Abstract
Mammalian genomes contain several billion base pairs of DNA which are packaged in chromatin fibers. At selected gene loci, cohesin complexes have been proposed to arrange these fibers into higher-order structures (1–7) but it is poorly understood how important this function is for determining overall chromosome architecture and how this process is regulated. Using conditional mutagenesis in the mouse, we show that depletion of the cohesin-associated protein Wapl (8, 9) stably locks cohesin on DNA, leads to clustering of cohesin in axial structures, and causes chromatin condensation in interphase. These findings reveal that the stability of cohesin-DNA interactions is an important determinant of chromatin structure and imply that cohesin has an architectural role in interphase chromosome territories. We further show that regulation of cohesin-DNA interactions by Wapl is important for embryonic development, expression of c-myc and other genes, and for cell cycle progression. In mitosis, Wapl mediated release of cohesin from DNA is essential for proper chromosome segregation and protects cohesin from cleavage by the protease separase, thus enabling mitotic exit in the presence of functional cohesin complexes.
Chromosome segregation depends on sister chromatid cohesion mediated by cohesin (reviewed in 10), whose subunits Smc1, Smc3 and Scc1 (also called Rad21 or Mcd1) embrace DNA strands as a ring (11). In mitotic prophase, cohesin is removed from chromosome arms by Wapl (8, 9), which might open the Smc3-Scc1 interface (10, 12, 13), and in metaphase from centromeres by separase, which opens the cohesin ring by cleaving Scc1 (14, 15). Separase is essential for sister chromatid separation (14, 16–18), but it is unknown if Wapl is also required for this process, despite the fact Wapl releases the bulk of cohesin from chromosomes.
Cohesin also associates with unreplicated DNA, presumably because cohesin has additional functions in gene regulation (reviewed in 19). These have been proposed to depend on long-range chromatin interactions which are mediated by cohesin together with the transcription factors CCCTC binding factor (CTCF) and Mediator (1–7). How important cohesin-mediated chromatin interactions are for the structural and functional organization of mammalian genomes, and how these interactions are regulated to allow changes in chromatin structure and gene expression, is poorly understood. We, therefore, analyzed if Wapl is required for releasing cohesin from DNA not only in mitosis but also in interphase, and if the regulation of cohesin-DNA interactions by Wapl is important for chromatin structure, gene regulation and chromosome segregation
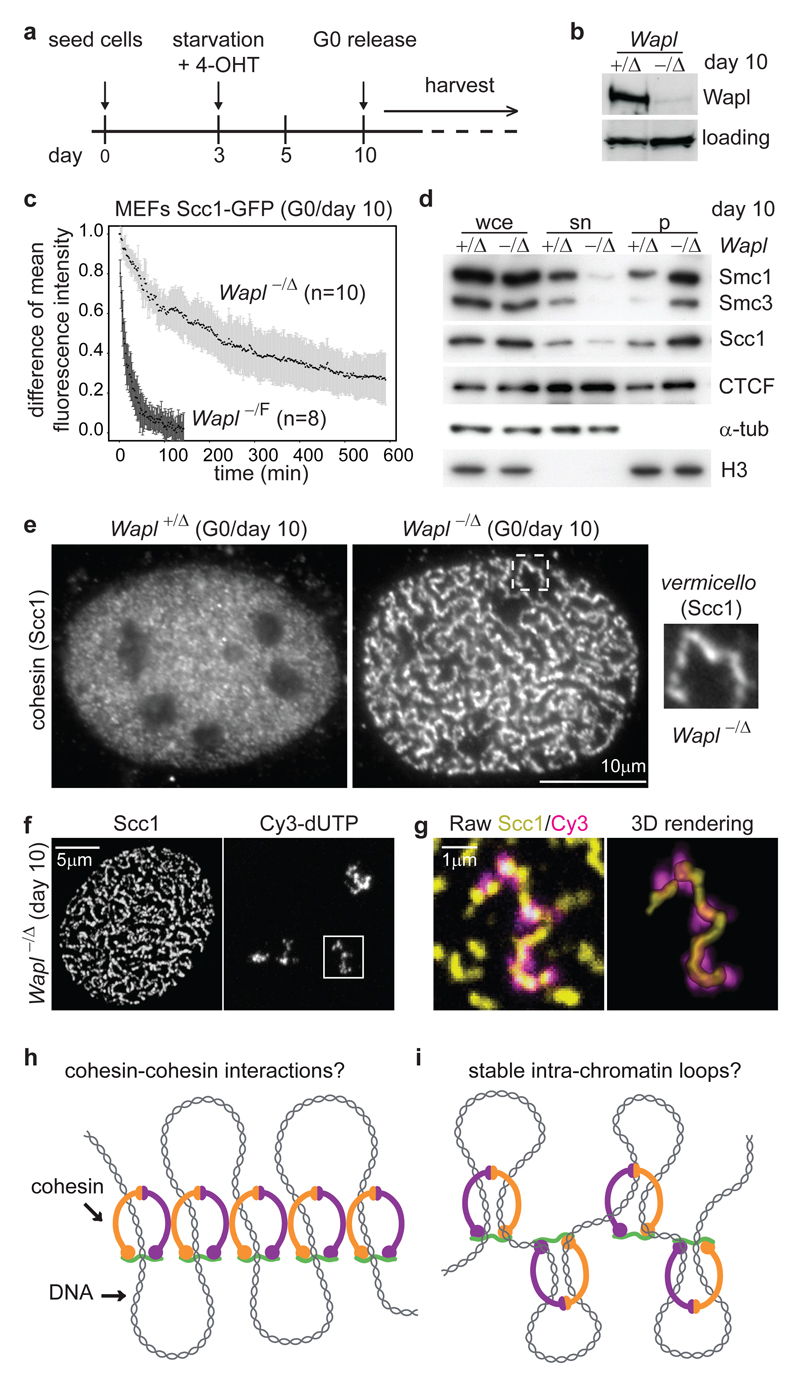
For this purpose we generated mice whose Wapl alleles (also called Wapal) were replaced by ‘floxed’ alleles (WaplF) which can be converted by Cre recombinase into WaplΔ ‘null’ alleles lacking exons 3 and 4 (called Wapl- following germline transmission; Supplementary Fig. 1a, b, and Methods Summary). No Wapl-/- mice were born (data not shown), indicating that Wapl is essential for development. We, therefore, used mouse embryonic fibroblasts (MEFs) in which WaplF is deleted following activation of Cre-estrogen receptor (Cre-ER(T2)) by 4-hydroxytamoxifen (4-OHT). When these cells, arrested in a quiescent state (G0) by serum starvation, were treated with 4-OHT, Wapl levels decreased over seven days (Fig. 1a, b and Supplementary Fig. 1c, d). Fluorescence recovery after photobleaching (FRAP) revealed that Wapl deletion increases cohesin’s chromatin-residence time more than twenty-fold: in quiescent Wapl-/F MEFs Scc1, tagged with green fluorescent protein (Scc1-GFP), bound to chromatin for 25±9 min (mean ± standard deviation (s.d.), n=8), but in Wapl-/Δ MEFs 73% ± 9% of Scc1-GFP had a chromatin-residence time of 540±240 min (n=10¸ Fig. 1c; Supplementary Fig. 2). Consequently, most cohesin accumulated on chromatin (Fig.1d). Wapl is, therefore, essential for proper release of cohesin from chromatin.
