Abstract
The anaphase promoting complex/cyclosome (APC/C) is a 1.2 MDa ubiquitin ligase complex with important functions in both proliferating and post-mitotic differentiated cells. In proliferating cells, APC/C controls cell cycle progression by targeting inhibitors of chromosome segregation and mitotic exit for degradation by the 26S proteasome. To understand how APC/C recruits and ubiquitylates its substrate proteins and how these processes are controlled, it is essential to analyse APC/C activity in vitro. In the past, such experiments have been limited by the fact that large quantities of purified APC/C were difficult to obtain and that mutated versions of the APC/C could not be easily generated. In this chapter we review recent advances in generating and purifying recombinant forms of the human APC/C and its co-activators, using methods that are scalable and compatible with mutagenesis. We also describe a method that allows the quantitative analysis of APC/C activity using fluorescently labelled substrate proteins.
Keywords: Enzyme activity, anaphase promoting complex/cyclosome, ubiquitin, mitosis
1. Introduction
The anaphase promoting complex/cyclosome (APC/C) is a ubiquitin ligase (E3 enzyme) which has essential functions in eukaryotes from yeast to man (reviewed in (1); (2)). In metaphase of mitosis, APC/C ubiquitylates securin, an inhibitor of the cohesin protease separase, and B-type cyclins, the activating subunits of cyclin dependent kinase 1 (CDK1). The APC/C recognises specific ‘degron’ sequences in these substrates, called the destruction box (D box) and the KEN box. The subsequent degradation of ubiquitylated securin and cyclin B by the 26S proteasome is essential for sister chromatid separation in anaphase and mitotic exit. APC/C also has functions in post-mitotic cells, for example during the terminal differentiation of cortical neurons (3, 4).
The APC/C is a 1.2 MDa protein complex composed of 14 protein subunits which are present in different stoichiometries (reviewed in (5)). To be able to recognise degron sequences in its substrate proteins, the APC/C has to associate with an additional co-activator protein, either with CDC20 in early mitosis, or with CDH1 during mitotic exit, G1-phase and in quiescent cells. CDC20 and CDH1 help to recruit substrates to the APC/C, presumably by directly binding their D and KEN boxes. Once associated with the APC/C, substrates are ubiquitylated by ubiquitin conjugating (E2) enzymes. These small monomeric enzymes are first trans-esterified with an activated ubiquitin residue by the ubiquitin activating (E1) enzyme, and subsequently transfer this ubiquitin residue to epsilon-amino side-chains of lysine residues in the APC/C substrate. The APC/C can interact with different E2s; UBCH5A, UBCH5B, UBCH5C (also known as UBE2D1, UBE2D2, UBE2D3, respectively), UBCH10 and UBE2S. UBCH10 is thought to be recruited to the APC/C via the RING finger domain of the APC/C subunit APC11. APC11 in turn is bound to the APC/C subunit APC2, which belongs to the cullin protein family. Cullin and RING finger subunits are also found in a number of other ubiquitin ligase complexes, implying that these use related catalytic mechanisms for substrate ubiquitylation reactions (6).
The activity of the APC/C is tightly controlled to enable proper progression through the cell cycle (reviewed in (7)). For example, securin and cyclin B are only recognised by APC/C-CDC20 once all chromosomes have been properly attached to both poles of the mitotic spindle. Before this state (metaphase) has been reached, APC/C’s ability to recruit securin and cyclin B is prevented by a four-subunit mitotic checkpoint complex (MCC). APC/C-CDH1 is also controlled by other mechanisms, for example the early mitotic inhibitor 1 (EMI1) (8).
Recent advances in single-particle electron microscopy have provided detailed insight into the structure of the APC/C (5). However, the catalytic mechanism of APC/C mediated substrate ubiquitylation reactions and their regulation remain incompletely understood. Studying these mechanisms requires the ability to measure the activity of APC/C using purified components, and structure-function analyses of these components via mutagenesis. For many years, these assays have utilised APC/C purified from cultured cells (or sometimes from tissues), typically using immuno-affinity purification techniques (reviewed in (9, 10)). Although these approaches have been instrumental for analysing APC/C, they were not easily scalable because the abundance of APC/C in cells is low and because the availability of APC/C antibodies needed for the affinity purification is often limited. These approaches were also not well suited for the analysis of APC/C loss-of-function mutants as these are difficult to generate in cells because APC/C is essential for cell viability.
To overcome these limitations, methods have been developed that enable the generation of recombinant forms of yeast and human APC/C, expressed in Baculovirus infected insect cells (11–13). These methods are more easily scalable and well suited for mutagenesis approaches. Here we describe one of these approaches for the generation of recombinant human APC/C. We also provide a brief description of how the other proteins can be purified that are required for the reconstitution of APC/C mediated substrate ubiquitylation reactions and describe how these reactions can be measured using a fluorescently labelled substrate.
In brief, APC/C activity can be measured by incubating the respective purified proteins, i.e., APC/C, an APC/C co-activator, E1, one or several of the E2s that cooperate with the APC/C, ubiquitin and a substrate protein in the presence of Mg2+ATP. The reaction is stopped by the addition of SDS sample buffer and analysed by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Ubiquitin-substrate conjugates can be detected on a fluorescence scanner if either a fluorescently labelled substrate or fluorescently labelled ubiquitin is used.
