Abstract
Objective:To develop automated classification methods for eligibility criteria in ClinicalTrials.gov to facilitate patient-trial matching for specific populations such as persons living with HIV or pregnant women.
Materials and Methods:We annotated 891 interventional cancer trials from ClinicalTrials.gov based on their eligibility for human immunodeficiency virus (HIV)-positive patients using their eligibility criteria. These annotations were used to develop classifiers based on regular expressions and machine learning (ML). After evaluating classification of cancer trials for eligibility of HIV-positive patients, we sought to evaluate the generalizability of our approach to more general diseases and conditions. We annotated the eligibility criteria for 1570 of the most recent interventional trials from ClinicalTrials.gov for HIV-positive and pregnancy eligibility, and the classifiers were retrained and reevaluated using these data.
Results:On the cancer-HIV dataset, the baseline regex model, the bag-of-words ML classifier, and the ML classifier with named entity recognition (NER) achieved macro-averaged F2 scores of 0.77, 0.87, and 0.87, respectively; the addition of NER did not result in a significant performance improvement. On the general dataset, ML + NER achieved macro-averaged F2 scores of 0.91 and 0.85 for HIV and pregnancy, respectively.
Discussion and Conclusion:The eligibility status of specific patient populations, such as persons living with HIV and pregnant women, for clinical trials is of interest to both patients and clinicians. We show that it is feasible to develop a high-performing, automated trial classification system for eligibility status that can be integrated into consumer-facing search engines as well as patient-trial matching systems.
Keywords: clinical trial screening, eligibility determination, machine learning, natural language processing, patient-trial matching
Objective
This paper describes the development of automated methods to classify clinical trials on the basis of their eligibility criteria in order to facilitate patient-trial matching for specific populations such as persons living with HIV (PLWH) or pregnant women. We describe and compare the implementation of several natural language processing–based and machine learning (ML) approaches and evaluate their performance using study records from ClinicalTrials.gov.
Background and significance
ClinicalTrials.gov, run under the auspices of the National Library of Medicine and the National Institutes of Health, is a public registry of clinical trials and, starting in 2008, also a repository and reporting mechanism for trial results. As of June 2016, over 218 000 studies were registered. The ClinicalTrials.gov database encompasses a wide variety of past and ongoing studies in terms of both funding source (public or private) and study type (interventional or observational). Each study record contains a large number of structured fields (eg, study ID, diseases/conditions studied, etc.) as well as semistructured or unstructured fields (eg, eligibility criteria). As a public resource, ClinicalTrials.gov is an important tool for patients attempting to find clinical trials for which they may be eligible, as well as for health care providers looking for experimental treatment options for their patients. Together, patients and health care providers account for more than one-third of the 170 million page views per month.1 Overall, randomized controlled trials are noted to have a positive effect on patient outcomes, even in the context of their inherently experimental nature,2 and inclusion in a clinical trial has been linked to higher survival rates in patients with cancer.3
However, the utility of ClinicalTrials.gov and other clinical trial registries to patients and physicians is limited by their ability to return relevant results in response to search queries, resulting in potential frustration and user dissatisfaction. The Essie search backend4 powering ClinicalTrials.gov performs well for general queries that require any or all of the search keywords and their related Unified Medical Language System (UMLS) concepts to appear in a study record, but due to the unstructured nature of fields such as eligibility criteria,5 it is unable to capture the semantic complexity and granularity of more sophisticated queries, such as “cancer trials that accept HIV+ patients.” Attempting to approximate this by searching for “cancer AND HIV” returns approximately 15 000 studies that mention both cancer and HIV, but manual review of the eligibility criteria from a random sample of these studies reveals that the majority of them (∼85% based on our estimate) explicitly exclude PLWH. This is not surprising, given the fact that conventional search engines do not implement negation-detection algorithms such as NegEx6 and therefore miss the negated concepts and phrases that are abundant in eligibility criteria. Although we do not wish to take a position on the validity of excluding HIV-positive patients from certain types of clinical trials,7 it is reasonable to infer that such exclusions make it more difficult for PLWH to find clinical trials for which they may be eligible. Pregnant women comprise another population that faces similar challenges when attempting to find clinical trials for which they may be eligible, as they are often excluded from interventional trials for various reasons.8 In the meantime, however, we believe that the automated classification methods presented in this paper have the potential to make it easier for traditionally excluded patient populations and their providers to locate clinical trials that may be of benefit.9,10
While the task of finding suitable clinical trials is addressed somewhat by existing solutions for patient-trial matching and eligibility screening, our classification framework differs in a couple of significant aspects. First, existing matching systems, eg, those by Ni et al.,11,12 Sahoo et al.,13 and Miotto and Weng,14 rely on data from clinical trial records and individual patient data in order to make an eligibility determination using methods such as cosine similarity and decision rules to determine the degree of match between specific patients and trials. Our framework relies on the corpus of clinical trial records only, which, when combined with a disease- or condition-specific training process, generates a model for classifying the trials themselves with regard to their eligibility status for the given disease/condition or patient population. While these systems are undoubtedly valuable for determining eligibility and recruiting patients at the point of care, where patient electronic health record data is readily accessible, the stand-alone nature of our classification framework is advantageous in terms of flexibility. For example, it could be implemented as a filter to enhance the usefulness and relevance of results returned from consumer-facing search engines powering clinical trial registries such as ClinicalTrials.gov, and as a provider of additional data points to the aforementioned patient-trial matching systems, thereby potentially improving their performance and utility to clinicians and other health care providers.