Figure 1. Wapl depletion reveals arrangement of cohesin in axial chromosomal domains.
a, Protocol for Wapl deletion by 4-OHT in G0 MEFs. b, Immunoblot of protein extracts from MEFs of indicated genotypes, treated as in (a). Loading, unidentified protein cross-reacting with Wapl antibodies. c, FRAP analysis of MEFs expressing Scc1-GFP. The difference in fluorescence intensity between bleached and unbleached regions is plotted over time. Black dots, mean values. Error bars, s.d.. d, Immunoblot of whole cell extracts (wce), supernatant (sn) and pellet (p) fractions from MEFs. α-tub, α -tubulin. e, Microscopy images of non-pre-extracted MEFs stained for Scc1. f, Maximum intensity projections of confocal z-stack images of MEFs in which chromosome territories were labeled with dUTP-Cy3, and stained for Scc1. Maximum projection was restricted to z-planes encompassing dUTP-Cy3 signals. g, Magnified images of regions boxed in (f). h, i Alternative models for how Wapl depletion might cause vermicelli formation and chromatin compaction.
Unexpectedly, immunofluorescence microscopy (IFM) experiments revealed that Wapl depletion changes cohesin localization: cohesin was located in most chromatin areas of quiescent Wapl+/Δ MEFs, but in 80% of Wapl-/Δ MEFs cohesin was enriched in elongated nuclear structures, which we call vermicelli (Italian for ‘little worms’; Fig. 1e, Supplementary Fig. 3). Vermicelli were also seen by live-cell imaging, ruling out fixation artefacts (Supplementary Fig. 4). CTCF was partially enriched in vermicelli, whereas histone H2B localization remained unchanged (Supplementary Fig. 5). Co-staining of chromosome 11 and cohesin by in situ hybridization (FISH) and IFM, respectively, suggested that one vermicello is present per chromosome territory (n=3) (Supplementary Fig. 6a, b). Labeling of individual chromosomes with dUTP confirmed this (n=31 chromosomes) (Fig. 1f, g and Supplementary Fig. 6c, d). In Wapl deficient cells, cohesin is, therefore, located in an axial chromosomal domain which might extend from one telomere to the other. Wapl depletion and stabilization of cohesin on DNA could either promote interactions between cohesin complexes, or could enhance an effect that cohesin might have on chromatin structure, such as intra-chromatid loop formation, and lead to cohesin clustering indirectly (Fig. 1h, i).
Chromatin immunoprecipitation and Solexa DNA sequencing (ChIP-seq) showed that most cohesin and CTCF sites remained unchanged following Wapl depletion (Supplementary Fig. 7), implying that vermicelli are composed of cohesin sites that also exist normally. We, therefore, analyzed if vermicelli are also present in wild-type cells. Although most cohesin had a fine-punctate distribution in chromatin, we observed short axial staining patterns in Wapl+/+ MEFs and HeLa cells. The axial cohesin staining overlapped only partially with H2B (data not shown), and became more prominent after Wapl RNAi (Supplementary Fig. 8) indicating that some cohesin is also located in axial domains in wild-type chromosomes.
