Abstract
To move beyond a select few genes/drugs, the successful adoption of pharmacogenomics into routine clinical care requires a curated and machine-readable database of pharmacogenomic knowledge suitable for use in an electronic health record (EHR) with clinical decision support (CDS). Recognizing that EHR vendors do not yet provide a standard set of CDS functions for pharmacogenetics, the Clinical Pharmacogenetics Implementation Consortium (CPIC) Informatics Working Group is developing and systematically incorporating a set of EHR-agnostic implementation resources into all CPIC guidelines. These resources illustrate how to integrate pharmacogenomic test results in clinical information systems with CDS to facilitate the use of patient genomic data at the point of care. Based on our collective experience creating existing CPIC resources and implementing pharmacogenomics at our practice sites, we outline principles to define the key features of future knowledge bases and discuss the importance of these knowledge resources for pharmacogenomics and ultimately precision medicine.
Keywords: pharmacogenetics, knowledge bases, electronic health records, clinical decision support systems, precision medicine
INTRODUCTION
Interest in tailored, effective treatments has prompted growing investment in the discovery and implementation of precision medicine.1 Precision medicine must leverage the electronic health record (EHR) as well as organize the vast body of evidence that supports translation of molecular markers to clinical care.2 The complexity and scale of molecular observations make knowledge resources essential to the delivery of clear, actionable interpretations. Early implementations of pharmacogenomics have demonstrated the ability to link discrete knowledge about genetic variants to point-of-care displays and clinical decision support (CDS).3–11 These programs have created their own local knowledge bases to perform tasks such as translating genotype to phenotype and constructing phenotype-based clinical recommendations, but such reinvention is difficult to sustain and inefficient for achieving national scale.12 The challenge is amplified when considering the effort required to deliver genomic results with consistent interpretations to all caregivers across a fragmented health care delivery system with limited EHR interoperability.13,14
Discovery efforts in precision medicine have been assisted by robust, nationally accessible reference databases that classify and annotate genomic variation eg, single nucleotide polymorphism database (dbSNP)/genomic structural variation database (dbVar), ClinVar, Online Mendelian Inheritance in Man (OMIM).15–18 However, few knowledge resources are attempting to cross the “omic chasm” of variant interpretation and delivery of evidenced-based clinical recommendations.19 Pharmacogenomics is leading the way as national knowledge resources, including the Clinical Pharmacogenetics Implementation Consortium (CPIC) and PharmGKB, are maturing to meet health care’s implementation needs. In this paper, we use pharmacogenomics as a prominent case example to consider current knowledge resources within precision medicine. We identify 5 principles based on those resources that will support the implementation of pharmacogenomics and precision medicine into EHRs and patient care.
CPIC Supports the Implementation of Pharmacogenomics into the EHR with CDS
Established in 2009, CPIC provides evidence-based, consensus clinical practice guidelines that enable the translation of genetic laboratory test results into actionable prescribing decisions for those genes/drugs with strong evidence for implementation. (For a detailed description of the CPIC guideline development process, see references.20) CPIC guidelines closely follow the National Academy of Medicine’s (formerly Institute of Medicine) Standards for Developing Trustworthy Clinical Practice guidelines and are freely available at cpicpgx.org/guidelines/ and www.guidelines.gov. CPIC members have expertise in various aspects of pharmacogenetics, and many are involved in implementing pharmacogenetics in clinical settings.