Second, we believe our proposed trial classification framework is the first to rely exclusively on unstructured input, ie, the free-text eligibility criteria of clinical trial records. By contrast, the existing systems mentioned above use structured data fields such as age, gender, vital signs, lab results, etc., in addition to or in lieu of free-text fields such as trial eligibility criteria or clinical notes. Notably, the oncology-oriented Trial Prospector platform of Sahoo et al.13 requires that the criteria of clinical trials be defined entirely in a structured format for the purpose of comparison with patient data. While the authors of these systems all reported good results within the constraints of their specific environments, we believe that the proposed classification framework offers additional portability and versatility. For example, it can be retrofitted into existing databases or environments that operate primarily with free-form text or where structured data may not be readily available or accessible.
MATERIALS AND METHODS
Acquisition of cancer-HIV trials from ClinicalTrials.gov
The 1000 most relevant study records from each of the search queries “cancer AND HIV” (representing study records that mention both cancer and HIV) and “cancer AND NOT HIV” (representing study records that mention cancer but not HIV) were downloaded from ClinicalTrials.gov in June 2016. The relaxation expansion feature in Essie4 was used so that records using synonymous or related terms such as “lymphoma” or “AIDS” would be included. Study types other than “Interventional” were excluded; however, to maximize the variety of writing and formatting styles in the eligibility criteria and the generalizability of our classification methods, no additional filtering was done based on study age or status. In all, 891 of the downloaded study records were randomly selected, and the eligibility criteria were manually reviewed in order to assign 1 of 3 annotations: “HIV-ineligible” or class 0, ie, studies that explicitly excluded HIV-positive patients from participation; “indeterminate” or class 1, ie, studies that made no explicit mention of HIV or HIV status; and “HIV-eligible” or class 2, ie, studies that specifically mentioned HIV and accepted HIV-positive patients. Some examples of HIV-related phrases found in the eligibility criteria are presented in Table 1. This process yielded a total of 626 HIV-ineligible annotations, 149 indeterminate annotations, and 116 HIV-eligible annotations.
Table 1.