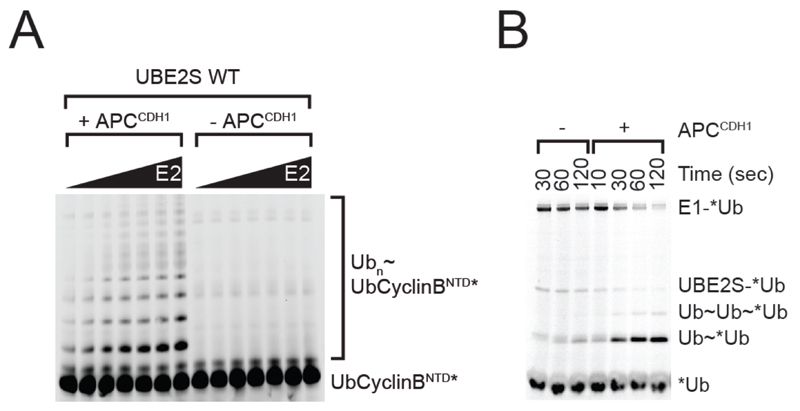
APC/C can ubiquitylate substrates at multiple lysine residues (14, 15) and, once the first ubiquitin residues have been linked to the substrate, also on lysine residues on the attached ubiquitin molecules (in the latter case APC/C typically modifies lysine residues 11, 48 and 63 on ubiquitin (16–18). APC/C mediated ubiquitylation reactions, therefore, result in the formation of complex mixtures of different substrate-ubiquitin conjugates. These are resolved by SDS-PAGE into a characteristic “ladder” of bands, each containing a different number of ubiquitin residues coupled to the substrate molecule (see Fig. 1A). SDS-PAGE analysis can therefore be used to estimate how many ubiquitin residues are coupled to a substrate by APC/C, but the identity of the modified lysines or whether linear or branched ubiquitin chains are formed can only be determined by mass spectrometric approaches (17). Alternatively, APC/C mediated ubiquitylation reactions can be performed in the presence of ubiquitin in which lysine residues have been chemically modified by methylation, so that ubiquitin itself cannot be ubiquitylated. Comparing reactions in the absence and presence of methyl-ubiquitin can, therefore, be used to determine if a substrate is modified by poly-ubiquitin chains, or by single ubiquitin residues which are attached to multiple lysine residues, or by a combination of both.
Figure 1.
(A) APC/C Drives PolyUbiquitin Chain Formation. SDS-PAGE gel illustrating that APC/C-CDH1 drives poly-ubiquitin chain formation. Visualised using a fluorescently labelled substrate. (20) (B) APC/C-CDH1-dependent formation of free Ub chains by UBE2S visualised using a fluorescently labelled donor ubiquitin, similar to previously described (20). For this experiment, the E1˜*Ub conjugate is formed by incubating 50 mM MgCl2ATP with Tube 2 containing E1 (10 uM) and fluorescent Ub (40 uM) for 10 min at room temperature. This reaction can be either left unquenched (shown) or, if desired, quenched by the addition of 50 mM Ethylenediaminetetraacetic acid (EDTA) and/or desalting. The contents of Tube 2 are diluted 10-fold into Tube 1 containing an unlabelled source of Ub (250 uM). Aliquots are then quenched by SDS sample buffer at the appropriate times and visualized by SDS-PAGE. In the experiment shown, the APC concentration used is 100 nM.
In most cases, APC/C activity is measured by using fluorescently labelled substrate, but the activity can also be analysed by using fluorescent labelled ubiquitin (see Fig. 1B). This permits not only the visualisation of substrate-ubiquitin conjugates, but also of a covalent E1-ubiquitin adenylate, and – if SDS-PAGE is performed in the absence of reducing agents – of E2-ubiquitin thioesters (which are unstable in the presence of reducing agents such as β-mercaptoethanol or dithiothreitol). A limitation of using fluorescently labelled ubiquitin is, however, that it cannot be determined to which protein the ubiquitin is conjugated (the substrate protein or to other proteins present in the reaction mixture).
When APC/C mediated ubiquitylation reactions are to be analysed in the presence of UBE2S it has to be kept in mind that this E2 preferentially ubiquitylates lysine residues (K11) in ubiquitin molecules, a property that enables it to elongate ubiquitin chains on substrate proteins (19). UBE2S activity is, therefore, typically analysed by using linear fluorescent ubiquitin-substrate fusion proteins, or by measuring the formation of fluorescent ubiquitin-ubiquitin conjugates (for an example, see (20)). Alternatively, UBE2S activity can also be analysed in the presence of either UBCH5 or UBCH10, which both efficiently ubiquitylate lysine residues in substrate proteins, thereby generating conjugates which can then serve as substrate for ubiquitin chain elongation by UBE2S.