Successful adoption of pharmacogenomics into routine clinical care requires a curated and machine-readable database of pharmacogenomic knowledge suitable for use in an EHR with CDS. In 2013, CPIC formed an Informatics Working Group to support the translation of CPIC’s recommendations into the clinical electronic environment. Recognizing that EHR vendors do not yet provide a standard set of CDS functions for pharmacogenetics, the CPIC Informatics Working Group is systematically incorporating a set of EHR-agnostic implementation resources into all CPIC guidelines (Table 1).21–26
Table 1:
Description and intended use of the implementation resources in the CPIC Guideline and Supplementa
| Name of Table in CPIC Guideline | Description | Intended Use |
|---|---|---|
| Translation of genotype test result into interpreted phenotype | Provides a crosswalk from genotype to interpreted phenotype. Includes diplotype in star allele nomenclature (if applicable). | Translates a laboratory result into a more clinically meaningful result. Phenotypes are helpful as discrete results in the EHR because they provide clinical context and can reduce the complexity needed in CDS rules. |
| Resources that demonstrate the genotypes that constitute the * alleles for gene X and their effect on X protein | Provides a crosswalk between pharmacogene star allele nomenclature, dbSNP identifier (rsID), variant nucleotide change, allele effect on protein. | Useful when evaluating limited published evidence to determine a potential phenotype and clinical recommendation. |
| Drugs that pertain to this guideline | Contains a list of the drugs covered in the guideline, referencing codes from standard terminologies (eg, RxNorm, DrugBank, ATC) and related databases (eg, PharmGKB). | Provides an unambiguous list of drugs that can be leveraged when creating CDS rules, using codes that are common in prescribing and pharmacy systems. |
| Genes that pertain to this guideline | Contains a list of genes covered in the guideline, referencing codes from standard nomenclatures and knowledge bases (eg, HGNC, NCBI, Ensembl, PharmGKB). | Useful when creating CDS rules; uses codes that can be cross-referenced to lab test results and used to look up data in knowledge databases. |
| Clinical implementation workflow for EHR | Contains the steps and decision flows needed to position a pharmacogenetic result in the EHR when a systematic CDS program is implemented. | Combine this workflow with the pharmacogenetic genotype/phenotype summary entries for appropriate results reporting. This workflow highlights where clinical care needs to be implemented for actionable results.b |
| Pharmacogenetic genotype/phenotype summary entries | Identifies required data to couple genetic result with an interpretation, including genotype, phenotype, EHR priority result notation, and example interpretation text. | Useful when reporting a genomic result to help clinicians understand the clinical relevance of genotype or phenotype information. It is important to have clinician input on the wording for these interpretations. |
| Point-of-care clinical decision support (table) | Describes the trigger conditions and example text for interruptive CDS alerts. | Useful when building the rules for interruptive CDS alerts. It is important to have clinician input on the final wording of these alerts. |
| Point-of-care clinical decision support (workflow) | Describes the evaluation criteria and decision flow needed to build rules for interruptive CDS alerts. | This workflow is combined with the point-of-care clinical decision support table to build interruptive CDS alerts. It should be customized to fit into local clinical workflows |
aCPIC Guideline and Supplement are available at https://cpicpgx.org/guidelines/. An example of tables can be found here: https://www.pharmgkb.org/guideline/PA166105005.
bAn actionable result is any result where a patient with that result and being prescribed a corresponding drug prompts a recommendation for a change in therapy. Also known as “priority” results in some settings.
PRINCIPLES OF PRECISION MEDICINE KNOWLEDGE RESOURCES BASED ON PHARMACOGENOMICS
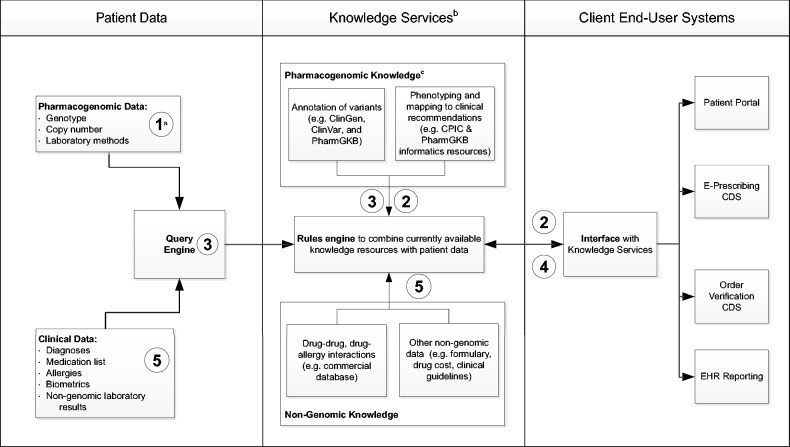
CPIC informatics resources are currently published in traditional journal formats with immediate posting on PharmGKB, but we recognize that static knowledge representations will not meet the ongoing needs of integrating precision medicine with the EHR.27–30 To maintain relevance and keep pace with rapid evidence generation, knowledge resources that are currently stored, maintained, and accessed locally19,31–33 will need to migrate to more service-oriented architectures that access consensus-based national resources. Centralized knowledge bases have their own challenges, but if designed properly, they would help meet many of the principles, or desiderata, already described for integrating molecular data with the EHR.34,35 In the following set of interlinked principles, we build upon our collective experience creating existing resources and implementing pharmacogenomics to define the key features of future knowledge bases (Box 1 and Figure 1). While not discussed in detail, all of these principles will require robust governance processes to maintain interoperability as knowledge resources evolve.