Examples of HIV-related phrases from study eligibility criteria
| CT.gov ID | Example text | Classification |
|---|---|---|
| NCT00393029 | Seronegative for human immunodeficiency virus (HIV) antibody. (The experimental treatment being evaluated in this protocol depends on an intact immune system. Patients who are HIV seropositive can have decreased immune competence and thus be less responsive to the experiment and more susceptible to its toxicities.) | 0: HIV-ineligible |
| NCT01143545 | Patients with active infections, including HIV, will be excluded, due to unknown effects of the vaccine on lymphoid precursors. | 0: HIV-ineligible |
| NCT01209520 | Patients with HIV infection (but not AIDS) are eligible for this trial. Therefore, no HIV testing will be required. | 2: HIV-eligible |
| NCT01434550 | No history of human immunodeficiency virus (HIV) or acquired immune deficiency syndrome (AIDS) or other immunosuppressive diseases. | 0: HIV-ineligible |
| NCT02365207 | Not be in an immunosuppressed state (eg, HIV+, use of chronic steroids >1 month). | 0: HIV-ineligible |
| NCT02818283 | [Inclusion Criteria] HIV-1 infection as documented by any licensed ELISA (enzyme-linked immunosorbent assay) test kit and confirmed by Western blot at any time prior to study entry. HIV-1 culture, HIV-1 antigen, plasma HIV-1 ribonucleic acid (RNA), or a second antibody test by a method other than ELISA is acceptable as an alternative confirmatory test. | 2: HIV-eligible |
Rule-based classification using regular expressions
Using the patterns we observed during annotation, we formulated rules and regular expressions to capture the boundaries between inclusion and exclusion criteria as well as recognize specific phrases and clauses in the study title, conditions studied, and eligibility criteria that mention HIV and/or immunodeficiency status (Box 1). The regular expressions we built for HIV status were ordered by specificity (to ensure that more specific regexes matched first over less specific ones) and can be grouped into 2 broad categories: positive (the pattern indicates PLWH are eligible) and negative (the pattern indicates PLWH are ineligible). The majority of our negative regular expressions were derived by prepending “no” and “not” to the positive regular expressions and/or changing “negative” to “positive” (or vice versa.) Next, we built a simple regex-based segmentation algorithm for the inclusion and exclusion criteria. If a line contained the word “criteria” or “characteristics,” it underwent additional matching to determine if it signified the start of inclusion criteria or exclusion criteria. We assigned different positive or negative integer point values depending on the context; eg, the phrase “history of HIV” can have completely opposite meanings depending on whether it is found under inclusion criteria or exclusion criteria. To minimize misclassification of the minority HIV-eligible class, we defined positive matches to be worth twice as much (+2) as negative matches (–1). If the text contained no occurrences of “HIV” or “human immunodeficiency virus,” it was classified as “indeterminate” (class 1).
Box 1.
Examples of regular expressions for HIV-positive status
| seropositive for (HIV|human immunodeficiency virus) |
| positiv.*?(HIV|human immunodeficiency virus).+?antibody |
| (documentation|evidence) of.+?(HIV|human immunodeficiency virus) |
| diagnosis of (HIV|human immunodeficiency virus) infection |
| test positive for.+?(HIV|human immunodeficiency virus) |
| suffering from[A-Z0‐9 -,/]+?(HIV|human immunodeficiency virus) |
| (known)?human immunodeficiency virus \(HIV\) infection |
| HIV-seropositive |
These 2 components were used to build a line-by-line classifier that assigned a total score for each study record. If a line matched multiple regular expressions, only the first was used. Studies with a total score ≥ 1 were labeled as HIV-eligible, studies with a total score ≤ −1 were labeled as HIV-ineligible, and studies with a total score = 0 were labeled as indeterminate.
Classification using machine learning
Next, we constructed a natural language processing pipeline to train and test a classifier using supervised ML techniques to see if we could improve upon our baseline regex-based method. For each study, we extracted the eligibility criteria and performed some light automated preprocessing for text cleanup, including adding additional line breaks where necessary and subsequently removing punctuation characters. The processed text was then tokenized into a composite unigram (bag of words) and bigram document term matrix using term frequency–inverse document frequency15 to represent the tokens. Using a chi-squared model, the top k features (k = 250) were selected. This feature representation, along with the corresponding class labels from our annotations, was used to train and test a one-vs-rest multiclass linear support vector machine (SVM) classifier using stratified 10-fold cross-validation. The scikit-learn16 toolkit along with LIBLINEAR17 were used to perform the text processing and construct the SVM. An outline of the pipeline is shown in Figure 1.
Figure 1.