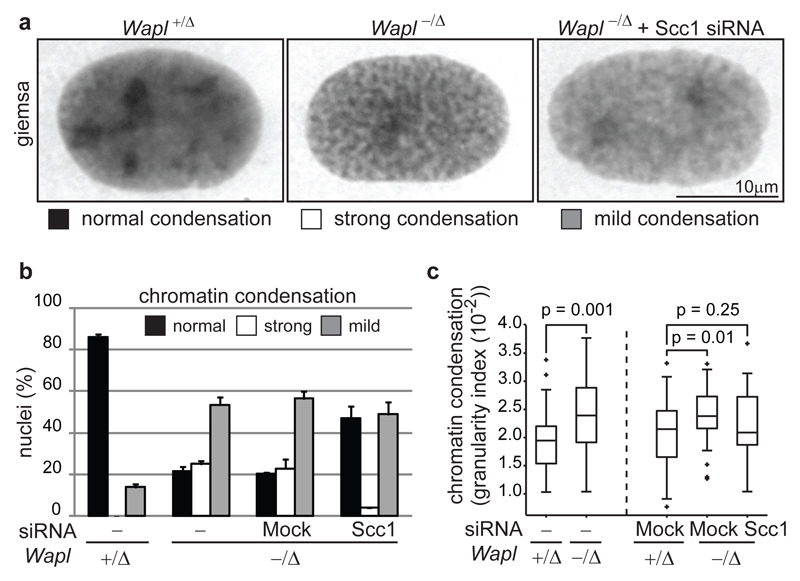
Wapl depletion also had major effects on chromatin structure: in FISH experiments, chromosome 11 occupied a significantly smaller median volume in Wapl-/Δ (7x102 voxel) than in Wapl+/Δ MEFs (8x102 voxel; Supplementary Fig. 6a, b). Giemsa staining showed that chromatin appeared more condensed in 78.5% of Wapl-/Δ MEFs (Fig. 2a, b). A ‘granularity index’ analysis of 4',6-diamidino-2-phenylindole (DAPI) stained cells (see Methods) confirmed this (Figure 2c and Supplemental Fig. 6e). In contrast, no changes in the distribution of heterochromatin protein 1a (HP1α), histone H3 trimethylated on K9 (H3-K9me3), condensin’s subunit Smc2 and topoisomerase-IIα and topoisomerase-β could be detected in IFM and immunoblotting experiments (data not shown). Partial cohesin depletion by Scc1 RNAi reduced chromatin condensation in Wapl-/Δ MEFs (Fig. 2a, b). These observations indicate that Wapl deficiency causes chromatin compaction by stabilizing cohesin on DNA, and not indirectly. The compaction state of interphase chromatin, therefore, depends on the stability of cohesin-DNA interactions, which is controlled by Wapl.
Figure 2. Wapl controls chromatin structure by regulating cohesin-DNA interactions.
a, Microscopy images of Giemsa stained interphase nuclei from MEFs of indicated genotypes. b, Quantification of nuclear morphologies in cells from (a), shown as mean and s.d. of two experiments (n=200/condition). c, Dense DAPI chromatin structures in MEFs were quantified by granularity index (n>40; p-values, Mann-Whitney U test). The boxes have lines at the lower quartile, median, and upper quartile values.
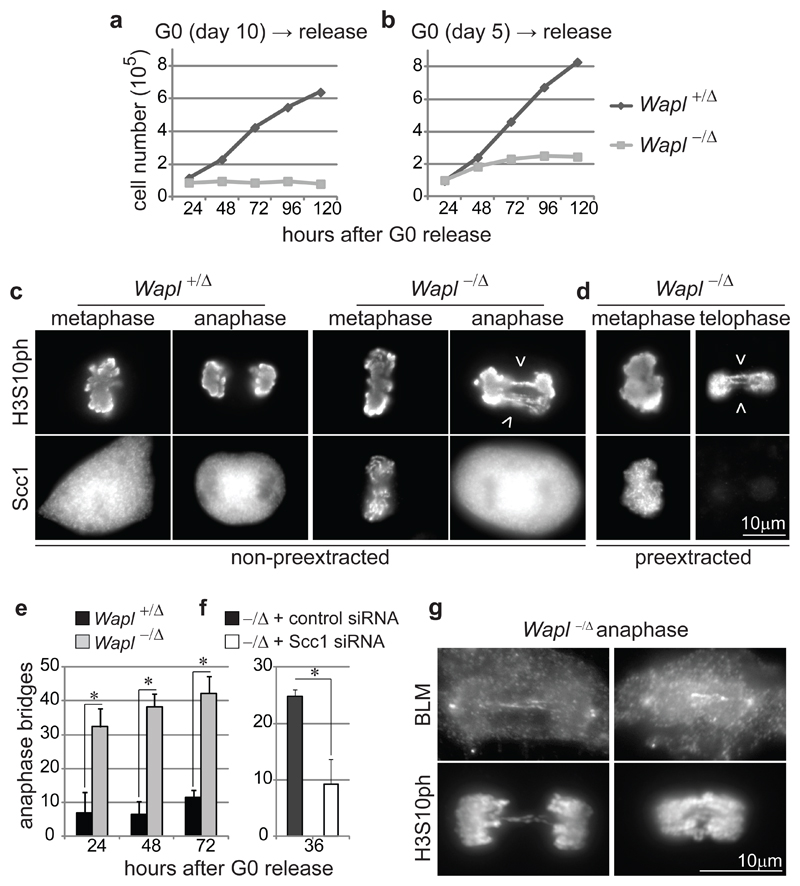
When stimulated with serum, quiescent Wapl-/Δ MEFs failed to proliferate (Fig. 3a). IFM experiments in which cells were released from quiescence in the continuous presence of bromodeoxyuridine (BrdU; Supplementary Fig. 9a, b) showed that fewer Wapl-/Δ than Wapl+/Δ MEFs incorporated BrdU. Wapl-/Δ cells which had been able to enter S or G2-phase also contained residual Wapl (data not shown), indicating that cells fully depleted of Wapl do not replicate DNA. Degradation of cyclin-dependent kinase inhibitor p27Kip1 and cyclin-A2 expression were reduced in Wapl-/Δ MEFs (Supplementary Fig. 9c), indicating that Wapl is required for G1-phase progression. To test if Wapl is also needed for DNA replication, we depleted Wapl from Xenopus egg extracts. This did not abrogate DNA replication (Supplementary Fig. 10), implying that Wapl deficiency prevents S-phase indirectly by interfering with exit form quiescence and/or G1 progression.
