Abstract
The development of modular and efficient methods to functionalize RNA with biophysical probes is very important in advancing the understanding of the structural and functional relevance of RNA in various cellular events. Herein, we demonstrate a two-step bioorthogonal chemical functionalization approach for the conjugation of multiple probes onto RNA transcripts using a 5-vinyl-modified uridine nucleotide analog (VUTP). VUTP, containing a structurally noninvasive and versatile chemoselective handle, was efficiently incorporated into RNA transcripts by in vitro transcription reactions. Furthermore, we show for the first time the use of a palladium-mediated oxidative Heck reaction in functionalizing RNA with fluorogenic probes by reacting vinyl-labeled RNA transcripts with appropriate boronic acid substrates. The vinyl label also permitted the post-transcriptional functionalization of RNA by a reagent-free inverse electron demand Diels–Alder (IEDDA) reaction in the presence of tetrazine substrates. Collectively, our results demonstrate that the incorporation of VUTP provides newer possibilities for the modular functionalization of RNA with variety of reporters.
Introduction
Functional modification of RNA in vitro and in cells have become necessary for understanding the regulation, structure, and function of RNA.1–3 While solid-phase oligonucleotide (ON) synthesis and enzymatic labeling methods using bacteriophage RNA polymerases are widely used in the generation of labeled RNA suitable for cell-free and in-cell analysis, analogous approaches to label endogenous RNA are less prevalent.4 Immunostaining of metabolically incorporated nucleoside analogs (e.g., BrU) using antibodies, structure-specific antibodies (e.g., G-quadruplexes), aptamers (e.g., spinach), and dyes have been employed to visualize cellular RNA structure and synthesis.5–10 However, limited permeability and selectivity of the antibodies and compromised function of aptamer-tagged RNA have been the downsides of these methods. Alternatively, bio-orthogonal chemical reactions provide an easy route to label RNA in vitro and cellular milieu for a variety of applications.11–15 In this approach, a reactive group is introduced into an RNA ON by either chemical, enzymatic (RNA polymerases), or chemo-enzymatic (transferases) means, and further bioconjugation to label RNA is achieved by performing a chemoselective reaction with a cognate reactive partner containing a desired biophysical reporter. Several chemoselective reactions including azide–alkyne cycloaddition (AAC), Staudinger ligation, and inverse electron demand Diels–Alder (IEDDA) reactions have been effectively utilized in labeling protein, glycan, lipid, and DNA.16–18 However, establishing RNA-labeling techniques under the conditions used in these reactions has always been a challenge due to the inherent instability of the RNA.15
Among the various bioorthogonal reactions, AAC reaction has been extensively used in labeling RNA postsynthetically.19–27 Typically, in this reaction, a copper(I) stabilizing ligand is used to alleviate the toxic effects of copper ions, which is known to produce species harmful to nucleic acids in the redox environment.14,28,29 In another strategy, the use of toxic copper has been circumvented by employing strain-promoted AAC (SPAAC) reaction, wherein strained alkynes (e.g., cyclooctynes) are used as one of the reactive counterparts.30–34 However, SPAAC reaction efficiency in labeling nucleic acids in cells has been limited by the poor permeability and reactivity of bulky cyclooctyne probes.33,34 Other concerns that have limited the application of AAC reactions are the involvement of alkyne substrates in side reactions such as homocoupling and thiol–yne addition and low stability of azide substrates in the chemical-labeling conditions.33,35,36
Analogous to azide and alkyne groups, the ability of alkene group to participate in a wide range of chemoselective reactions has been elegantly utilized in devising several bioconjugation strategies. Bioconjugation methods based on ene-thiol and ene-tetrazole reactions have been put to good use in cross-linking and functionalizing biomolecules, especially proteins.37–39 However, the use of UV radiation and nonspecific reaction with cellular thiols has hampered their applications in in-cell analysis. Recently, protein labeling has been achieved by using oxidative Heck reaction between protein-bound alkenes and boronic acid reporters.40–42 This reaction is particularly interesting because it can be effectively performed in aqueous buffer under near physiological conditions (~pH 7, room temperature and oxygen atmosphere) as compared to other analogous Pd-assisted biomolecular labeling reactions.43,44 Very recently, base-modified 2′-deoxynucleosides, containing a minimally perturbing vinyl group as a dienophile, have been successfully incorporated in vitro into DNA ONs and metabolically into replicating DNA in cells.45,46 Although the reaction rates are significantly lower as compared to strained dienophiles, vinyl-labeled DNA did enable the labeling as well as the visualization of DNA in cell-free and cellular environments by using IEDDA reactions.
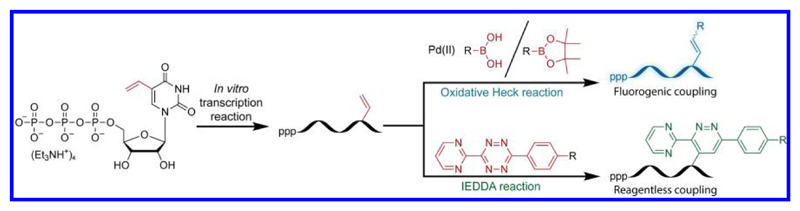
Coupling reactions have been employed to attach heterocyclic aryl moieties via an alkene to natural nucleobases. Such conjugations have produced microenvironment-sensitive nucleoside probes, which have been used in studying the conformation of the G-quadruplex structure.47 Similarly, fluorogenic reporters have been developed to specifically detect epigenetic and epitranscriptomic nucleobase modifications (e.g., 5-formylcytosine and 5-formyluracil) by using a chemoselective reaction between the base and trimethylindole derivatives.48 Taking advantage of this knowledge, we rationalized that enzymatic incorporation of a minimally perturbing vinyl-modified ribonucleotide into RNA would facilitate the generation of microenvironment-sensitive fluorogenic RNA reporters by post-transcriptional oxidative Heck reaction with appropriate boronic acid derivatives (Figure 1). Also, we envisioned that vinyl-labeled RNA transcript would open up possibilities for investigating its potential to functionalize RNA by reagentless IEDDA reaction. Despite the successes with protein and DNA,40,49 the utility of vinyl group as a versatile chemoselective handle in developing RNA-labeling protocols has not been well-explored.50 In this context, here we describe the synthesis of 5-vinyl-modified UTP (VUTP) analog and its effective incorporation into RNA ONs by in vitro transcription reactions. Furthermore, we illustrate the post-transcriptional chemical functionalization of vinyl-labeled RNA transcripts with a variety of biophysical reporters by using oxidative Heck and IEDDA reactions. Notably, this is the first report on the bioconjugation of an RNA ON by employing Pd-mediated oxidative Heck reactions.