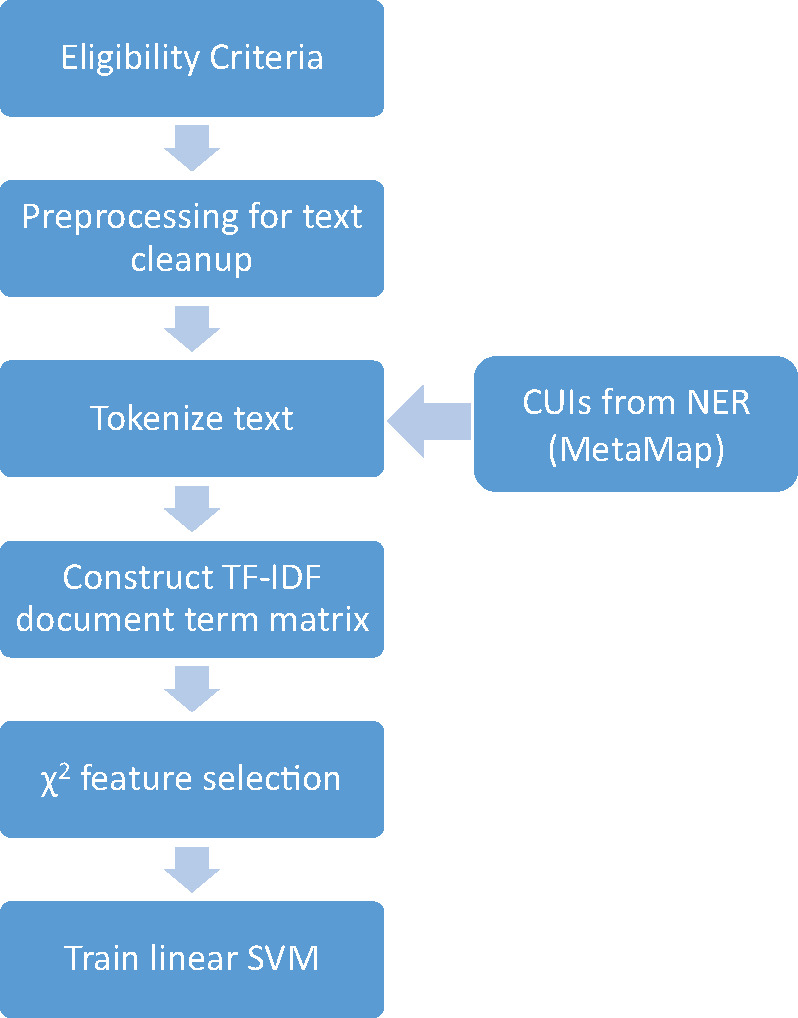
An architectural diagram of the machine learning–based classification framework.
Adding additional features using named entity recognition
Due to the highly variable nature of natural language and our observation of the many variations in which a simple concept like “HIV positive” or “HIV negative” might be expressed, we hypothesized that using named entity recognition (NER) to preprocess the text and annotate variant phrases with standardized terminology might increase the performance of our SVM classifier. We used MetaMap18 to recognize concepts and transform them into concept-unique identifiers (CUIs); negated CUIs detected by the NegEx6 functionality of MetaMap were denoted by prepending an “N” (eg, NC0002965). These CUIs were then appended to the term frequency–inverse document frequency matrix as additional features. The same vectorization and SVM classification approach as described above was used.
Evaluation metrics
To evaluate classification performance, we calculated macro-averaged values and 95% confidence intervals for precision, recall, F2 score, and area under the precision-recall (PR) curve, and plotted PR curves for each label class; due to the inherently imbalanced nature of the label classes in the cancer-HIV data, computing PR curves instead of receiver operating characteristic curves gives us a more complete picture of classification performance.19–21 We opted to use the F2 metric over the more commonly used F-measure because in this particular case, recall is arguably more important than precision; for instance, misclassifying a study that accepts HIV-positive patients and subsequently missing a trial-patient match (false negative) is much more costly than having to review an HIV-ineligible study erroneously classified as HIV-eligible (false positive). The F2 score is calculated using the generic formula for with as follows22:
Workload reduction
Under a manual approach, ie, without the aid of any automated classification system, a clinician or patient would need to review all 891 studies in our collection to determine eligibility status for PLWH. Using our automated classifiers, the clinician or patient would only need to review all studies classified as either HIV-eligible or indeterminate. Since one of the main use cases for automated trial classification algorithms and patient-trial matching systems is minimizing the need for time-consuming human review, we used the above definitions to calculate a mean “workload reduction” metric12 for each of our classifiers.
Generalizing to other diseases and conditions
After finding that our ML classifiers were able to achieve good performance on the cancer-HIV dataset (see below), we broadened our scope to include all studies regardless of disease/context, and also selected a different condition (pregnancy) to evaluate the generalizability of our approach. We downloaded eligibility criteria for all interventional study records with a status of recruiting (as of July 2016) from ClinicalTrials.gov. Standardized classification guidelines devised by the authors were given to a group of 12 volunteers recruited among National Library of Medicine informatics program trainees, who annotated the eligibility criteria for 1660 of the most recent studies (as determined by start date) for both HIV-positive and pregnancy eligibility, under the same ineligible/indeterminate/eligible scheme. The volunteers were instructed in a training session during which the provided annotation guidelines were discussed. Pregnancy annotation was skipped for all studies whose eligibility was restricted to males only. Pairwise Cohen’s kappa scores were computed to measure interannotator agreement. The ML classifiers were then retrained and tested using the same methodology described above. For the HIV annotation, we also tested both ML classifiers (1) without retraining and (2) retrained and tested on a merged version of both datasets.
