Abstract
The discovery of organic ligands that bind specifically to proteins is a central problem in chemistry, biology and biomedical sciences. The encoding of individual organic molecules with distinctive DNA tags, serving as amplifiable identification barcodes, allows the construction and screening of combinatorial libraries of unprecedented size, thus facilitating the discovery of ligands to many different protein targets. Fundamentally, one links powers of genetics and chemical synthesis. After the initial description of DNA-encoded chemical libraries in 1992, several experimental embodiments of the technology have been reduced to practice. This review provides an historical account on important milestones for the development of DNA-encoded chemical libraries, a survey of relevant on-going research activities and a glimpse into the future.
Keywords: DNA-encoded chemical libraries, combinatorial chemistry, drug discovery
Ligand discovery: a central problem in chemistry, biology and biomedical sciences
The discovery of specific ligands that bind to protein targets of interest represents an activity of fundamental importance both for basic research and for industrial applications. Many drug discovery programs start with the search for a molecule that interacts with a validated protein target (1–3). Similarly, the deciphering of complex biochemical processes in basic research often relies on the availability of specific reagents, which bind (or even block) macromolecular structures of interest, thus allowing their visualization, quantification or functional investigation (4–6).
The advent of monoclonal antibodies by hybridoma technology has revolutionized many areas of scientific research. This innovation (for which Cesar Milstein and Georges Köhler were awarded the Nobel Prize in Medicine and Physiology in 1984) has made it possible to detect individual macromolecular structures with an unprecedented level of precision, using affinity reagents of exquisite specificity (7). The formidable advances of the last few decades in the molecular characterization of biochemical and cellular processes would have been unthinkable without the use of monoclonal antibody reagents. It would be highly desirable if, in addition to monoclonal antibodies, small organic ligands to protein targets of interest could be readily available.
The value of small organic ligands capable of high-affinity binding to a cognate protein is illustrated by the versatility of the biotin / (strept)avidin system (8). Derivatives of biotin are recognized by avidin or streptavidin with dissociation constants in the sub-picomolar range. These tight-binding complexes have found numerous applications not only for the development of specific reagents in biochemistry and immunology, but also in areas as diverse as chemical synthesis (9), nuclear medicine (10), and material sciences (11), to name just a few.
For many years, the discovery of small organic ligands to protein targets has been performed by screening very large sets of organic molecules (termed “chemical libraries”), one by one (1–3, 12). Large pharmaceutical companies typically construct and assay chemical libraries comprising one million organic molecules or more, using high-throughput screening procedures. It has been estimated that the assembly of such libraries may cost between 0.4 and 2 billion U.S. dollars (i.e., approximately 1,000 $ per compound, times 1 million compounds) (13). While the value of high-throughput library screening has been demonstrated in various pharmaceutical applications, it is not uncommon that binding molecules of sufficient affinity and specificity (called “hits”) cannot be discovered using conventional screening campaigns (14).
In light of these considerations, considerable efforts have been devoted and continue to be devoted to the discovery and development of methods which facilitate the identification of specific binding molecules to macromolecular targets and proteins in particular. DNA-encoded chemical library technology enables the construction and screening of compound sets of unprecedented size and, as a consequence, the discovery of small organic ligands. When library size grows, the concentration of individual library members decreases, to an extent that those molecules may no longer be detectable even with the most sophisticated analytical methods. However, DNA tags allow the amplification, identification and relative quantification of molecules in very large libraries.
From encoded libraries of polypeptides to DNA-encoded chemical libraries
The advent of encoded combinatorial libraries of polypeptides has not only played an important role for the engineering of proteins with novel properties, with applications in many research fields, but has also been conceptually instrumental for the genesis of DNA-encoded chemical libraries. For this reason, it is convenient to briefly discuss a few milestones in this research area.
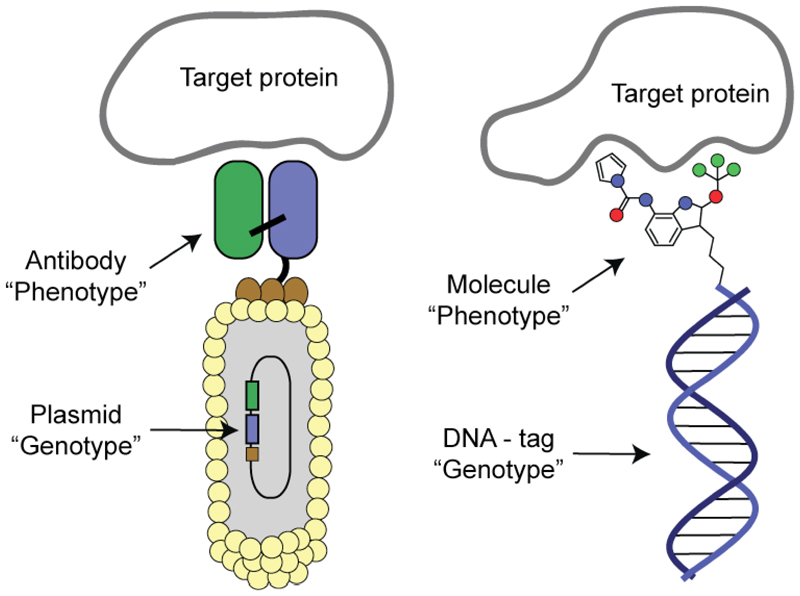
In 1985, George P. Smith proposed the use of filamentous phage as tools for the “display” of polypeptides on the surface of these bacterial viruses (15). In a popular implementation, peptides or proteins would be genetically fused at the N-terminal end of the minor coat protein pIII of filamentous phage. The resulting viral particle features a potential functional property (e.g., a binding “phenotype”, embodied by the polypeptide on the phage surface), while simultaneously bearing the corresponding genetic information (i.e., “genotype”) as part of the modified phage genome [Figure 1]. The central idea is that genotype and phenotype are linked in a single phage package. Combinatorial phage display libraries could be generated by inserting a degenerate DNA sequence upstream of gene III (coding for the minor coat protein pIII, downstream of the leader peptide). This simple cloning procedure enables the generation and interrogation of billions of different peptides. Initially, combinatorial phage display libraries of peptides were used to identify the sequence of linear epitopes, recognized by monoclonal antibodies (16). Soon afterwards, however, the groups of Sir Gregory Winter (17) and of one of us (R.A.L; 18) realized that very large combinatorial libraries of antibodies, rather than of short peptides, could be created and that phage display could be used to amplify and isolate rare binding specificities within those libraries (19,20). In this process, “man-made antibodies” could be created by interrogating large combinatorial antibody libraries against virtually any target antigen of interest, immobilized on a solid support. The procedure bears logical and functional similarities to the clonal expansion of antigen-specific B cells in the immune system. Indeed, a B lymphocyte with an immunoglobulin on its surface constitutes a cellular entity, which simultaneously features a binding phenotype and the corresponding genotype, in full analogy to an antibody fragment displayed on the surface of a filamentous phage (21). Thus, in B-cells genotype and phenotype are linked albeit at the cellular level.
Figure 1.
Schematic representation of antibody phage display libraries and of DNA-encoded chemical libraries. In antibody phage display libraries, an antibody fragment (e.g., a scFv fragment) is displayed on the surface of filamentous phage and could potentially bind to a cognate protein target. The genetic information coding for the recombinant antibody is contained in the genome of the bacteriophage. In full analogy, small organic molecules can be attached to DNA-fragments, serving as amplifiable identification barcodes.
The impact of antibody phage technology in basic research and in pharmaceutical sciences has been substantial. For example, the fully-human antibody Humira™ (a TNF blocker and one of the best-selling pharmaceuticals of all times) was generated thanks to antibody phage technology (22). Antibody phage libraries have also taught us important lessons. It has been formally demonstrated that larger libraries are much more likely to yield numerous and high-affinity antibodies, compared to smaller aliquots of the same library. For example, a library comprising 65 billion antibody clones allowed the isolation of numerous antibodies with a dissociation constant in the low nanomolar range, while a portion of the same library, comprising only 10 million antibodies, yielded only few binders, with Kd values in the 0.8 - 12 µM range, confirming that not only molecular design by also “library size” matters (23). Encoded combinatorial libraries of antibodies and of other polypeptides have provided an inspiration for DNA-encoded chemical libraries: a general technology that could be broadly applied to the world of organic molecules. Phage display, yeast display and ribosome display are some of the most popular selection systems which enable a physical connection between a phenotype and the corresponding genetic information, exploiting the biosynthetic linkage between nucleic acids and proteins (24,25). This biosynthetic dependence is not needed in DNA-encoded chemical libraries, which use DNA tags only as amplifiable identification barcodes.
In 1992, one of us (R.A.L.) together with Sidney Brenner postulated that it should be possible to “encode” chemistry with DNA (26). The authors envisaged the possibility to simultaneously synthesize distinctive polypeptide and oligonucleotide sequences on beads, using orthogonal chemistry and split-and-pool procedures. The synthesis of oligonucleotides on beads would not be limited to the stepwise assembly of bases, since other assembly strategies (e.g., the stepwise ligation of DNA fragments) could also be considered. The 1992 landmark paper revealed that oligonucleotides could serve as amplifiable barcodes for the corresponding peptide displayed on the same bead. The barcode would only act as “identifier” for the corresponding peptide structure and would not act as genotype in a biosynthetic sense. Indeed, the original paper anticipated that the bead could be replaced by a generic chemical linker and the connection between binding “phenotype” and the corresponding barcode would be preserved [Figure 1]. Shortly afterwards, Needels and colleagues, as well as Janda and colleagues, exemplified the Brenner and Lerner concept with the synthesis of peptide libraries, successfully using bead-based libraries to retrieve known antibody epitopes (27,28). In 2004, three groups reported on the construction of DNA-encoded combinatorial libraries of organic molecules and on the selection of specific binders (29–31), using libraries devoid of beads and affinity-capture procedures, which are similar to the ones previously used for the panning of phage display libraries [Figure 2]. Several DNA-encoded chemical libraries have been synthesized and screened afterwards, as we will see in the following sections.
Figure 2.
