Abstract
Kif15 is a kinesin-12 that contributes critically to bipolar spindle assembly in humans. Here we use force-ramp experiments in an optical trap to probe the mechanics of single Kif15 molecules under hindering or assisting loads and in a variety of nucleotide states. While unloaded Kif15 is established to be highly processive, we find that under hindering loads, Kif15 takes <∼10 steps. As hindering load is increased, Kif15 forestep:backstep ratio decreases exponentially, with stall occurring at 6 pN. In contrast, under assisting loads, Kif15 detaches readily and rapidly, even from its AMPPNP state. Kif15 mechanics thus depend markedly on the loading direction. Kif15 interacts with a binding partner, Tpx2, and we show that Tpx2 locks Kif15 to microtubules under both hindering and assisting loads. Overall, our data predict that Kif15 in the central spindle will act as a mechanical ratchet, supporting spindle extension but resisting spindle compression.
INTRODUCTION
Kinesin molecular motors play pivotal roles in the assembly and maintenance of bipolar spindles. By transducing the energy freed by ATP hydrolysis, kinesins generate impulses of force and movement that are essential for spindle self-organization (Cross and McAinsh, 2014). Kif15 (hKlp2) is a kinesin-12 whose role in human spindles is partially redundant with that of the kinesin-5, Eg5 (Tanenbaum et al., 2009; Gayek and Ohi, 2014). Eg5 is essential for spindle pole separation in humans, but dispensable for maintaining spindle bipolarity in several human cell lines, provided Kif15 is functional (Gayek and Ohi, 2014). Eg5 and Kif15 both move toward microtubule (MT) plus ends and counteract the actions of the minus end–directed motors dynein and kinesin-14 (van Heesbeen et al., 2014). Kif15 can support spindle assembly in the absence of Eg5, either when overexpressed or in the absence of dynein (Tanenbaum et al., 2009; van Heesbeen et al., 2014). The apparent functional overlap between Kif15 and Eg5 has led the field to suspect that these two kinesins may have similar mechanisms and make equivalent contributions to spindle assembly (Tanenbaum et al., 2009). However recent in vitro reconstitution work argued otherwise, by revealing Kif15-specific behavior (Drechsler et al., 2014; Drechsler and McAinsh, 2016; Mann et al., 2017). In such assays, Kif15 molecules display a combination of paused, diffusive, and motile behaviors. At low loads, Kif15 is highly processive, being able to move >1 μm along a microtubule before detaching. Additionally, Kif15 reduces the catastrophe rate of dynamic MTs, cross-links pairs of MTs, and promotes the assembly of parallel bundles of microtubules (Drechsler and McAinsh, 2016), consistent with its proposed in vivo role in stabilizing parallel K-fibers (Sturgill and Ohi, 2013). The localization of Kif15 to K-fibers (and interpolar MTs) is dependent on Tpx2 (Tanenbaum et al., 2009). In vitro, Tpx2 causes both Eg5 and Kif15 to bind statically to MTs (Drechsler et al., 2014; Balchand et al., 2015; Mann et al., 2017).
Whether the functional form of Kif15 is a dimer or a tetramer has been controversial (Drechsler et al., 2014; Drechsler and McAinsh, 2016; Sturgill et al., 2014). Kif15 motors within mitotic or interphase extracts were recently confirmed to be tetrameric by single-molecule imaging (Mann et al., 2017). However, the arrangement of the primary chains in functional Kif15 remains to be established. It is also clear that Kif15 can autoinhibit, via a C-terminal subdomain (Sturgill et al., 2014), though again, the exact arrangement of the primary chains remains unclear.
The critical role of Kif15 in bipolar spindle assembly and function makes it especially important to find out its mechanochemical mechanism. This problem is also important for practical reasons—for example, Kif15-based spindle assembly could allow cells to develop resistance to Eg5-inhibiting antiproliferatives (Rath and Kozielski, 2012). Kif15 is essential for the development of such resistance in HeLa cells (Sturgill et al., 2016).
Here we report measurements of the single-molecule mechanics of Kif15 under calibrated loads. We find that while Kif15 walks processively for long distances at zero load, it slows and reduces its processivity under hindering load and detaches readily under assisting load. The tendency for single Kif15 molecules to release rapidly under forward loads, even from their strongly binding AMPPNP state, suggests that Kif15 motors have “stepping rules” that differ from those of any other kinesin so far characterized.
Recent optical trapping work from the Lang lab also measured Kif15 single-molecule mechanics, but using a truncated construct and different analyses (Reinemann et al., 2017). Reinemann et al. ‘s work also measured the steps and sliding forces generated by Kif15 between parallel and antiparallel microtubules in bundles. Our own data set on the mechanics of single Kif15 molecules is independent and complementary, and we discuss the implications of our own results in the light of this related work.
RESULTS
Full-length Kif15 steps processively against hindering loads
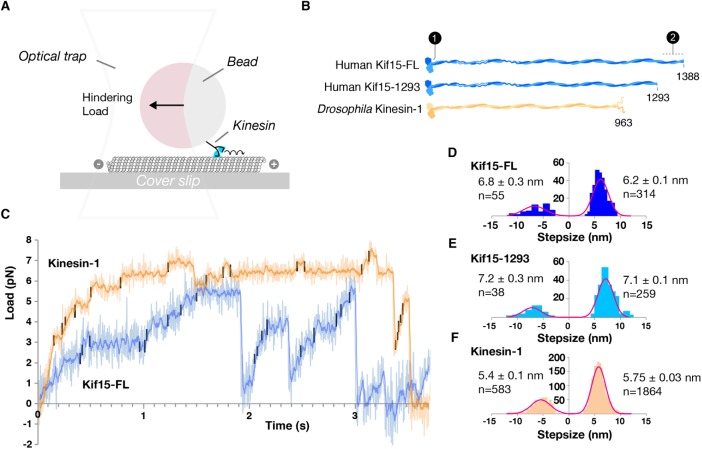
We measured the mechanics of a full-length Kif15 construct, Kif15-FL, and a truncated construct, Kif15-1293, which lacks the C-terminal 95 aa region of the tail containing the regions required for autoinhibition and Tpx2 interaction (Figure 1; Tanenbaum et al., 2009; Sturgill et al., 2014). We bound these to polystyrene beads, at a dilution designed to ensure one motor molecule per bead (see Materials and Methods), and observed the stepping behavior of the bead-attached motors as they walked along MTs at 1 mM ATP under loads exerted by a parked optical trap (Figure 1A). At zero load (unpublished data), we saw frequent runs of more than 1 bead diameter (560 nm), indicating that Kif15 is highly processive when unloaded, consistent with earlier single-molecule TIRF experiments done at zero external load (Drechsler et al., 2014). In contrast, under hindering loads applied by the optical trap, single Kif15-FL molecules (Figure 1C, blue trace) still step processively, but run lengths are reduced compared with those at zero load, and stepping is slower. To control for instrumental effects, we made parallel measurements with kinesin-1, whose-single molecule stepping we previously characterized (Carter and Cross, 2005). Compared with the stepping of full-length kinesin-1 molecules (Figure 1C, orange trace), Kif15-FL stepping is slower, run lengths are shorter, and the motor takes fewer steps at high loads. Step size distributions for both Kif15-FL and Kif15-1293, measured using an automated step detection algorithm, were centered on ±∼8 nm, with no substeps apparent, as were those for control kinesin-1 dimers (Figure 1, D–F).