RESULTS
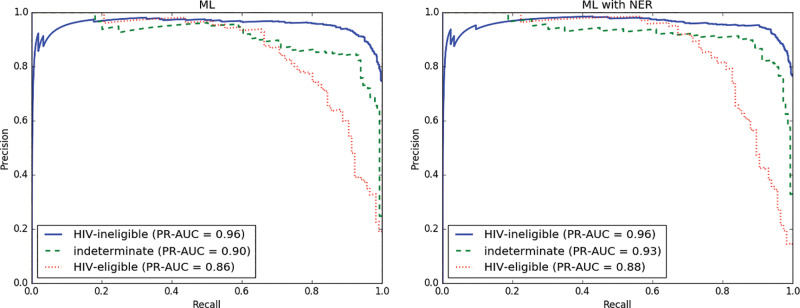
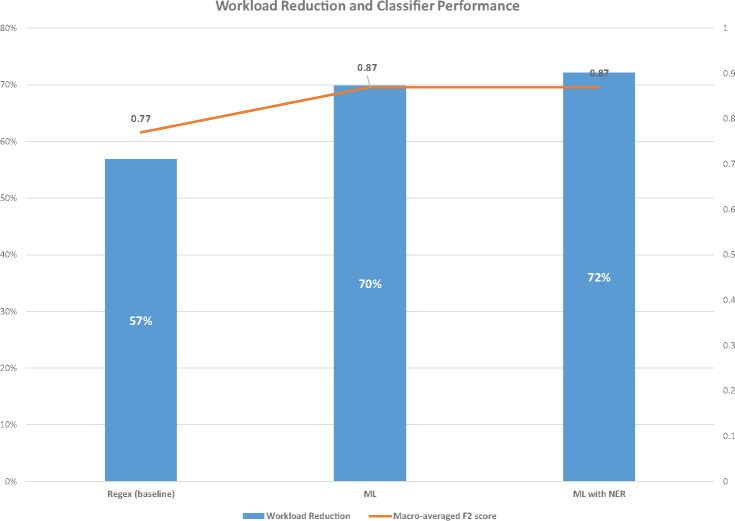
The evaluation metrics for the baseline (regex) classifier, the ML classifier, and the ML with NER classifier are shown in Table 2 and Figure 2. Using the fold-matched macro-averaged F2 scores as a metric, both the ML and ML + NER classifiers significantly outperformed the baseline regex classifier (P < .001). The results of our experiments indicate that both the ML and ML + NER methods are able to effectively distinguish between the 3 eligibility classes we defined, with the ML + NER method achieving 90% precision (95% confidence interval, 0.86–0.94, Table 2). However, the addition of NER to the ML model did not result in a significant performance benefit when using the fold-matched macro-averaged F2 scores as a metric. Both ML methods also achieved a substantial workload reduction of 70% or more (Figure 3). Taking into account both classification performance and workload reduction, ML + NER performed best overall.
Table 2.
Evaluation metrics for classifier performance on the cancer-HIV dataset
| Classifier | Precision | Recall | F2 score |
|---|---|---|---|
| Baseline (regex) | 0.70 (0.67–0.74) | 0.81 (0.78–0.85) | 0.77 (0.74–0.81) |
| ML | 0.87 (0.83–0.91) | 0.87 (0.83–0.91) | 0.87 (0.83–0.90) |
| ML + NER | 0.90 (0.86–0.94) | 0.86 (0.82–0.91) | 0.87 (0.83–0.91) |
Figure 2.
Per-class precision-recall curves and their corresponding areas under the curve for the ML classifier (left) and ML + NER classifier (right) on the cancer-HIV dataset.
Figure 3.
Workload reduction percentages and macro-averaged F2 scores for each of the 3 classifiers on the cancer-HIV dataset.