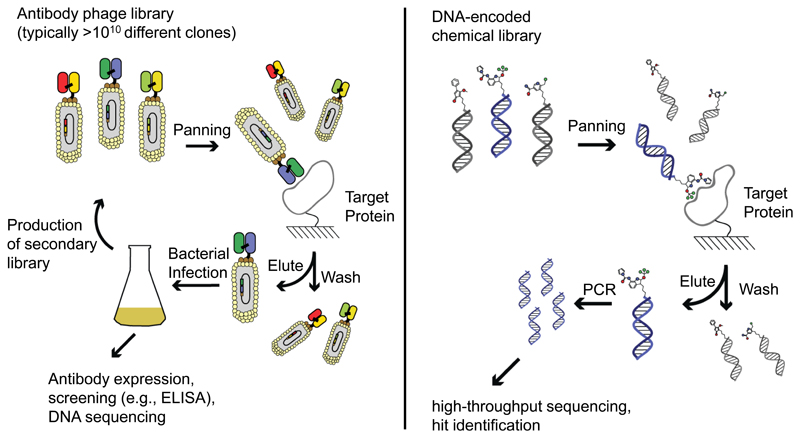
Schematic representation of biopanning procedures with antibody phage display libraries and with DNA-encoded chemical libraries. In the first case, a large library of phage antibodies is incubated with the target protein of interest, immobilized on a solid support. Selective binders are captured on the affinity support, while the majority of other phage antibodies (which do not bind toia the target) can be washed away. Selected phage particles can be amplified by infecting bacteria, leading to an amplification step and to the generation of more phage particles, which can be submitted to a second round of panning. Alternatively, infected bacteria can be plated onto selective plates and individual colonies correspond to distinct monoclonal antibody clones. Similarly, large collections of organic molecules (individually tagged with DNA barcodes) can be interrogated using an affinity capture procedure. Preferentially enriched molecules can be identified by PCR amplification of the DNA tags, followed by high-throughput DNA sequencing.
Many reviews have covered the field of DNA-encoded chemistry, some of which are listed here (32–42). In this manuscript, we wish to provide essential information not only about the origin of the technology, but also discuss strategies which will continue to shape future research activities in the field.
Types of DNA-encoded chemical libraries
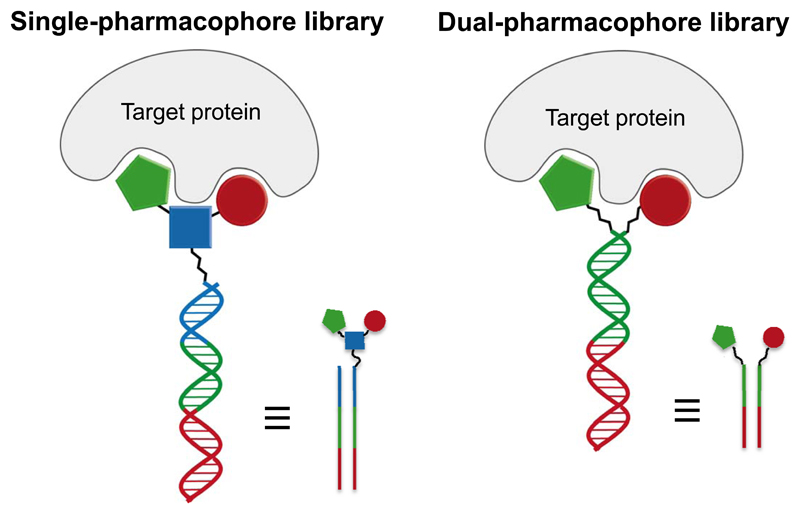
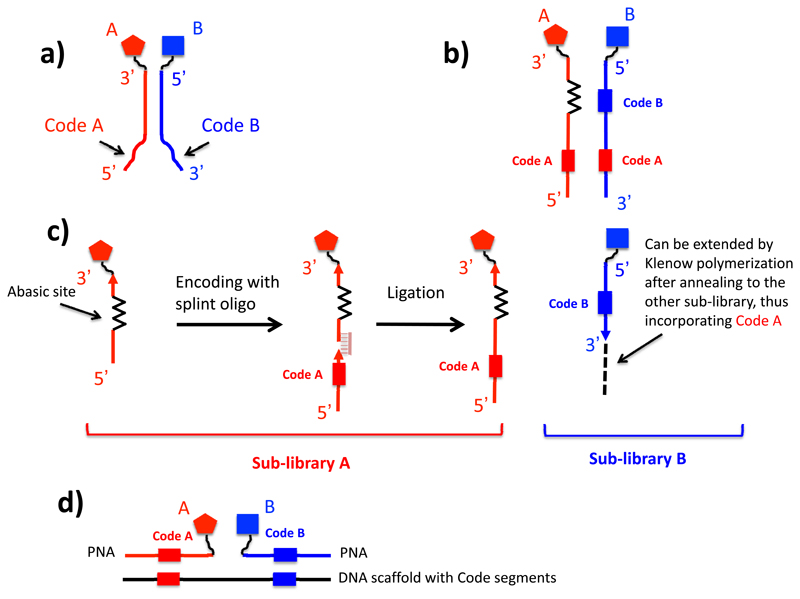
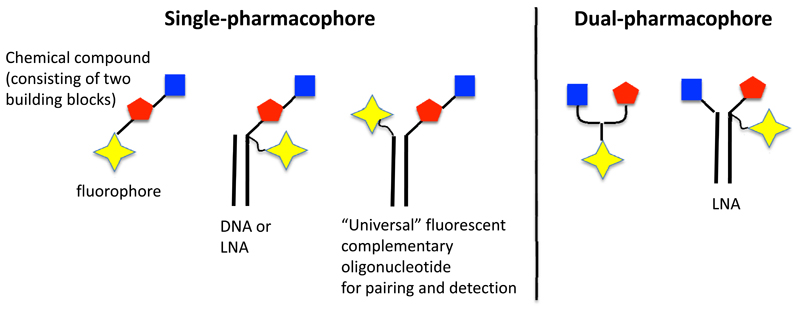
One can distinguish between “single pharmacophore” and “dual pharmacophore” DNA-encoded chemical libraries [Figure 3]. In the first case, individual molecules (no matter how complex) are attached to distinctive DNA fragments. In most instances, those molecules are coupled to one of the two complementary DNA-strands. However, researchers at Praecis (now GSK) had introduced a technology featuring the use of a chemical linker (43), which would provide a covalent connection between complementary strands and, at the same time, a site for the stepwise growth of organic molecular structures [Figure 3].
Figure 3.
Schematic representation of “single-pharmacophore” and “dual-pharmacophore” DNA-encoded chemical libraries. Next to the double-helix representation of the DNA structure, a linear schematic representation is also displayed, as this graphical representation is used in subsequent slides, describing encoding procedures.
By contrast, dual pharmacophore chemical libraries can be produced by coupling pairs of organic molecules at the extremity of complementary DNA strands [Figure 3]. If two sub-libraries are constructed, featuring two sets of partially complementary oligonucleotides (each modified with organic molecules), these two sub-libraries may then be reassembled (i.e., hybridized) to create a large combinatorial diversity. This technology is sometimes referred to as “encoded self-assembling chemical (ESAC) library technology” (30). Details of their construction and specific aspects of the technology are described in more detail in the following section.
Encoding and library synthesis strategies
Over the last two decades, various techniques have been used for the tagging of organic molecules with DNA and, consequently, for the construction of encoded combinatorial libraries.
Let us consider the construction of a library comprising 1 billion (109) different molecules. Obviously, it would be highly unpractical (and prohibitively expensive) to couple 1 billion organic molecules to 1 billion different oligonucleotides (or DNA fragments) one at a time. Luckily, split-and-pool synthesis procedures greatly facilitate the facile and economic assembly of single-pharmacophore chemical libraries. Alternatively, ESAC library technology can also be used to create large molecular repertoires, capitalizing on the combinatorial self-assembly of sublibraries.
Single-pharmacophore chemical libraries
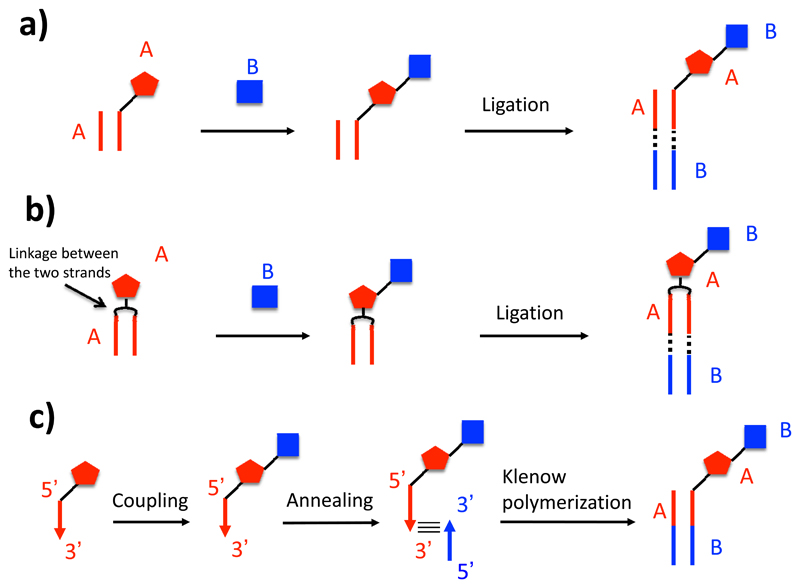
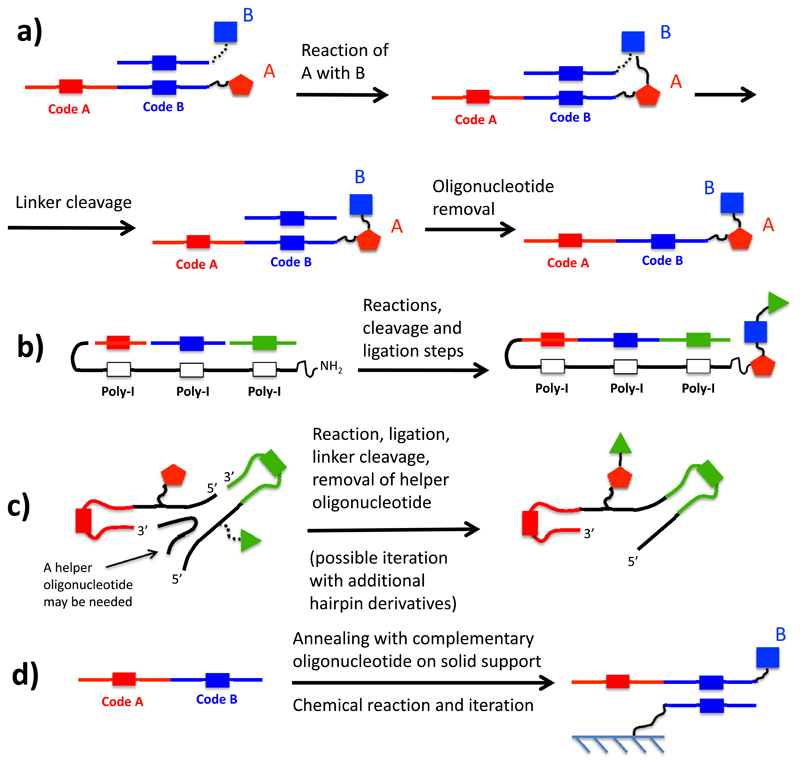
The most commonly used method for the encoding and construction of single-pharmacophore DNA-encoded chemical libraries relies on “DNA-recorded” (sometimes also referred to as “DNA encoded”) synthesis. From a conceptual viewpoint, the simplest way to construct a library relies on the stepwise ligation of double-stranded DNA-fragments, which “record” the identity of individual chemical building blocks [Figure 4a]. As an example, let us consider a set of 100 different organic molecules, each of which is coupled to a distinctive double-stranded DNA fragment. The set of 100 organic molecules coupled to the cognate DNA fragments can be pooled, as the DNA sequences allow an unambiguous identification of the corresponding building blocks. The resulting pool can be split into individual reaction vessels (for example 100 vessels), each of which is allowed to react with a new chemical moiety. The identity of each building block, which is being added to a nascent molecular structure through a suitable chemical reaction, can then be encoded in a ligation step with a double-stranded DNA fragment. In other words, the use of 2 x 100 building blocks (i.e., 200 organic molecules) and of 2 x 100 DNA fragments (i.e, 200 DNA fragments) allows the construction of an encoded library, comprising 10’000 organic molecules. These molecules originate from the encoded combination of 100 x 100 building blocks. If the process is re-iterated, larger libraries can be constructed (e.g., 1-million compound library from 3 sets of 100 building blocks each). A variation on the theme features the use of linkers which connect the two complementary strands and enable the stepwise growth of a molecular structure [Figure 4b] (43). It is worth noting that the molecular mass of the DNA-encoded molecules increases at each round of synthesis (with potentially deleterious effects for pharmaceutical applications). Furthermore, the increase in number of reaction steps may lead to a worsening of library purity.