2. Materials
2.1. Bacterial protein expression
Bacterial cells: E.coli BL21 (DE3) CodonPlus-RIL (see Note 1)
Lysogeny broth - Miller (LB): 10 g tryptone, 5 g yeast extract, 10 g NaCl per litre
0.6 M Isopropyl β-D-1-thiogalactopyranoside (IPTG)
2.2. Baculovirus-insect cell expression
2.3. Expression and purification of recombinant APC/C
Low and high speed centrifuges
Phosphate buffered saline (PBS)
APC/C lysis buffer: 50 mM Hepes pH 8, 250 mM NaCl, 5% glycerol, 2 mM Dithiothreitol (DTT), 2 mM benzamidine, 5 units/mL benzonase, 10 μg/mL aprotnin, 10 μg/uL leupeptin 5 μg/μL avidin and 1 Complete™ EDTA-free protease inhibitor tablet (per 50 mL buffer)
Sonicator
Streptactin resin
APC/C wash buffer: 50 mM Hepes pH 8, 250 mM NaCl, 5% glycerol, 2 mM DTT
Gravity flow column
APC/C elution buffer: 50 mM Hepes pH 8, 250 mM NaCl, 5% glycerol, 2.5 mM desthiobiotin, 2 mM DTT
Bradford reagent
SDS-PAGE gels and gel running equipment
APC/C buffer A: 50 mM Hepes pH 8, 5% glycerol, 2 mM DTT
APC/C buffer B: 50 mM Hepes pH 8, 1 M NaCl, 5% glycerol, 2 mM DTT
AKTA protein purification system
POROS HQ 50 μm anion exchange column
Superose 6 size exclusion column
APC/C size exclusion buffer: 20 mM Hepes pH 8, 200 mM NaCl, 1 mM DTT
2.4. Expression and purification of the co-activators - CDC20 and CDH1
Low and high speed centrifuges
PBS
Co-activator lysis buffer: 20 mM Hepes pH 7, 100 mM (NH4)SO4, 20 mM Imidazole, 5% glycerol, 1 mM DTT, 1 Complete™ EDTA-free protease inhibitor tablet (per 50 mL buffer)
Dounce homogenisors
Ni-NTA resin
Co-activator wash buffer: 20 mM Hepes pH 7, 300 mM (NH4)SO4, 20 mM Imidazole, 2.5% glycerol, 1 mM DTT
Gravity flow column
Co-activator elution buffer: 20 mM Hepes pH 7, 300 mM (NH4)SO4, 250 mM Imidazole, 2.5% glycerol, 250 mM imidazole, 1 mM DTT
Bradford reagent
SDS-PAGE gels and gel running equipment
AKTA protein purification system
Superdex 200 26/60 size exclusion column
Co-activator size exclusion buffer: 20 mM Hepes pH 7, 300 mM NaCl, 2.5 % glycerol, 1 mM DTT.
2.5. Expression and purification of Substrate-Cyclin B 1-95* and Ub-Cyclin B 1-95*
Low and high speed centrifuges
PBS
Substrate lysis buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT, 0.25 μg/mL Lysozyme, 1 Complete™ EDTA-free protease inhibitor tablet (per 50 mL buffer)
Sonicator
Glutathione Sepharose
Substrate wash buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT
Tobacco Etch Virus protease
Gravity flow column
SDS-PAGE gels and gel running equipment
Ni-NTA
Substrate elution buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 250 mM Imidazole, 1 mM DTT
Bradford reagent
Amicon Ultra-15 centrifugal concentrator – 3kDa
DTT
PD-10 columns
Substrate pre-label buffer: 50 mM Hepes pH 7, 150 mM NaCl buffer (do not include DTT!)
Fluorescein-5-maleimide (F5M) in Dimethyl sulfoxide (DMSO)
Substrate post-label buffer: 50 mM Hepes pH 7, 150 mM NaCl, 1 mM DTT
AKTA protein purification system
Superdex 75 16/60 column
Substrate size exclusion buffer: 20 mM Hepes pH 8, 200 mM NaCl, 1 mM DTT
2.6. Expression and purification of E1 - UBA1
Low and high speed centrifuges
PBS
E1 lysis buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT, 0.25 μg/mL Lysozyme, 1 Complete™ EDTA-free protease inhibitor tablet (per 50 mL buffer)
Sonicator
Glutathione Sepharose
E1 wash buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT
Gravity flow column
E1 elution buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 20 mM reduced glutathione, 1 mM DTT
Bradford reagent
SDS-PAGE gels and gel running equipment
AKTA protein purification system
Resource Q anion exchange column
E1 buffer A: 20 mM Tris pH 7.6, 1 mM DTT
E1 buffer B: 20 mM Tris pH 7.6, 1M NaCl, 1 mM DTT
Superdex 200 16/60 size exclusion column
E1 size exclusion buffer: 20 mM Hepes pH 8, 200 mM NaCl, 1 mM DTT buffer
2.6. Expression and purification of E2s - UBCH10 & UBE2S
Low and high speed centrifuges
PBS
E2 lysis buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT, 0.25 ug/mL Lysozyme, 1 Complete™ EDTA-free protease inhibitor tablet (1 tablet per 50 mL buffer)
Sonicator
Ni-NTA resin
E2 wash buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT
Gravity flow column
E2 elution buffer: 20 mM Tris pH 7.6, 150 mM NaCl, 20 mM reduced glutathione, 1 mM DTT
Bradford reagent
SDS-PAGE gels and gel running equipment
AKTA protein purification system
Superdex 200 16/60 size exclusion column
E2 size exclusion buffer: 20 mM Hepes pH 8, 200 mM NaCl, 1 mM DTT buffer