Figure 1.
Diagram illustrating the post-transcriptional chemical labeling of RNA by using oxidative Heck and IEDDA reactions. Incorporation of VUTP into RNA transcripts, followed by oxidative Heck reaction with boronic acid derivatives and IEDDA reaction with tetrazole derivatives, enabled the synthesis of RNA conjugated to various reporters and tags.
Results and Discussion
Synthesis and Enzymatic Incorporation of 5-Vinyluridine Triphosphate 3
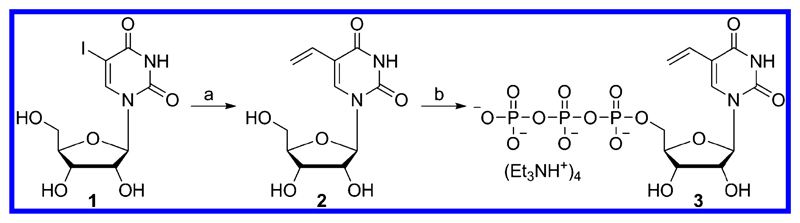
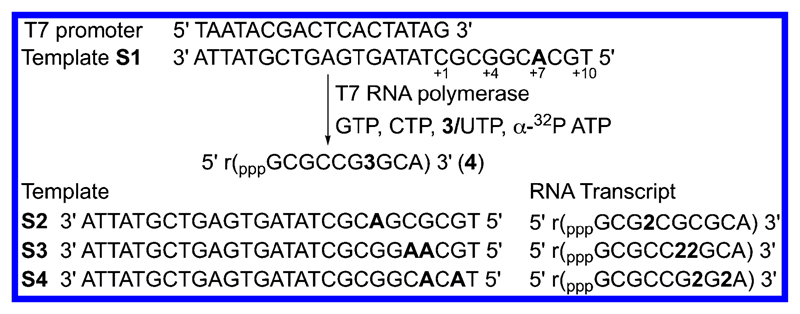
VUTP 3 was synthesized in simple steps starting from 5-iodouridine 1 (Scheme 1). 5-Vinyluridine (VU) 2 was synthesized by Stille cross-coupling reaction using vinyltributylstannane and a palladium catalyst, tris-(dibenzylideneacetone)dipalladium(0).51 VU was further phosphorylated in the presence of POCl3 and bis-tributylammonium pyrophosphate to afford VUTP. The ability of VUTP to serve as a UTP analog for RNA polymerase was studied by performing in vitro transcription reactions in the presence of a small series of DNA templates S1−S4 (Figure 2). The templates contained one or two dA residues in the coding region to guide the insertion of monophosphate of VUTP into RNA during the polymerization reaction. The 5′-end of each template ended with a dT residue to introduce a radiolabeled α-32P adenosine at the 3′-end of the full-length transcript. Transcription was performed using GTP, CTP, UTP/3, and α-32P ATP in the presence of T7 RNA polymerase. The transcription products were then electrophoresed on an analytical polyacrylamide gel and phosphor-imaged.
Scheme 1. Synthesis of VUTP 3a.
a(a) Pd2(dba)3, P(Fur)3, vinyltributylstannane, DMF, 60 °C, 3 h, 53%. (b) (i) POCl3, proton sponge, trimethylphosphate, ~4 °C, 30 min and (ii) bis-tributylammonium pyrophosphate, tributylamine, ~4 °C, 30 min, 21%.
Figure 2.
Incorporation of the vinyl group into RNA ONs by in vitro transcription reaction using DNA templates S1−S4 and VUTP 3. “r” preceding the sequence in parentheses represents RNA ON transcripts.
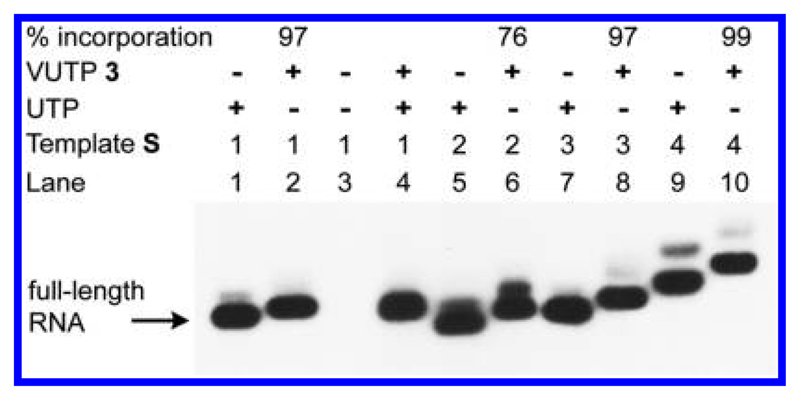
The incorporation of VUTP 3 in the presence of template S1 proceeded with very good efficiency at the +7 position, yielding a 10 mer full-length RNA transcript 4 (Figure 3, lane 2). Slower migration of the transcription product 4 in lane 2 hinted at the incorporation of 3 into the transcript (compare lanes 1 and 2). While a control reaction in the absence of UTP and 3 did not yield full-length RNA transcript, at equimolar concentrations, both natural UTP and 3 got incorporated into RNA (Figure 3 lanes 3 and 4, respectively). Templates S2–S4 incorporated modified UTP near the promoter region and at more than one site with good to excellent efficiency (Figure 3 lanes 6, 8, and 10).
Figure 3.
Phosphor image of transcripts obtained by transcription of templates S1−S4 in the presence of UTP and 3. “The percent of incorporation of 3 is reported relative to a control transcription with UTP. For a complete gel picture, see Figure S1.
Transcript 4 was scaled up by performing large-scale reactions with S1 in the presence of cold NTPs. Typically, a reaction in the presence of 75 pmol of the template gave nearly 15 nmol of the modified transcript after gel electrophoretic purification, which was further used for post-transcriptional chemical labeling experiments. The presence of VU in transcript 4 was confirmed by mass analysis and enzymatic digestion assay (Figures S2 and S3 and Table S1).