FIGURE 1:
Kif15 takes 8-nm steps toward microtubule plus ends under hindering loads of up to 6 pN. (A) Schematic of the single-molecule optical trapping assay. (B) Constructs used in these experiments: full-length hKif15 with N-terminal His-tag (Kif15-FL), C-terminally truncated Kif15 lacking the terminal 95 a.a. that includes the Tpx2 binding region (Kif15-1293), and full-length Drosophila kinesin-1 (Kinesin-1). 1 = 6-his tag; 2 = C-terminal 95 residue section. (C) Example Kif15-FL (blue trace) and kinesin-1 (orange trace) processive runs under load at saturation with ATP (1 mM). In these example traces, both the Kif15-FL and kinesin-1 motors slow to a stall at around 6 pN. Walking Kif15-FL typically reaches lower loads and steps more slowly than walking kinesin-1. Both traces show clear 8-nm steps in both the forward and backward directions. Trap stiffnesses were 0.095 and 0.081 pN/nm for Kif15-FL and kinesin-1, respectively. Steps were detected by processing the raw data using a t test–based algorithm (see Materials and Methods) and are marked in black. (D–F) Step-size distributions for Kif15-FL, Kif15-1293, and kinesin-1. Values given are mean ± SD. Step-size values shown are not compliance-corrected, so they are slightly less than 8 nm.
C-terminal antibodies, or a C-terminal truncation, block entry to a Kif15 lock state
For Kif15-FL, only 27% of motor–bead complexes moved processively. The remainder bound statically to the MT lattice (Figure 2A). Applying a load to these “locked” motors, in either direction, did not provoke stepping. Autoinhibition of Kif15 has been reported previously (Sturgill et al., 2014) and shown to require a C-terminal section of the Kif15 tail. We speculate that our locked motors are autoinhibited. Consistent with this, treatment of bead–motor complexes with antibodies that target the C-terminus of Kif15 increased the motile fraction of MT-binding bead–motor complexes to over 60% (Figure 2A). Truncating the Kif15 tail to remove the C-terminal 95 amino acids (Kif15-1293) was even more effective, increasing the motile fraction to 84% (Figure 2A), while having a negligible effect on its velocity (Figure 2B). Here we have worked not only with Kif15-FL, but also, given the favorable effects of truncation, with Kif15-1293.
FIGURE 2:
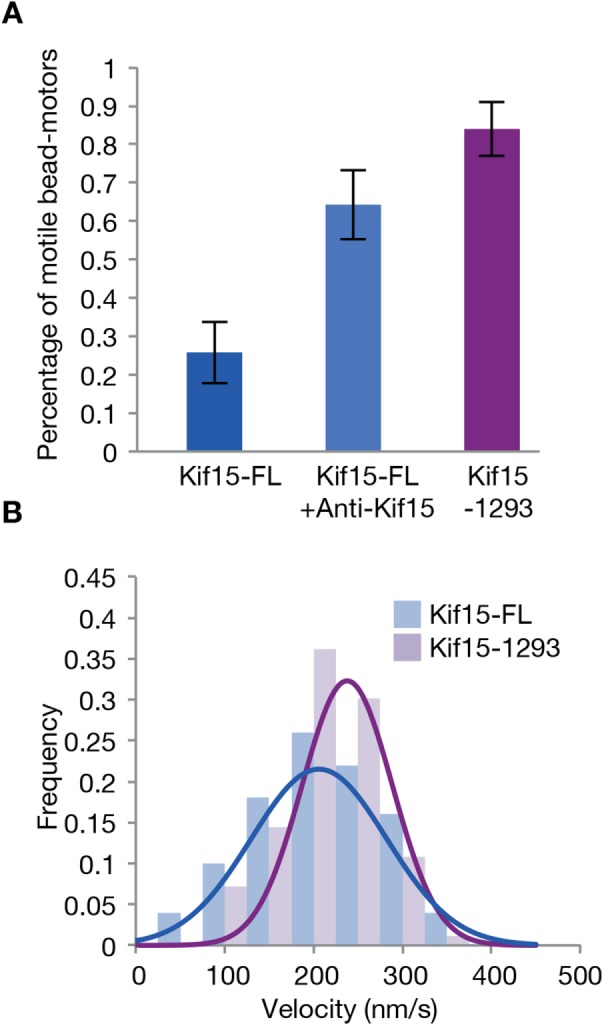
C-terminal antibody and C-terminal truncation increase the motile fraction of Kif15. (A) Motile fraction of all beads that bind the MT for Kif15-FL (n = 31), for Kif15-FL in the presence of 1 mg/ml anti-Kif15 antibody (n = 28), and for Kif15-1293 (n = 25). (B) Unloaded velocities of Kif15-FL motors (blue) and Kif15-1293 motors (purple). The velocity of Kif15-1293 is 238 ± 6 nm/s (n = 83), and that of Kif15-FL is 206 ± 10 nm/s (n = 50) mean and SE.