Figure 4.
Schematic representation of encoding strategies for DNA-recorded chemical libraries. (a) In the simplest implementation of this technology, each building block in the synthesis procedure is encoded (i.e., identified) by a distinctive double-stranded DNA fragment. After each chemical reaction, the identity of the newly introduced building block is provided by an additional DNA fragment, which is ligated to the nascent DNA structure. (b) In a variation of the procedure described before, the complementary DNA strands are connected by a linker, which also supports the growth of the nascent molecule. (c) In a different encoding procedure, a first building block is attached at the 5’ end of an oligonucleotide, which contains a suitable identification code. After a second building block has been added to the nascent molecule by chemical reaction, its identity is encoded by the annealing of a partially complementary oligonucleotide, followed by a Klenow polymerization procedure.
The procedure described above allows the construction of libraries with relatively short DNA barcodes, which are easy to sequence using modern high-throughput procedures. However, the encoding of n building blocks through ligation of DNA fragments requires 2n oligonucleotides, due to the heterodimeric nature of DNA. The group of one of us (D.N.) has described an encoding procedure, which makes use of a set of only n oligonucleotides for the encoding of n building blocks [Figure 4c]. Let us consider a set of 100 building blocks, each of which is coupled at the 5’ end of a corresponding oligonucleotide (e.g., through the functionalization of an appended linker terminating with a primary amine). The oligonucleotides will be designed to have an identical sequence, except for a short central portion, which is distinctive for each building block and acts as barcode. The 100 oligonucleotide conjugates can be purified (e.g., by HPLC) and can be pooled, as each building block can be identified by the barcode to which it is associated. At this stage, splitting of the mixture in individual reaction vessels (e.g., 100 vessels) allows the execution of reactions with a second set of building blocks (e.g, 100 building blocks). The identity of each reaction can be “recorded” by annealing with a partially complementary oligonucleotide, followed by a Klenow polymerization step (44–49). The procedure can be expanded to ≥3 sets of building blocks by means of “splint” oligonucleotides and ligation procedures (50,51).
An alternative way of encoding single-pharmacophore chemical libraries has been pioneered by the group of David Liu and is often referred to as “DNA-templated” (or “DNA-directed”) synthesis [Figure 5a]. In this approach, long pre-formed oligonucleotides are used to mediate a stepwise series of annealing steps with shorter complementary oligonucleotides, carrying chemical moieties that can react with organic structures displayed on the longer oligonucleotide templates (29). These reaction steps may benefit from the high effective molarity advantage caused by the hetero-dimerization of the complementary oligonucleotides. Indeed, the Liu group has demonstrated the usefulness of DNA-templated methods for the execution of bimolecular chemical reactions, which would otherwise be inefficient in water solution and in the presence of DNA (52). The transfer of building blocks onto a nascent molecular structure needs to be followed by a suitable cleavage step, in order to allow for the next round of coupling to proceed. The procedure appears to be particularly suited for the synthesis of complex macrocyclic structures (53). In principle, the requirement for a high-fidelity annealing between the long oligonucleotide template and the shorter complementary oligonucleotides could represent a limitation for the DNA-directed synthesis methodology. However, Xiaoyu Li and coworkers have described an ingenious optimization of the encoding method, featuring the use of an oligonucleotide containing a nascent organic molecule and a poly-inosine segment, serving as promiscuous hybridization stretch for short oligonucleotides, carrying transferrable chemical moieties (54) [Figure 5b].
Figure 5.
Schematic representation of encoding strategies for DNA-templated synthesis of chemical libraries. (a) An oligonucleotide template (here depicted for the construction of a molecule based on two building blocks) is annealed with a chemically modified oligonucleotide, which transfers a chemical moiety to the nascent molecule. At the end of the coupling step, the cleavable connection between the building block B and the corresponding oligonucleotide (depicted with a dotted line). (b) In a modified procedure, a general template (containing poly-I stretches for annealing with various coding segments) is sequentially hybridized with oligonucleotide derivatives, which transfer building blocks onto a nascent molecule. (c) In YoctoReactor™ technology, hairpin structures (containing a coding sequence and a building block, are used to mediate successive cycles of DNA ligation and of chemical reactions. Cleavable linkers are indicated with a dotted line. (d) In DNA routing strategies, long oligonucleotides (containing multiple coding sequences) are sequentially hybridized to columns carrying complementary oligonucleotides. Individual column hybridization steps dictate which chemical reaction (i.e., which building block addition) is performed on a given nascent molecule.
Two further variations of the DNA-templated synthesis method have been described. The Danish company Vipergen has developed an elegant methodology, called YoctoReactor™ technology, featuring the annealing and subsequent ligation of hairpin loops, carrying suitable reactive moieties (55). The end-result of a series of hybridization, building block transfer and cleavage events is a set of double-stranded DNA products, containing the sequence information which is needed to unambiguously identify the molecular entities, assembled through proximity-enhanced chemical reactions [Figure 5c]. A second technology (described as early as in 2004 by the Harbury group and sometimes called “DNA routing”) makes use of long preformed oligonucleotides, which are sequentially annealed to complementary oligonucleotides on solid supports. These hybridization steps allow the transfer of different building blocks in correspondence of given oligonucleotide hybridization events, thus providing a direct linkage between DNA sequence of the oligonucleotide templates and the resulting chemical structure of the stepwise synthesized molecules [Figure 5d]. The procedure depends on the fidelity of the hybridization steps and may be complex to perform in practice, if serial (rather than parallel) affinity-based routing steps are performed, in order to direct chemical synthesis. The authors have reported the successful synthesis of a peptide library, from which a known ligand could be retrieved (31,56–58)
Dual-pharmacophore chemical libraries
From the beginning, dual-pharmacophore chemical libraries were designed to be compatible with either microarray-based or with DNA-sequencing-based decoding procedures (30,51,59). In the simplest implementation, two sets of oligonucleotides would be used for library construction, featuring a conserved portion (essential for the self-assembly of the library) and a variable portion (distinctive for each chemical compound) (30). The resulting ESAC libraries can be “decoded” using microarrays coated with sets of complementary oligonucleotides, but this procedure does not provide information about the enrichment factors of individual pairs of building blocks, since the barcodes are present in different DNA strands [Figure 6a].
Figure 6.
Encoding strategies for dual-pharmacophore encoded chemical libraries. (a) General structure of ESAC library members, which are decoded by hybridization onto oligonucleotide microarrays. (b) In a more powerful procedure, ESAC library members are encoded with a methodology, that leads to the simultaneous presence of two codes (identifying building blocks A and B) onto one of the two complementary strands. (c) Encoding strategy, compatible with high-throughput sequencing decoding procedures. Members of the “red” sublibrary are constructed by the coupling of building blocks to a general oligonucleotide carrying an abasic site, followed by encoding by ligation assisted by a splint degradable oligo. By contrast, members of the “blue” sublibrary are created by coupling building blocks to the 5’ extremity of suitable oligonucleotides, carrying an identification code. The two sublibraries can then be annealed followed by a Klenow polymerization step. (c) In one of the possible implementations of the technology, chemically-modified PNAs are hybridized to DNA templates, which carry suitable complementary coding sequences.
A more elaborate procedure allows the unambiguous identification of pairs of building blocks in ESAC libraries and (as will be described a later section) and a library that can be decoded by high-throughput DNA sequencing [Figure 6b]. In this setting, a single oligonucleotide containing an abasic site is coupled to various chemical building blocks at the 3’ end and these reactions are subsequently encoded by ligation with suitable oligonucleotide barcodes (51). In parallel, complementary oligonucleotides can be modified at the 5’ end, generating a second sub-library that can be used for self-assembly procedures. Conveniently, the base segment that identifies molecular entities at the 5’ end of one sub-library corresponds to the abasic site in the sub-library modified at the 3’ end. A Klenow polymerization step allows the incorporation of pairs of coding segments into a single oligonucleotide, thus permitting the unambiguous identification of all building block combinations within the library [Figure 6c].