Oxidative Heck Reaction on Vinyl-Labeled RNA Transcript
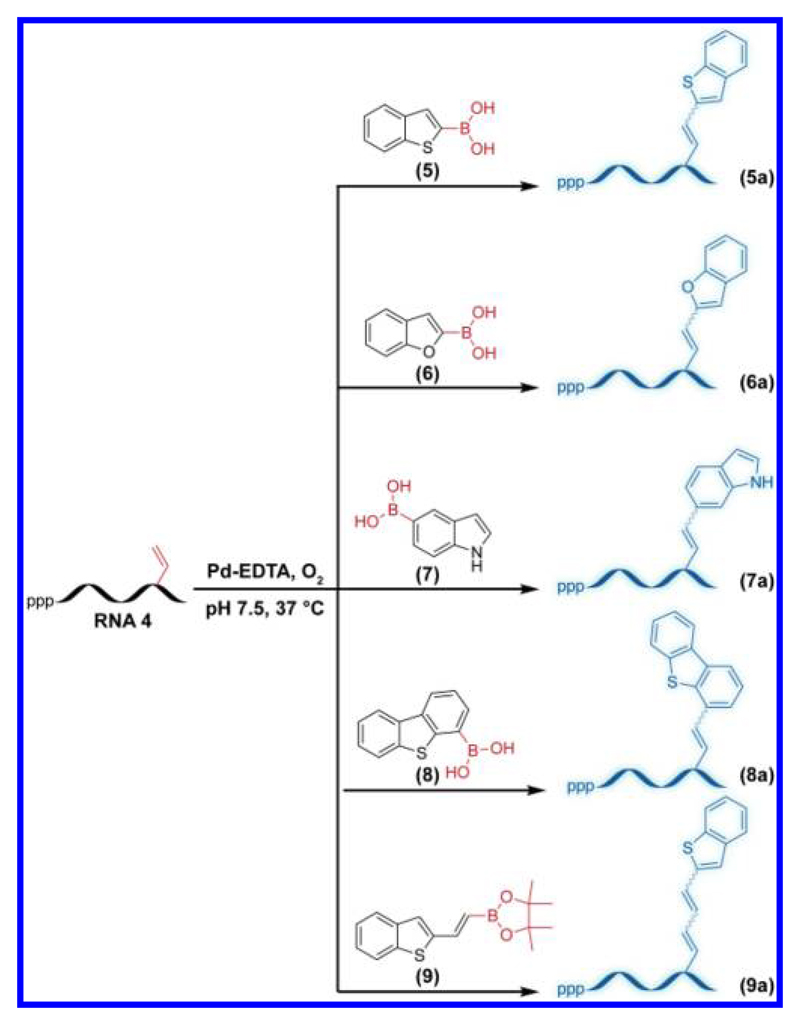
Electronically unbiased terminal alkene (e.g., allylic) substrates have been reported to undergo oxidative Heck reaction with boronic acids in the presence of water-soluble Pd-EDTA complex.41 However, the feasibility of such a reaction on RNA has not been explored. In this regard, we sought to investigate the possibility of performing oxidative Heck reaction on vinyl-modified RNA with suitable boronic acid and ester substrates. For this purpose we chose a range of heterobicycles, namely benzothiophene-2- (5), benzofuran-2- (6), indole-5- (7), dibenzothiophene-4-boronic acid, (8) and a previously reported pinacol boronic ester, benzothiophene-2-vinyl boronic ester (9),47 which, upon conjugation to a uracil ring, could impart fluorescence to otherwise nonemissive nucleobase (Figure 4). Fluorescent nucleosides have been developed by attaching heterocycles to pyrimidine and purine bases, respectively, and such analogs incorporated into ONs have been used as probes in various nucleic acid studies.2,52–57
Figure 4.
Oxidative Heck reaction between vinyl-modified RNA ON 4 and boronic acid substrates 5−8 or boronic ester 9 in the presence of Pd−EDTA complex.
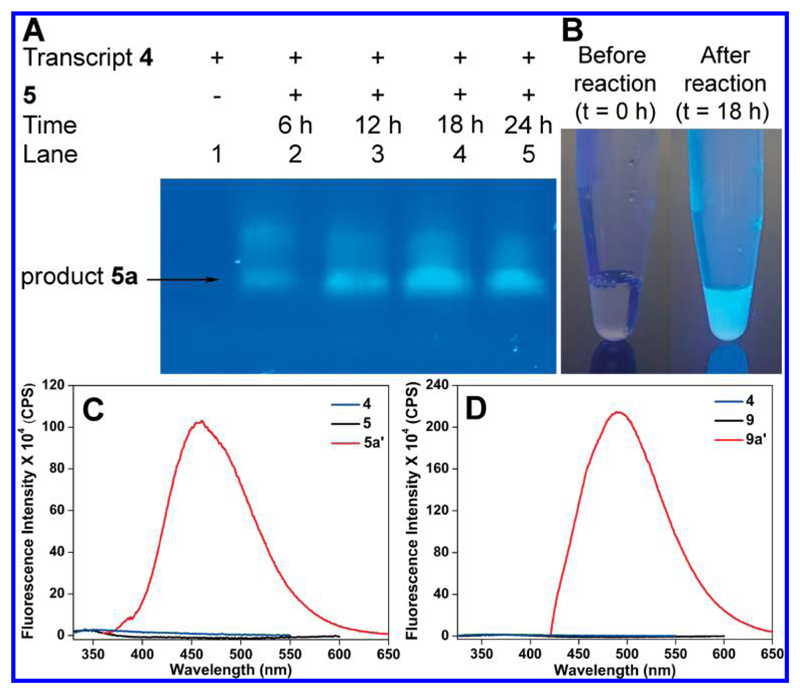
For performing oxidative Heck reaction on RNA, transcript 4 was incubated with boronic acid 5 in the presence of Pd–EDTA complex at 37 °C. Aliquots of reaction mixture at various time intervals were resolved by PAGE under denaturing conditions. Almost complete conversion was observed at 18 h, and rewardingly, we observed an intense fluorescent band corresponding to the coupled product along with a minor band (Figures 5 and S4). Reactions with other substrates (6–8) also proceeded well (Figure S5). Further, products from large-scale reactions between transcript 4 and boronic acid 5–8 or boronic ester 9 were analyzed by reverse-phase high-performance liquid chromatography (RP-HPLC) at 260 and 338 nm (conjugation of heterocycles to pyrimidine bases results in a strong absorption band near 338 nm).2,58 Mass analysis of HPLC fractions corresponding to both 260 and 338 nm absorption bands revealed the formation of the oxidative Heck products 5a–9a (Figures S6 and S7 and Table S2). Reactions performed at a 5 nmole scale of the RNA ON 4 provided 0.5 to 1.1 nmole of the coupled products (Table S3).
Figure 5.