Figure 3. Wapl is essential for cell cycle progression and proper chromosome segregation.
a,b Proliferation curves of MEFs obtained as in Fig. 1a. c,d IFM images of metaphase, anaphase and telophase MEFs 72 hours after G0 release as in (b), either pre-extracted (d) or not (c) and co-stained for Scc1 and histone H3 phosphorylated on serine 10 (H3S10ph). Arrowheads indicate chromosome bridges. e, Quantification of chromosome bridge frequency in MEFs from (c, d), shown as mean and s.d. of three experiments (n≥ 100/condition; asterisk, P< 0.05, two-tailed Student’s test). f, Quantification of chromosome bridge frequency in Wapl -/Δ MEFs treated with Scc1 or control siRNAs, shown as mean and s.d. of three experiments (n≥ 156/condition; P< 0.05). g, IFM images of Wapl -/Δ anaphase MEFs 48 hours after G0 release (b) co-stained for BLM and H3S10ph.
Because cell cycle progression depends on transcription, we tested if Wapl is required for gene regulation. As serum stimulates proliferation in Wapl proficient but not in Wapl deficient cells, we analyzed quiescent cells to avoid indirect cell cycle effects (Supplementary Fig. 3). When we compared the levels of 21,169 RefSeq mRNAs between Wapl+/Δ and Wapl-/Δ MEFs using DNA microarrays, we found 1,152 differentially expressed genes (DEGs; 469 up-regulated, 683 down-regulated;), including the cell cycle transcription factor c-myc whose mRNA was reduced 4-fold (Supplementary Fig. 11). Wapl-/Δ MEFs in which c-myc was expressed ectopically did still not proliferate, but entered S and G2-phase more frequently (Supplementary Fig. 12), indicating that Wapl enables cell cycle progression in part by promoting c-myc expression. Wapl depletion increased the mean Smc3-ChIP read density at the transcription start site (TSS) of many down-regulated genes, including c-myc, and at non-TSS cohesin sites (Supplementary Fig. 13), implying that Wapl depletion down-regulates some genes by stabilizing cohesin on DNA. However, Wapl depletion decreased the mean Smc3-ChIP read density at the TSSs of most up-regulated genes (Supplementary Fig. 14). Although it remains to be understood how Wapl affects cohesin at these genes and increases their expression, our results raise the possibility that changes in gene expression depend on regulation of cohesin-DNA interactions by Wapl. Precedence for the regulatability of cohesin-DNA interactions comes from the observation that sister chromatid cohesion depends on inhibition of Wapl by sororin (20), which stabilizes cohesin on DNA (21), and from the finding that Wapl-mediated cohesin release is activated in prophase (9).
To analyze if Wapl is required for mitosis, we analyzed MEFs containing residual Wapl sufficient for one round of cell division (Fig. 3b and Supplementary Fig. 15a-d). In these cells, chromosome bridges occurred frequently in anaphase and telophase and many cells became binucleated (Fig. 3c-e and Supplementary Fig. 15e-g). Although partial Scc1 depletion by RNAi reduced the chromosome bridge frequency (Fig. 3f) we could not detect Scc1 on them (Fig. 3d and Supplementary Fig.16a, c). The chromosome bridge phenotype does, therefore, depend on cohesin, but may not be caused by an inability of Wapl depleted cells to remove cohesin from chromosomes at anaphase onset. Instead, Wapl depletion might cause chromosome bridges by preventing release of cohesin in early mitosis, which might be required for de-catenation of sister chromatids (22,23). Consistently, at least one chromosome bridge in 54.5±14.9% of Wapl-deficient MEFs contained the Bloom Syndrome helicase (BLM; Fig. 3g; triplicate experiments; n cells with bridges=86), which has been implicated in de-catenation (24; for further characterization of these bridges see Supplementary Fig. 17). Proper sister chromatid separation in anaphase, therefore, depends on early mitotic release of cohesin from chromosomes by Wapl.
In Wapl-/Δ MEFs from which soluble proteins had not been pre-extracted, Scc1 was located on chromosomes in metaphase but cytoplasmically in anaphase (n=200), implying that Wapl deficiency delays cohesin release from prophase to anaphase onset (Fig. 3c, d). To test if separase is required for releasing cohesin from chromosomes in Wapl-depleted cells we analyzed MEFs in which both Wapl and separase genes were deleted. Chromosome spread analysis confirmed that Wapl and separase deficiency prevent chromosome arm separation in prometaphase (8,9) and centromere separation in anaphase, respectively, leading to diplochromosome formation due to mitotic exit with unseparated sister chromatids in separase-depleted cells (17,18). Prometaphase Wapl-/Δ separase-/Δ cells showed a combination of both phenotypes, including diplo-chromosomes with un-separated arms (Fig. 4a and Supplementary Fig. 18a), indicative of defects in arm and centromere separation. IFM revealed that in Wapl-/Δ separase-/Δ MEFs sister chromatids remained connected and cohesin persisted on chromosomes even when cells exited mitosis, as measured by a decrease in cyclin-B1 levels and re-localization of Aurora-B from centromeres to mid-spindles (Fig. 4b and Supplementary Fig. 18b-d). Separase is, therefore, required for releasing cohesin from both centromeres and chromosome arms in Wapl depleted cells.