Figure 1:
Idealized information flow for querying pharmacogenomic knowledge resources and return to an EHR with CDS.
aNumbers correspond with relevant principles. bEach knowledge component may be used alone or in combination depending on the clinical scenario. cAll resources listed are freely available (ClinGen, ClinVar, PharmGKB, and CPIC).
1. Pharmacogenomic knowledge resources must support traceability between interrogated variants, primary results, and clinical interpretations.
Knowledge resources must connect the specific variants tested with the clinical interpretations being used in patient care. Genomic data are expected to have persistent, potentially lifelong value, but for the foreseeable future, many rapid, inexpensive laboratory methods will identify only a limited subset of variants. Within pharmacogenomics, misclassification of a drug phenotype is possible when only a subset of variants is measured. For example, when CYP2C19 is assayed for *17 (resulting in increased enzyme function) but does not interrogate for *4B, which includes both *17 and a variant rendering the encoded enzyme nonfunctional, the interpretation of increased enzyme function could be erroneous because the assay has not interrogated a key indicator of decreased metabolizer phenotype, leading to the wrong drug recommendation. A second common misclassification occurs when normal metabolizer status is predicted based on a lack of detected variants, when loss-of-function or gain-of-function may be present in regions that were not tested by the assay. Thus, since laboratory methods contain significant inherent limitations, the downstream interpretation and knowledge frameworks need to be able to transmit these limitations to the point of care along with recommendations.
2. Pharmacogenomic knowledge resources must rate level of evidence for each variant as well as for the overall recommendation
Level of evidence is a common feature of clinical guidelines, where individual recommendations are rated based on the rigor of original scientific research. CPIC and PharmGKB have established rating scales, which are currently used to prioritize the clinical implementation of drug-gene interactions, or used by individual clinicians to decide on clinical actionability.20,36–38 The degree of scientific support when interpreting phenotypes can hinge on a combination of the rarity, type, and clinical data supporting the biological impact of the underlying variants. For example, rare variants encoding stop codons have an almost certain detrimental impact on protein function, but due to relatively low allele frequency, there is often limited in vivo evidence to confirm their effect. When reporting the interpretation of drug-specific genotype results, knowledge frameworks must account for these different attributes, integrating across reported variants when determining an overall level of evidence. With the transition to next-generation sequencing, the number of identified variants relevant to drug metabolism or effect is expected to grow exponentially, making it even more critical to report the level of evidence at the variant level.