Kif15 run length under hindering load depends on trap stiffness
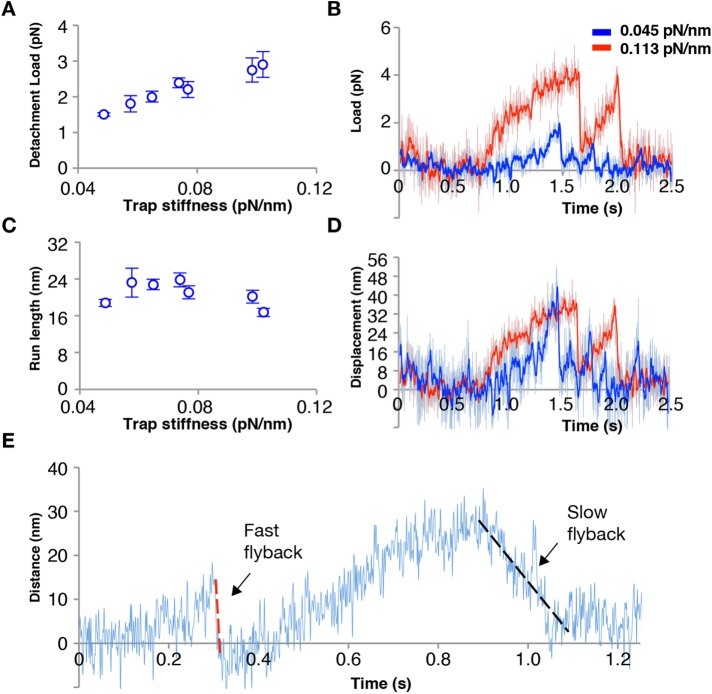
Figure 3 plots the influence of hindering load on Kif15 processivity. At low trap stiffnesses, Kif15 makes short processive runs of steps and typically detaches at low (1–3 pN) loads, as recently reported by Reinemann and colleagues for their C-terminally truncated Kif15 construct (Reinemann et al., 2017; N700, lacking the C-terminal ∼2/3 of the tail—see Discussion). At higher trap stiffnesses, Kif15 reaches higher loads before detaching (Figure 3A), with no change in run length (Figure 3C), showing that progress away from the trap center is limited by the intrinsic run length of the motor and not by an inability to step against higher loads. Detachments typically lead to rapid flyback of the bead–motor complex to the trap center, but on occasion the flyback was much slower (Figure 3E).
FIGURE 3:
Average detachment loads (A) and run lengths (C) for a sequence of records obtained from the same Kif15-FL motor measured at different trap stiffnesses. The average detachment load increases with increasing trap stiffness, while run length is invariant. (B, D) Example Kif15-FL traces for the same bead taken at trap stiffnesses of 0.045 pN/nm (blue) and 0.113 pN/nm (red), with the two traces scaled together according to load, B, or travel distance, D. (E) Example Kif15-FL trace showing that on detachment, Kif15-FL returns either rapidly (red broken line) or slowly (black broken line) to the trap center, suggesting that on some occasions, the motor slips progressively backward while still engaged with the MT rather than fully detaching. This behavior is seen for both Kif15-FL and Kif15-1293.
Kif15 run length under hindering load is the same under dimer and tetramer conditions
At ionic strengths in the physiological range, both Kif15-FL and Kif15-1293 are tetrameric, as judged by nondenaturing PAGE (Figure 4A). To look for functional differences between Kif15-FL dimers and tetramers, we preincubated the motors at high ionic strength before attaching them to beads and testing the beads in the optical trap. We previously showed that this treatment dissociates Kif15 tetramers into dimers (Drechsler et al., 2014). Preincubation at high ionic strength had no effect on the measured mechanical characteristics of Kif15-FL (Figure 4, B and C), suggesting that under the conditions of our assay, only one pair of Kif15 motor domains interacts with the MT. This is consistent with 8-nm steps (Figure 1); if both head-pairs of the Kif15-FL tetramer were to interact with the MT, we would expect step size to vary, especially at higher loads, since the load would then be distributed variably between the two engaged head pairs.
FIGURE 4:
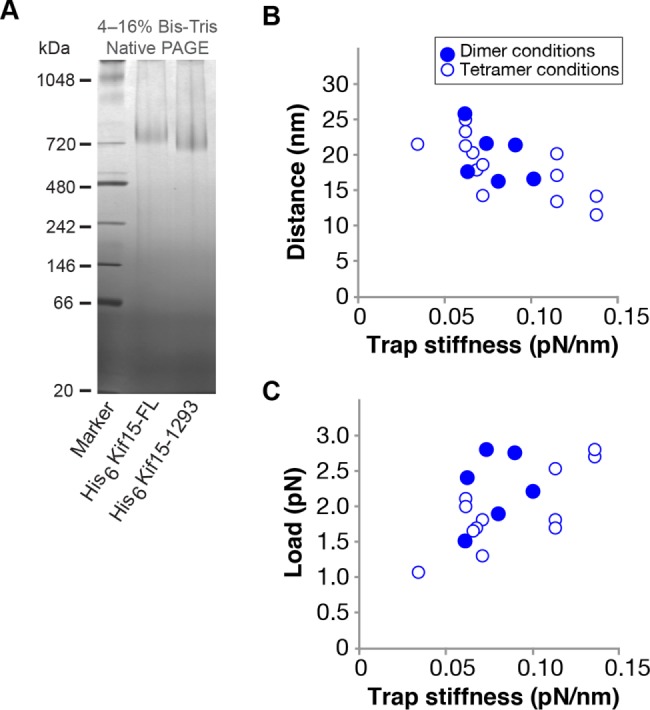
Shifting from dimer to tetramer conditions does not affect the stepping mechanics of bead-attached Kif15-FL. (A) Non-denaturing PAGE of Kif15-FL and Kif15-1293. Both constructs run as tetramers. (B) Mean distances at detachment and (C) mean loads at detachment vs. trap stiffness for walking Kif15-FL–bead complexes prepared under dimer-favoring (filled symbols, assay buffer + 380 mM NaCl) or tetramer-favoring (open symbols, assay buffer alone) incubation conditions.