Resource S cation exchange columns
E2 buffer A: 20 mM Tris pH 7.6, 1 mM DTT
E2 buffer B: 20 mM Tris pH 7.6, 1M NaCl, 1 mM DTT
TEV protease
Amicon Ultra-15 centrifugal concentrator – 10 kDa
2.7. Expression and purification of Ubiquitin
Low and high speed centrifuges
PBS
Ub lysis buffer 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT, 0.25 μg/mL Lysozyme and 1 Complete™ EDTA-free protease inhibitor tablet (per 50 mL buffer)
Glacial acetic acid
SDS-PAGE gels and gel running equipment
Dialysis tubing - 3 kDa cutoff
Ub buffer A: 25 mM NaC2H3O2pH 4.5
AKTA protein purification system
Resource S cation exchange column
Ub buffer B: 25 mM NaC2H3O2pH 4.5, 1 M NaCl
Amicon Ultra-15 centrifugal concentrator – 3 kDa
Superdex 75 16/60 size exclusion column
Ub size exclusion buffer: 20 mM Hepes pH 8, 200 mM NaCl buffer
2.7. APC/C ubiquitylation assay
10 mg/mL BSA (Applichem) stock solution
50 mM MgCl2+ATP pH7 solution
4x SDS sample buffer: 250 mM Tris pH 6.8, 8% SDS, 40% glycerol, 400 mM DTT and 0.04% bromophenolblue
SDS-PAGE gels and gel running equipment
Fluorescence scanner
3. Methods
3.1. Expression and purification of recombinant APC/C
Expression of recombinant APC/C is performed in insect cells. To date, published methods utilise co-expression techniques ranging from co-expressing each individual subunit from Baculoviruses each containing the open reading frame of an individual APC/C subunit (12) to co-expression of the APC/C subunits using two Baculoviruses containing a distribution of the individual subunits (13). Here we briefly describe the expression and purification protocol we used to purify recombinant human APC/C which has been characterised in (8, 12, 20).
Co-infect Sf9 cells with the respective Baculoviruses at a cell density of 1.0 x 106 and incubate at 27°C at 100 rpm for 72 h to allow protein expression.
Harvest cells by centrifugation at 500 g at 4°C for 15 min, wash once with phosphate buffered saline (PBS) before re-suspending in APC/C lysis buffer.
Sonicate cells for 15 sec, repeat 3x, before centrifuging at 150,000 g at 4°C for 30 min.
Incubate the supernatant with Streptactin resin on a rotor at 4°C for 1 h (APC4 contains a C-terminal twin Strep tag).
After binding, centrifuge the slurry at 1,000 g at 4°C for 5 min, re-suspend the Streptactin resin in APC/C wash buffer, transfer to a gravity flow column and wash with 10 column volumes (CV) of wash buffer.
Elution of recombinant APC/C from the Streptactin resin is performed over 5 CV using APC/C elution buffer. Follow the elution by analysing fractions using the Bradford assay.
Analyse the fractions by SDS-PAGE.
The relevant fractions can then be pooled, and diluted with APC/C buffer A to a final concentration of 100 mM NaCl and loaded onto a pre equilibrated POROS HQ anion exchange column. The bound recombinant APC/C is eluted using a gradient of APC/C buffer B.
Fractions should be analysed by SDS-PAGE with the appropriate fractions pooled, concentrated and loaded onto a Superose 6 size exclusion column equilibrated in APC/C size exclusion buffer (see Note 3).
The resulting fractions from size exclusion should then be analysed by SDS-PAGE with respective fractions pooled and concentrated to 1 mg/mL, aliquoted, flash frozen and stored at -80 °C.
3.2. Expression and purification of the co-activators - CDC20 and CDH1
Working with a recombinant form of the human APC/C allows great control over which co-factors in vitro experiments are performed with. As such the co-activators need to be purified separately. We briefly describe this below.
Infect Sf9 cells at a cell density of 1.0 x 106 with a Baculovirus containing either the full length reading frame of CDC20 or CDH1 with an N-terminal 3myc-his6 and incubate at 100 rpm at 27°C for 72 h.
Following expression, harvest cells by centrifugation at 500 g at 4°C for 15 min, wash once with PBS before re-suspending in co-activator lysis buffer.
Dounce-homogenise cells 5x, repeat 3x, before centrifuging at 150,000 g at 4°C for 30 min.
Incubate the supernatant with equilibrated Ni-NTA resin on a rotor at 4°C for 1 h.
After binding, centrifuge the slurry at 1,000 g for 5 min at 4°C, re-suspend in co-activator wash buffer, transfer to a gravity flow column and wash with 10 CV of co-activator wash buffer.
Perform the elution over 5 CV using co-activator elution buffer. Follow the elution by analysing fractions using the Bradford assay.