(A) UV-transilluminator image (364 nm) of the gel of a reaction between RNA 4 and 5. See Figure S4 for the UV-shadow image of the gel at 254 nm. (B) Fluorescence of RNA transcript before and after the reaction between RNA 4 and boronic acid 5 under UV lamp (364 nm). (C) Fluorescence spectra (0.2 μM) of RNA transcript 4, boronic acid 5, and RNA product 5a (HPLC fraction corresponding to 5a′). Samples were excited at 289, 310, and 340 nm, respectively, with an excitation and emission slit width of 8 and 9 nm. (D) Fluorescence spectra (0.2 μM) of RNA transcript 4, boronic ester 9, and RNA product 9a (HPLC fraction corresponding to 9a′). Samples were excited at 289, 310, and 372 nm, respectively, with an excitation and emission slit width of 7 and 8 nm. See Figure S6 for HPLC profiles.
An oxidative Heck reaction between an electronically unbiased terminal alkene (e.g., allylic system) and arylboronic acid substrates typically gives linear and branched cross-coupled products.40 However, reactions with electronically biased aryl vinyl systems (e.g., styrene) give predominantly linear transcoupled product.59,60 Formation of branched regioisomer product has also been reported using Pd in the presence of certain bulky ligands (e. g., 2,9-dimethyl-1,10-phenanthroline). 60 Although it is not a major concern in most bioconjugation strategies, we sought to determine the formation of different isomers in the oxidative Heck reaction of vinyl-labeled RNA transcript. First, we carried out a reaction between free vinyl-modified uridine 2 and boronic acid 5 under similar conditions used for RNA ligation. Interestingly, at the ribonucleoside level, a very low conversion was observed even after prolonged incubation time. Varying the Pd–ligand complex and oxidant did not give a better yield (data not shown). However, a fluorescent product was isolated in analytical quantities, and further characterization by NMR revealed the formation of a linear trans-isomer product 2a (Figure S8). Further, the coupled product gave a unique longer wavelength absorption band at ~338 nm as compared to native nucleoside and nucleic acid, which absorbs at 260 nm (Figure S8B). Next, the major HPLC fraction corresponding to the RNA products (designated as 5a′) of a reaction between vinyl-labeled RNA ON 4 and boronic acid 5 was subjected to enzymatic digestion in the presence of calf intestinal alkaline phosphatase, RNase A, RNase T1 and snake venom phosphodiesterase. Enzymatic digestion of 5a′ gave individual ribonucleosides as well as Heck-coupled ribonucleosides, whose identity was determined by comparing the HPLC chromatogram of the digested sample and oxidative Heck reaction product 2a obtained from a reaction between uridine 2 and boronic acid 5 (Figure S9). The chromatogram of the digest revealed the presence of native ribonucleosides (rC, rG, and rA) and two peaks corresponding to coupled products absorbing at 338 nm. One of the peaks clearly matched with the trans-isomer product 2a, whereas the chemical structure of the peak X could not be determined. However, both 2a and X gave the same mass, indicating that a mixture of isomers (X may be the cis isomer) are formed in these reactions. Formation of a mixture of isomers, which is an inherent “limitation” of this reaction, has also been observed in protein ligation experiments.40,41 Although, at the present, this reaction may not be viable for introducing a specific type of conformation-sensitive probe into RNA, development in chelate-control strategies could vastly improve the selective of the reaction.60
We next studied the fluorogenic nature of the RNA products by steady-state fluorescence spectroscopy. Benzothiophene-coupled RNA product 5a (HPLC fraction corresponding to 5a′) showed a ~40-fold enhancement in fluorescence intensity as compared to the boronic acid substrate 5 (Figures 4, 5C, and S6). Interestingly, benzothiophene-alkene-coupled RNA product 9a (HPLC fraction corresponding to 9a′) showed a remarkable ~170-fold enhancement in fluorescence intensity as compared to substrate 9. Furthermore, the emission profile of 9a showed a significant bathochromic shift compared to RNA 5a as a direct result of increased conjugation between the uracil and benzothiophene rings (Figures 4, 5D, and S6). Reactions with benzofuran (6), indole (7), and dibenzothiophene (8) boronic acid substrates also produced fluorescent RNA products (Figure S10). While benzofuran- and dibenzothiophene-conjugated RNA products were reasonably fluorescent, indole-conjugated RNA was found to be weakly fluorescent. Taken together, this approach of functionalizing vinyl-labeled RNA transcripts by fluorogenic Heck-type coupling reaction with appropriate boronic acid/boronic ester substrates could provide direct access to RNA functionalized with fluorescent reporters.48,61,62
IEDDA Reaction on Vinyl-Labeled RNA Transcripts
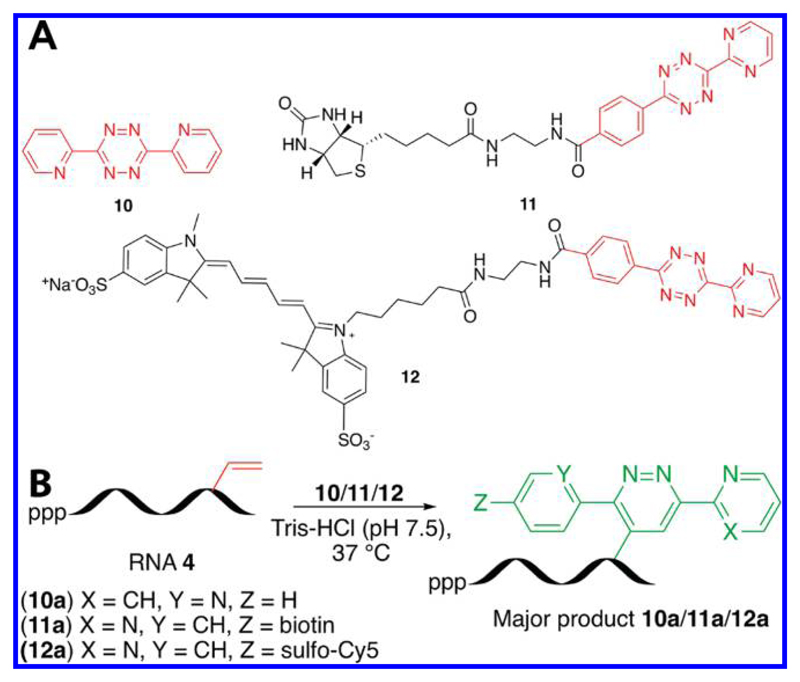
Analogous to AAC chemistry, IEDDA reaction between an electron-rich dienophile and electron-deficient tetrazine is gaining particular attention as a useful biomacromolecular labeling strategy because this reaction is reagent-free, reasonably fast, highly selective, and biocompatible.63–65 Bioconjugation strategies based on IEDDA reaction commonly use ONs labeled with reactive dienophiles such as norbornene, trans-cyclooctene, and cyclopropene groups.66–73 While the reaction rates of strained dienophiles with tetrazine derivatives are generally fast, such bulky dienophile substituents on nucleoside are particularly not suitable for labeling cellular nucleic acids as they may not serve as good substrates for endogenous polymerases. Despite low reactivity as compared to that of strained dienophiles, vinylated 2′-deoxynucleosides have been used as chemical reporters to label and visualize DNA in cells by IEDDA reaction.45 Encouraged by these results, we decided to study the reactivity of vinyl label of transcript 4 toward diene counterpart in IEDDA reaction. For this purpose, we chose two tetrazine cores, which had reportedly shown a good reactivity against an electron-rich strained alkene.66–73 The reactivity of transcript 4 was tested by using a commercially available tetrazine 10 and biotinylated (11) and Cy5-conjugated (12) tetrazines, which were synthesized by following an analogous literature procedure (Figure 6 and Scheme S1).64
Figure 6.