Additional classifier variants
We also explored some variations on the ML classifiers described above. As the indeterminate (class 1) and HIV-eligible (class 2) classes are of greatest interest to HIV-positive patients, we tried merging these 2 classes and constructing a binary classifier instead using the ML + NER model. The performance of this classifier turned out to be worse (F2 = 0.87 for the HIV-ineligible class and F2 = 0.81 for the merged class), so we did not investigate this method further. Next, we tried a cascade approach, in which we trained 2 binary classifiers, the first to separate studies that mentioned HIV from studies that did not mention HIV, ie, “indeterminate” (class 1 vs rest), and the second on the HIV-mentioning studies from the first classifier to further distinguish between HIV-ineligible and HIV-eligible (class 0 vs 2). While the first classifier achieved reasonably good performance (F2 = 0.97 for the HIV-mentioning class), the second classifier performed quite poorly on the HIV-eligible class (recall = 0.63 and F2 = 0.67). A possible explanation for this is propagation of misclassification errors from the first classifier to the second, which is an inherent limitation of multistage approaches.
Generalizing to other diseases and conditions
The interannotator agreement on the general dataset was good for both the HIV and pregnancy annotations, with mean pairwise Cohen’s kappa scores of 0.74 and 0.73, respectively. Due to the comparatively poor performance of the baseline regex method on the cancer-HIV dataset and the additional time investment required to compose domain-specific rules and regular expressions, we tested only the 2 ML classifiers. However, because the number of studies in the set that were labeled as HIV-eligible or pregnancy-eligible (class 2) was very small ( for both), we merged it with indeterminate (class 1), which effectively yielded binary classifiers for excludes/does not exclude HIV/pregnancy. The evaluation metrics are shown in Table 3. As with the cancer-HIV dataset, ML + NER did not significantly outperform the standard ML model for either HIV or pregnancy in terms of macro-averaged F2 score.
Table 3.
Evaluation metrics for ML and ML + NER classifiers on general dataset of recent studies: HIV and pregnancy
| Classifier/dataset | Precision | Recall | F2 score |
|---|---|---|---|
| ML (HIV, cancer-HIV model with no retraining) | 0.35 (0.28–0.43) | 0.51 (0.47–0.54) | 0.29 (0.28–0.31) |
| ML + NER (HIV, cancer-HIV model with no retraining) | 0.34 (0.33–0.36) | 0.50 (0.46–0.55) | 0.35 (0.33–0.36) |
| ML (HIV) | 0.94 (0.93–0.95) | 0.90 (0.88–0.91) | 0.91 (0.89–0.92) |
| ML + NER (HIV) | 0.95 (0.93–0.96) | 0.90 (0.89–0.92) | 0.91 (0.90–0.92) |
| ML (HIV, merged dataset) | 0.90 (0.88–0.91) | 0.88 (0.86–0.89) | 0.88 (0.87–0.89) |
| ML + NER (HIV, merged dataset) | 0.91 (0.89–0.92) | 0.89 (0.87–0.90) | 0.89 (0.88–0.90) |
| ML (pregnancy) | 0.79 (0.77–0.82) | 0.88 (0.85–0.9) | 0.86 (0.84–0.87) |
| ML + NER (pregnancy) | 0.78 (0.76–0.81) | 0.87 (0.84–0.9) | 0.85 (0.83–0.87) |
Discussion
The results above demonstrate that a simple supervised ML approach is effective at classifying trial eligibility criteria on the basis of inclusion or exclusion of specific patient populations, such as PLWH or pregnant women.
We observed during the annotation process that the concept of HIV positivity (or lack thereof) can be expressed in a multitude of ways: “seropositive for HIV,” “known diagnosis of HIV infection,” “test positive for HIV,” “positive HIV antibody,” “documentation of HIV infection,” etc. Thus, while regular expression-based methods are simple to understand and easy to implement, they are insufficient to fully capture the semantics of free-form language. Furthermore, the contextual location of HIV-related phrases is significant, as they can be found under an “inclusion criteria” heading or an “exclusion criteria” heading (and sometimes both). Although we attempted to account for this by developing regular expressions to segment the inclusion and exclusion criteria, this process was imperfect due to variations in punctuation and wording. As evidenced by the significantly better performance of our supervised ML-based methods, these methods are able to overcome some limitations and minimize the need to manually recognize and define patterns. Although human annotation is still needed to generate labeled training data, such a task is preferable to the laborious and nonscalable process of defining rules and regular expressions for each problem domain.