Kif15 stepping rate and forestep:backstep ratio both depend on load
Kif15-1293, which lacks the C-terminal 95 residues of the Kif15 tail, does not autoinhibit, consistent with Sturgill et al. (2014), allowing us to collect stepping data more readily over a range of loads. Under a hindering load, Kif15-1293 takes both forward and backward steps. Figure 5 plots the average dwell time (the time interval preceding each step) for stepping of Kif15-1293 under load. For this plot, we pooled the measured dwell times for forward and backward steps and divided the data into 1-pN bins. Fitting exponentials to the substall parts of the data in Figure 5A yields dwell = 0.013 e(0.41 × load) seconds for kinesin-1 and dwell = 0.027 e(0.49 × load) seconds for Kif15-1293. The exponential factors describing the load dependence are thus similar for kinesin-1 and Kif15, but the projected zero-load velocities are different, corresponding to ∼615 nm s−1 for kinesin-1 (measured at 670 nm s−1) and 296 nm s−1 for Kif15 (measured at 238 nm s−1). Figure 5B plots the ratio {number of forward steps}/{number of back steps} versus load, again using 1-pN bins. The motor stall force is defined as the hindering load at which the motor makes an equal number of forward and backward steps and so makes no net progress. Kif15-1293 stall force is 5.6 ± 0.8 pN, while kinesin-1 control stall force is 6.8 ± 0.7 pN, in quantitative agreement with previous data (Carter and Cross, 2005). Above stall force (superstall), Kif15-1293 can take several successive backward steps. Figure 5C compares force–velocity curves for Kif15-1293 and kinesin-1.
FIGURE 5:
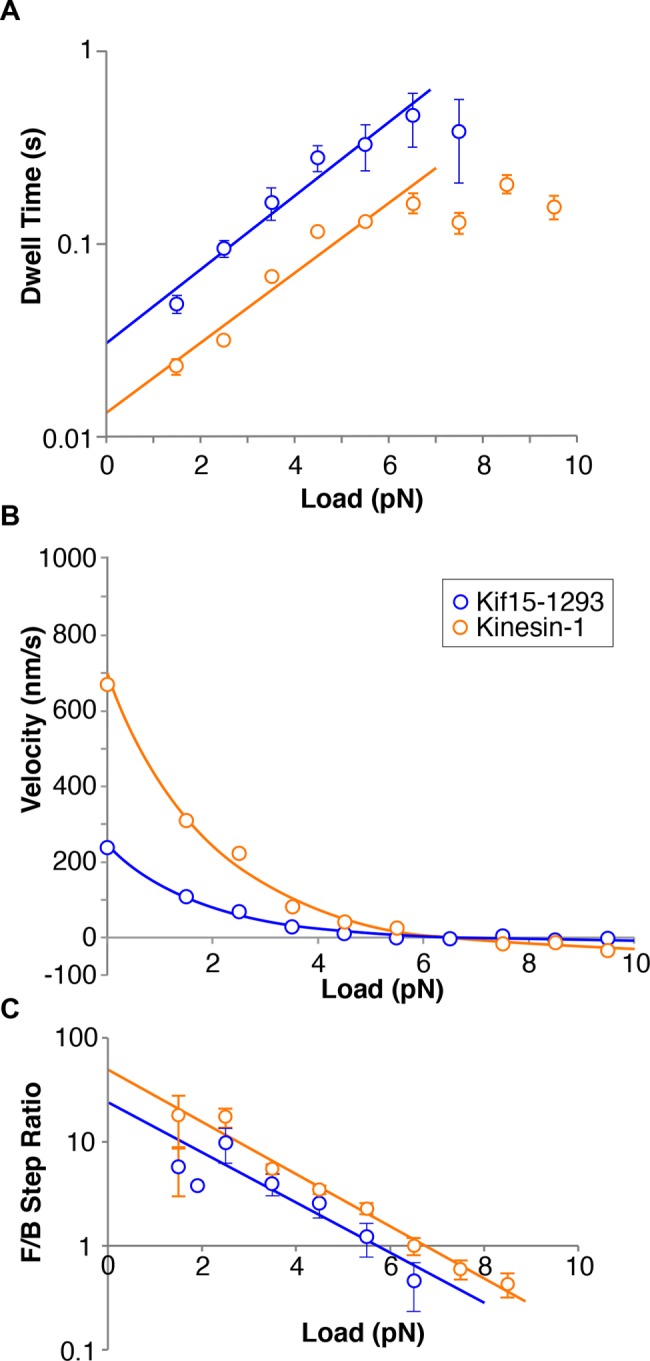
Load dependence of Kif15 stepping rate and forestep:backstep ratio. (A) Dwell time distributions for Kif15-1293 (blue symbols, n = 355) and kinesin-1 control (orange symbols, n = 2446) in 1 mM ATP. Dwell time is the time that the motor waits between steps. The average dwell time for forward plus backward steps increases sharply with increasing hindering load. (B) Forward:backward stepping ratios for Kif15-1293 (blue) and kinesin-1 (orange). Data were divided into 1-pN bins. Solid lines are exponentials, fitted using least squares. Stall forces (the force at which the ratio of the numbers of forward and backward steps is unity) are 5.6 ± 0.8 pN for Kif15-1293 and 6.8 ± 0.8 pN for kinesin-1. Errors are calculated from the standard error in the slopes and intercepts of the least-squares line fits. (C) Dependence of velocity on hindering load for Kif15-1293 (blue) and kinesin-1 (orange). Velocity is calculated from dwell time and forward:backward stepping ratio at each load, assuming a step size of 8.1 nm for both motors. Zero-load data points are from unloaded bead velocities. Kif15-1293 and kinesin-1 dwell time distributions are fitted with exponentials. For kinesin1, 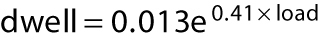 ; for Kif15-1293,
; for Kif15-1293, 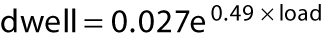 .
.