(A) Tetrazine substrates 10−12 used in this study. (B) IEDDA reaction between RNA ON 4 and tetrazine substrates. 10a−12a are RNA products obtained from a reaction between RNA ON 4 and tetrazines 10−12, respectively.
Initially, the IEDDA reaction was performed using the free nucleoside 2 and a tetrazine substrate 10. The reaction proceeded well, and almost complete conversion was observed after 21 h (Scheme S2 and Figure S11). The reaction resulted in a mixture of ligated products (2b and 2c), which were isolated and characterized. 2a was found to be oxidized pyridazine product, whereas 2c corresponded to the dihydropyridazine product, which underwent slow oxidation to form 2b. Formation of a mixture of products and slow oxidation event has been documented earlier in DNA conjugation experiments.45,74
Next, IEDDA reactions were performed between RNA ON 4 and tetrazine substrates in Tris-HCl buffer (pH 7.5) at 37 °C, and aliquots of reaction mixture at various time intervals were analyzed by analytical PAGE under denaturing conditions. A reaction with tetrazine 10 was found to be almost complete in 12 h, whereas with substrates 11 and 12, the reaction was partially complete even after 15 h (Figure S12). Reactions at elevated temperatures (45 and 55 °C) did not result in noticeable improvement in reaction efficiency (data not shown). This is in agreement with reports of less reactivity of the tetrazine core of 11 and 12 as compared to a tetrazine core of 10.65
Large-scale reactions were then performed to isolate and characterize the RNA products. Reaction with substrates 10−12 produced major and minor (could not be isolated) bands (Figure S13). Mass analysis of major bands confirmed the formation of pyridazine products 10a−12a (Figure S14 and Table S2), similar to previous reports.72 UV–vis profile of purified products was distinguishingly different from the profile of transcript 4, and further fluorescence analysis also confirmed the bioconjugation by IEDDA reaction (Figure S15). While HPLC analysis of the major band obtained from a reaction with symmetrical tetrazine 10 gave a single peak corresponding to the pyridazine 10a, reactions with asymmetrical tetrazines 11 and 12 afforded mixture of pyridazine isomers (Figure S16). The formation of mixture of isomeric products is consistent with literature reports.45,75
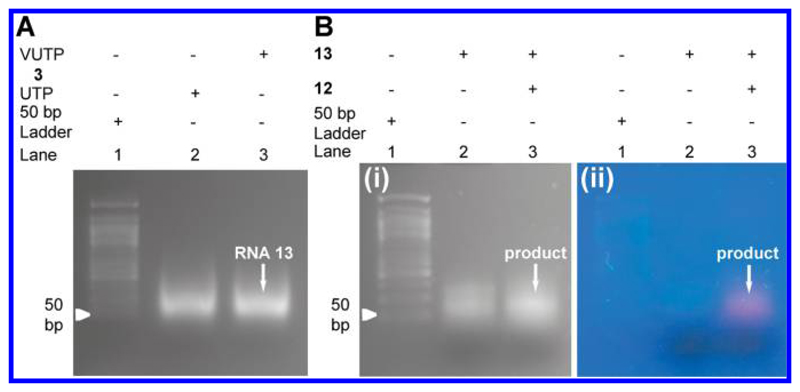
High-density labeling of VUTP into a longer RNA transcript was studied by using a DNA template that would generate a 59 mer RNA containing nine modifications. T7 RNA polymerase effectively incorporated the vinyl analog into the 59 mer RNA 13 with efficiency comparable to that of natural UTP (Figure 7A). Transcript 13 was subsequently reacted with sulfo-Cy5 labeled tetrazine 12, and the reaction product was resolved on a 2% agarose gel. The image captured using UV-transilluminator (364 nm) clearly revealed the fluorescence labeling of the longer RNA transcript (Figure 7B).
Figure 7.
(A) Agarose gel picture of 59 mer control (lane 2) and VU-modified RNA transcript 13 (lane 3). Lane 1: 50 bp DNA ladder. (B) Agarose gel picture of IEDDA reaction between RNA 13 and Cy5-tetrazine 12. UV-transilluminator image at (i) 254 nm and (ii) 364 nm. Lane 1: 50 bp DNA ladder.
Conclusions
We have successfully incorporated the vinyl functionality into RNA by in vitro transcription reaction using VUTP 3. The results demonstrate that VU incorporated into RNA transcripts can serve as a useful handle for chemoselective functionalization of RNA in a modular fashion by oxidative Heck and IEDDA reactions. In particular, the generation of fluorogenic RNA by oxidative Heck reaction with boronic acid and ester is advantageous. It suggests that a screening reaction with appropriate boronic acid and ester substrates could provide direct access to RNA emitting at different wavelengths. Even though conjugation by utilizing oxidative Heck reaction resulted in a mixture of isomers, this study represents a promising initial step on further research on oxidative Heck reaction for the design of made-to-order biologically compatible regioselective and stereoselective ligands for palladium catalysis.60,76 Such studies would pave the way for selective incorporation of desired biophysical probes (for example, microenvironment-sensing probes onto RNA). Furthermore, metabolic labeling of VU followed by post-transcriptional functionalization by these methods could potentially enable the imaging of RNA in cells. Taken together, the studies presented here are expected to complement other bioorthogonal RNA labeling strategies by providing alternative access to RNA labeled with biophysical probes and tags.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.7b00169.
Experimental procedure, characterization data, and additional experimental figures and tables. (PDF)
Acknowledgments
S.G.S. thanks DST, India (EMR/2014/000419) and Wellcome Trust-DBT India Alliance (IA/S/16/1/502360) for the research grants. J.T.G. is grateful to IISER, Pune for graduate research fellowship. We thank Manisha Walunj for providing a boronic ester substrate.
Footnotes
Orcid
Seergazhi G. Srivatsan: 0000-0001-5765-3967
Notes
The authors declare no competing financial interest.