Analyse the fractions by SDS-PAGE.
Pool the appropriate fractions, concentrate and load onto a Superdex 200 26/60 size exclusion column equilibrated in co-activator size exclusion buffer.
Fractions should be analysed by SDS-PAGE with the respective fractions concentrated to ˜1 mg/mL and aliquoted, flash frozen and stored at -80 °C (see Note 4).
3.3. Expression and purification of Substrate - Cyclin B 1-95* and Ub-Cyclin B 1-95*
This substrate construct is an iteration of previous versions of the N-terminus of Cyclin B (15).
Substrate is bacterially produced and is expressed in the E.coli strain BL21 (DE3) CodonPlus-RIL. Bacteria are grown to an OD of 0.8, protein expression is induced by treatment with 0.6 M Isopropyl β-D-1-thiogalactopyranoside (IPTG) and the culture is grown overnight at 16°C.
Harvest cells by centrifugation at 4,500 g at 4°C for 15 min, wash once with PBS before re-suspending in substrate lysis buffer.
Incubate cells rotating at 4°C for 30 min before sonicating for 15 sec, repeat 4x and then centrifuge at 25,000 g at 4°C for 30 min.
Incubate the supernatant with equilibrated Glutathione Sepharose resin on a rotor at 4°C for 1 h.
After binding, centrifuge the slurry at 1,000 g at 4°C for 5 min and wash with 5 CV substrate wash buffer.
Re-suspend the resin in 1 CV substrate wash buffer supplemented with tobacco etch virus (TEV) protease and incubate on a rotor overnight at 4°C.
The next morning transfer the slurry to a gravity flow column and collect the flow through. Add a further 1 CV pulse of substrate wash buffer and collect the flow through.
Analyse the flowthrough by SDS-PAGE.
Incubate the flowthrough with equilibrated Ni-NTA resin on a rotor at 4°C for 1 h.
After binding, centrifuge the slurry at 1,000 g at 4°C for 5 min, re-suspend in substrate wash buffer, transfer to a gravity flow column and wash with 10 CV of substrate wash buffer.
Elute the protein over 5 CV using substrate elution buffer. Follow the elution by analysing fractions using the Bradford assay.
Analyse the flowthrough by SDS-PAGE.
Concentrate the flow through containing substrate to ˜2.5 mL and continue to label the substrate of interest.
3.4. Fluorescent labelling of the Substrate of Interest
Reduce the substrate by incubating with 20 mM DTT for 20 min.
De-salt the reduced substrate twice using PD-10 columns into substrate pre-label buffer.
Dissolve the Fluorescein-5-maeimide (F5M) in Dimethyl sulfoxide (DMSO).
The dissolved F5M is mixed with the substrate at a 10x higher concentration and incubated at room temperature for 3 h (see Note 5).
Quench the chemical reaction between the maleimide group and the cysteine in the substrate by the addition of a final concentration of 10 mM DTT.
De-salt the labelled substrate a further 2x using PD-10 columns using substrate post-label buffer and further purify by size exclusion chromatography using a Superdex 75 16/60 column equilibrated in substrate size exclusion buffer.
Fractions should be analysed by SDS-PAGE with the appropriate fractions concentrated to ˜5 mg/mL and aliquoted, flash frozen and stored at -80°C (20).
3.5. Expression and purification of E1 - UBA1
The following method has been previously published (22).
GST-UBA1 (E1; a kind gift from Cynthia Wolberger) is bacterially produced and is expressed in the E.coli strain BL21 (DE3) CodonPlus-RIL. Bacteria are grown to an OD of 0.8, protein expression is induced by treatment with 0.6 M IPTG and the culture is grown overnight at 16°C.
Harvest cells by centrifugation at 4,500 g at 4°C for 15 min, wash once with PBS and resuspend in E1 lysis buffer.
Incubate cells by rotating for 30 min at 4°C, before sonicating for 15 sec, repeat 4x, and centrifuge at 25,000 g at 4°C for 30 min.
Incubate the supernatant with equilibrated Glutathione Sepharose resin on a rotor at 4°C for 1 h.
Following binding, centrifuge the resin at 1,000 g at 4°C for 5 min, re-suspend in E1 wash buffer, transfer to a gravity flow column and wash with 10 CV of E1 wash buffer.
Elute the protein over 5 CV using E1 elution buffer. Follow the elution by analysing fractions using the Bradford assay.
Analyse the fractions by SDS-PAGE.
The respective fractions are then pooled, and diluted with E1 buffer A to 50 mM NaCl and loaded onto an equilibrated anion exchange column. The bound UBA1 is eluted using a gradient of E1 buffer B.
Analyse fractions by SDS-PAGE and pool the appropriate fractions, concentrate and load onto a Superdex 200 16/60 size exclusion column equilibrated in E1 size exclusion buffer.
Fractions should be analysed by SDS-PAGE with the respective fractions concentrated to ˜5 mg/mL and aliquoted, flash frozen and stored at -80°C.
3.6. Expression and purification of E2s - UBCH10
UBCH10-His6 is bacterially produced and is expressed in the E.coli strain BL21 (DE3) CodonPlus-RIL. Bacteria are grown to an OD of 0.8, protein expression is induced by treatment with 0.6 M IPTG and the culture is grown overnight at 23°C.