Figure 4. The prophase pathway of cohesin dissociation protects cohesin from cleavage by separase.
a, Microscopy images of Giemsa stained prometaphase chromosomes from MEFs of indicated genotypes. Diplo, diplochromosomes (n prometaphase cells=100). b, IFM images of metaphase and anaphase MEFs co-stained for DAPI, Scc1 and Cyclin-B1. c, Immunoblot of extracts from MEFs expressing Scc1-9myc synchronized in G0, prometaphase (0’) or anaphase (15’). White and black arrow-heads and the asterisk indicate full length Scc1-9myc, an anaphase-specific cleavage product of Scc1-9myc, and a cross-reacting band, respectively. d, IFM images of preextracted early-G1 MEFs co-stained for DAPI, Aurora B and Scc1. e, Quantification of Scc1 immunofluorescence signals from MEFs analyzed as in (d). Magenta bars, means of three experiments. Dots, single data points (n>37/condition;asterisk, P< 0.0001, two-tailed Student’s test).
We noticed that separase cleaved more Scc1 in Wapl depleted cells than normally, presumably because separase cleaves chromosome-bound but not soluble cohesin (15,25) (Fig. 4c; note that these cells also contained un-cleaved Scc1, possibly because cells had been treated only for two days with 4-OHT and thus contained residual Wapl, and/or because in these experiments only 35% of MEFs entered anaphase after release from the prometaphase arrest, Supplementary Fig. 16a-d). This implied that Wapl deficient cells might exit mitosis with less intact cohesin and can, therefore, not load as much cohesin onto chromatin as normally in the next cell cycle. Indeed, IFM revealed that significantly less chromatin-bound Scc1 was detectable in Wapl-/Δ than in Wapl+/Δ MEFs in telophase and G1 (Fig. 4d, e and Supplementary Fig. 16e; note that in this experiment Wapl was depleted more efficiently than in Fig. 4c by treating cells for 7 days with 4-OHT, presumably causing more Scc1 cleavage than in Fig. 4c). The prophase pathway of cohesin, therefore, allows mitotic exit in the presence of functional cohesin, enabling it to function early in the next cell cycle.
Our findings that Wapl depletion ‘locks’ cohesin on chromatin in a stably bound state and alters chromatin compaction to a degree that can be seen by light microscopy, indicate that the stability of cohesin-DNA interactions is an important determinant of chromatin structure, perhaps because cohesin’s residence time on DNA determines if and for how long intra-chromatid loops can be formed (Fig. 1h, i). Wapl’s role in chromatin structure may be evolutionary conserved as alterations in chromosome morphology have been observed in fruit flies and yeast Wapl mutants (26–28). Our observation that cohesin complexes form axial structures (vermicelli) in interphase chromosomes of Wapl depleted cells and to a lesser degree in wild type cells, implies that cohesin has an architectural role in the organization of interphase chromosome territories. Similar roles have been proposed for cohesin and the related condensin complexes in meiotic and mitotic chromosomes, respectively (29,30). Because cohesin-related complexes exist in all kingdoms of life, the architectural role of cohesin that we propose here for interphase chromosomes may represent an ancient function which may have helped to organize DNA in chromosomes before nucleosomes existed.
Methods Summary
Generation of conditional Wapl allele
The conditional Wapl allele was generated by inserting loxP sites into introns 2 and 4 to allow Cre-mediated excision of exons 3 and 4 (Supplementary Fig. 1). Correctly targeted ES cell clones were identified by Southern blotting of EcoRI and SacI digested genomic DNA to confirm the presence of 5’ and 3’ loxP sites, respectively (data not shown).
Cell culture
Immortalized MEFs (iMEFs) were used for the fractionation experiments in Fig. 1d, the nucleotide labeling experiment in Fig. 1f, and for experiments in which tagged versions of Scc1 were stably expressed. All the other experiments were performed in primary MEFs from E13.5 embryos. MEFs and iMEFs were cultured as described (18).
To synchronize MEFs or iMEFs in quiescence and to activate CreER(T2), cells were first cultured to confluence and then serum starved for seven or two days (corresponding to day 10 and 5 in Fig. 1a, respectively) with optiMEM media (Invitrogen) supplemented with 2% charcoal/dextran-treated serum (Hyclone), 100 unitis/ml penicillin (Sigma), 100 μg/ml streptomycin (Sigma), 0.5 μM 4-OHT (Sigma, 10 mM stock in ethanol). Cells were split and released in fresh medium (10% serum) for cell cycle analysis.
Cells were enriched in prometaphase of mitosis by nocodazole (Sigma) treatment for five hours at 300 ng/ml (Fig 4a) or at 200 ng/ml (Fig 4c). Cells were either continuously labeled (Supplementary Fig. 9a) or pulsed (2 hours) (Supplementary Fig. 12f) with 40 μM BrdU (Sigma).