3. Knowledge resources must use standards to facilitate information exchange and enable interoperability among disparate systems
Several aspects of pharmacogenomics must be standardized to provide common semantics among disparate systems. Key data elements include which genetic variants were interrogated, phenotype terms, and medications involved in the gene-drug interaction. Standards are also needed to represent pharmacogenomic knowledge, including translation tables, clinical recommendations, and levels of evidence, as well as the evolution of that knowledge over time. Standardization of these elements is required not only to build and maintain knowledge resources, but also (and more importantly) to facilitate the exchange of pharmacogenomic knowledge across sites. Ultimately, standardization will assist in moving beyond local, fragmented knowledge resources to consensus-based national resources. A single publicly available knowledge resource may avoid many of the challenges of commercial knowledge bases used for CDS.39
4. Pharmacogenomic knowledge resources must support long-term reinterpretation of results
A challenge to implementing pharmacogenomics, which also applies to many areas of precision medicine, is accounting for the rapidly expanding scientific knowledge base. New genomic knowledge may prompt new clinical recommendations for previously reported variants, new findings that supersede prior recommendations, or completely new variants. Procedures to manage reconciliation of new and old interpretations and to maintain a robust system of knowledge versioning need to be established. To support frequent reinterpretation, the relationship between the knowledge consumer and the knowledge resource will need to be significantly re-engineered. At least 2 models are possible: one allows the knowledge resource to store all previous requests for interpretation of molecular data and push revised interpretations to subscribing clients; the second has a knowledge resource that might need to be queried routinely across all assayed variants. A local implementation of the first model has been described.40 Regardless, the CDS system should maintain transparency to the original result, but as illustrated in Figure 1, it is not necessary for the result data to reside in the knowledge base.
5. Pharmacogenomic knowledge resources must be positioned to be integrated with other knowledge at the point of care
Current pharmacogenomic implementation efforts and knowledge resources from CPIC are focused on highly penetrant single or 2-gene interactions with a drug or set of drugs. However, genotype results are only one of the many important patient data types that influence prescribing. Most knowledge resources do not account for more complex genomic interactions, other classes of omic data (eg, proteomics), other categories of drug knowledge (eg, interacting medications), or patient context (eg, known drug levels) when generating recommendations. For molecular knowledge to translate to improved patient care, the impact of other types of knowledge and patient data must be anticipated and eventually incorporated into guidelines. As precision medicine evolves, the exponential increase in combinatorial effects is a major challenge to developing a comprehensive knowledge resource. One solution is to frame patient recommendations relative to a consensus standard instead of in absolute terms, which allows multiple recommendations to be integrated at the point of care. Another approach is to foster the development of well-calibrated algorithms to explicitly integrate quantitative genomic and non-genomic predictors of drug response. More research is needed on methods to synthesize knowledge at the point of care to improve pharmacotherapy.
DISCUSSION
A longstanding goal of precision medicine is to leverage large curated repositories of molecular evidence and other knowledge resources to electronically communicate with patients and providers the ideal selection and dosing of pharmacotherapy, thereby optimizing patient outcomes. A growing number of health systems are successfully deploying customized genomic CDS through the EHR.4–6,8,41–43 However, these initial efforts have largely addressed single gene effects by relying on local versions of national guidelines with manual knowledge maintenance processes. To move from local to national implementation, CPIC believes these principles are critical to the design and implementation of future resources.
Other Efforts to Develop Knowledge Resources and Implement Genomics in the EHR
Other national efforts linked with CPIC are also making strides toward developing a comprehensive repository of pharmacogenomic knowledge. The Clinical Genome Resource (ClinGen) is an emerging NIH National Human Genome Research Institute (NHGRI)–funded project dedicated to building an authoritative central repository that defines the clinical relevance of genomic variants for use in precision medicine and research.44 A pharmacogenomics working group has convened within ClinGen, which is initially focusing on the published CPIC guidelines. This group is working in conjunction with CPIC and PharmGKB to represent the variants associated with each guideline and create knowledge for clinical decisions aligned with the predicted phenotype derived from the allele-level variant calls. Once fully operational, it is expected that ClinGen will be a definitive source for clinical care. While creating ClinGen represents significant progress, simply having a resource does not mean the information will be used for clinical care. To interface with clinical information systems, ClinGen supports an EHR working group focused on a variety of projects, including accessing the ClinGen resource through e-resource and infobutton standards proposed in Meaningful Use Stage 3.45
