current guidelines recommend adjuvant chemotherapy for high-risk patients with stage II CRC. The risk of a poor outcome or relapse in stage II disease has been clinically identified using the following criteria: fewer than 12 lymph nodes analyzed after surgery; poorly differentiated histology (exclusive to those that are MSI-H); lymphatic/vascular invasion; bowel obstruction; perineural invasion; localized perforation; and close, indeterminate, or positive margins [4]. However, these clinicopathological risk factors cannot clearly identify the high-risk patients who may benefit from chemotherapy [4]. Accordingly, the two nomograms, which incorporate multiple prognostic parameters into the current staging system, might assist in identifying patients with poor odds of survival who are likely to benefit from adjuvant chemotherapy.
Substantial efforts have been made toward identification of molecular signatures to predict survival in patients with CRC, including gene signatures, and microRNAs [2], [49]. However, these gene-based signatures have not been widely introduced into clinical practice as initially expected due to the variability of measurements in microarray assays, inconsistencies in assay platforms, and the requirement for analytical expertise [49]. IHC not only provides a semi-quantitative assessment of protein abundance but also defines the cellular localization of their expression. Therefore, identification of biomarkers with IHC, which has been widely applied in clinical diagnosis, is found to serve as promising alternative strategies for the molecular profiling of tumors [16], [22], [50].
However, this study has some limitations. First, the study is a retrospective study that relied exclusively on a single-institutional database. Validation by external cohorts is necessary for the prognostic value of CSN2and the generalized use of the nomograms as the basis for postoperative treatment recommendations. Further studies of the biological function of CSN2 expression in cell lines and animal models should be performed in the future. Other prognostic and/or predictive variables may be integrated to enhance the accuracy of the nomograms. In addition, the application of the nomograms requires several IHC analyses and pathologic variables that are available only after surgery, e.g., the pT and pN stages. Hence, the nomograms will have a limited impact on alternative treatments prior to surgery, including neoadjuvant chemotherapy.
In conclusion, this study is the first to reveal that CRC patients with lowCSN2 expression had a higher risk of recurrence/metastasis and poorer survival than patients with high CSN2 expression. Our results demonstrated that CSN2 expression in CRC was an independent predictor of patient outcomes. The two nomograms were developed and validated for predicting the probability of 1-, 3-, and 5-year DFS and OS. The model might facilitate both clinician and patient counseling and individualized adjuvant treatment decision-making, as well as follow-up scheduling.
Acknowledgments
Acknowledgements
This study was supported by the main project of the natural science fund of Bengbu Medical College (BYKY1632ZD) and the National Natural Science Foundation of China (81603155).
Authors' Contributions
Conception and design: Bing Zhu, Hao Liu and Jun Fu.
Development of methodology: Bing Zhu and Jun Fu.
Acquisition of data (acquired and managed patients, provided facilities, etc.): Bing Zhu, Pei Zhang, Mulin Liu, Congqiao Jiang, and Jun Fu.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Bing Zhu, Pei Zhang, Mulin Liu, and Congqiao Jiang.
Writing, review, and/or revision of the manuscript: Bing Zhu, Pei Zhang, and Jun Fu.
Administrative, technical, or material support (i.e., reporting, organizing data):Bing Zhu, Pei Zhang, Mulin Liu, Congqiao Jiang, Hao Liu and Jun Fu.
Study supervision: Hao Liu and Jun Fu.
Contributed with sampling of tissue and clinical information: Bing Zhu, Pei Zhang, Mulin Liu, Hao Liu and Jun Fu.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.07.006.
Contributor Information
Bing Zhu, Email: bbmczhubing@163.com.