Methods
Generation of conditional Wapl allele
The conditional Wapl allele was generated by inserting loxP sites into introns 2 and 4 to allow Cre-mediated excision of exons 3 and 4 (Supplementary Fig. 1). Correctly targeted ES cell clones were identified by Southern blotting of EcoRI and SacI digested genomic DNA to confirm the presence of 5’ and 3’ loxP sites, respectively (data not shown).
Mice
The conditional and null Wapl mice were generated by crossing with FLPe (31) and MORE mice (32), respectively. Separase mice (18), Rosa26CreER(T2) mice (33), and H2B-GFP transgenic mice (34) have been described. Mice were hybrids between C57BL/6 and 129 strains.
Cell culture
Immortalized MEFs (iMEFs) were used for the fractionation experiments in Fig. 1d, the nucleotide labeling experiment in Fig. 1f, and for experiments in which tagged versions of Scc1 were stably expressed. All the other experiments were performed in primary MEFs from E13.5 embryos. MEFs and iMEFs were cultured as described (18). Wapl -/F Rosa26 ERCre (T2)/+ iMEFs stably expressing mouse Scc1 (cDNA) tagged with nine myc epitopes (Scc1-9myc) or EGFP (Scc1-EGFP) or Dronpa (Scc1-Dronpa) (18) or mouse Scc1 (BAC RP23-375K15) tagged with LAP (35) were generated as described.
To synchronize MEFs or iMEFs in quiescence and to activate CreER(T2), cells were first cultured to confluence and then serum starved for seven or two days (corresponding to day 10 and 5 in Fig. 1a, respectively) with optiMEM media (Invitrogen) supplemented with 2% charcoal/dextran-treated serum (Hyclone), 100 unitis/ml penicillin (Sigma), 100 μg/ml streptomycin (Sigma), 0.5 μM 4-OHT (Sigma, 10 mM stock in ethanol). Cells were split and released in fresh medium (10% serum) for cell cycle analysis.
Cells were enriched in prometaphase of mitosis by nocodazole (Sigma) treatment for five hours at 300 ng/ml (Fig 4a) or at 200 ng/ml (Fig 4c). Cells were either continuously labeled (Supplementary Fig. 9a) or pulsed (2 hours) (Supplementary Fig. 12f) with 40 μM BrdU (Sigma).
MEFs used in Fig. 4a were infected with AdCre expressing virus (18) and collected 48 hours after infection for chromosome spread and Giemsa staining (36) as described. Plasmids for c-myc rescue experiments (Supplementary Fig. 12) have been described in (37).
Nucleotide labeling of interphase chromosome territories
To incorporate Cy3-dUTP in iMEFs, cells were grown on Lab-tek chambered coverglass and synchronized at the G1/S transition by two days serum starvation followed by 42 hours incubation in growing medium containing 1ug/mL aphidicolin (Sigma-Aldrich). Immediately after aphidicolin release, the cell layer, bathed with growing medium containing 10 µM of Cy3-dUTP (GE Healthcare), was scratched using the tip of a syringe needle to allow nucleotide loading (38) and integration to the DNA during replication. After 4 days in growing medium and due to the successive cell divisions, each nucleus was containing only a few labeled chromosomes, each of them appearing as a dense group of fluorescent spots by light microscopy as described (39). Cells were then serum starved in presence of 4-hydroxy-tamoxifen for seven days and imaged.
Antibodies
Most of the antibodies have been used at 1 μg/ml, except where otherwise indicated.
Immunofluorescence: rabbit anti-myc (myc (527), ID A957) (15), mouse anti-myc (myc (9E10), ID A668) (15), mouse anti-BrdU (BD 347580), mouse anti-BrdU (Caltag MD5110, 1:500), rabbit anti-Wapl (AfR266-3L, a gift from T. Hirano, 1:250), chicken anti-GFP (Abcam ab13970), rabbit anti-CTCF (CTCF-35, ID A992), rabbit anti-cyclinB1(Cell Signaling 4138S), mouse anti-AuroraB (BD 611083), mouse anti-Scc1 (UPSTATE 05-908), rabbit anti-Scc1 (Scc1 (623) ID A900) (15), rabbit anti-H3S10ph (Millipore 06570, 1:500), mouse anti-H3S10ph (Cell Signaling 9706L, 1:100), rabbit anti-BLM (Abcam ab476, 1:200), rat anti-RPA32 (Cell Signaling 4EA, 1:200), Alexa fluorophore (488,568,633) conjugated antibodies (Molecular Probes, 1:1000).
Western Blotting: rabbit anti-Wapl (hWapl (4748), IDs A960; hWapl (4749), A961) (9), goat anti-H3 (Santa Cruz sc-8654), rabbit anti-cyclinA (Santa Cruz sc-596), mouse anti-Scc1 (UPSTATE 05-908), rabbit anti-Smc3 (Bethyl A300-060A), rabbit anti-Smc1 (Bethyl A300-055A), mouse anti-tubulin (Sigma B512), rabbit anti-p27 (a gift from M. Barbacid, 1:250), rabbit anti-CTCF (CTCF-35, ID A992).
Chromatin IP (ChIP): rabbit anti-Smc3 (Bethyl A300-060A) (40) and rabbit anti-CTCF (UPSTATE 07-729) (4,41) have been described. Methods for subcellular fractionation and Western blotting (9) have been described.
Immunofluorescence and immuno-FISH
Cells were either grown on 22 mm glass coverslips (Menzel), fibronectin coated slides (BD Biosciences), Lab-tek chambered coverglass (Nunc), or spun onto glass slides (Menzel) using a Cytospin centrifuge (Shandon), fixed with 4% PFA and stained as described (36). Where indicated, cells were extracted using 0.1% Triton X-100 for 2 min before fixation. BrdU staining was performed as described (42) except that cells were fixed with 4% PFA instead of 70% ethanol. RPA32 and BLM staining was performed as described (43).
