Abstract
MetaMap is a widely used named entity recognition tool that identifies concepts from the Unified Medical Language System Metathesaurus in text. This study presents MetaMap Lite, an implementation of some of the basic MetaMap functions in Java. On several collections of biomedical literature and clinical text, MetaMap Lite demonstrated real-time speed and precision, recall, and F1 scores comparable to or exceeding those of MetaMap and other popular biomedical text processing tools, clinical Text Analysis and Knowledge Extraction System (cTAKES) and DNorm.
Keywords: natural language processing, algorithms, software design, software validation, unified medical language system
BACKGROUND AND SIGNIFICANCE
MetaMap, a software program for finding Unified Medical Language System (UMLS) Metathesaurus1 concepts in biomedical text, was developed in 1994.2 The original Prolog implementation evolved into a sophisticated UMLS-based named entity recognition tool with many options.3 MetaMap is widely used both as a service for remote file processing and as a downloadable tool. In 2015, MetaMap software was downloaded 2174 times. In addition, MetaMap was accessed 1 391 751 times through its Web and application program interface facilities to process 90 429 494 documents. Reviews from MetaMap users communicated directly to the development team and in the annual UMLS users’ survey are mostly positive, with some concerns expressed about processing speed and finding the best combination of many options to be used for a given task. In addition, many developers who would like to modify MetaMap for local use expressed a strong preference for a Java implementation. A previous Java implementation, MetaMap Transfer (MMTx), an open-source downloadable version of MetaMap, addressed the need to comply with privacy issues and the needs of developers, while preserving the rich set of processing options.4 With improvements to the Prolog implementation, MMTx became slower than MetaMap and the team focused on making MetaMap available for downloads, rather than maintaining 2 versions of the software. Regretfully, some old unsupported versions of MMTx are still used,5 which, in addition to the need for real-time processing, motivated the development of MetaMap Lite as a replacement for MMTx.
In MetaMap Lite, we focus on real-time processing speed and start with a limited basic set of functions, such as longest term match and negation detection. In this paper, we present MetaMap Lite and evaluate its performance compared to the current Prolog implementation of MetaMap3 and other widely used medical named entity recognition tools, clinical Text Analysis and Knowledge Extraction System (cTAKES)6 and DNorm,7 using the ShARe corpus used in SemEval/CLEF 2013–2015 evaluations,8 the National Center for Biotechnology Information (NCBI) Disease Corpus,9 the BioScope Corpus,10 the 2010 i2b2 collection,11 and the Indexing Initiative collection of biological and clinical journal abstracts (Lister Hill Center [LHC] test collection) that is being released concurrently with this publication.
MetaMap and MetaMap Lite use the same UMLS-based lookup thesauri and rely on the UMLS to provide meta-information about the terms identified in the text, normalizing them in the process, ie, mapping the identified named entities to UMLS unique concept identifiers (CUIs). Both tools allow use of customized dictionaries and either focus on a specific domain or provide broad coverage of text types and semantic types. The other 2 tools that we use in the evaluation were intended for clinical text processing or disorder extraction and combine knowledge-based and machine learning methods or rely solely on machine learning.
cTAKES is a general-purpose clinical NLP system built within the Unstructured Information Management Architecture framework. Its pipeline components, many of which are trained on clinical data, are as follows: (1) a sentence splitter, (2) a context-sensitive tokenizer, (3) an OpenNLP12-based part-of-speech tagger, (4) an OpenNLP-based shallow parser, (5) 2 implementations of an entity recognizer and ontology mapper (cTAKES dictionary lookup and fast dictionary), (6) a negation detector that implements NegEx,13 (7) an uncertainty detector inspired by NegEx, (8) an OpenNLP-based constituency parser, (9) a dependency parser, (10) a semantic role labeler, (11) a coreference resolver, (12) a relation extractor, (13) a CLEAR-TK–based event recognizer, (14) a CLEAR-TK–based temporal expression recognizer, and (15) a CLEAR-TK–based temporal relation extractor. Modules 1 through 7 are used for named entity and attribute recognition. cTAKES is widely used in clinical informatics and has inspired development of many extensions.14–16
DNorm was originally built using the BANNER named entity recognizer, the NCBI disease corpus, and pairwise learning to rank to normalize the identified terms to MEDIC,17 a disease lexicon for indexing diseases in biomedical literature that merges the Online Mendelian Inheritance in Man and the “Diseases” branch of the National Library of Medicine’s Medical Subject Headings.7 Later, DNorm was adapted to process clinical notes.18
OBJECTIVE
Our objective was to introduce a new real-time open-source UMLS-based named entity recognition tool and evaluate its performance compared to state-of-the-art, widely used, publicly available biomedical named entity recognition tools.