References
- (1).Wachowius F, Höbartner C. Chemical RNA modifications for studies of RNA structure and dynamics. ChemBioChem. 2010;11:469–480. doi: 10.1002/cbic.200900697. [DOI] [PubMed] [Google Scholar]
- (2).Sinkeldam RW, Greco NJ, Tor Y. Fluorescent analogs of biomolecular building blocks: design, properties, and applications. Chem Rev. 2010;110:2579–2619. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Holstein JM, Rentmeister A. Current covalent modification methods for detecting RNA in fixed and living cells. Methods. 2016;98:18–25. doi: 10.1016/j.ymeth.2015.11.016. [DOI] [PubMed] [Google Scholar]
- (4).Baker M. RNA imaging in situ. Nat Methods. 2012;9:787–790. [Google Scholar]
- (5).Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu X-D, van Driel R, Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem. 2013;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wlodkowic D, Skommer J, Darzynkiewicz Z. SYTO probes in the cytometry of tumor cell death. Cytometry, Part A. 2008;73A:496–507. doi: 10.1002/cyto.a.20535. [DOI] [PubMed] [Google Scholar]
- (8).Xu Y, Suzuki Y, Ito K, Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proc Natl Acad Sci U S A. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Laguerre A, Hukezalie K, Winckler P, Katranji F, Chanteloup G, Pirrotta M, Perrier-Cornet J-M, Wong JMY, Monchaud D. Visualization of RNA-quadruplexes in live cells. J Am Chem Soc. 2015;137:8521–8525. doi: 10.1021/jacs.5b03413. [DOI] [PubMed] [Google Scholar]
- (11).Gramlich PME, Wirges CT, Manetto A, Carell T. Postsynthetic DNA modification through the copper-catalyzed azide–alkyne cycloaddition reaction. Angew Chem, Int Ed. 2008;47:8350–8358. doi: 10.1002/anie.200802077. [DOI] [PubMed] [Google Scholar]
- (12).Weisbrod SH, Marx A. Novel strategies for the site-specific covalent labelling of nucleic acids. Chem Commun. 2008:5675–5685. doi: 10.1039/b809528k. [DOI] [PubMed] [Google Scholar]
- (13).Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).El-Sagheer AH, Brown T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc Natl Acad Sci U S A. 2010;107:15329–15334. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).George JT, Srivatsan SG. Posttranscriptional chemical labeling of RNA by using bioorthogonal chemistry. Methods. 2017 doi: 10.1016/j.ymeth.2017.02.004. [DOI] [PubMed] [Google Scholar]
- (16).El-Sagheer AH, Brown T. Click nucleic acid ligation: Applications in biology and nanotechnology. Acc Chem Res. 2012;45:1258–1267. doi: 10.1021/ar200321n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).van Berkel SS, van Eldijk MB, van Hest JCM. Staudinger ligation as a method for bioconjugation. Angew Chem, Int Ed. 2011;50:8806–8827. doi: 10.1002/anie.201008102. [DOI] [PubMed] [Google Scholar]
- (18).Wu H, Devaraj NK. Inverse electron-demand Diels–Alder bioorthogonal reactions. Top Curr Chem. 2016;374:3. doi: 10.1007/s41061-015-0005-z. [DOI] [PubMed] [Google Scholar]
- (19).Jayaprakash KN, Peng CG, Butler D, Varghese JP, Maier MA, Rajeev KG, Manoharan M. Non-nucleoside building blocks for copper-assisted and copper-free click chemistry for the efficient synthesis of RNA conjugates. Org Lett. 2010;12:5410–5413. doi: 10.1021/ol102205j. [DOI] [PubMed] [Google Scholar]
- (20).Ishizuka T, Kimoto M, Sato A, Hirao I. Site-specific functionalization of RNA molecules by an unnatural base pair transcription system via click chemistry. Chem Commun. 2012;48:10835–10837. doi: 10.1039/c2cc36293g. [DOI] [PubMed] [Google Scholar]
- (21).Rao H, Tanpure AA, Sawant AA, Srivatsan SG. Enzymatic incorporation of an azide-modified UTP analog into oligoribonucleotides for post-transcriptional chemical functionalization. Nat Protoc. 2012;7:1097–1112. doi: 10.1038/nprot.2012.046. [DOI] [PubMed] [Google Scholar]
- (22).Curanovic D, Cohen M, Singh I, Slagle CE, Leslie CS, Jaffrey SR. Global profiling of stimulus-induced polyadenylation in cells using a poly(A) trap. Nat Chem Biol. 2013;9:671–673. doi: 10.1038/nchembio.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Holstein JM, Schulz D, Rentmeister A. Bioorthogonal site-specific labeling of the 5′-cap structure in eukaryotic mRNAs. Chem Commun. 2014;50:4478–4481. doi: 10.1039/c4cc01549e. [DOI] [PubMed] [Google Scholar]
- (24).Phelps KJ, Ibarra-Soza JM, Tran K, Fisher AJ, Beal PA. Click modification of RNA at adenosine: structure and reactivity of 7-ethynyl- and 7-triazolyl-8-aza-7-deazaadenosine in RNA. ACS Chem Biol. 2014;9:1780–1787. doi: 10.1021/cb500270x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Schmid K, Adobes-Vidal M, Helm M. Alkyne-functionalized coumarin compound for analytic and preparative 4-thiouridine labeling. Bioconjugate Chem. 2017 doi: 10.1021/acs.bioconj-chem.7b00035. [DOI] [PubMed] [Google Scholar]
- (26).Cahova H, Winz M-L, Hofer K, Nubel G, Jaschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2014;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- (27).Sawant AA, Mukherjee PP, Jangid RK, Galande S, Srivatsan SG. A clickable UTP analog for the posttranscriptional chemical labeling and imaging of RNA. Org Biomol Chem. 2016;14:5832–5842. doi: 10.1039/c6ob00576d. [DOI] [PubMed] [Google Scholar]
- (28).Paredes E, Das SR. Click chemistry for rapid labeling and ligation of RNA. ChemBioChem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- (29).Kislukhin AA, Hong VP, Breitenkamp KE, Finn MG. Relative performance of alkynes in copper-catalyzed azide–alkyne cycloaddition. Bioconjugate Chem. 2013;24:684–689. doi: 10.1021/bc300672b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).van Delft P, Meeuwenoord NJ, Hoogendoorn S, Dinkelaar J, Overkleeft HS, van der Marel GA, Filippov DV. Synthesis of oligoribonucleic acid conjugates using a cyclooctyne phosphoramidite. Org Lett. 2010;12:5486–5489. doi: 10.1021/ol102357u. [DOI] [PubMed] [Google Scholar]