Post expression, harvest cells by centrifugation at 4,500 g at 4°C for 15 min, wash once with PBS before being re-suspending in E2 lysis buffer.
Incubate rotating for 30 min at 4°C before sonicating for 15 sec, repeat 4x and centrifuge at 25,000 g at 4°C for 30 min.
Incubate the supernatant with equilibrated Ni-NTA resin on a rotor at 4°C for 1h.
After binding, centrifuge the slurry at 1,000 g at 4°C for 5 min, resuspend in E2 wash buffer, transfer to a gravity flow column and wash with 10 CV of E2 wash buffer.
Conduct elution over 5 CV using E2 elution. Follow the elution by analysing fractions using the Bradford assay.
Analyse fractions by SDS page.
Pool the appropriate fractions, concentrate and load onto a Superdex 200 16/60 size exclusion column equilibrated in E2 size exclusion buffer.
Fractions from size exclusion should be analysed by SDS-PAGE with respective fractions pooled, concentrated to ˜5 mg/mL, aliquoted, flash frozen and stored at -80°C.
3.7. Expression and purification of E2s – UBE2S
His6-TEV-Flag-PS-UBE2S is bacterially produced and is expressed in the E.coli strain BL21 (DE3) CodonPlus-RIL. Bacteria are grown to an OD of 0.8, protein expression is induced by treatment with 0.6 M IPTG and the culture is grown overnight at 23°C.
Harvest cells by centrifugation at 4,500 g at 4°C for 15 min, wash once with PBS before being re-suspending in E2 lysis buffer.
Incubate cells rotating at 4°C for 30 min before sonicating for 15 sec, repeat 4x and centrifuge at 25,000 g at 4°C for 30 min.
Incubate the supernatant with equilibrated Ni-NTA resin rotating at 4°C for 1 h.
After binding, centrifuge the slurry at 1,000 g at 4°C for 5 min and wash with 5 CV E2 wash buffer.
Re-suspend the resin in 1 CV E2 wash buffer supplemented with TEV protease and incubate on a rotor overnight at 4°C.
The following morning transfer the slurry to a gravity flow column and collect the flow through. Pulse with a further 1 CV E2 wash buffer and collect the flow through.
Concentrate the flow through to ˜2.5 mL and dilute with E2 buffer A to a final concentration of 50 mM NaCl and load onto an equilibrated cation exchange column. The bound UBE2S is eluted using a gradient of E2 buffer B.
Analyse fractions by SDS-PAGE and pool the appropriate fractions, concentrate and load onto a Superdex 200 16/60 size exclusion column equilibrated in E2 size exclusion buffer.
Fractions from size exclusion should be analysed by SDS-PAGE with respective fractions pooled, concentrated to ˜5 mg/mL, aliquoted, flash frozen and stored at -80°C.
3.8. Expression and purification of Ubiquitin
The following method has been previously published (23).
Ubiquitin is bacterially produced and is expressed in the E.coli strain BL21 (DE3) CodonPlus-RIL. Bacteria are grown to an OD of 0.8, protein expression is induced by treatment with 0.6 M IPTG and the culture is grown overnight at 16°C.
Harvest cells by centrifugation at 4,500 g at 4°C for 15 min, wash once with PBS before resuspending in Ub lysis buffer.
Incubate cells rotating at 4°C for 30 min before sonicating for 15 sec, repeat 4x and centrifuge at 25,000 g at 4°C for 30 min.
Acidify the supernatant with glacial acetic acid to approximately pH 4-4.5 (see Note 6).
Centrifuge the supernatant again at 25,000 g at 4°C for 30 min.
Analyse the supernatant by SDS-PAGE.
Dialyse the supernatant in 3 kDa dialysis tubing at 4°C overnight in Ub buffer A.
Centrifuge the supernatant again at 25,000 g at 4°C for 30 min and load onto an equilibrated cation exchange column. The bound ubiquitin is eluted using a gradient of Ub buffer B.
Analyse fractions by SDS-PAGE and pool the appropriate fractions, concentrate and load onto a Superdex 7516/60 size exclusion column equilibrated in Ub size exclusion buffer.
Fractions from size exclusion should be analysed by SDS-PAGE with respective fractions pooled, concentrated to ˜10 mg/mL, aliquoted, flash frozen and stored at -80°C.