Unbinding load of Kif15 depends acutely on loading direction
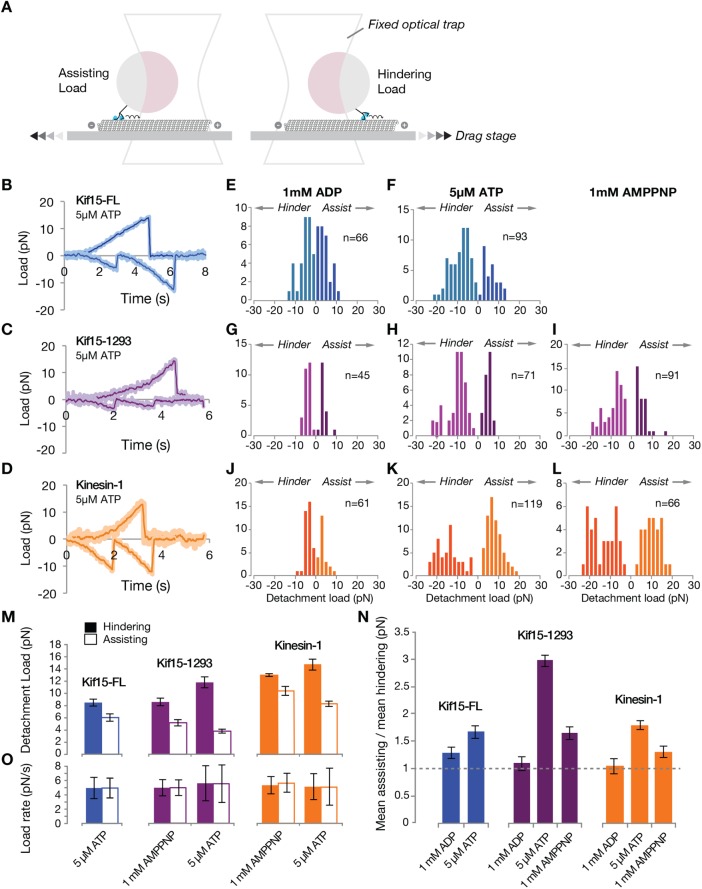
To examine the load dependence of Kif15 detachment in different nucleotide states, we performed drag-to-detachment (force-ramp) experiments. In these experiments, which were similar to those of Uemura et al. (2002), a bead–motor complex is first caught in the optical trap and the motor allowed to bind to a MT. Slowly transporting the microscope stage then progressively increases the load on the motor–MT connection (Figure 6A). Eventually the motor unbinds from the MT and the bead-attached motor returns to the trap center. Example bead position recordings are shown in Figure 6, B–D. By recording the loads at which the motor–bead complex detached from the MT under assisting (plus end–directed) or hindering (minus end–directed) loads, we were able to quantitate the dependence of the unbinding rate on load, for plus and minus end–directed loads, for both Kif15-1293 and for control kinesin-1, under both stepping (ATP) and nonstepping (ADP, AMPPNP) conditions.
FIGURE 6:
Unbinding of Kif15 from MTs is highly sensitive to loading direction. (A) Experimental setup for applying assisting (negative) and hindering (positive) loads. With the motor bound to the MT, the stage is moved at a defined rate (black chevrons) to drag the MT relative to the bead-attached motor in the trap center. The bead-attached motor is thereby steadily translated out of the trap center and experiences a steadily increasing restoring force (black arrows). The position of the bead is tracked continuously and the position and load at which the motor detaches from the MT are recorded. (B, C) Example traces for (B) Kif15-FL (blue) and (C) Kif15-1293 (magenta) in the presence of 5 µM ATP. Positive values indicate hindering loads and negative values indicate assisting loads. (D) Example traces for kinesin-1 (orange) in the presence of 5 µM ATP. (E–L) Detachment load distributions. (M) Mean hindering loads at detachment (filled bars) and mean assisting loads at detachment (unfilled bars) for Kif15-FL, Kif15-1293, and kinesin-1. Detachments under hindering loads occur at higher forces than those under assisting loads for all motors and nucleotide conditions. (N) Ratios of mean assisting load at detachment to mean hindering load at detachment. Kif15-1293 detaches at a threefold lesser force under assisting load compared with hindering load. (O) Loading rates for the data sets shown above, calculated from the slopes of individual traces. Loading rates were 3.6 ± 1.1 pN/s for ADP data.
In ADP, Kif15-FL detaches at low loads in both hindering and assisting directions (Figure 6E), consistent with the ADP state of Kif15 being the weakest binding state. More interestingly, for conditions under which Kif15-FL is stepping only very slowly (5 µM ATP), a clear difference appears between the effects of hindering and assisting loads—the motor detaches at low assisting loads, but resists detachment under hindering loads (Figure 6F). This pattern is even clearer for Kif15-1293, which lacks autoinhibition. In ADP, Kif15-1293 detaches at very low assisting and hindering loads (Figure 6G). In 5-μM ATP, unloaded Kif15-1293 steps at ∼36 nm/s, corresponding to 4.5 steps/s. Under these conditions, while stepping actively but slowly, Kif15-1293 detaches at very low assisting loads (Figure 5H), but resists detachment under hindering loads. In AMPPNP, a nonhydrolyzable ATP analogue, similar sensitivity to loading direction is seen (Figure 6I), showing that even when trapped in a putatively ATP-like state, Kif15 has a ratchet-like quality, tending to grip when pulled backward and release when pulled forward. In ATP, this acute sensitivity of the unbinding reaction to loading direction is dramatic, with threefold more hindering load than assisting load needed, on the average, to detach Kif15-1293 (Figure 6, M and N).
For comparison, we also did drag-to-detachment experiments with kinesin-1. Kinesin-1 run length is strongly dependent on loading direction (Milic et al., 2014). Figure 6, J and L, shows the unbinding loads for kinesin-1 in ADP and AMPNP and closely resembles the earlier results of Uemura et al. (2002). As noted by Uemura et al., unbinding loads of kinesin-1 in AMPPNP show a bimodal distribution, thought to reflect motors unbinding from either a single or double head–bound state. The high force component is less prominent for Kif15 (Figure 6I) than for kinesin-1 (Figure 6L), suggesting that in AMP-PNP Kif15 spends more time bound to the MT by a single motor head than does kinesin-1. Data in 5 µM ATP hint at a bimodal distribution of detachment loads for all three motors (Figure 6, F, H, and K). Under assisting load, kinesin-1 has a much shorter run length than under hindering load (Milic et al., 2014), consistent with assisting loads disrupting the coordination of kinesin-1 stepping. Our data show that Kif15 and kinesin-1 share a ratchetlike quality, whereby the motor attaches stably under backward load but slips under forward load, with Kif15 being if anything even more ratchetlike than kinesin-1.