- (31).Someya T, Ando A, Kimoto M, Hirao I. Site-specific labeling of RNA by combining genetic alphabet expansion transcription and copper-free click chemistry. Nucleic Acids Res. 2015;43:6665–6676. doi: 10.1093/nar/gkv638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Santner T, Hartl M, Bister K, Micura R. Efficient access to 3′-terminal azide-modified RNA for inverse click-labeling patterns. Bioconjugate Chem. 2014;25:188–195. doi: 10.1021/bc400513z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Neef AB, Luedtke NW. An azide-modified nucleoside for metabolic labeling of DNA. ChemBioChem. 2014;15:789–793. doi: 10.1002/cbic.201400037. [DOI] [PubMed] [Google Scholar]
- (34).Sawant AA, Tanpure AA, Mukherjee PP, Athavale S, Kelkar A, Galande S, Srivatsan SG. A versatile toolbox for posttranscriptional chemical labeling and imaging of RNA. Nucleic Acids Res. 2016;44:e16. doi: 10.1093/nar/gkv903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ourailidou ME, Zwinderman MRH, Dekker FJ. Bioorthogonal metabolic labelling with acyl-CoA reporters: targeting protein acylation. MedChemComm. 2016;7:399–408. [Google Scholar]
- (36).van Geel R, Pruijn GJM, van Delft FL, Boelens WC. Preventing thiol-yne addition improves the specificity of strain-promoted azide–alkyne cycloaddition. Bioconjugate Chem. 2012;23:392–398. doi: 10.1021/bc200365k. [DOI] [PubMed] [Google Scholar]
- (37).Yu Z, Pan Y, Wang Z, Wang J, Lin Q. Genetically encoded cyclopropene directs rapid, photoclick chemistry-mediated protein labeling in mammalian cells. Angew Chem, Int Ed. 2012;51:10600–10604. doi: 10.1002/anie.201205352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Li Q-F, Yang Y, Maleckis A, Otting G, Su X-C. Thiol-ene reaction: a versatile tool in site-specific labelling of proteins with chemically inert tags for paramagnetic NMR. Chem Commun. 2012;48:2704–2706. doi: 10.1039/c2cc17900h. [DOI] [PubMed] [Google Scholar]
- (39).Li Y, Yang M, Huang Y, Song X, Liu L, Chen PR. Genetically encoded alkenyl-pyrrolysine analogues for thiolene reaction mediated site-specific protein labeling. Chem Sci. 2012;3:2766–2770. [Google Scholar]
- (40).Ourailidou ME, van der Meer J-Y, Baas B-J, Jeronimus-Stratingh M, Gottumukkala AL, Poelarends GJ, Minnaard AJ, Dekker FJ. Aqueous oxidative Heck reaction as a protein-labeling strategy. ChemBioChem. 2014;15:209–212. doi: 10.1002/cbic.201300714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ourailidou ME, Dockerty P, Witte M, Poelarends GJ, Dekker FJ. Metabolic alkene labeling and in vitro detection of histone acylation via the aqueous oxidative Heck reaction. Org Biomol Chem. 2015;13:3648–3653. doi: 10.1039/c4ob02502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhang Y, Pan Y, Liu W, Zhou YJ, Wang K, Wang L, Sohail M, Ye M, Zou H, Zhao ZK. In vivo protein allylation to capture protein methylation candidates. Chem Commun. 2016;52:6689–6692. doi: 10.1039/c6cc02386j. [DOI] [PubMed] [Google Scholar]
- (43).Omumi A, Beach DG, Baker M, Gabryelski W, Manderville RA. Post-synthetic guanine arylation of DNA by Suzuki-Miyaura cross-coupling. J Am Chem Soc. 2011;133:42–50. doi: 10.1021/ja106158b. [DOI] [PubMed] [Google Scholar]
- (44).Lercher L, McGouran JF, Kessler BM, Schofield CJ, Davis BG. DNA modification under mild conditions by Suzuki–Miyaura cross-coupling for the generation of functional probes. Angew Chem, Int Ed. 2013;52:10553–10558. doi: 10.1002/anie.201304038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Rieder U, Luedtke NW. Alkene–tetrazine ligation for imaging cellular DNA. Angew Chem, Int Ed. 2014;53:9168–9172. doi: 10.1002/anie.201403580. [DOI] [PubMed] [Google Scholar]
- (46).Batroff E, Niederwieser A, Abdel-Rahman OS, Winter RF, Wittmann V, Marx A, Bußkamp H. Efficient labelling of enzymatically synthesized vinyl-modified DNA by an inverse-electron-demand Diels-Alder reaction. Chem Commun. 2014;50:10827–10829. doi: 10.1039/c4cc04332d. [DOI] [PubMed] [Google Scholar]
- (47).Blanchard DJM, Fadock KL, Sproviero M, Deore PS, Cservenyi TZ, Manderville RA, Sharma P, Wetmore SD. Photophysical properties of push-pull 8-aryl-deoxyguanosine probes within duplex and G-quadruplex structures. J Mater Chem C. 2016;4:2915–2924. [Google Scholar]
- (48).Samanta B, Seikowski J, Höbartner C. Fluorogenic Labeling of 5-Formylpyrimidine Nucleotides in DNA and RNA. Angew Chem, Int Ed. 2016;55:1912–1916. doi: 10.1002/anie.201508893. [DOI] [PubMed] [Google Scholar]
- (49).Dadová J, Orság P, Pohl R, Brázdová M, Fojta M, Hocek M. Vinylsulfonamide and acrylamide modification of DNA for cross-linking with proteins. Angew Chem, Int Ed. 2013;52:10515–10518. doi: 10.1002/anie.201303577. [DOI] [PubMed] [Google Scholar]
- (50).Holstein JM, Stummer D, Rentmeister A. Enzymatic modification of 5′-capped RNA with a 4-vinylbenzyl group provides a platform for photoclick and inverse electron-demand Diels-Alder reaction. Chem Sci. 2015;6:1362–1369. doi: 10.1039/c4sc03182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wicke L, Engels JW. Postsynthetic on column RNA labeling via Stille coupling. Bioconjugate Chem. 2012;23:627–642. doi: 10.1021/bc200659j. [DOI] [PubMed] [Google Scholar]
- (52).Srivatsan SG, Tor Y. Fluorescent pyrimidine ribonucleotide: synthesis, enzymatic incorporation, and utilization. J Am Chem Soc. 2007;129:2044–2053. doi: 10.1021/ja066455r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Pawar MG, Nuthanakanti A, Srivatsan SG. Heavy atom containing fluorescent ribonucleoside analog probe for the fluorescence detection of RNA-ligand binding. Bioconjugate Chem. 2013;24:1367–1377. doi: 10.1021/bc400194g. [DOI] [PubMed] [Google Scholar]