Heterodimeric and multimeric self-assembling structures can also be generated using DNA templates and peptide-nucleic acids (PNAs) [Figure 6d]. Winssinger and colleagues have extensively used this technology to isolate binders to various molecular targets (37,60). Importantly, PNA-encoded chemical libraries can also be encoded by the stepwise chemical coupling of PNA fragments (61). In most experimental conditions, PNAs are more stable than the corresponding DNA-based oligonucleotides and may therefore be compatible with reaction conditions which are not possible in the presence of DNA (37). However, unlike DNA, PNA structures cannot be amplified by a suitable polymerase procedure (e.g., PCR). The DNA moiety that directs the assembly of chemically-functionalized PNAs allows a PCR amplification step after selection on target proteins, thus facilitating subsequent decoding procedures.
Selection methodologies and the impact of selection conditions
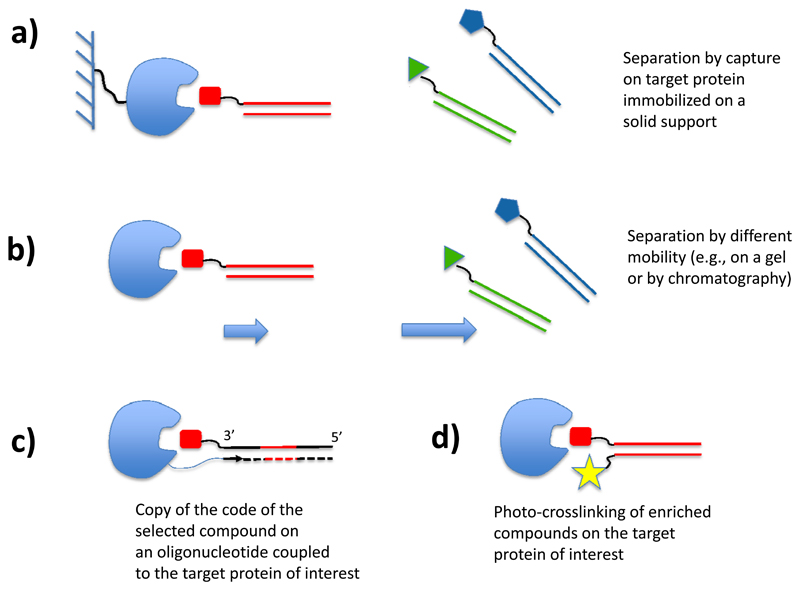
Figure 7 schematically summarizes methods that can be used for the screening of DNA-encoded chemical libraries. In full analogy to phage display libraries (17–20,23) or ribosome display libraries (25), specific binders can be identified from DNA-encoded chemical libraries by performing an affinity capture step on the target protein of interest, immobilized on a solid support [Figure 7a]. The use of magnetic beads (46,47), of streptavidin-coated tips (43) and of CNBr-activated sepharose (after suitable modification step with the target protein; 49,62) have been described and reviewed (63). Certain experimental parameters (e.g., washing conditions, concentration of detergents, protein coating procedures) have been shown to have a profound impact both on the absolute recovery and on the selectivity of selection fingerprints (47,63). The use of quantitative PCR methodologies has recently been introduced as a tool for the experimental determination of selection performance, especially when using a mixture of DNA-tagged ligands of known biochemical properties and compounds of irrelevant specificity used as negative controls (64,65).
Figure 7.
Selection methods for DNA-encoded chemical libraries. (a) Library members are captured on a target protein, immobilized on a solid support. (b) DNA derivatives, capable of binding to a target protein with sufficient stability in given experimental conditions, are separated from non-binding library members by chromatography or by electrophoresis. (c) Library members encoded by single-stranded DNA molecules, capable of binding to a target protein of interest equipped with a suitable oligonucleotide primer, can be identified by a DNA polymerization step, followed by PCR amplification. (d) Capture of preferentially-binding library members by photo-crosslinking.
Capillary electrophoresis techniques can also be considered for the separation of protein-bound DNA derivatives and the rest of the library (66) [Figure 7b]. Alternatively, a selective PCR amplification of the DNA tag associated with preferential binders can be used, if the protein target of interest is equipped with a suitable oligonucleotide primer [Figure 7c] (67,68). This methodology has been implemented in the context of water-in-oil emulsions, allowing a preferential productive PCR amplification step of those coding sequences associated with protein binders (42).
The use of photo-crosslinking methods [Figure 7d] for the screening of DNA-encoded libraries has also been proposed, with encouraging experimental results (69–70). These approaches can be performed in solution and may avoid certain artifacts, that could potentially arise from the immobilization of target proteins on solid supports.
Decoding strategies
The composition of a DNA-encoded chemical library can be characterized by DNA sequencing before and after protein selections, thus providing an experimental determination of relative enrichment factors for all compounds in the library (45,46,48,53,71). We assume that the frequency at which a certain DNA sequence is found corresponds to the relative abundance of the corresponding DNA fragment (and hence the associated compound) in the library. Since each base can have four different possibilities (A, C, G and T), a stretch of 10 bases can encode 410 = 220 = 1,048,576 different events (41). Considering the iterative nature of most library construction steps, the coding sequences that identify the individual molecules are often found in variable blocks flanked by constant sequences, rather than in a single continuous stretch (differing across library members). The use of constant sequence segments facilitates PCR amplification procedures, as well as the implementation of certain encoding methods [Figures 4-6].
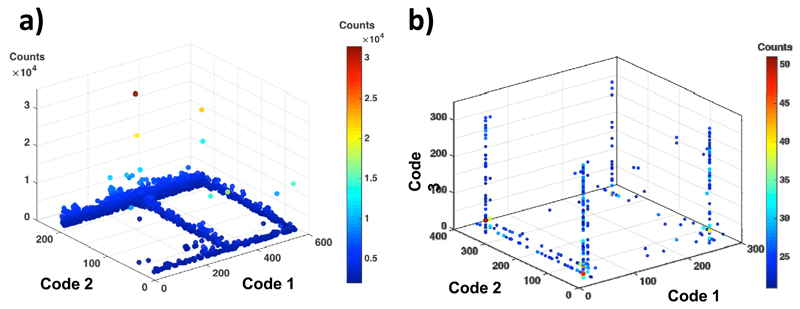
The advent of high-throughput DNA sequencing technologies (e.g., 454 technology or Illumina sequencing; 45,46,72) has revolutionized the field of DNA-encoded chemistry, permitting the screening of very large compound collections. The relative abundance of compounds in library consisting of two sets of building blocks can be depicted as a cube, in which two dimensions are used to determine the identity of pairs of building blocks, while the third dimension corresponds to the relative compound frequency (i.e., to the number of times individual sequence tags are read in a high-throughput sequencing experiment) [Figure 8a]. A library based on three sets of building blocks will need four dimensions, in order to display the identity of all compounds and their relative frequency. In most instances, spheres of different radius or color are used, in order to graphically represent this pseudo-four dimensional space [Figure 8b]. The use of suitable cut-off filters in frequency counts allows the display of the most common molecules in a collection (e.g., the most enriched compounds, after a selection procedure). Lines and planes of enriched compounds in libraries based on three sets of building blocks correspond to set of molecules, for which two or one building blocks (respectively) dominate the selection procedure. It is indeed possible, as we will see in a successive section, that not all building blocks equally contribute to binding affinity.
Figure 8.
Illustrative fingerprints of selection results, performed with a library consisting of two (a) or three (b) sets of building blocks.
Library design, chemical reactions and hit compounds
Many different chemical strategies can be considered, in order to construct an encoded library, using multi-step synthesis procedures. Some approaches have been inspired by previous combinatorial chemistry approach, making use of “DNA-friendly” transformations. Compilation of chemical reactions which can be performed on DNA and which are potentially compatible with library construction have been described (52,73). However, not all building blocks are equally reactive even for the most straightforward transformations (e.g., amide bond formation) (74) and it is recommended to investigate reaction conditions for individual compounds prior to library synthesis.
Members of single-pharmacophore chemical libraries, produced using the DNA-recorded method, are routinely purified after the first step of synthesis. However, all subsequent transformations are performed with pools of compounds and, as a consequence, are not purified and are difficult to characterize using analytical methodologies. “Catch and cap” technologies have been proposed as an avenue to achieve higher library purity, but the additional chemical steps may lead to loss in DNA quantities (75).
Dual-pharmacophore chemical libraries (such as ESAC libraries) are generated by the combinatorial self-assembly of sub-libraries of very high quality, whose members are individually purified by HPLC and checked by mass spectrometry.
Recent surveys of selection hits have been published, which describe publications with hit compounds isolated from DNA-encoded chemical libraries (32,40,41). Here, we highlight some notable hit discovery procedures, detailing how the corresponding libraries had been constructed and which building blocks mostly contributed to the experimentally-observed affinity constants.