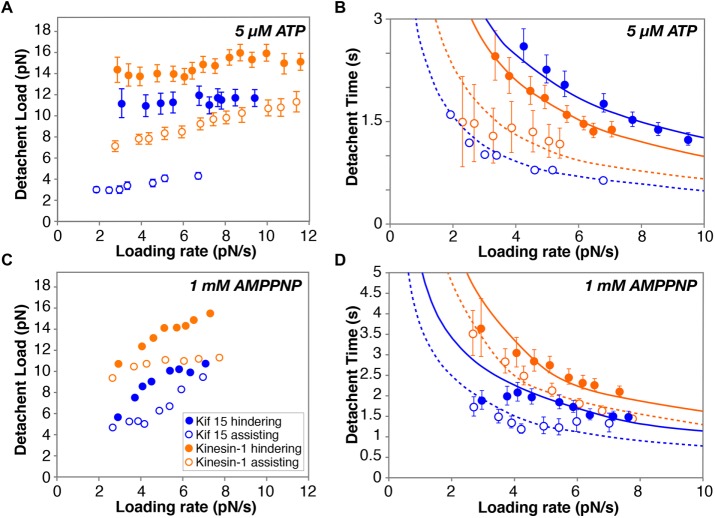
Figure 7 displays the loading rate versus time to detachment for Kif15-1293 and control kinesin-1 in 5-µM ATP and in 1-mM AMPPNP. In 5-µM ATP (Figure 7A), there is only slight loading rate dependence of detachment loads for both Kif15 and kinesin-1. In contrast, in AMPPNP (Figure 7C), loading rate dependence was obvious for both motors.
FIGURE 7:
Loading rate dependence of detachment. (A) In 5-µM ATP, there is little loading rate dependence of the detachment load for either Kif15 or kinesin-1. (B) For both motors, the loading rate dependence of the residence time is well fitted by a model in which the probability of detachment is related only to the instantaneous load (see the text). (C) In 1-mM AMPPNP, the mean detachment load shows an obvious dependence on loading rate and the fits in D are correspondingly poorer. Open symbols, assisting load; filled symbols, hindering load. Fitted values are given in the main text.
Pursuing the analysis originally applied by Uemura and colleagues (Uemura et al., 2002; Kawaguchi et al., 2003), we can apply a model in which the tethered motor must diffuse over an activation energy barrier in order to detach. In this case,
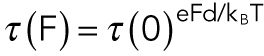 |
where τ(F) is the average time to unbinding at load F, τ(0) is the average time to unbinding at zero load, kB is the Boltzmann constant, T is the absolute temperature, and d is a characteristic distance. We can fit this model to the data by setting 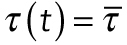 , the average time to detachment at load F = ατ, where α is the loading rate:
, the average time to detachment at load F = ατ, where α is the loading rate:
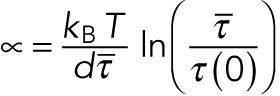 |
In the presence of 5-µM ATP, fitting this equation by least squares using τ(0) = 100 s for kinesin-1 and 30 s for Kif15-1293 gives a good fit and run lengths of 5 and 1 µm for kinesin-1 and Kif15-1293 at zero load, in broad agreement with published data. The values of d for kinesin-1 in 5 µM ATP are then dassisting = 3.33 nm and dhindering = 1.85 nm. For Kif15-1293 in 5-µM ATP the values are dassisting = 3.8 nm and dhindering = 1.0 nm (Figure 7B). The direction dependence of d is evident for both motors but greater for Kif15-1293, emphasizing its ratchetlike properties.
For dimeric Kif15, τ(0) in 1-mM AMP-PNP has been experimentally measured to be 12.2 s (Sturgill et al., 2014). Fitted characteristic distances for Kif15-1293 are then dassisting = 1.7 nm and dhindering = 1 nm for plus and minus end–directed loading, respectively; τ(0) is 19 s. For kinesin-1 in AMPPNP, using τ(0) = 950 s gives dassisting = 2.3 nm and dhindering = 1.7 nm (Figure 7D). In this case the fit is poorer, potentially due to the loading rate dependence of the detachment load in AMPPNP. One of the features of the fitted model is that there is no such dependence.
Overall, this analysis confirms substantial differences in the average residence time at particular loads, depending on the direction of the load. This asymmetry, expressed as dassisting/dhindering, is greater for Kif15-1293 than for kinesin-1, especially in 5-µM ATP.
Tpx2 augments the load resistance of MT-bound Kif15-FL, but not Kif15-1293
How does Tpx2 influence the mechanics of Kif15? Our data argue that the interaction of Kif15-FL with Tpx2 occurs via its C-terminal 95 residues. The motor activity of Kif15-1293, which is missing this section of the tail (Figure 1B), is relatively unaffected by Tpx2 (Figure 8, A and B), whereas Kif15-FL is consistently arrested by Tpx2 (Figure 8A), indicating also that bead-bound Kif15-FL exposes its Tpx2 binding site. Because Kif15-FL and Tpx2 do not interact tightly in solution (Drechsler et al., 2014), we infer that Tpx2 binds appreciably only to the Kif15-FL-MT complex.
FIGURE 8:
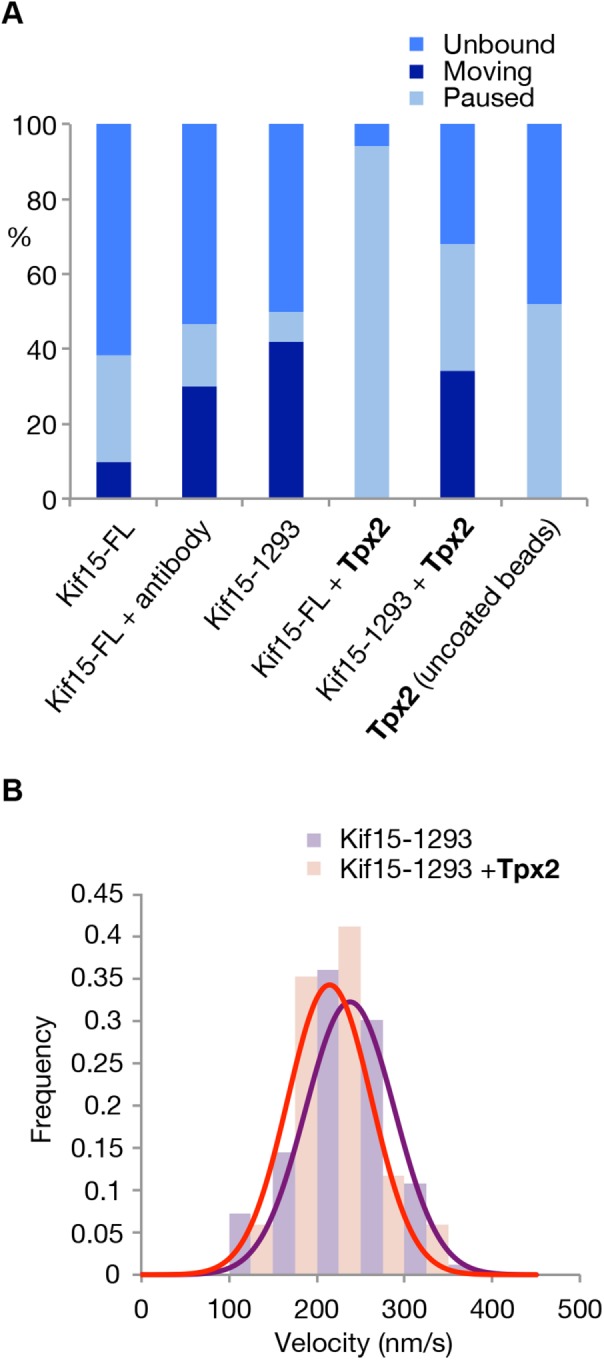
Tpx2 arrests Kif15-FL, but not Kif15-1293. (A) Motile fraction of all beads that bind the MT for Kif15-FL (n = 31), for Kif15-FL in the presence of 1 mg/ml anti-Kif15 antibody (n = 28), and for Kif15-1293 (n = 25). Error bars show region of 95% certainty. (B) Unloaded velocities of Kif15-1293 motors and Kif15-FL motors. The velocity of Kif15-1293 is 238 ± 6 nm/s (n = 83), and that of Kif15-FL is 206 ± 10 nm/s (n = 50) mean and SE.