- (54).Tanpure AA, Srivatsan SG. Conformation-sensitive nucleoside analogues as topology-specific fluorescence turn-on probes for DNA and RNA G-quadruplexes. Nucleic Acids Res. 2015;43:e149. doi: 10.1093/nar/gkv743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sproviero M, Fadock KL, Witham AA, Manderville RA. Positional impact of fluorescently modified G-tetrads within polymorphic human telomeric G-quadruplex structures. ACS Chem Biol. 2015;10:1311–1318. doi: 10.1021/cb5009926. [DOI] [PubMed] [Google Scholar]
- (56).Suzuki A, Yanaba T, Saito I, Saito Y. Molecular design of an environmentally sensitive fluorescent nucleoside, 3-deaza-2'-deoxyadenosine derivative: distinguishing thymine by probing the DNA minor groove. ChemBioChem. 2014;15:1638–1644. doi: 10.1002/cbic.201402078. [DOI] [PubMed] [Google Scholar]
- (57).Kanamori T, Ohzeki H, Masaki Y, Ohkubo A, Takahashi M, Tsuda K, Ito T, Shirouzu M, Kuwasako K, Muto Y, Sekine M, et al. Controlling the fluorescence of benzofuran-modified uracil residues in oligonucleotides by triple-helix formation. ChemBioChem. 2015;16:167–176. doi: 10.1002/cbic.201402346. [DOI] [PubMed] [Google Scholar]
- (58).Tanpure AA, Pawar MG, Srivatsan SG. Fluorescent nucleoside analogs: probes for investigating nucleic acid structure and function. Isr J Chem. 2013;53:366–378. [Google Scholar]
- (59).Shaikh TM, Hong F-E. Palladium(II)-catalyzed Heck reaction of aryl halides and arylboronic acids with olefins under mild conditions. Beilstein J Org Chem. 2013;9:1578–1588. doi: 10.3762/bjoc.9.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Zheng C, Stahl SS. Regioselective aerobic oxidative Heck reactions with electronically unbiased alkenes: efficient access to α-alkyl vinylarenes. Chem Commun. 2015;51:12771–12774. doi: 10.1039/c5cc05312a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Furukawa K, Abe H, Hibino K, Sako Y, Tsuneda S, Ito Y. Reduction-triggered fluorescent amplification probe for the detection of endogenous RNAs in living human cells. Bioconjugate Chem. 2009;20:1026–1036. doi: 10.1021/bc900040t. [DOI] [PubMed] [Google Scholar]
- (62).He Z, Chen Y, Wang Y, Wang J, Mo J, Fu B, Wang Z, Du Y, Zhou X. A rapidly photo-activatable light-up fluorescent nucleoside and its application in DNA base variation sensing. Chem Commun. 2016;52:8545–8548. doi: 10.1039/c6cc03098j. [DOI] [PubMed] [Google Scholar]
- (63).Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Bioorthogonal turn-on probes for imaging small molecules inside living cells. Angew Chem, Int Ed. 2010;49:2869–2872. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Knall A-C, Slugovc C. Inverse electron demand Diels-Alder (iEDDA)-initiated conjugation: a (high) potential click chemistry scheme. Chem Soc Rev. 2013;42:5131–5142. doi: 10.1039/c3cs60049a. [DOI] [PubMed] [Google Scholar]
- (66).Schoch J, Wiessler M, Jäschke A. Post-synthetic modification of DNA by inverse-electron-demand Diels–Alder reaction. J Am Chem Soc. 2010;132:8846–8847. doi: 10.1021/ja102871p. [DOI] [PubMed] [Google Scholar]
- (67).Šečkutė J, Yang J, Devaraj NK. Rapid oligonucleotide-templated fluorogenic tetrazine ligations. Nucleic Acids Res. 2013;41:e148–e148. doi: 10.1093/nar/gkt540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Asare-Okai PN, Agustin E, Fabris D, Royzen M. Site-specific fluorescence labelling of RNA using bioorthogonal reaction of trans-cyclooctene and tetrazine. Chem Commun. 2014;50:7844–7847. doi: 10.1039/c4cc02435d. [DOI] [PubMed] [Google Scholar]
- (69).Ameta S, Becker J, Jäschke A. RNA-peptide conjugate synthesis by inverse-electron demand Diels-Alder reaction. Org Biomol Chem. 2014;12:4701–4707. doi: 10.1039/c4ob00076e. [DOI] [PubMed] [Google Scholar]
- (70).Pyka AM, Domnick C, Braun F, Kath-Schorr S. Diels–Alder cycloadditions on synthetic RNA in mammalian cells. Bioconjugate Chem. 2014;25:1438–1443. doi: 10.1021/bc500302y. [DOI] [PubMed] [Google Scholar]
- (71).Domnick C, Eggert F, Kath-Schorr S. Site-specific enzymatic introduction of a norbornene modified unnatural base into RNA and application in post-transcriptional labeling. Chem Commun. 2015;51:8253–8256. doi: 10.1039/c5cc01765c. [DOI] [PubMed] [Google Scholar]
- (72).Eggert F, Kath-Schorr S. A cyclopropene-modified nucleotide for site-specific RNA labeling using genetic alphabet expansion transcription. Chem Commun. 2016;52:7284–7287. doi: 10.1039/c6cc02321e. [DOI] [PubMed] [Google Scholar]
- (73).Lorenz DA, Garner AL. A click chemistry-based microRNA maturation assay optimized for high-throughput screening. Chem Commun. 2016;52:8267–8270. doi: 10.1039/c6cc02894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Kore AR, Yang B, Srinivasan B. Synthesis of 5-[3,6-di(pyridin-2-yl)pyridazine-4-yl]-2′-deoxyuridine-5′-O-triphosphate a potential probe for fluorescence detection and imaging DNA. Tetrahedron Lett. 2015;56:808–811. [Google Scholar]
- (75).Niederwieser A, Späte A-K, Nguyen LD, Jüngst C, Reutter W, Wittmann V. Two-color glycan labeling of live cells by a combination of Diels–Alder and click chemistry. Angew Chem, Int Ed. 2013;52:4265–4268. doi: 10.1002/anie.201208991. [DOI] [PubMed] [Google Scholar]
- (76).Zhang L, Dong C, Ding C, Chen J, Tang W, Li H, Xu L, Xiao J. Palladium-catalyzed regioselective and stereoselective oxidative Heck arylation of allylamines with arylboronic acids. Adv Synth Catal. 2013;355:1570–1578. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.