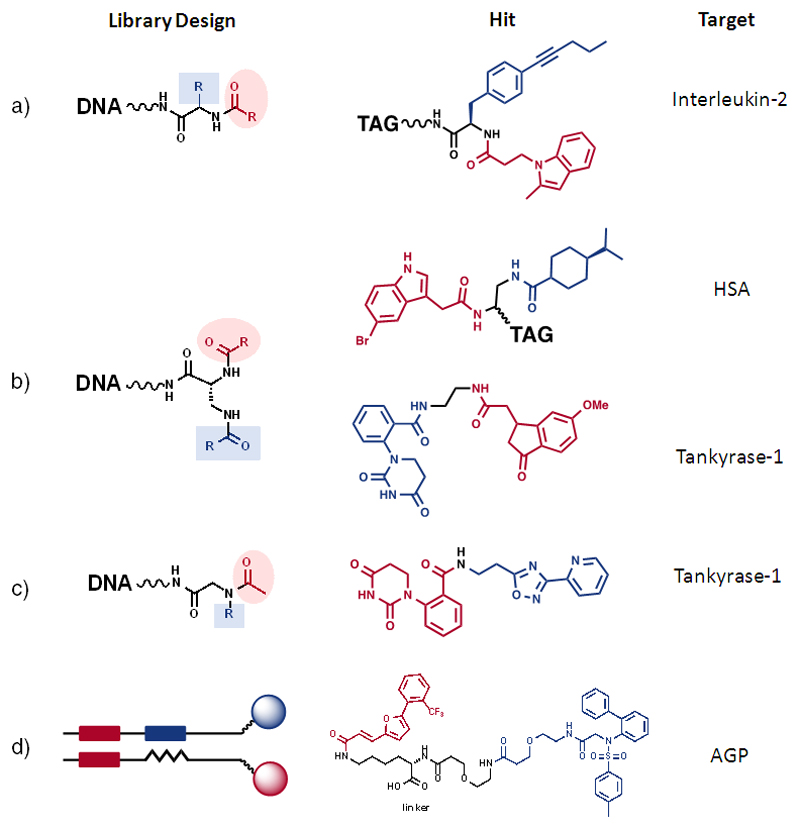
A few interesting hits, isolated from DNA-encoded chemical libraries based on two sets of building blocks, have been described. For example, Leimbacher et al. described the synthesis of a combinatorial library, featuring a set of amino acids which were subsequently reacted with a set of carboxylic acids, from which specific interleukin-2 inhibitors could be isolated [Figure 9a]. In those compounds, a methyl indole derivative played a crucial role for the molecular recognition of the cytokine target (47). In this library and in many other applications, it was convenient to substitute the DNA moiety with a fluorophore, in order to facilitate the determination of dissociation constants in solution, using fluorescence polarization methodologies (see also next section).
Figure 9.
Selected examples of binding molecules, isolated from DNA-encoded chemical libraries. The individual examples show the general library designs, which were used for the selections. The color-code of the building blocks is retained when displaying the chemical structure of the hit molecule, facilitating the detection of synthetic strategies and of modular structures.
A library based on a central scaffold featuring two primary amino groups, which were sequentially modified with two sets of carboxylic acids, yielded high-affinity binders against various target proteins, including selective albumin binders (71,76) and tankyrase 1 inhibitors (71) [Figure 9b]. The latter protein could also be drugged using compact structures, generated through the acylation of secondary amines (77) [Figure 9c].
ESAC libraries have allowed the isolation of high-affinity alpha-1 acid glycoprotein binders [Figure 9d]. Interestingly, the combination of two building blocks yielded binders with a dissociation constant in the nanomolar range, while the individual building blocks did not exhibit any detectable binding in fluorescence polarization assays or in isothermal titration calorimetry (51). This observation suggests that a chelate binding effect may be exploited for molecular recognition applications, when low affinity ligands recognizing adjacent pockets on the target protein of interest are joined together. The chemical nature of the linker connecting two building blocks can also strongly contribute to the resulting binding affinity (61,78).
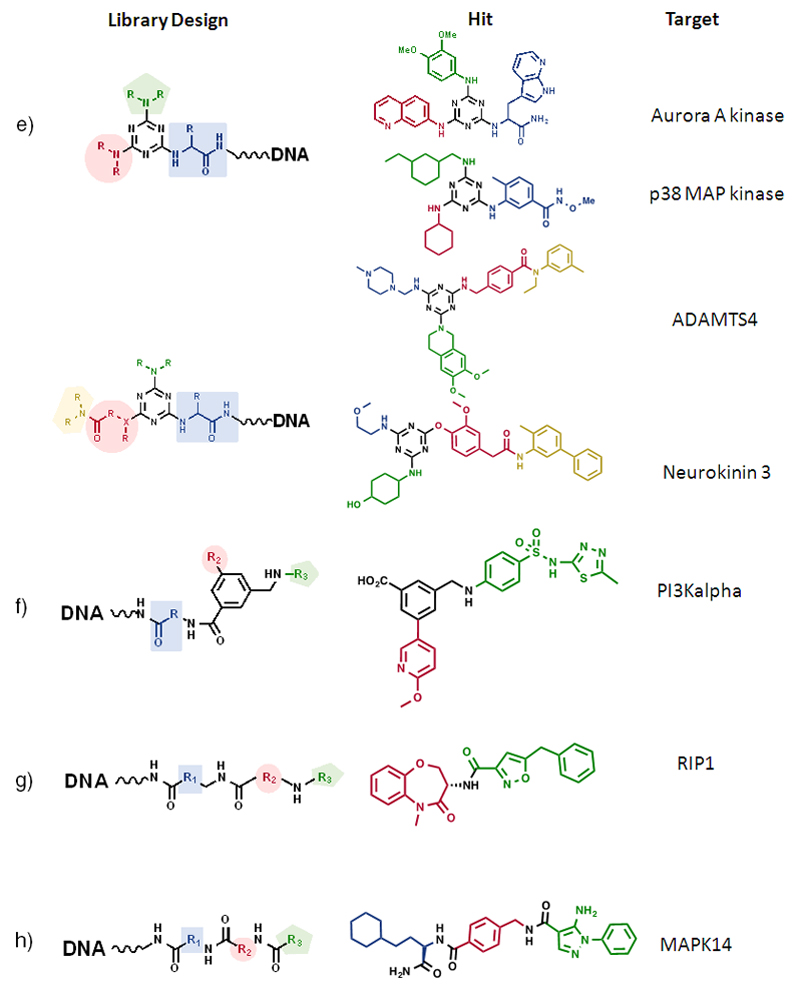
Libraries based on three or more sets of scaffolds have yielded interesting binders against targets of pharmaceutical interest. For example, scientists at GSK described in 2009 novel libraries, including one which contained >800 million substituted triazines, resulting from the combinatorial assembly of four sets of building blocks [Figure 9e]. The authors isolated inhibitors for Aurora A kinase and p38 MAP kinase, some of which had potency in the low nanomolar concentration range (43). Triazine-based libraries have also yielded high affinity ligands towards other classes of protein targets, including a 2 nM inhibitor of neurokinin 3 (79) and a 10 nM binder to ADAMTS4 (80), indicating the versatility of triazines for library design [Figure 9e]. The neurokinin 3 hit discovery activities were remarkable, since encoded library technology was used to find inhibitors of a cell surface target, using selection conditions that mimic the cellular environment (79).
The GSK group has reported the isolation of phosphoinositide 3-kinase-α (PI3Kα) inhibitors (IC50 = 10 nM), using a library of 3.5 million compounds, assembled by the sequential reaction of 191 aminoacids with an aromatic multifunctional scaffold, which was subsequently reacted with 96 boronates and 196 amines (81) [Figure 9f]. Suzuki- and Sonogashira-coupling methodologies are particularly attractive for the formation of carbon-carbon bonds in the presence of DNA (73,82). Scientists at GSK also reported the successful isolation of potent inhibitors of the Receptor Interacting Protein 1 (RIP1) kinase, from a library of 2.6 billion compounds, obtained by the sequential reaction of three sets of compounds (632 aminoacids, 632 aminoacids and 6594 amine capping reagents, such as carboxylic acids) [Figure 9g]. Interestingly, the most potent inhibitors (a set of substituted benzodiazepines) relied only on the chemical structure of the second and third building block, while the first set of building blocks could be removed without loss of inhibitory potency (83). This observation suggests that the same inhibitors could have been isolated from libraries based on two sets of building blocks, for example having the design of Figure 9a.
Using YoctoReactor™ technology, scientists at Vipergen have described the synthesis of a linear library containing >12 million compounds, resulting from three series of acylation, reductive amination and urea formation steps. The library allowed the isolation of a MAPK14 inhibitor with potency in the single-digit nanomolar range in cellular assays (84) [Figure 9h]
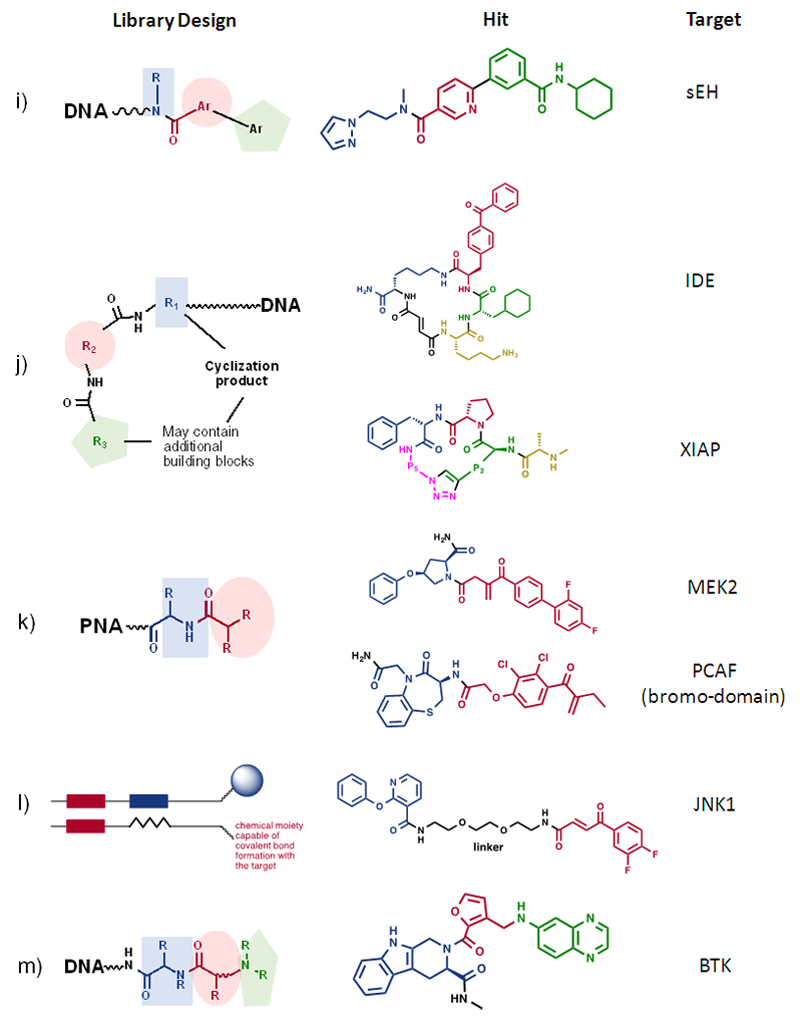
Scientists at X-Chem have been very active in the field of DNA-encoded chemical libraries and have reported, among other results, the synthesis of a library containing > 300 million compounds, resulting from the sequential reaction of 2,259 primary amines, 666 bromoaryl acids and 667 boronic acids [Figure 9i]. Fingerprints of selections performed against soluble epoxide hydrolase enabled the synthesis and confirmation of a hit with 2 nM potency and drug-like properties [MW = 431 Dalton; CLogP = 3.1] (85)
DNA-encoded chemistry has also been used to generate libraries of peptide macrocycles, from which interesting binders against “difficult” target proteins have been isolated. For example, David Liu and collaborators reported the isolation of a 50 nM inhibitor of Insulin Degrading Enzyme (IDE), from a library of 13,824 macrocycles, constructed using a DNA-templated synthesis approach (86) [Figure 9j]. A similar approach has been used by scientists at Ensemble to isolate XIAP inhibitors with IC50 = 140 nM from a library of 160’000 cyclic peptides (87) [Figure 9j].