DISCUSSION
Kif15 stall force is 6 pN
While Kif15 is highly processive at zero load and while hindering loads dramatically reduce its processivity, our data show that Kif15 has a stall force of 6 pN, approaching that of kinesin-1. This value is higher than the 3 pN recently reported by Reinemann et al. (2017). The constructs used for the Reinemann experiments lacked the C-terminal 688 residues of the Kif15 tail, whereas we have worked with Kif15-FL or with Kif15-1293, which lacks only the C-terminal 95 residues. But we suspect that the discrepancy in our measured stall forces may have more to do with our two different techniques. We obtained the stall force by measuring the forestep:backstep ratio over a range of loads and extrapolating to unity. The stall force quoted by Reinemann et al. (2017) is their highest accessible hindering load. In earlier work (Drechsler et al., 2014), we obtained a similar value. In a compliant trap this value can be limited by the processivity of the motor. In our current work, we counted foresteps and backsteps and obtained stall force by extrapolation (Figure 5B). Accordingly, our two sets of experiments are, we believe, fully consistent and complementary.
Kif15 slips under assisting load
Under assisting load, we find that Kif15 detaches rapidly, even at low loads and even from its “strong” state. In this respect Kif15 resembles kinesin-1, whose run length is markedly dependent on loading direction, albeit the asymmetric response of Kif15 to loading direction is even more acute.
Kif15 waits between steps in a one head–attached state
Strikingly, our force-ramp data point to two different attached populations of Kif15 in AMP-PNP (Figure 6), which we attribute, by analogy to kinesin-1 (Uemura et al., 2002), to one head–attached and two -head–attached states. Whether the functional form of Kif15 is a dimer or a tetramer has been controversial. We confirmed tetramerization of Kif15-FL and Kif15-1293 in low-salt buffer (Figure 4A), but we find that the mechanics of Kif15 under dimer conditions (high-salt buffer) are indistinguishable from those under tetramer conditions, suggesting that tetramers, when attached to beads, function mechanically as twin-headed dimers—in other words, that bead attachment allows only one Kif15 head pair to engage the MT. Our experiments in ATP show two classes of attached state, for both kinesin-1 and Kif15, but with the putatively one head–attached state dominating. Force-ramp experiments in ATP have not previously been reported for any kinesin. Our data argue that Kif15 dwells between steps predominantly in the one head–attached state, implying that the acute dependence of detachment rate on loading direction is an intrinsic property of each Kif15 head.
A Kif15 refractory (lock) state is superstabilized by Tpx2
We find that in addition to its processive stepping mode, Kif15 can bind statically to microtubules, in a stably refractory (lock) state. In our hands, this state never arose during a processive run under load. Rather, a typical pattern was that following a processive run in the laser trap, the motor would detach and then reattach, either in an active state that underwent a further processive run or in an inactive lock state that bound nucleotide-independently and resisted both hindering and assisting loads. This state appears to be further stabilized by Tpx2. In its Tpx2-stabilized autoinhibited state, detachment of Kif15 from the MT requires loads >35 pN in either direction. The C-terminal 95aa region of the Kif15 tail that is required for the lock state includes the Tpx2 binding region. Tpx2 binds via its C-terminus (Mann et al., 2017) to this C-terminal region of Kif15. Deletion of this region dramatically reduces the effect of Tpx2 on Kif15 (Figure 8A), so that velocities of motile Kif15-1293 motors are essentially unaffected by the presence of Tpx2 (Figure 8B). Tpx2 has previously been shown to cause Kif15 to pause on the MT lattice for extended periods of time (Drechsler et al., 2014). Figure 9A summarizes these properties of the Kif15 motor domain as a mechanical cartoon.