Jun Fu, Email: fujun3186682@163.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 3.Weiser MR, Gonen M, Chou JF, Kattan MW, Schrag D. Predicting survival after curative colectomy for cancer: individualizing colon cancer staging. J Clin Oncol. 2011;29:4796–4802. doi: 10.1200/JCO.2011.36.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu MX, Qu H, Bu ZD, Chen DL, Jiang BH, Cui M, Xing JD, Yang H, Wang ZZ, Di JB. Validation of the Memorial Sloan-Kettering Cancer Center nomogram to predict overall survival after curative colectomy in a Chinese colon cancer Population. Ann Surg Oncol. 2015;22:3881–3887. doi: 10.1245/s10434-015-4495-2. [DOI] [PubMed] [Google Scholar]
- 5.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JH, Powell AG, Roxburgh CS, Horgan PG, McMillan DC, Edwards J. Mismatch repair status in patients with primary operable colorectal cancer: associations with the local and systemic tumour environment. Br J Cancer. 2016;114:562–570. doi: 10.1038/bjc.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Menon S, Lykke-Andersen K, Tsuge T, Di X, Wang X, Rodriguez-Suarez RJ, Zhang H, Wei N. The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr Biol. 2002;12:667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 8.Fang LK, Lu WS, Choi HH, Yeung SCJ, Tung JY, Hsiao CD, Fuentes-Mattei E, Menter D, Chen CQ, Wang L. ERK2-dependent phosphorylation of CSN6 is critical in colorectal cancer development. Cancer Cell. 2015;28:183–197. doi: 10.1016/j.ccell.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10:3057–3066. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Zheng JN, Pei DS. The emerging roles of Jab1/CSN5 in cancer. Med Oncol. 2016;33:90. doi: 10.1007/s12032-016-0805-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Ni P, Mou C, Zhang Y, Guo H, Zhao T, Loh YH, Chen L. Cops2 promotes pluripotency maintenance by Stabilizing Nanog Protein and Repressing Transcription. Sci Rep. 2016;6 doi: 10.1038/srep26804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong W, Li J, Wang B, Chen L, Niu W, Yao Z, Baniahmad A. Epigenetic involvement of Alien/ESET complex in thyroid hormone-mediated repression of E2F1 gene expression and cell proliferation. Biochem Biophys Res Commun. 2011;415:650–655. doi: 10.1016/j.bbrc.2011.10.130. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Wang J, Li J, Zhang H, Guo S, Yan M, Zhu Z, Lan B, Ding Y, Xu M. Identification of serum biomarkers for gastric cancer diagnosis using a human proteome microarray. Mol Cell Proteomics. 2016;15:614–623. doi: 10.1074/mcp.M115.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leal JF, Fominaya J, Cascon A, Guijarro MV, Blanco-Aparicio C, Lleonart M, Castro ME, Ramon YCS, Robledo M, Beach DH. Cellular senescence bypass screen identifies new putative tumor suppressor genes. Oncogene. 2008;27:1961–1970. doi: 10.1038/sj.onc.1210846. [DOI] [PubMed] [Google Scholar]
- 15.Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol. 2013;25:261–267. doi: 10.1016/j.coi.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C. Towards the introduction of the 'Immunoscore' in the classification of malignant tumours. J Pathol. 2014;232:199–209. doi: 10.1002/path.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 18.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 19.Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, Pommerencke T, Doeberitz MV, Folprecht G, Luber B. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. 2011;71:5670–5677. doi: 10.1158/0008-5472.CAN-11-0268. [DOI] [PubMed] [Google Scholar]
- 20.Sinicrope FA. DNA mismatch repair and adjuvant chemotherapy in sporadic colon cancer. Nat Rev Clin Oncol. 2010;7:174–177. doi: 10.1038/nrclinonc.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li TJ, Jiang YM, Hu YF, Huang L, Yu J, Zhao LY, Deng HJ, Mou TY, Liu H, Yang Y. Interleukin-17-producing neutrophils link inflammatory stimuli to disease progression by promoting angiogenesis in gastric cancer. Clin Cancer Res. 2017;23:1575–1585. doi: 10.1158/1078-0432.CCR-16-0617. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Zhang Q, Hu Y, Li T, Yu J, Zhao L, Ye G, Deng H, Mou T, Cai S. ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann Surg. 2018;267:504–513. doi: 10.1097/SLA.0000000000002116. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Zhang Q, Jiang Y, Yu J, Hu Y, Mou T, Chen G, Li G. Gastric cancer cells inhibit natural killer cell proliferation and induce apoptosis via prostaglandin E2. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2015.1069936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Detre S, Saclani Jotti G, Dowsett M. A "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350 doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 26.Liang WH, Zhang L, Jiang GN, Wang Q, Liu LX, Liu DR, Wang Z, Zhu ZH, Deng QH, Xiong XG. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol. 2015;33:861–869. doi: 10.1200/JCO.2014.56.6661. [DOI] [PubMed] [Google Scholar]