In addition, 2 recent efforts are actively addressing the generalizability of genomic CDS. In 2014, the NHGRI focused the Genomic Medicine 7 meeting on genomic CDS. The meeting’s purpose was to define both the current and ideal states of GCDS and develop a plan to move from the current to the ideal state, including a prioritized research agenda.46 The second effort is under the auspices of the National Academy of Medicine, the Displaying and Integrating Genetic Information Through the EHR (DIGITizE) project.47 This project engages key stakeholder populations to adopt and extend standards in order to improve patient health and maximize the knowledge that could be gained if genomic information were successfully integrated into the EHR in a format that would allow for research. DIGITizE is working on 2 pharmacogenetic use cases from conceptualization to implementation in a generalizable, standards-based EHR. The complexity of such a task is illustrated in the conceptual use model.48
Finally, the rendering of clinical guidelines as structured, coded knowledge sources will facilitate their adoption and implementation as CDS interventions by reducing the amount of expertise and effort required to extract and transform the knowledge from human-readable forms into technical specifications for programming teams. This will allow adopting institutions to focus more on vetting and gaining consensus on the rules to be implemented locally, and less on the informatics and knowledge engineering tasks. In conclusion, we have highlighted the important role of knowledge resources for pharmacogenomics and ultimately precision medicine. Before CPIC’s contributions, freely available knowledge resources to support the implementation of pharmacogenomics in the EHR with CDS were limited. CPIC resources give examples of how to integrate pharmacogenomic test results in clinical information systems with CDS to facilitate the application of patient genomic data at the point of care. To expand on this foundation, we have provided a set of principles of precision medicine knowledge resources based on pharmacogenomics. Putting these principles into practice will facilitate the development of the infrastructure needed to implement pharmacogenomics and precision medicine.
ACKNOWLEDGMENTS
The members of CPIC and the CPIC Informatics Working Group are acknowledged for their support in continuing to develop resources to support the implementation of pharmacogenomics in the EHR. All members are listed here: https://cpicpgx.org/members/. We thank Mary Relling, PharmD, and Jonathan Burlison, PhD, for their contributions to this paper.
CONTRIBUTORS
All authors are members of the CPIC Informatics Working Group. All authors contributed to designing the implementation resources in CPIC guidelines and developing the principles for the development of knowledge resources to support precision medicine described in this paper. James M Hoffman and Josh F Peterson drafted the manuscript. All authors contributed revisions to the manuscript. All authors critically reviewed and approved the final version of the manuscript.
COMPETING INTERESTS
All authors are members of the Clinical Pharmacogenetics Implementation Consortium Informatics Working Group. The authors have no competing interests to declare.
FUNDING
This work was supported by ALSAC and NIH grants CA 21765, R24GM115264-01, U01 HL105198, and U01 GM92666 (J.M.H. and K.E.C.); R24 GM61374 and R24GM115264-01 (M.W.C. and T.E.K.); U19 GM61388 (R.R.F.); and 1U01HG007253 (J.F.P.).
REFERENCES
- 1. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jameson JL, Longo DL. Precision medicine: personalized, problematic, and promising. N Engl J Med. 2015;372(23):2229–2234. [DOI] [PubMed] [Google Scholar]
- 3. Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Ann Rev Pharmcol Toxicol. 2015;55:89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharm Ther. 2012;92(5):563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peterson JF, Bowton E, Field JR, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med. 2013;15(10):833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166C(1):45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shuldiner AR, Relling MV, Peterson JF, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clin Pharm Ther. 2013;94(2):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2014;21(e1):e93–e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Driest SL, Shi Y, Bowton EA, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharm Ther. 2014;95(4):423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharm Ther. 2012;92(1):87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williams MS. Genomic medicine implementation: learning by example. Am J Med Genet C Semin Med Genet. 2014;166C(1):8–14. [DOI] [PubMed] [Google Scholar]
- 13. Payne TH, Corley S, Cullen TA, et al. Report of the AMIA EHR 2020 task force on the status and future direction of EHRs. J Am Med Inform Assoc. 2015;pii:ocv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei WQ, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J Am Med Inform Assoc. 2012;19(2):219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Online Mendelian Inheritance in Man. http://omim.org/. Accessed August 28, 2015.