For immuno-FISH experiments MEFs were fixed with 2% PFA and stained with a mouse anti-Scc1 antibody (UPSTATE 05-908) and Alexa 488 secondary antibody as described (36). MEFs were then fixed with 1% PFA and hybridized with mouse paint specific for mouse chromosome 11 labeled with a red emitting fluorochrome (Metasystems) according to manufacturer’s instruction. DNA was counterstained with DAPI (Roche Diagnostics) at 2 μg/ml. Slides were mounted using Vectashield Mounting medium (Vector Laboratories).
Microscopy and image analysis
Epifluorescence Microscopy was performed with an Axioplan 2 microscope (Zeiss) using a CoolSnap HQ camera (Photometrics) and images were processed with MetaMorph (Universal Imaging).
FRAP imaging and analysis was carried out as described (45). Cells were imaged with a Zeiss 780 confocal microscope using a Zeiss Fluar 40x/1.3 Oil objective. After capture of a pre-bleach image, half of the nucleus was bleached followed by time-lapse imaging with 2 min time resolution over 2 hrs (control cells) and 10 hrs (Wapl depleted cells). For each time-point measurements were taken in user-defined regions.
Single point confocal laser scanning microscopy was performed with Zeiss 510 (Supplementary Fig. 5) and Zeiss LSM780 (Fig. 1 f, g) with a plan-apochromat 63x/1.4 oil objective. Different fluorescence signals were collected in multitrack mode set up for three channels to fit with the fluorescence spectrum of the respective dyes. Colocalization of Scc1 and H2B or CTCF was performed with ImageJ (44). The association of cohesin vermicelli with dUTP-Cy3 labeled DNA is described in Supplementary Information. Confocal spinning disk microscopy (Supplementary Fig. 6a) was performed using Ultraview run under Velocity software with a plan-apochromat 63x/1.4 oil objective (Perkin Elmer). To avoid cross-talk of fluorophors, channels were acquired sequentially using bandpass filters covering respective emission spectra of used fluorophors. Deconvolution was performed using Huygens Professional (SVI, Netherland) software using classic maximum likelihood estimation (CMLE) algorithm.
Live-cell imaging was performed in a Zeiss TIRF 3 microscope used in widefield mode at 37 °C using CO2 independent medium (Invitrogen). To image Scc1-Dronpa and H2B-PAmCherry, cells were illuminated with a 405 nm laser for 5-10 seconds to activate its fluorescent state. Afterwards, cells could be imaged using a 488 nm and 561 nm laser for Scc1-Dronpa and H2B-PAmCherry, respectively. Hoechst and Scc1-EGFP were imaged using a 405 nm and 488 nm laser, respectively.
RNA interference
Transfection of control GL2 siRNA (Ambion), human Wapl siRNA (Ambion) (9), and mouse Scc1 siRNA (Ambion, ID s72658) was performed with Lipofectamine RNAi MAX (Invitrogen) according to manufacturer’s instruction. All siRNAs were used at a final concentration of 100 nM. MEFs in Fig. 2a-c were released from quiescent (day 10 in Fig. 1a) into medium containing the indicated siRNA. Cells were then analyzed 48 hours after transfection (Scc1 levels measured by quantitative IFM were reduced to 57±13.4% (mean and s.d. of two experiments)).
Chromatin immunoprecipitation and ChIP-seq data analysis
Cohesin and CTCF ChIP was performed as described (41), except that samples were sonicated for 12 x 60 second pulses with a Covaris sonicator, and the DNA was purified with Phase lock gel tubes (5PRIME). ChIP-seq sample preparation, sequencing and data analysis are described in Supplementary Information.
Transcriptome microarray analysis
RNAs were extracted from MEFs with RNeasy Mini Kit (QIAGEN). Microarray hybridization and data analysis are described in Supplementary Information. Methods for RT-qPCR (41) have been described.
Statistics
Statistical analyses were carried out using the GraphPad Prism software package (GraphPad Software, Inc.). Statistical analyses for Transcriptome and ChIP-seq experiments are described in Supplementary Information.
Supplementary Material
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We dedicate this paper to the memory of Dr. Beate Peters who performed the first experiments on Wapl in our laboratory. We are grateful to K. Aumayr, O.F.-Capetillo, T. Hoffmann, M.E. Idarraga-Amado, S. Kueng, T. Kulcsar, M. Leeb, P. Pasierbek, D. Santamaría, G. Schmauss, A. Souabni, H. Tkadletz, K. Wendt and members of the Peters laboratory for discussions and assistance, J. Hutchins for suggesting the term vermicelli, K. Nasmyth for the separase mouse model, and M. Barbacid, T. Cremer, T. Hirano, T. Jenuwein, M. Malumbres, J. Zuber for reagents. T.N. was supported by the European Molecular Biology Organization (EMBO) and the Japanese Society for the Promotion of Science (JSPS). S.H. was supported by funds from the Agence National de la Recherche (JCJC-SVSE2-2011, ChromaTranscript project) and from the European Union (FP7-PEOPLE-2011-CIG, ChromaTranscript project). D.A.C. was supported by MFPL VIPS Programm (BMWF and City of Vienna). Research in the laboratory of J.-M.P is supported by Boehringer Ingelheim, the Austrian Science Fund (FWF special research program SFB F34 ‘Chromosome Dynamics’, and Wittgenstein award Z196-B20), the Austrian Research Promotion Agency (FFG, Laura Bassi Center for Optimized Structural Studies), the Vienna Science and Technology Fund (WWTF LS09-13), and the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 241548 (MitoSys).