DNA-encoded chemical libraries for the discovery of covalent inhibitors
Most applications of DNA-encoded chemical libraries relate to the isolation of non-covalent inhibitors, but the technology is also ideally suited for the discovery of covalent binders. Winssinger and colleagues used a library of 10’000 compounds, based on chemically-modified PNAs and featuring suitable Michael acceptors, for the discovery of irreversible covalent binders of MEK2 (88) [Figure 9k]. The same group reported the use of PNA-encoded chemical libraries for the discovery of two small molecules that form a covalent bond with cysteine residues conserved across the bromodomain family (epigenetic readers that are important for the regulation of transcriptional programs; 89) [Figure 9l].
Zimmermann et al. have recently described the use of ESAC libraries, containing building blocks suitable for the chemical modification of protein targets, to discover a covalent inhibitor of JNK1 kinase, a protein containing 8 cysteine residues (of which one in close proximity of the active site). Interestingly, the covalent protein modification was highly selective compared to other kinases with cysteine residues in the active site (90) and proceeded with 1:1 stoichiometry, even in the presence of >1000-fold excess of glutathione, used to simulate the thiol-rich intracellular reducing milieu [Figure 9m].
Clark and colleagues reported the construction of an encoded library, containing 27 million acrylamide derivatives, from which potent irreversible covalent inhibitors of Bruton Tyrosine Kinase (BTK, a validated target for the treatment of certain forms of lymphoma, with a cysteine residue in the active site) could be isolated (91) [Figure 9n]. Liu and coworkers have recently used interaction-dependent screening methodologies with libraries of DNA-encoded bioactive compounds and mixtures of DNA-tagged human kinases to identify ligand:protein binding partners out of 32’096 possible combinations in a single solution-phase panning experiment. The results confirmed known small molecule:protein interactions and also revealed that ethacrynic acid is a novel ligand and inhibitor of MAP2K6 kinase. Ethacrynic acid inhibits MAP2K6, in part, through alkylation of a non-conserved cysteine residue (92).
“On-DNA” and “off-DNA” hit validation strategies
DNA is an invaluable tag for the encoding and decoding of chemical libraries. However, for most applications, it is desirable to use chemical compounds in the absence of DNA. In many cases, the confirmation of hits from library selections proceeds through the re-synthesis of the most enriched compounds “off-DNA”, followed by a characterization of their binding properties using biochemical or biophysical assays. However, the synthesis and purification of organic molecules can be labor-intensive, depending on the structure of the compounds and the number of hits which need to be measured. Furthermore, when the target is not amenable to biochemical characterization (e.g., using an enzyme inhibition assay), it can be difficult to measure binding affinities in the absence of a suitable tag. Surface plasmon resonance methodologies (e.g., BIAcore) may be complex to perform with proteins that lose activity upon immobilization on a solid support or during microsensor chip regeneration procedures. Isothermal titration calorimetry may allow the determination of dissociation constants in solution, but the procedure poses certain requirements on the availability, buffer composition and solubility for both binding partners. For these reasons, it is often convenient to resynthesize hit compounds with a fluorophore moiety, at the position which was originally occupied by the DNA tag, and to perform fluorescence polarization measurements in solution (46). In an initial phase of the hit validation process, it is practical to re-synthesize compounds “on-DNA” (51,93). Zimmermann and colleagues have recently described a general and versatile methodology, based on the hybridization of oligonucleotide conjugates with complementary strands, labeled with a suitable fluorophore and/or additional chemical moieties (93) [Figure 10]. The technology benefits from the fact that “on-DNA” synthesis procedures are available from the previously performed library construction steps. Furthermore, oligonucleotides facilitate the purification of the conjugates and their analytical characterization by mass spectrometry, while also contributing to the solubilization of lipophilic molecules.
Figure 10.
Methods for the fluorescent labeling of library members, enabling fluorescence polatization measurement procedures. For single-pharmacophore library members, hits can be resynthesized as fluorescently-labeled compounds, or on oligonucleotides (DNA or LNA) which carry the fluorophore on the same strand or on a complementary strand. Similarly, hits from dual-pharmacophore chemical libraries can be re-synthesized as fluorescent molecules (with a suitable linker connecting the two building blocks) or on complementary oligonucleotides (one of which is fluorescently labeled).
The chemistry of large numbers
DNA-encoded chemical libraries are often referred to as “the chemistry of large numbers”. What is meant by this is that most chemical procedures are about the reaction between molecules, but usually one at a time. However, if one has a strategy to de-convolute reactions in a mixture, DNA encoding allows one to carry out vast numbers of reactions at once. Current encoded libraries can be used to find ligands for many protein targets, but not (yet) for all (94). This raises the question of how large should these libraries be. A simple answer is that, when one builds diversity systems, the incorporation of more diversity is never a bad idea, so long as one has a robust strategy for the de-convolution of binding events. In our experience, when screening libraries with conventional affinity capture methodologies, it is convenient to ensure that individual molecules are present in a selection experiment with least 105 copies each (64,65,95,96). In practice, this prerequisite may constrain the size of encoded libraries that can be efficiently selected. In most cases, we aim at perturbing the biological world either by construction of a ligand that mimics the physiological binding event or by generating a new binding event such as an allosteric effector. Thus, it is useful to ask what does nature use as components of ligands, because it is this that we are trying to duplicate. Actually, nature uses relatively few chemical interactions (i.e. hydrogen bonds, pi stacking, hydrophobic interactions, etc.) but can achieve enormous binding energy by using them in concert in a relatively anhydrous, confined space, such as the active site of an enzyme.
Some of the major reactions to generate DNA-encoded chemical libraries include amide bond formation and reductive amination, that can be used with various molecular scaffolds bearing reactive moieties similar to those used in nature. However, natural ligands are a product of evolution where the best chemical entities and their organization in space were selected over vast periods of time. The central concept is that nature took advantage of what was available or could be biologically constructed (such as by incorporating cofactors co-opted from the environment) but used selection over time to optimize the organization of the individual components of a ligand. In the case of DNA-encoded chemical libraries, the large numbers are the equivalent of evolutionary time because one hopes that somewhere in the large number of molecules is one or a few that are at least as good as the one that was the product of evolution. There are, however, other significant directions being pursued in the generation of DNA-encoded chemical libraries. Importantly, the use of enzymes in the synthesis of organic ligands is only beginning to be explored (97). Additionally, organic chemists are inventing new reactions that are compatible with DNA and water solutions. This allows DNA-encoded chemical libraries to incorporate adducts that were not available to natural selection. These new adducts may give components or scaffolds that are beyond the chemistry available to nature.
Thus, in the end the numbers question needs to be rephrased as a diversity question. As for numbers, one simply asks how many unique members are there in the system, whereas diversity concerns how different are the members from each other? To achieve both large numbers and maximum diversity, it would seem that one should link simple amide based chemistries to more sophisticated chemical transformations. In the end, in an amalgamation of genetics and chemical synthesis, new exciting diversity systems will probably result from the sequential use of simple and complex organic transformations, exploring a chemical space that may contain a solution to biological problems which cannot be addressed by other means. These diversity systems, and the DNA-encoded chemical libraries described in this review, are new to organic chemistry, because they endow organic molecules with information capable of replication thereby linking phenotype to genotype in single molecules.
Acknowledgements
D.N. acknowledges financial support from ETH Zürich, the Swiss National Science Foundation (CRSII2_160699 and 310030B_163479/1), of the ERC Advanced Grant “ZAUBERKUGEL” and of Philochem AG.
R.A.L. acknowledges financial support from the J.P.B. Foundation.
We are grateful to Gerry Joyce, Ian Wilson, Raphael Franzini and Jörg Scheuermann for having read the manuscript and for useful insights. We also thank Nicholas Favalli, Gabriele Bassi and Jörg Scheuermann for help in the preparation of Figure 9.