MATERIALS AND METHODS
MetaMap Lite provides the longest concept match for UMLS semantic types defined by users and uses ConText19 or NegEx13 for negation detection. The processing pipeline consists of 7 steps, presented in Figure 1 and described below.
Figure 1.
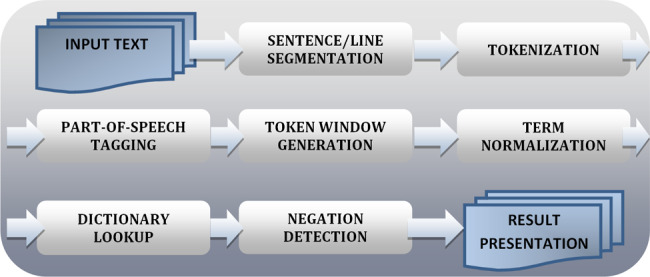
MetaMap Lite processing steps.
Sentence/line segmentation uses the default OpenNLP’s sentence segmenter12 or its own blank line segmenter. Tokenization is based on the original MetaMap tokenization algorithm. Part-of-speech tagging, which is optional, is performed by the default OpenNLP part-of-speech tagger. Token window is predefined to be the length of the sentence if the OpenNLP segmenter is used, or a nonoverlapping window of 15 tokens for the blank line segmenter. No additional chunking is performed. Term normalization is based on MetaMap string normalization with slight modifications. The following operations are applied to a term before the dictionary lookup: (1) removal of parentheticals, (2) syntactic un-inversion, (3) conversion to lowercase, and (4) stripping of possessives.
Dictionary lookup is the most time-consuming part of MetaMap Lite processing. To speed it up, we experimented with a publicly available Lucene search engine,20 for which multiple requests needed for named entity recognition were also time-consuming. This experiment showed that we need to optimize dictionary lookup. The underlying dictionary lookup uses an implementation of inverted files,21 in which the dictionary is divided into several partitions with each partition containing only terms of the same length.22 The implementation uses the Java NIO class java.nio.MappedByteBuffer to access the system’s virtual memory facilities to improve I/O performance.
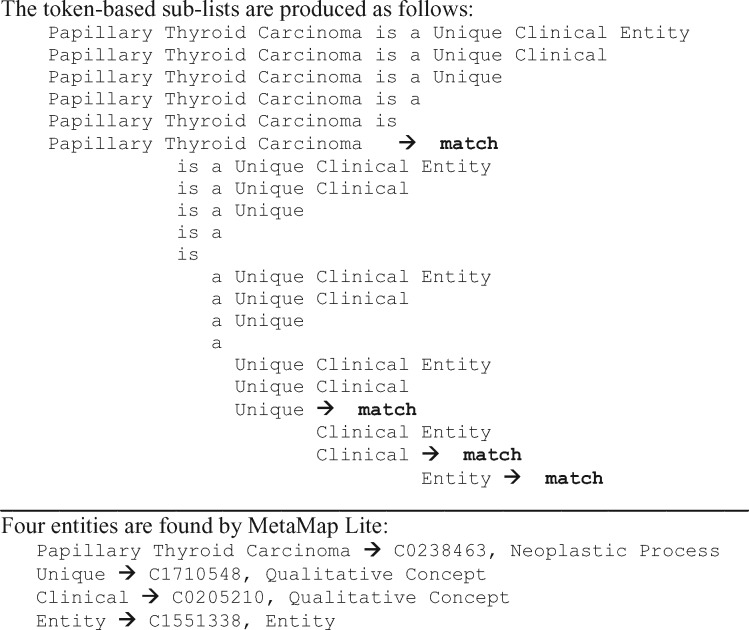
Mapping is done for sentence- or line-based chunks dynamically divided into sublists during processing, as shown in Figure 2. Each chunk is normalized and then looked up in a dictionary. Any match found in the dictionary that is subsumed by a longer match is discarded.