- 18. Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Starren J, Williams MS, Bottinger EP. Crossing the omic chasm: A time for omic ancillary systems. JAMA. 2013;309(12):1237–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15(2):209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin MA, Hoffman JM, Freimuth RR, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA-B Genotype and Abacavir Dosing: 2014 update. Clin Pharm Ther. 2014;95(5):499–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saito Y, Stamp LK, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA-B) genotype and allopurinol dosing: 2015 update. Clin Pharm Ther. 2015. [Epub ahead of print, doi: 10.1002/cpt.161]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caudle KE, Rettie AE, Whirl-Carrillo M, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clin Pharm Ther. 2014;96(5):542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramsey LB, Johnson SG, Caudle KE, et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharm Ther. 2014;96(4):423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharm Ther. 2015;98(2):127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Birdwell KA, Decker B, Barbarino JM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin Pharm Ther. 2015;98(1):19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoffman MA, Williams MS. Electronic medical records and personalized medicine. Hum Genet. 2011;130(1):33–39. [DOI] [PubMed] [Google Scholar]
- 28. Kannry JL, Williams MS. Integration of genomics into the electronic health record: mapping terra incognita. Genet Med. 2013;15(10):757–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shoenbill K, Fost N, Tachinardi U, Mendonca EA. Genetic data and electronic health records: a discussion of ethical, logistical and technological considerations. J Am Med Inform Assoc. 2014;21(1):171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chute CG, Kohane IS. Genomic medicine, health information technology, and patient care. JAMA. 2013;309(14):1467–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramos EM, Din-Lovinescu C, Berg JS, et al. Characterizing genetic variants for clinical action. Am J Med Genet C Semin Med Genet. 2014;166C(1):93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kullo IJ, Jarvik GP, Manolio TA, Williams MS, Roden DM. Leveraging the electronic health record to implement genomic medicine. Genet Med. 2012;15(4):270–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Masys DR, Jarvik GP, Abernethy NF, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inform. 2012;45(3):419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Welch BM, Eilbeck K, Del Fiol G, Meyer LJ, Kawamoto K. Technical desiderata for the integration of genomic data with clinical decision support. J Biomed Inform. 2014;51:3–7. [DOI] [PubMed] [Google Scholar]
- 36. Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenomics knowledge base. Methods Mol Bio. 2013;1015:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whirl-Carrillo M, McDonagh E, Hebert J, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharm Ther. 2012;92(4):414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clinical Pharmacogenetics Implementation Consortium(CPIC). CPIC Genes/Drugs Legend. https://cpicpgx.org/genes-drugs/. Accessed January 20, 2016.
- 39. Kuperman GJ, Reichley RM, Bailey TC. Using commercial knowledge bases for clinical decision support: opportunities, hurdles, and recommendations. J Am Med Inform Assoc. 2006;13(4):369–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilcox AR, Neri PM, Volk LA, et al. A novel clinician interface to improve clinician access to up-to-date genetic results. J Am Med Inform Assoc. 2014;21(e1):e117–e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shirts BH, Salama JS, Aronson SJ, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc. 2015:pii: ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldspiel BR, Flegel WA, DiPatrizio G, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J Am Med Inform Assoc. 2014;21(3):522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharm Ther. 2014;96:482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rehm HL, Berg JS, Brooks LD, et al. ClinGen: The Clinical Genome Resource. N Engl J Med. 2015;372(23):2235–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. ClinGen EHR WG. http://clinicalgenome.org/about/working-groups/ehr/ Accessed August 31, 2015.
- 46. NHGRI. Genomic Medicine Meeting VII: Genomic Clinical Decision Support: Developing Solutions for Clinical and Research Implementation. https://www.genome.gov/27558904. Accessed August 28, 2015.
- 47. National Academy of Medicine. DIGITizE: Displaying and Integrating Genetic Information Through the EHR http://iom.nationalacademies.org/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/EHR.aspx. Accessed August 28, 2105.
- 48. National Academy of Medicine. IOM Use Models Pharmacogenomic Use Pilot for TPMT (Thiopurine) and CYP2C19 (Plavix) http://iom.nationalacademies.org/∼/media/Files/Activity%20Files/Research/GenomicBasedResearch/Action%20Collaboratives/Pharmacogenetics%20Standards%20Model.pdf?la=en. Accessed August 28, 2015.