References
- [1].Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov. 2016;15:605–19. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- [2].Folmer RH. Integrating biophysics with HTS-driven drug discovery projects. Drug Discov Today. 2016;21:491–8. doi: 10.1016/j.drudis.2016.01.011. [DOI] [PubMed] [Google Scholar]
- [3].Nielsen TE, Schreiber SL. Towards the optimal screening collection: a synthesis strategy. Angew Chem Int Ed Engl. 2008;47:48–56. doi: 10.1002/anie.200703073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schreiber SL, Kotz JD, Li M, Aubé J, Austin CP, et al. Advancing Biological Understanding and Therapeutics Discovery with Small-Molecule Probes. Cell. 2015;161:1252–65. doi: 10.1016/j.cell.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, et al. The promise and peril of chemical probes. Nat Chem Biol. 2015;11:536–41. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–95. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- [7].Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–7. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- [8].Wilchek M, Bayer EA, Livnah O. Essentials of biorecognition: the (strept)avidin-biotin system as a model for protein-protein and protein-ligand interaction. Immunol Lett. 2006;103:27–32. doi: 10.1016/j.imlet.2005.10.022. [DOI] [PubMed] [Google Scholar]
- [9].Heinisch T, Ward TR. Artificial Metalloenzymes Based on the Biotin-Streptavidin Technology: Challenges and Opportunities. Acc Chem Res. 2016;49:1711–21. doi: 10.1021/acs.accounts.6b00235. [DOI] [PubMed] [Google Scholar]
- [10].Paganelli G, Pervez S, Siccardi AG, Rowlinson G, Deleide G, et al. Intraperitoneal radio-localization of tumors pre-targeted by biotinylated monoclonal antibodies. Int J Cancer. 1990;45:1184–9. doi: 10.1002/ijc.2910450632. [DOI] [PubMed] [Google Scholar]
- [11].Sapsford KE, Algar WR, Berti L, Gemmill KB, Casey BJ, et al. Functionalizing nanoparticles with biological molecules: developing chemistries that facilitate nanotechnology. Chem Rev. 2013;113:1904–2074. doi: 10.1021/cr300143v. [DOI] [PubMed] [Google Scholar]
- [12].Halai R, Cooper MA. Using label-free screening technology to improve efficiency in drug discovery. Expert Opin Drug Discov. 2012;7:123–31. doi: 10.1517/17460441.2012.651121. [DOI] [PubMed] [Google Scholar]
- [13].Goodnow RAJ. In: A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space. Goodnow RAJ, editor. Wiley & Sons; 2014. pp. 417–426. [Google Scholar]
- [14].Stark JL, Powers R. Application of NMR and molecular docking in structure-based drug discovery. Top Curr Chem. 2012;326:1–34. doi: 10.1007/128_2011_213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–7. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- [16].Clackson T, Wells JA. In vitro selection from protein and peptide libraries. Trends Biotechnol. 1994;12:173–84. doi: 10.1016/0167-7799(94)90079-5. [DOI] [PubMed] [Google Scholar]
- [17].McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–4. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- [18].Huse WD, Sastry L, Iverson SA, Kang AS, Alting-Mees M, et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246:1275–81. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- [19].Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, et al. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–97. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- [20].Kang AS, Barbas CF, Janda KD, Benkovic SJ, Lerner RA. Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. Proc Natl Acad Sci U S A. 1991;88:4363–6. doi: 10.1073/pnas.88.10.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Winter G, Milstein C. Man-made antibodies. Nature. 1991;349:293–9. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- [22].Jespers LS, Roberts A, Mahler SM, Winter G, Hoogenboom HR. Guiding the selection of human antibodies from phage display repertoires to a single epitope of an antigen. Biotechnology (N Y) 1994;12:899–903. doi: 10.1038/nbt0994-899. [DOI] [PubMed] [Google Scholar]
- [23].Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 13:3245–60. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–73. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Plückthun A. Ribosome display: a perspective. Methods Mol Biol. 2012;805:3–28. doi: 10.1007/978-1-61779-379-0_1. [DOI] [PubMed] [Google Scholar]
- [26].Brenner S, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci U S A. 1992;89:5381–3. doi: 10.1073/pnas.89.12.5381. [Earliest proposal of using DNA tags to encode chemical libraries] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Needels MC, Jones DG, Tate EH, Heinkel GL, Kochersperger LM, et al. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc Natl Acad Sci U S A. 1993;90:10700–4. doi: 10.1073/pnas.90.22.10700. [Earliest demonstration that DNA-encoded peptides of known specificity to a cognate antibody can be retrieved from a large mixture of compounds] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nielsen J, Brenner S, Janda KD. Synthetic methods for the implementation of encoded combinatorial chemistry. J Am Chem Soc. 1993;115:9812. [Earliest demonstration that DNA-encoded peptides of known specificity to a cognate antibody can be retrieved from a large mixture of compounds] [Google Scholar]
- [29].Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–5. doi: 10.1126/science.1102629. [Earliest proposal and implementation of a DNA-templated chemical library] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded self-assembling chemical libraries. Nat Biotechnol. 2004;22:568–74. doi: 10.1038/nbt961. [Earliest proposal and implementation of dual-pharmacophore DNA-encoded chemical libraries] [DOI] [PubMed] [Google Scholar]
- [31].Halpin DR, Harbury PB. DNA display I. Sequence-encoded routing of DNA populations. PLoS Biol. 2004;2:E173. doi: 10.1371/journal.pbio.0020173. [Earliest proposal of encoding by DNA routing] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Franzini RM, Neri D, Scheuermann J. DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries. Acc Chem Res. 2014;47:1247–55. doi: 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]
- [33].Kleiner RE, Dumelin CE, Liu DR. Small-molecule discovery from DNA-encoded chemical libraries. Chem Soc Rev. 40:5707–17. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zimmermann G, Neri D. DNA-encoded chemical libraries: foundations and applications in lead discovery. Drug Discov Today. 2016;21:1828–1834. doi: 10.1016/j.drudis.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Franzini RM, Randolph C. Chemical Space of DNA-Encoded Libraries. J Med Chem. 2016;59:6629–44. doi: 10.1021/acs.jmedchem.5b01874. [DOI] [PubMed] [Google Scholar]
- [36].Clark MA. Selecting chemicals: the emerging utility of DNA-encoded libraries. Curr Opin Chem Biol. 2010;14:396–403. doi: 10.1016/j.cbpa.2010.02.017. [DOI] [PubMed] [Google Scholar]
- [37].Zambaldo C, Barluenga S, Winssinger N. PNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- [38].Shi B, Zhou Y, Huang Y, Zhang J, Li X. Recent advances on the encoding and selection methods of DNA-encoded chemical library. Bioorg Med Chem Lett. 2017;27:361–369. doi: 10.1016/j.bmcl.2016.12.025. [DOI] [PubMed] [Google Scholar]
- [39].Keefe AD, Clark MA, Hupp CD, Litovchick A, Zhang Y. Chemical ligation methods for the tagging of DNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:80–8. doi: 10.1016/j.cbpa.2015.02.015. [DOI] [PubMed] [Google Scholar]
- [40].Salamon H, Klika Škopić M, Jung K, Bugain, Brunschweiger A. Chemical Biology Probes from Advanced DNA-encoded Libraries. ACS Chem Biol. 2016;11:296–307. doi: 10.1021/acschembio.5b00981. [DOI] [PubMed] [Google Scholar]
- [41].Goodnow RA, Jr, Dumelin CE, Keefe AD. DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat Rev Drug Discov. 2017;16:131–147. doi: 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- [42].Blakskjaer P, Heitner T, Hansen NJ. Fidelity by design: Yoctoreactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery. Curr Opin Chem Biol. 2015;26:62–71. doi: 10.1016/j.cbpa.2015.02.003. [DOI] [PubMed] [Google Scholar]
- [43].Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–54. doi: 10.1038/nchembio.211. [One of the earliest examples for the synthesis and successful screening of a very large DNA-encoded chemical library] [DOI] [PubMed] [Google Scholar]
- [44].Buller F, Mannocci L, Zhang Y, Dumelin CE, Scheuermann J, Neri D. Design and synthesis of a novel DNA-encoded chemical library using Diels-Alder cycloadditions. Bioorg Med Chem Lett. 2008;18:5926–31. doi: 10.1016/j.bmcl.2008.07.038. [DOI] [PubMed] [Google Scholar]
- [45].Buller F, Zhang Y, Scheuermann J, Schäfer J, Bühlmann P, et al. Discovery of TNF inhibitors from a DNA-encoded chemical library based on diels-alder cycloaddition. Chem Biol. 2009;16:1075–86. doi: 10.1016/j.chembiol.2009.09.011. [DOI] [PubMed] [Google Scholar]
- [46].Mannocci L, Zhang Y, Scheuermann J, Leimbacher M, De Bellis G, et al. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc Natl Acad Sci U S A. 2008;105:17670–5. doi: 10.1073/pnas.0805130105. [Earliest publication on DNA-recorded chemical library and on the use of high-throughput DNA sequencing for library decoding] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Leimbacher M, Zhang Y, Mannocci L, Stravs M, Geppert T, et al. Discovery of small-molecule interleukin-2 inhibitors from a DNA-encoded chemical library. Chemistry. 2012;18:7729–37. doi: 10.1002/chem.201200952. [DOI] [PubMed] [Google Scholar]
- [48].Buller F, Steiner M, Frey K, Mircsof D, Scheuermann J, et al. Selection of Carbonic Anhydrase IX Inhibitors from One Million DNA-Encoded Compounds. ACS Chem Biol. 2011;6:336–44. doi: 10.1021/cb1003477. [DOI] [PubMed] [Google Scholar]
- [49].Mannocci L, Melkko S, Buller F, Molnàr I, Bianké JP, et al. Isolation of potent and specific trypsin inhibitors from a DNA-encoded chemical library. Bioconjug Chem. 2010;21:1836–41. doi: 10.1021/bc100198x. [DOI] [PubMed] [Google Scholar]
- [50].Bain JD, Switzer C. Regioselective ligation of oligoribonucleotides using DNA splints. Nucleic Acids Res. 1992;20:4372. doi: 10.1093/nar/20.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, et al. Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat Chem. 2015;7:241–9. doi: 10.1038/nchem.2158. [Earliest publication on the use of high-throughput DNA sequencing for the decoding of dual-pharmacophore chemical libraries] [DOI] [PubMed] [Google Scholar]
- [52].Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature. 2004;431:545–9. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kleiner RE, Dumelin CE, Tiu GC, Sakurai K, Liu DR. In vitro selection of a DNA-templated small-molecule library reveals a class of macrocyclic kinase inhibitors. J Am Chem Soc. 2010;132:11779–91. doi: 10.1021/ja104903x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li Y, Zhao P, Zhang M, Zhao X, Li X. Multistep DNA-templated synthesis using a universal template. J Am Chem Soc. 2013;135:17727–30. doi: 10.1021/ja409936r. [DOI] [PubMed] [Google Scholar]