Figure 2.
An example of UMLS concept recognition in a sentence.
Figure 2 presents an example of sentence-level named entity recognition in which a token list for the sentence “Papillary thyroid carcinoma is a unique clinical entity” is generated and processed.
For dictionary lookup, MetaMap Lite currently uses 3 dictionaries originally created for MetaMap: (1) cuiconcept, which maps CUIs to concept preferred names; (2) cuisourceinfo, the primary dictionary that contains the UMLS CUI, the UMLS string identifier, a sequence number, the source-derived string, the source abbreviation, and the source term type; and (3) cuist, which maps CUIs to semantic types. Examples of dictionary entries and more details about dictionary file organization are provided in Supplementary Appendix A.
Finally, optional Negation detection relies on ConText19 or our implementation of NegEx12; the modules are interchangeable and the preference to use one or the other is set in the preferences file or in the command line options at run time.
Evaluation
We used the following collections to evaluate MetaMap Lite concept extraction and negation detection:
BioScope, which contains 1954 clinical notes, 9 full-text articles, and 1273 abstracts in which 3032 negation cues and 4611 speculation cues with 7643 linguistic scopes are annotated.
NCBI disease corpus, which contains 793 PubMed abstracts, in which 6120 disease mentions are mapped to 682 distinct UMLS concepts.
i2b2 2010 collection of 871 clinical notes, which provides annotations for 30 518 problems, 20 852 treatments, 22 060 tests, several assertion types, and 8 types of relations. In this evaluation we used only problems and 6144 negation annotations.
ShARe corpus, which contains 300 clinical notes with 12 095 annotated disorders and their attributes.
LHC test collection, which contains 150 clinically oriented PubMed abstracts and 150 biology-oriented abstracts in which 2242 disorders are annotated and normalized to their 2015 AA UMLS CUIs.
We included in the evaluation all annotated entities and restricted MetaMap, MetaMap Lite, and other tool processing to the semantic types in the UMLS semantic group Disorders.
RESULTS
Table 1 presents the time it took each tool to run on each collection. Table 2 presents recall, precision, and F1 scores for each tool on each collection that we were able to process using the tool. Negation detection is not a native MetaMap Lite feature; therefore, we compared it only to the MetaMap implementation of NegEx, to verify that we maintained its level of performance. Due to difficulties in obtaining meaningful offsets, we were not able to evaluate DNorm on 2 collections.
Table 1.
Total time (in minutes) to annotate test collections
| BioScope (negation) | NCBI disease | ShARe corpus | i2b2 2010 | LHC test | |
|---|---|---|---|---|---|
| MetaMap | 24 m 2 s | 29 m 58 s | 42 m 36 s | 52 m 15 s | 17 m 8 s |
| cTAKES (DL) | No evaluation | 25 m 14.04 s | 35 m 74 s | 35 m 13 s | 10 m 1 s |
| DNorm | No evaluation | 1 m 59.800 s | Failed to run | Failed to run | 2 m 54 s |
| MetaMap Lite | 4 m 41.995 s | 1 m 35.906 s | 2 m 50.54 s | 2 m 33.06 s | 1 m 15.62 s |
Note that only the cTAKES dictionary lookup (cTAKES DL) model was used in our experiments. Independent experiments indicate that this model is 30 times slower than the fast lookup model.5
Table 2.