- 27.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Localio AR, Goodman S. Beyond the usual prediction accuracy metrics: reporting results for clinical decision making. Ann Intern Med. 2012;157:294–295. doi: 10.7326/0003-4819-157-4-201208210-00014. [DOI] [PubMed] [Google Scholar]
- 29.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 30.de Groen FL, Timmer LM, Menezes RX, Diosdado B, Hooijberg E, Meijer GA, Steenbergen RD, Carvalho B. Oncogenic role of miR-15a-3p in 13q amplicon-driven colorectal adenoma-to-carcinoma progression. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: A meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Ohrling K, Edler D, Hallstrom M, Ragnhammar P. Mismatch repair protein expression is an independent prognostic factor in sporadic colorectal cancer. Acta Oncol. 2010;49:797–804. doi: 10.3109/02841861003705786. [DOI] [PubMed] [Google Scholar]
- 33.Lanza G, Gafa R, Santini A, Maestri I, Guerzoni L, Cavazzini L. Immunohistochemical test for MLH1 and MSH2 expression predicts clinical outcome in stage II and III colorectal cancer patients. J Clin Oncol. 2006;24:2359–2367. doi: 10.1200/JCO.2005.03.2433. [DOI] [PubMed] [Google Scholar]
- 34.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sargent DJ, Marsoni S, Thibodeau SN, Labianca R, Hamilton SR, Torri V, Monges G, Ribic C, Grothey A, Gallinger S. Confirmation of deficient mismatch repair (dMMR) as a predictive marker for lack of benefit from 5-FU based chemotherapy in stage II and III colon cancer (CC): A pooled molecular reanalysis of randomized chemotherapy trials. J Clin Oncol. 2008;26:431–436. [Google Scholar]
- 36.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel M, Arnold CA, Miyai K. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 37.Benatti P, Gafa R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 38.Tougeron D, Mouillet G, Trouilloud I, Lecomte T, Coriat R, Aparicio T, Des Guetz G, Lecaille C, Artru P, Sickersen G. Efficacy of adjuvant chemotherapy in colon cancer with microsatellite instability: A large multicenter AGEO study. J Natl Cancer Inst. 2016;108:djv438. doi: 10.1093/jnci/djv438. [DOI] [PubMed] [Google Scholar]
- 39.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 40.Lee V, Murphy A, Le DT, Diaz LA., Jr. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. 2016;21:1200–1211. doi: 10.1634/theoncologist.2016-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prall F, Duhrkop T, Weirich V, Ostwald C, Lenz P, Nizze H, Barten M. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Hum Pathol. 2004;35:808–816. doi: 10.1016/j.humpath.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 42.Timmermann B, Kerick M, Roehr C, Fischer A, Isau M, Boerno ST, Wunderlich A, Barmeyer C, Seemann P, Koenig J. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zlobec I, Terracciano LM, Lugli A. Local recurrence in mismatch repair-proficient colon cancer predicted by an infiltrative tumor border and lack of CD8+ tumor-infiltrating lymphocytes. Clin Cancer Res. 2008;14:3792–3797. doi: 10.1158/1078-0432.CCR-08-0048. [DOI] [PubMed] [Google Scholar]
- 44.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gatalica Z, Snyder C, Maney T, Ghazalpour A, Holterman DA, Xiao NQ, Overberg P, Rose I, Basu GD, Vranic S. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev. 2014;23:2965–2970. doi: 10.1158/1055-9965.EPI-14-0654. [DOI] [PubMed] [Google Scholar]
- 47.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 48.Weiser MR, Landmann RG, Kattan MW, Gonen M, Shia J, Chou J, Paty PB, Guillem JG, Temple LK, Schrag D. Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol. 2008;26:380–385. doi: 10.1200/JCO.2007.14.1291. [DOI] [PubMed] [Google Scholar]
- 49.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 50.Jiang Y, Liu W, Li T, Hu Y, Chen S, Xi S, Wen Y, Huang L, Zhao L, Xiao C. Prognostic and predictive value of p21-activated kinase 6 associated support vector machine classifier in gastric cancer treated by 5-fluorouracil/oxaliplatin chemotherapy. EBioMedicine. 2017;22:78–88. doi: 10.1016/j.ebiom.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material