- [55].Hansen MH, Blakskjaer P, Petersen LK, Hansen TH, Højfeldt JW, et al. A yoctoliter-scale DNA reactor for small-molecule evolution. J Am Chem Soc. 2009;131:1322–7. doi: 10.1021/ja808558a. [Earliest report on Yoctoreactor™ encoding technology] [DOI] [PubMed] [Google Scholar]
- [56].Halpin DR, Harbury PB. DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol. 2004;2:E174. doi: 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Halpin DR, Lee JA, Wrenn SJ, Harbury PB. DNA display III. Solid-phase organic synthesis on unprotected DNA. PLoS Biol. 2004;2:E175. doi: 10.1371/journal.pbio.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. Synthetic ligands discovered by in vitro selection. J Am Chem Soc. 2007;129:13137–43. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Neri D, Melkko S. Encoded self-assembling chemical libraries. Patent number WO03/076943. 2002
- [60].Gorska K, Huang KT, Chaloin O, Winssinger N. DNA-templated homo- and heterodimerization of peptide nucleic acid encoded oligosaccharides that mimick the carbohydrate epitope of HIV. Angew Chem Int Ed Engl. 2009;48:7695–700. doi: 10.1002/anie.200903328. [DOI] [PubMed] [Google Scholar]
- [61].Dagauer JP, Zambaldo C, Ciobanu M, Morieux P, Barluenga S, et al. DNA display of fragment pairs as a tool for the discovery of novel biologically active small molecules. Chem Sci. 2015;6:739–44. doi: 10.1039/c4sc01654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dumelin CE, Trüssel S, Buller F, Trachsel E, Bootz F, et al. A portable albumin binder from a DNA-encoded chemical library. Angew Chem Int Ed Engl. 2008;47:3196–201. doi: 10.1002/anie.200704936. [DOI] [PubMed] [Google Scholar]
- [63].Decurtins W, Wichert M, Franzini RM, Buller F, Stravs MA, et al. Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat Protoc. 2016;11:764–80. doi: 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li Y, Zimmermann G, Scheuermann J, Neri D. Quantitative PCR is a valuable tool to monitor the performance of DNA-encoded chemical library selections. ChemBioChem. 2017;18:848–852. doi: 10.1002/cbic.201600626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Belyanskaya SL, Ding Y, Callahan JF, Lazaar AL, Israel DI. Discovering Drugs with DNA-Encoded Library Technology: From Concept to Clinic with an Inhibitor of Soluble Epoxide Hydrolase. ChemBioChem. 2017;18:837–42. doi: 10.1002/cbic.201700014. [DOI] [PubMed] [Google Scholar]
- [66].Bao J, Krylova SM, Cherney LT, Hale RL, Belyanskaya SL, et al. Predicting Electrophoretic Mobility of Protein-Ligand Complexes for Ligands from DNA-Encoded Libraries of Small Molecules. Anal Chem. 2015;88:5498–506. doi: 10.1021/acs.analchem.6b00980. [DOI] [PubMed] [Google Scholar]
- [67].McGregor LM, Gorin DJ, Dumelin CE, Liu DR. Interaction-dependent PCR: identification of ligand-target pairs from libraries of ligands and libraries of targets in a single solution-phase experiment. J Am Chem Soc. 2010;132:15522–4. doi: 10.1021/ja107677q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].McGregor LM, Jain T, Liu DR. Identification of ligand-target pairs from combined libraries of small molecules and unpurified protein targets in cell lysates. J Am Chem Soc. 2014;136:3264–70. doi: 10.1021/ja412934t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Denton KE, Krusemark CJ. Crosslinking of DNA-linked ligands to target proteins for enrichment from DNA-encoded libraries. MedChemComm. 2016;7:2020–7. doi: 10.1039/C6MD00288A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shi B, Deng Y, Zhao P, Li X. Selecting a DNA-Encoded Chemical Library against Non-immobilized Proteins Using a "Ligate-Cross-Link-Purify" Strategy. Bioconj Chem. 2017;28:2293. doi: 10.1021/acs.bioconjchem.7b00343. [DOI] [PubMed] [Google Scholar]
- [71].Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, et al. Identification of structure-activity relationships from screening a structurally compact DNA-encoded chemical library. Angew Chem Int Ed Engl. 2015;54:3927–31. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- [72].Buller F, Steiner M, Scheuermann J, Mannocci L, Nissen I, et al. High-throughput sequencing for the identification of binding molecules from DNA-encoded chemical libraries. Bioorg Med Chem Lett. 2010;20:4188–92. doi: 10.1016/j.bmcl.2010.05.053. [DOI] [PubMed] [Google Scholar]
- [73].Satz AL, Cai J, Chen Y, Goodnow R, Gruber F, et al. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjug Chem. 2015;26:1623–32. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- [74].Li Y, Gabriele E, Samain F, Favalli N, Sladojevich F, et al. Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb Sci. 2016;18:438–43. doi: 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Franzini RM, Biendl S, Mikutis G, Samain F, Scheuermann J, et al. "Cap-and-Catch" Purification for Enhancing the Quality of Libraries of DNA Conjugates. ACS Comb Sci. 2015;17:393–8. doi: 10.1021/acscombsci.5b00072. [DOI] [PubMed] [Google Scholar]
- [76].Franzini RM, Nauer A, Scheuermann J, Neri D. Interrogating target-specificity by parallel screening of a DNA-encoded chemical library against closely related proteins. Chem Commun (Camb) 51:8014–6. doi: 10.1039/c5cc01230a. [DOI] [PubMed] [Google Scholar]
- [77].Samain F, Ekblad T, Mikutis G, Zhong N, Zimmermann M, et al. Tankyrase 1 Inhibitors with Drug-like Properties Identified by Screening a DNA-Encoded Chemical Library. J Med Chem. 2015;58:5143–9. doi: 10.1021/acs.jmedchem.5b00432. [DOI] [PubMed] [Google Scholar]
- [78].Melkko S, Zhang Y, Dumelin CE, Scheuermann J, Neri D. Isolation of high-affinity trypsin inhibitors from a DNA-encoded chemical library. Angew Chem Int Ed Engl. 2007;46:4671–4. doi: 10.1002/anie.200700654. [DOI] [PubMed] [Google Scholar]
- [79].Wu Z, Graybill TL, Zeng X, Platchek M, Zhang J, et al. Cell-Based Selection Expands the Utility of DNA-Encoded Small-Molecule Library Technology to Cell Surface Drug Targets: Identification of Novel Antagonists of the NK3 Tachykinin Receptor. ACS Comb Sci. 2015;17:722–731. doi: 10.1021/acscombsci.5b00124. [DOI] [PubMed] [Google Scholar]
- [80].Ding Y, O'Keefe H, DeLorey JL, Israel DI, Messer JA, et al. Discovery of Potent and Selective Inhibitors for ADAMTS-4 through DNA-Encoded Library Technology (ELT) ACS Med Chem Lett. 2015;6:888–93. doi: 10.1021/acsmedchemlett.5b00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yang H, Medeiros PF, Raha K, Elkins P, Lind KE, et al. Discovery of a Potent Class of PI3Kα Inhibitors with Unique Binding Mode via Encoded Library Technology (ELT) ACS Med Chem Lett. 2015;6:531–6. doi: 10.1021/acsmedchemlett.5b00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Malone ML, Paegel BM. What is a "DNA-Compatible" Reaction? ACS Comb Sci. 2016;18:182–7. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Harris PA, King BW, Bandyopadhyay D, Berger SB, Campobasso N, et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J Med Chem. 2016;59:2163–78. doi: 10.1021/acs.jmedchem.5b01898. [DOI] [PubMed] [Google Scholar]
- [84].Petersen LK, Blakskjaer P, Chaikuad A, Christensen AB, Dietvorst J, et al. Novel p38α MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library. MedChemComm. 2016;7:1332–9. [Google Scholar]
- [85].Litovchick A, Dumelin CE, Habeshian S, Gikunju D, Gujé MA, et al. Encoded library synthesis using chemical ligation and the discovery of sEH inhibitors from a 334-million member library. Sci Rep. 2015;5 doi: 10.1038/srep10916. 10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Maianti JP, McFedries A, Foda ZH, Kleiner RE, Du XQ, et al. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature. 2014;511:94–8. doi: 10.1038/nature13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Seigal BA, Connors WH, Fraley A, Borzilleri RM, Carter PH, et al. The discovery of macrocyclic XIAP antagonists from a DNA-programmed chemistry library, and their optimization to give lead compounds with in vivo antitumor activity. J Med Chem. 2015;58:2855–61. doi: 10.1021/jm501892g. [DOI] [PubMed] [Google Scholar]
- [88].Zambaldo C, Daguer JP, Saarbach J, Barluenga S, Winssinger N. Screening for covalent inhibitors using DNA-display of small molecule libraries functionalized with cysteine reactive moieties. MedChemComm. 2016;7:1340–1351. [Google Scholar]
- [89].Daguer JP, Zambaldo C, Abegg D, Barluenga S, Tallant C, et al. Identification of covalent bromodomain binders through DNA display of small molecules. Angew Chem Int Ed Engl. 2015;54:6057–61. doi: 10.1002/anie.201412276. [DOI] [PubMed] [Google Scholar]
- [90].Zimmermann G, Rieder U, Bajic D, Vanetti S, Chaikuad A, et al. A specific and covalent JNK-1 ligand selected from an encoded self-assembling chemical library. Chemistry. 2017;23:8152–55. doi: 10.1002/chem.201701644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Cuozzo JW, Centrella PA, Gikunju D, Habeshian S, Hupp CD, et al. Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library. Chembiochem. 2017;18:864–871. doi: 10.1002/cbic.201600573. [DOI] [PubMed] [Google Scholar]
- [92].Chan AI, McGregor LM, Jain T, Liu DR. Discovery of a covalent kinase inhibitor from a DNA-encoded small-molecule library x protein selection. J Am Chem Soc. 2017;139:10192–5. doi: 10.1021/jacs.7b04880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zimmermann G, Li Y, Rieder U, Mattarella M, Neri D, et al. Hit-validation methodologies for ligands isolated from DNA-encoded chemical libraries. Chembiochem. 2017;18:853–857. doi: 10.1002/cbic.201600637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Machutta CA, Kollmann CS, Lind KE, Bai X, Chan PF, et al. Prioritizing multiple therapeutic targets in parallel using automated DNA-encoded library screening. Nat Commun. 2017;8:16081. doi: 10.1038/ncomms16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Satz AL, Hochstrasser R, Petersen AC. Analysis of current DNA encoded chemical library screening data indicates higher false negative rates for numerically larger libraries. ACS Comb Sci. 2017;19:234. doi: 10.1021/acscombsci.7b00023. [DOI] [PubMed] [Google Scholar]
- [96].Li Y, De Luca R, Cazzamalli S, Pretto F, Bajic D, et al. Versatile protein recognition by the encoded display of multiple chemical elements on a constant macrocyclic scaffold. Nat Chem. 2018 doi: 10.1038/s41557-018-0017-8. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Thomas B, Lu X, Birmingham WR, Huang K, Both P, et al. Applications of biocatalysis to on-DNA carbohydrate library synthesis. ChemBioChem. 2017;18:858. doi: 10.1002/cbic.201600678. [DOI] [PubMed] [Google Scholar]