Entity recognition and negation detection results at the mention level
| Collection/Tool | MetaMap |
cTAKES (DL) |
DNorm |
MetaMap Lite |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F-1 | P | R | F-1 | P | R | F-1 | P | R | F-1 | |
| BioScope (negation) | 43.7 | 34.4 | 38.5 | 85.2 | 37.9 | 52.4 | ||||||
| NCBI disease | 60.3 | 68.3 | 64.1 | 47.0 | 53.8 | 47.4 | 74.1 | 67.6 | 70.7 | 73.1 | 71.9 | 72.5 |
| ShARe (entities) | 59.5 | 48.1 | 53.2 | 46.3 | 46.2 | 46.2 | N/A | N/A | N/A | 74.2 | 42.1 | 53.8 |
| i2b2 2010 (entities) | 38.1 | 35.7 | 36.8 | 31.9 | 34.1 | 32.9 | N/A | N/A | N/A | 47.0 | 31.9 | 38.0 |
| i2b2 2010 (negation) | 40.2 | 32.2 | 38.3 | 53.8 | 38.0 | 44.6 | ||||||
| LHC clinical articles | 58.8 | 77.2 | 66.8 | 42.6 | 59.9 | 49.8 | 71.5 | 58.2 | 64.2 | 69.4 | 74.9 | 70.0 |
| LHC biological articles | 46.8 | 75.6 | 57.8 | 47.1 | 60.6 | 53.0 | 67.7 | 62.8 | 65.2 | 67.5 | 77.9 | 72.4 |
Grayed-out cells indicate that negation detection was tested only for MetaMap and MetaMap Lite. We were not able to run DNorm on the ShARe and i2b2 2010 collections. Note that only the cTAKES dictionary lookup (cTAKES DL) model was used in our experiments. Independent experiments indicate that this model has lower F1 scores than the fast lookup model.5
The results were obtained using a Dell T5500n with 16 Xeon processors running at 2.66 GHz with 24 GB of RAM running Red Hat Enterprise Linux 6. All of the systems were evaluated using a single thread.
DISCUSSION
MetaMap Lite achieved the intended aim of significantly speeding up text processing while maintaining and somewhat exceeding the default MetaMap level of performance. Our evaluation is limited to the UMLS semantic group Disorders, primarily due to the availability of the test collections and tools that are heavily skewed toward that group, likely because of its importance in clinical text processing and downstream applications, such as extraction of phenotypes, adverse reactions to drugs, and question answering, to name just a few. MetaMap Lite’s speed of processing is approached only by that of DNorm; however, we were not able to obtain mention-level results with the version of DNorm available to us on the i2b2 2010 and ShARe collections. In general, we exerted nontrivial efforts to install, run, and obtain mention-level offsets for the third-party tools. We are fairly confident that we have achieved results attainable to average end users of the versions of these tools available at the time we conducted our experiments. MetaMap Lite achieved F1 scores higher than all other tools on all collections. The absolute scores obtained in our experiments may differ from those reported elsewhere (eg5), which could be explained by the versions of the tools and the choice of their settings in our experiments. All our installations of the tools, the test collections that can be freely distributed, and the evaluation scripts are available upon request.
MetaMap Lite has several limitations. First, it implements very few of the rich set of MetaMap options. We have only compared MetaMap Lite to the default MetaMap settings, and we can therefore only suggest that its performance is comparable to out-of-the-box MetaMap settings with no tuning for a given task. Rather than implementing all of the options, we will continue to introduce features as they are requested by users. Second, MetaMap Lite’s processing speed depends on the options: the fastest processing is achieved without part-of-speech tagging, with an insignificant reduction in the F1 score. Third, MetaMap Lite does not yet implement any word-sense disambiguation modules and will provide all senses of a given term available in the UMLS; as a result, given the sentence “The steroid will be kept for now and tapered at a later date on follow-up with Dr Coma,” it maps “Coma” to the UMLS concept [C0009421] Comatose. As with MetaMap, some mappings that are always wrong could be added to an exclude list; alternatively, a custom-built data file could be used instead of the standard dictionary files provided with the tools. Finally, an important limitation of the evaluation is that it covers only disorders and negation. We are planning to extend annotation of the LHC collection to cover other important semantic groups in the future.