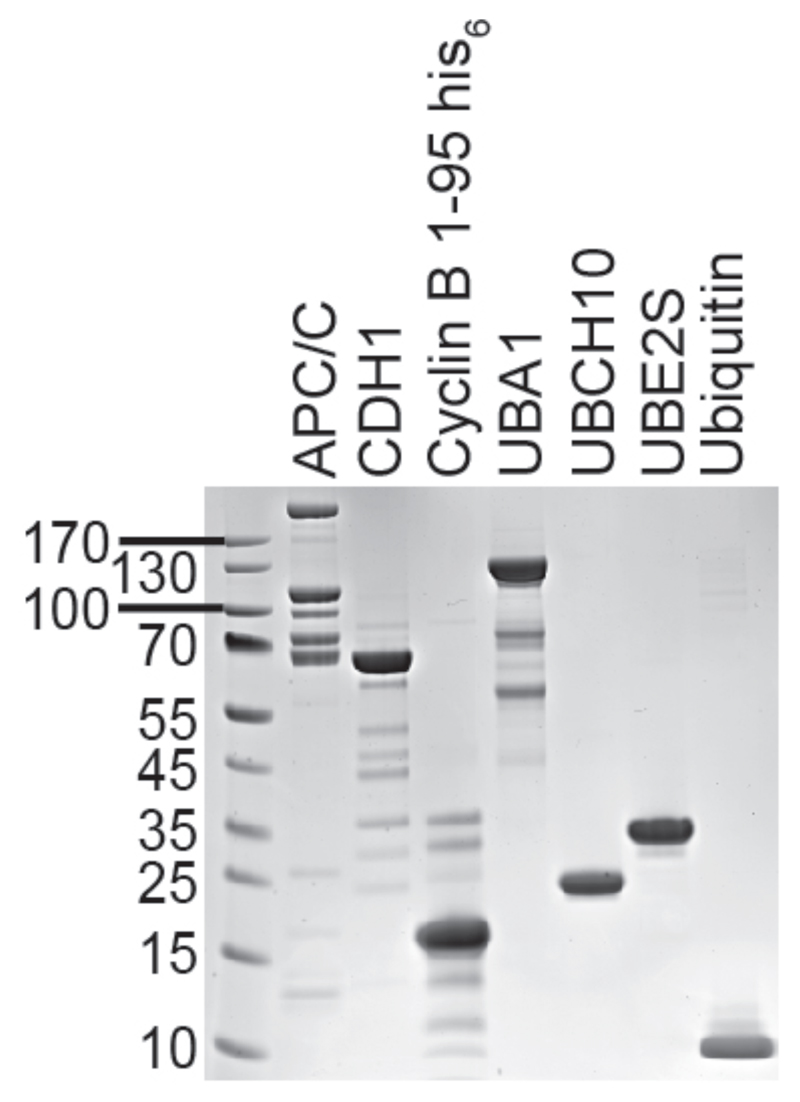
Fig 2. Illustrates the purity to which the above described individual proteins can be purified.
Figure 2.
SDS-PAGE gel of the individual purified protein components described in this chapter. From left to right: APC/C, CDH1, Cyclin B 1-95, UBA1, UBCH10, UBE2S, Ubiquitin.
3.9. APC/C ubiquitylation assay
A typical APC/C ubiquitylation assay is conducted at room temperature in a volume of 20 uL. Below we describe the basic procedure of such an assay. Note that the components in the assay can be varied depending on the purpose of the experiment and the pipetted volumes may change depending on the stock concentrations of the purified components used (see Note 7).
The individual purified components (APC/C, co-activator, substrate, E2s) are mixed together, on ice, to reach the appropriate final concentrations. In a second tube, E1 and ubiquitin are mixed together.
The mixtures are then equilibrated to room temperature for 10 min.
The ubiquitylation reaction is initiated by adding the volume in tube 1 to that of tube 2, with the assay allowed to proceed for 5-15 min at room temperature.
The reaction is stopped by the addition of 6 uL 4x SDS sample buffer to the reaction volume followed by a 5 minute incubation at 95°C.
The reaction products are then resolved in the dark by SDS-PAGE using a NuPAGE 4-12% Bis-Tris, 8 cm x 8 cm, 1.0 mm thick, 15 well protein gel according to the manufacturers recommendations. Alternatively, homemade polyacrylamide gels can also be used. The results are subsequently visualised by use of a fluorescence scanner set for detecting Alexa Fluor 488 (we use a Typhoon laser scanner in fluorescence scanning mode, with a voltage of 750V (see Note 8).
Tube 1.
| Purified Component | Concentration of stock | Volume | Final concentration in assay |
|---|---|---|---|
| BSA | 10 mg/mL | 0.5 ul | 0.25 mg/mL |
| MgCl2ATP | 50 mM | 2 ul | 5 mM |
| Substrate | 10 uM | 1 uL | 500 nM |
| UBCH10 | 2.5 uM | 2 ul | 250 nM |
| UBE2S | 2.5 uM | 2 ul | 250 nM |
| APC/C | 150 nM | 2 ul | 15 nM |
| Cdh1 | 20 uM | 1 ul | 1 uM |
| Buffer | 1x | 5.5 ul | - |
| Total volume: 16 ul | |||
Tube 2.
| Purified Component | Concentration of stock | Volume | Final concentration in assay |
|---|---|---|---|
| E1 | 1 uM | 2 ul | 100 nM |
| Ubiquitin | 1 mM | 2 ul | 100 uM |
| Total volume: 4 ul | |||
3.8. Quantification
This assay allows assaying for a wide-range of parameters regarding APC/C activities with a range of different substrates, E2 combinations, and inhibitors, as have been published (8, 20). We describe one typical scenario here for quantifying the apparent Km for the E2, UBE2S.
We determined assay conditions in the initial velocity range for 10 min reactions at room temperature to use 10 nM APC/C, 0.5 μM Ub-CyclinBNTD*.