Footnotes
Author Contributions Experiments were designed and data interpreted by A.T., G.W., S.H., M.J., J.E. and J.-M.P. A.T. generated Wapl floxed ES cells, established and maintained the mouse colony required for this study, and carried out chromosome segregation, transcriptome, and ChIP-seq studies. A.T. and G.W. analyzed cohesin localization by IFM, and cleavage of Scc1 in anaphase MEFs. G.W. generated MEFs expressing Scc1-LAP and Scc1-9myc, and carried out cohesin IFM experiments in telophase and G1 MEFs. G.W. and V.B. performed chromatin fractionation experiments. G.W. and T.N. performed cleavage assays of Scc1 with Xenopus egg extracts. A.T., W.G. and W.T analyzed cell cycle progression of MEFs. S.H. performed DNA granularity analysis and dUTP-Cy3 labeling of DNA. M.J. carried out bioinformatic analyses. A.Wuensche performed cohesin FRAP experiments. E.S. performed FISH experiments. A.Wutz supervised work of A.T. with ES cells and participated in designing Wapl targeting strategy. I.F.D performed replication assays with Xenopus egg extracts. D.A.C. performed live imaging of cohesin vermicelli under guidance of A.V. A.T. and J.-M.P. wrote the manuscript.
Author Information ChiP-seq and microarray data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under accession code GSE41603. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishiro T, et al. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. Embo J. 2009;28(9):1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nativio R, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5(11):e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seitan VC, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2012;476(7361):467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demare LE, et al. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013 doi: 10.1101/gr.156570.113. Advance online article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips-Cremins JE, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153(6):1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16(24):2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kueng S, et al. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127(5):955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 10.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22(22):3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 11.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454(7202):297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 12.Chan KL, et al. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150(5):961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buheitel J, Stemmann O. Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3-Scc1 gate. Embo J. 2013;32(5):666–676. doi: 10.1038/emboj.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400(6739):37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 15.Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103(3):399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 16.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293(5533):1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 17.Kumada K, et al. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. J Cell Biol. 2006;172(6):835–846. doi: 10.1083/jcb.200511126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirth KG, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172(6):847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seitan VC, Merkenschlager M. Cohesin and chromatin organisation. Curr Opin Genet Dev. 2012;22(2):93–100. doi: 10.1016/j.gde.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama T, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143(5):737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz J, Watrin E, Lenart P, Mechtler K, Peters JM. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol. 2007;17(7):630–636. doi: 10.1016/j.cub.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 22.Wang LH, Mayer B, Stemmann O, Nigg EA. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J Cell Sci. 2010;123(Pt 5):806–813. doi: 10.1242/jcs.058255. [DOI] [PubMed] [Google Scholar]
- 23.Farcas AM, Uluocak P, Helmhart W, Nasmyth K. Cohesin's concatenation of sister DNAs maintains their intertwining. Mol Cell. 2011;44(1):97–107. doi: 10.1016/j.molcel.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol. 2011;22(8):906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, et al. Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner. Cell. 2009;137(1):123–132. doi: 10.1016/j.cell.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verni F, Gandhi R, Goldberg ML, Gatti M. Genetic and molecular analysis of wings apart-like (wapl), a gene controlling heterochromatin organization in Drosophila melanogaster. Genetics. 2000;154(4):1693–1710. doi: 10.1093/genetics/154.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guacci V, Koshland D. Cohesin-independent segregation of sister chromatids in budding yeast. Mol Biol Cell. 2012;23(4):729–739. doi: 10.1091/mbc.E11-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F. Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol. 2012;23(1):64–69. doi: 10.1016/j.cub.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 29.Pezzi N, et al. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams- Beuren syndrome deletion. Faseb J. 2000;14(3):581–592. doi: 10.1096/fasebj.14.3.581. [DOI] [PubMed] [Google Scholar]
- 30.Ono T, et al. Differential Contributions of Condensin I and Condensin II to Mitotic Chromosome Architecture in Vertebrate Cells. Cell. 2003;115(1):109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25(2):139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 32.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26(2):113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Seibler J, et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31(4):e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadjantonakis AK, Papaioannou VE. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. Journal of Cell Biology. 2003;161(2):281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schermelleh L, Solovei I, Zink D, Cremer T. Two-color fluorescence labeling of early and mid-to-late replicating chromatin in living cells. Chromosome Res. 2001;9(1):77–80. doi: 10.1023/a:1026799818566. [DOI] [PubMed] [Google Scholar]
- 39.Zink D, et al. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102(2):241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]
- 40.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. Embo J. 2008;27(4):654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 42.Bugler B, Schmitt E, Aressy B, Ducommun B. Unscheduled expression of CDC25B in S-phase leads to replicative stress and DNA damage. Mol Cancer. 2010;9:29. doi: 10.1186/1476-4598-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerqueira A, et al. Overall Cdk activity modulates the DNA damage response in mammalian cells. J Cell Biol. 2009;187(6):773–780. doi: 10.1083/jcb.200903033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-Cell Imaging Reveals a Stable Cohesin-Chromatin Interaction after but Not before DNA Replication. Curr Biol. 2006;16(15):1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.