CONCLUSION
This paper presents MetaMap Lite, a lightweight Java implementation of MetaMap, one of the most-used named entity recognition tools for identification of UMLS Metathesaurus concepts in biomedical text. This tool is meant for applications that emphasize processing speed and ease of use. The tool is modular and publicly available, which we hope will advance its development through requests submitted by end users and contributions of additional modules by the developers.
The software is downloadable from https://metamap.nlm.nih.gov/download/new/public_mm_lite_2016_3.0_SNAPSHOT.tar.bz2.
Download requires a valid UMLS license.
FUNDING
This work was supported by the intramural research program at the US National Library of Medicine, National Institutes of Health.
COMPETING INTERESTS
There are no competing interests.
CONTRIBUTOR
DDF and ARA planned and guided the development of the tool and the evaluation. WJR developed the tool and conducted the evaluation. DDF and WJR authored the manuscript. All authors read and commented on the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material are available at Journal of the American Medical Informatics Association online.
Supplementary Material
REFERENCES
- 1. Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aronson AR, Rindflesch TC, Browne AC. Exploiting a large thesaurus for information retrieval. Proc RIAO. 1994;94:197–216. [Google Scholar]
- 3. Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;173:229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Divita G, Tse T, Roth L, Pt C. Failure analysis of MetaMap Transfer (MMTx). Stud Health Technol Inform. 2004;107():763–67. [PubMed] [Google Scholar]
- 5. Tseytlin E, Mitchell K, Legowski E et al. . NOBLE: Flexible concept recognition for large-scale biomedical natural language processing. BMC Bioinformatics. 2016;171:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Savova GK, Masanz JJ, Ogren PV et al. . Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;175:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leaman R, Islamaj Dogan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics. 2013;2922:2909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pradhan S, Elhadad N, Chapman W, Manandhar S, Savova G. SemEval-2014 Task 7: Analysis of Clinical Text. Notes. 2014;19999:133. [Google Scholar]
- 9. Doğan RI, Leaman R, Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inform. 2014;47:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vincze V, Szarvas G, Farkas R, Móra G, Csirik J. The BioScope corpus: biomedical texts annotated for uncertainty, negation and their scopes. BMC Bioinformatics. 2008;9 (Suppl 11):S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;185:552–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Apache OpenNLP. Online: https://opennlp.apache.org/. Accessed July 2016.
- 13. Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;345:301–10. [DOI] [PubMed] [Google Scholar]
- 14. Garla V, Lo Re V 3rd, Dorey-Stein Z et al. . The Yale cTAKES extensions for document classification: architecture and application. J Am Med Inform Assoc. 2011;185:614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu J, Wu Y, Zhang Y, Wang J, Lee HJ, Xu H. CD-REST: a system for extracting chemical-induced disease relation in literature. Database (Oxford). 2016;2016:baw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Velupillai S, Mowery DL, Abdelrahman S, Christensen L, Chapman WW. Towards a generalizable time expression model for temporal reasoning in clinical notes. AMIA Annu Symp Proc. 2015;2015:1252–59. eCollection 2015. [PMC free article] [PubMed] [Google Scholar]
- 17. Davis AP, Wiegers TC, Rosenstein MC, Mattingly CJ. MEDIC: a practical disease vocabulary used at the Comparative Toxicogenomics Database. Database. 2012;2012:bar065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leaman R, Khare R, Lu Z. Challenges in clinical natural language processing for automated disorder normalization. J Biomed Inform. 2015;57:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;425:839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Apache Lucene. Online: https://lucene.apache.org/. Accessed July 2016.
- 21. Zhang J, Long X, Suel T. Performance of compressed inverted list caching in search engines. In Proceedings of the 17th International Conference on World Wide Web. 2008:387–96. ACM. [Google Scholar]
- 22. Rogers W, Candela G, Harman D. Space and time improvements for indexing in information retrieval. In: Proceedings of the Annual Symposium on Document Analysis and Information Retrieval (SDAIR-95). Las Vegas, NV, US, April 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.