Because UBE2S processively generates poly-ubiquitin chains, reaction products were quantified by determining the intensity of all ubiquitinated species (individual bands) using ImageQuant software, corrected for background by subtracting the sum of intensities in the corresponding lane for control reactions lacking APC/C.
The values for ½ Vmax and Km values can be calculated by fitting the initial velocity to a hyperbolic curve according to Michaelis-Menton kinetics using the equation: v = Vmax[UBE2S]/(Km +[UBE2S]). Prism 6 software (GraphPad) is suitable for performing curve-fitting.
4. Notes
E.coli offers a fast and effective system for protein expression. There are many different strains of E.coli offering various advantages such as the co-expression of rare codon encoding t-RNAs (e.g., BL21 (DE3) CodonPlus-RIL), tight control of protein expression suitable for toxic proteins (e.g., BL21 (DE3) pLysS) and induction of proper protein folding (e.g., Origami (DE3)).
The Baculovirus-insect cell expression system provides a mode of production that yields large quantities of recombinant proteins that require eukaryotic chaperones or the need to be post-translationally modified by eukaryotic enzymes. There are various insect cell lines available such as Sf9 and High Five, as well as many different types of media including Grace’s media (Sigma-Aldrich), Hyclone SFX (GE Lifesciences), Sf-900 III (Life technologies) and ESF 921 (Expression Systems) offering many solutions in recombinant protein production.
Be conservative with respect to the fractions that you pool for size exclusion chromatography. Focus on fractions that contain nice stoichiometric APC/C.
Do not concentrate the co-activators to higher concentrations than is necessary because these proteins tend to form aggregates and to precipitate.
Ensure that less than 5% of the total volume contains DMSO as to limit the susceptibility of the substrate to DMSO.
Don’t be concerned that a lot of protein is precipitating, these are bacterial host proteins!
We recommend making small working aliquots to avoid repeated freeze thaw cycles of these enzymes. APC/C, the co-activators and E1 are particularly sensitive to freeze-thaw cycles.
When setting up this assay it is worth to test different scan voltages to determine at which settings the best images are obtained.
References
- 1.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nature Reviews Molecular Cell Biology. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 2.Primorac I, Musacchio A. Panta rhei: The APC/C at steady state. The Journal of Cell Biology. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delgado-Esteban M, García-Higuera I, Maestre C, et al. APC/C-Cdh1 coordinates neurogenesis and cortical size during development. Nature Communications. 2013;4 doi: 10.1038/ncomms3879. [DOI] [PubMed] [Google Scholar]
- 4.Eguren M, Porlan E, Manchado E, et al. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nature Communications. 2013;4 doi: 10.1038/ncomms3880. [DOI] [PubMed] [Google Scholar]
- 5.Chang L, Barford D. Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Current Opinion in Structural Biology. 2014;29:1–9. doi: 10.1016/j.sbi.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Duda DM, Scott DC, Calabrese MF, et al. Structural regulation of cullin-RING ubiquitin ligase complexes. Current Opinion in Structural Biology. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craney A, Rape M. Dynamic regulation of ubiquitin-dependent cell cycle control. Current Opinion in Cell Biology. 2013;25:704–710. doi: 10.1016/j.ceb.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Frye JJ, Brown NG, Petzold G, et al. Electron microscopy structure of human APC/CCDH1–EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nature Structural & Molecular Biology. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herzog F, Peters J. Large-Scale Purification of the Vertebrate Anaphase-Promoting Complex/Cyclosome. Methods in Enzymology. 2005:175–195. doi: 10.1016/S0076-6879(05)98016-6. Elsevier. [DOI] [PubMed] [Google Scholar]
- 10.Kraft C, Gmachl M, Peters JM. Methods to measure ubiquitin-dependent proteolysis mediated by the anaphase-promoting complex. Methods. 2006;38:39–51. doi: 10.1016/j.ymeth.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber A, Stengel F, Zhang Z, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 12.Uzunova K, Dye BT, Schutz H, et al. APC15 mediates CDC20 autoubiquitylation by APC/CMCC and disassembly of the mitotic checkpoint complex. Nature Structural & Molecular Biology. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Yang J, Kong EH, et al. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) Biochemical Journal. 2013;449:365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 14.Dimova NV, Hathaway NA, Lee B-H, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nature Cell Biology. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Molecular Biology of the Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin L, Williamson A, Banerjee S, et al. Mechanism of Ubiquitin-Chain Formation by the Human Anaphase-Promoting Complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkpatrick DS, Hathaway NA, Hanna J, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nature Cell Biology. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 18.Meyer H-J, Rape M. Enhanced Protein Degradation by Branched Ubiquitin Chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu T, Merbl Y, Huo Y, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the Anaphase-Promoting Complex. Proceedings of the National Academy of Sciences. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown NG, Watson ER, Weissmann F, et al. Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly. Molecular Cell. 2014;56:246–260. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang L, Zhang Z, Yang J, et al. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eletr ZM, Huang DT, Duda DM, et al. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nature Structural & Molecular Biology. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 23.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods in Enzymology. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]