We would also like to briefly note the relative class imbalance in the cancer-HIV dataset; eg, 626 of the 891 trials were annotated as HIV-ineligible. We speculate that, given the cancer domain–specific nature of the dataset, the investigators in these types of trials might prefer to err on the side of caution and exclude any factors or variables that are potentially confounding with regard to outcome, such as positive HIV status.7
Generalizing to other diseases and conditions
As mentioned previously, there are other conditions, such as pregnancy, that are often excluded from clinical trials for various reasons, and this provided the motivation to investigate the usefulness and performance of our ML + NER classifier in a general context. The classifier performed well for both HIV and pregnancy (Table 3). In the case of HIV, neither classifier performed well on the general dataset without retraining (Table 3), indicating that there might be linguistic differences between cancer trials and other types of trials when discussing HIV status. More generally, this implies that domain adaptation is necessary to achieve high classification performance. Additionally, it is worth pointing out the relative differences in class frequency between HIV and pregnancy; specifically, of the 1660 trials we looked at, many more trials explicitly excluded pregnant women (n = 949) than PLWH (n = 375). Our classifier is thus more useful in some situations than others in terms of workload reduction, depending on whether or not the majority class consists of negative (ineligible) samples. Nevertheless, the good performance in both scenarios indicates that our approach should be generalizable to other commonly excluded conditions, eg, hypertension or hepatitis infection.
Error analysis
We performed error analysis on the classification of the cancer-HIV dataset. Of the 116 cancer studies in our set that accepted PLWH, the ML + NER classifier correctly classified 94 of them, with 19 misclassified as HIV-ineligible and 3 misclassified as indeterminate. We manually reviewed the eligibility criteria of the 19 studies misclassified as HIV-ineligible to see if there were any linguistic commonalities that could account for classification error. Four studies (21%) contained HIV drug-related language (protease inhibitors and/or highly active antiretroviral therapy), 2 studies (11%) mentioned AIDS in addition to HIV, another 2 contained phrases stating that HIV-positive patients would be excluded at the investigator’s discretion, and 1 was a complex study with multiple arms seeking both HIV-positive and HIV-negative patients. There was no discernable pattern among the remaining 9 misclassified studies. The misclassifications involving HIV-related drugs and AIDS could potentially be explained by the presence of several studies that used these types of phrases yet still excluded all HIV-positive patients.
Limitations and future work
One difficulty that we encountered while constructing the regex-based classifier was reliably segmenting the inclusion and exclusion sections from the eligibility criteria. This proved to be cumbersome enough that we did not incorporate this step into either of the ML classifiers. In theory, however, adding inclusion/exclusion context to the set of features would be beneficial to classifier performance, due to its highly significant nature in terms of semantics. In addition, potentially there is room for improvement by employing more sophisticated text-processing techniques such as sentence segmentation and part-of-speech tagging.
Second, it is worth noting that our ML classifiers are SVM-based and thus a form of supervised learning; there is an up-front time investment required to perform annotation for each new problem domain. Since our small team of annotators was able to produce almost 1600 annotators in about a week’s time that were of high enough quality to effectively train the classifiers, we do not view this as a major limitation.
Finally, the results and conclusions from this research were generated in an offline, retrospective environment. In the future, it would be interesting to study the feasibility of using the aforementioned classifiers in a real-world environment, in consumer-facing search engines for clinical trial registries and/or patient-trial matching systems, and to quantify their benefit to clinicians and patients who are searching for clinical trials. Furthermore, we would like to further investigate the generalizability of our approach to other common diseases/exclusions, eg, diabetes, hypertension, hepatitis, tuberculosis, etc.
CONCLUSION
The eligibility status of specific patient populations such as PLWH and pregnant women for clinical trials are of interest to both patients and clinicians. However, conventional search engines are not able to fully leverage the information contained within the eligibility criteria. We developed an automated trial classification system for specific diseases/conditions using a supervised ML approach and show that it is effective in terms of both classification performance and workload reduction, as well as generalizable to other diseases or conditions commonly mentioned in trial eligibility criteria. The proposed methods have immediate real-world usability in that they can be implemented as filters within consumer-facing search engines for clinical trial registries, or used as additional input into existing patient-trial matching systems.
ACKNOWLEDGMENTS
The authors would like to thank Nicholas Ide and Russell Loane for their support in helping to acquire the data, Dr Clem McDonald for his mentorship and comments on the manuscript, Russell Loane for his comments on the manuscript, and the National Library of Medicine informatics program trainees for their assistance with annotations.
Funding
This work was supported by the intramural research program at the US National Library of Medicine, National Institutes of Health.
Competing Interests
The authors have no competing interests to declare.
Contributors
DDF conceived the original idea for the study, provided guidance and suggestions on methodology and analysis, and contributed to the manuscript. KZ acquired the data, implemented the classifiers and ran the experiments and data analysis, and wrote the manuscript.
References
- 1. Williams RJ. ClinicalTrials.gov: Policy Updates in Trial Registration and Results Reporting. https://clinicaltrials.gov/ct2/about-site/for-media: ClinicalTrials.gov; 2016. [Google Scholar]
- 2. Braunholtz DA, Edwards SJL, Lilford RJ. Are randomized clinical trials good for us (in the short term)? Evidence for a ‘trial effect.’ J Clin Epidemiol. 2001;54:217–24. [DOI] [PubMed] [Google Scholar]
- 3. Stiller CA. Centralised treatment, entry to trials and survival. Br J Cancer. 1994;70:352–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ide NC, Loane RF, Demner-Fushman D et al. . Essie: a concept-based search engine for structured biomedical text. J Am Med Inform Assoc. 2007;14:253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhattacharya S, Cantor MN. Analysis of eligibility criteria representation in industry-standard clinical trial protocols. J Biomed Inform. 2013;46:805–13. [DOI] [PubMed] [Google Scholar]
- 6. Chapman WW, Bridewell W, Hanbury P et al. . A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. J Biomed Inform. 2001;34:301–10. [DOI] [PubMed] [Google Scholar]
- 7. Persad GC, Little RF, Grady C. Including persons with HIV infection in cancer clinical trials. J Clin Oncol. 2008;26:1027–32. [DOI] [PubMed] [Google Scholar]
- 8. Foulkes MA, Grady C, Spong CY et al. . Clinical research enrolling pregnant women: a workshop summary. J Womens Health (Larchmt). 2011;20:1429–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thadani SR, Weng C, Bigger JT et al. . Electronic screening improves efficiency in clinical trial recruitment. J Am Med Inform Assoc. 2009;16:869–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Penberthy LT, Dahman BA, Petkov VI et al. . Effort required in eligibility screening for clinical trials. J Oncol Pract. 2012;8:365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ni Y, Wright J, Perentesis J et al. . Increasing the efficiency of trial-patient matching: automated clinical trial eligibility Pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak. 2015;15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ni Y, Kennebeck S, Dexheimer JW et al. . Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc. 2015;22:166–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sahoo SS, Tao S, Parchman A et al. . Trial prospector: Matching patients with cancer research studies using an automated and scalable approach. Cancer Inform. 2014;13:157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miotto R, Weng C, Hersh W et al. . Case-based reasoning using electronic health records efficiently identifies eligible patients for clinical trials. J Am Med Inform Assoc. 2015;22:e141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li-Ping Jing L-P, Hou-Kuan Huang H-K, Hong-Bo Shi H-B. Improved feature selection approach TFIDF in text mining. In: Proceedings, International Conference on Machine Learning and Cybernetics. IEEE 2002: 944–46. [Google Scholar]
- 16. Pedregosa F, Varoquaux G, Gramfort A et al. . Scikit-learn: Machine Learning in Python. Mach Learn. 2012;12:2825–30. [Google Scholar]
- 17. Fan R-E, Chang K-W, Hsieh C-J et al. . LIBLINEAR: A Library for Large Linear Classification. J Mach Learn Res. 2008;9:1871–74. [Google Scholar]
- 18. Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davis J, Goadrich M. The relationship between Precision-Recall and ROC curves. In: Proceedings of the 23rd International Conference on Machine Learning 2006. New York, NY: ACM Press; 2006: 233–40. [Google Scholar]
- 20. Keilwagen J, Grosse I, Grau J. Area under precision-recall curves for weighted and unweighted data. PLoS One. 2014;9:e92209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lobo JM, Jiménez-Valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008;17:145–51. [Google Scholar]
- 22. Hripcsak G, Rothschild AS, Hersh W et al. . Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc. 2005;12:296–98. [DOI] [PMC free article] [PubMed] [Google Scholar]