FIGURE 9:
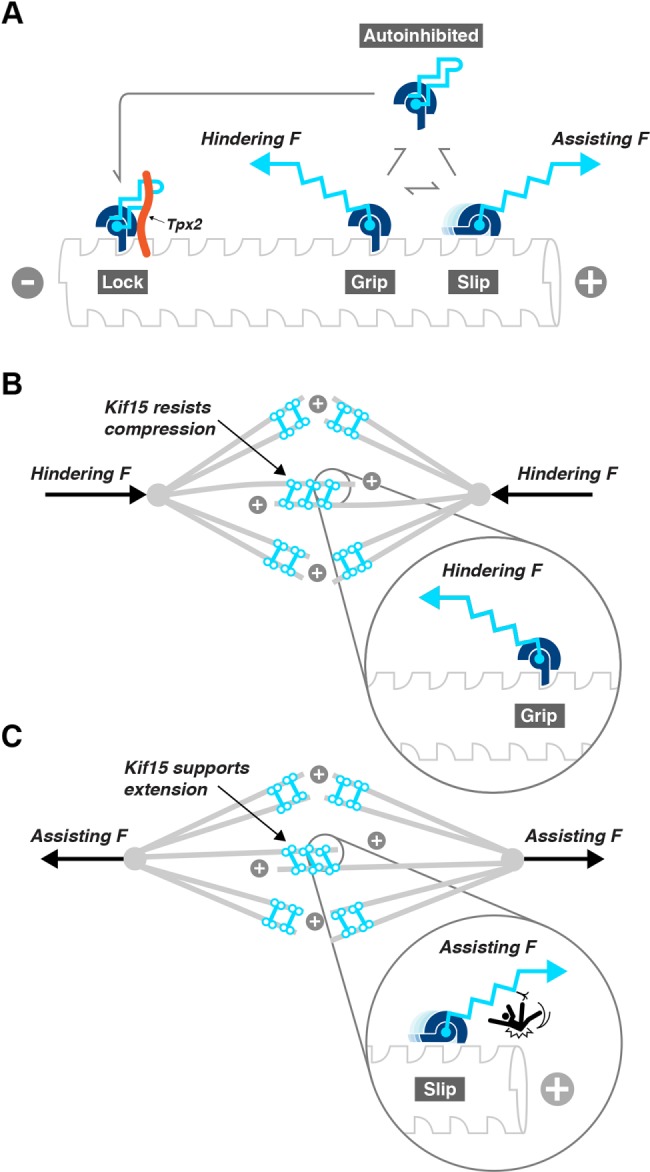
(A) Ratchet and pawl cartoon depicting the load direction–dependent single-molecule mechanics of Kif15. The model focuses on an individual Kif15 motor domain, which is represented as a ratchet and pawl that slip (Slip state) under assisting load but grip (Grip state) under hindering load. A further Lock state arises when the motor binds the microtubule stably and nucleotide-independently. Tpx2 (orange) stabilizes this state. (B, C) How Kif15 might contribute to force balance in spindles. Speculative model showing possible implications of Kif15 single-molecule mechanics for spindle function. For spindles under axial compression, B, Kif15 in the overlap region of the spindle will tend to lock the sliding of MTs in this zone. In contrast, for spindles subject to extensile forces tending to drive the poles apart, C, Kif15 in the overlap region (antiparallel MTs) will experience assisting load and will slip, allowing the spindle to elongate. Kif15 in the K-fibers (parallel MTs) functions to organize and stabilize.
Implications of our data for the in vivo roles of Kif15
There is now considerable evidence that Kif15 in spindles binds both in the overlap zone and to K-fibers. Our mechanical data support a model in which Kif15 can shift between gripping and walking under hindering load and slipping under assisting load. In addition, and especially when interacting with Tpx2, Kif15 can lock passively to the microtubule in a state that resists load in either direction (Figure 9A). With Kif15 in the latter state, it will resist sliding driven by other force generators (van Heesbeen et al., 2014). Fluxing of MTs in K-fibers might, in this situation, drive spindle pole separation. Under hindering load, walking Kif15 will grip and resist compressive forces that would otherwise drive the spindle poles toward one another (Figure 9B), whereas under assisting load, arising when the spindle poles are driven apart by other force generators, Kif15 will slip, ensuring minimal drag and supporting pole separation (Figure 9C).
Implications for the partial in vivo redundancy of Kif15 with Eg5
The cell-biological role of Kif15 is partially functionally redundant with that of Eg5, and this has been taken as evidence that the two motors might have similar mechanisms. Our data show the contrary. Kif15, like Eg5, Kif15, has a short run length and tends to detach from the MT before reaching stall (Reinemann et al., 2017). But Eg5 is directionally insensitive—its low detachment rate under both hindering and assisting loads allows it to restrict the sliding apart of overlapping MTs (Shimamoto et al., 2015) so that it both drives microtubule sliding and acts as a drag brake (Saunders et al., 2007; Shimamoto et al., 2015; Muretta et al., 2018) that limits the rate of anaphase spindle elongation. By contrast, Kif15 is an active ratchet, stepping slowly and securely under hindering load, and slipping under assisting load. Kif15 and Eg5 thus support spindle bipolarity by fundamentally different molecular (and cell-biological) mechanisms, albeit both bind Tpx2, suggesting the possibility of linked regulation. We suspect that the active ratchet mechanism of Kif15 (Figure 9) would allow it to lock and resist spindle collapse in prometaphase, consistent with evidence that Kif15 supplements and complements the mitotic activities of a range of other force generators (van Heesbeen et al., 2014), including dynamic microtubules and potentially including nonmotor MAPs such as Ase 1/Prc1, which can produce sliding force by entropic expansion of overlaps (Rincón et al., 2017). Kif15 is not involved in anaphase spindle elongation (Saunders et al., 2007).
In conclusion, we have found that the single-molecule mechanics of Kif15 allows it to function as an active mechanical ratchet that walks rapidly toward microtubule plus ends when unloaded, slows down and become less processive under hindering load, and detaches rapidly under assisting load. We predict that Kif15’s ability to unbind and slip under assisting load will allow it to function effectively in teams involving other plus end–directed steppers. Further work on the spatial regulation of Kif15 by Tpx2, Ki67, Aurora A, or other mitotic regulators in cells will be needed to understand the in-context contribution of these intrinsic properties of Kif15 motor domains to spindle assembly and maintenance.
MATERIALS AND METHODS
Preparation of bead–motor complexes
Sample preparation was as described in Drechsler et al. (2014) and instrumentation as described in Carter and Cross (2005). Motors were nonspecifically bound to polystyrene beads (550 nm, NIST calibration size standards; Polysciences, USA) by incubation in a solution of 80 mM K.PIPES (piperazine-N,N’-bis(2-ethanesulfonic acid)), 2 mM MgSO4, 1 mM ethylene glycol-bis(b-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA), 1 mM 1,4-dithiothreitol (DTT), 3 mg/ml d-glucose, 0.2 mg/ml casein, and 1 μM ATP at pH 7.0. Incubation was performed for at least 1 h before addition to the flow cell. Motor concentration was reduced until no more than one-third of beads showed any microtubule binding. A bead-binding event was defined as a bead that remained attached to the microtubule for at least 2 s after the laser trap was turned off. Each bead was tested for binding using at least two different microtubules. For measurements using preincubation at high ionic strength, motors were incubated in assay buffer plus 380 mM NaCl for 30 min prior to incubation with beads, and beads were preincubated in this same solution for at least 1 h. Bead–motor complexes were then formed by rapid dilution as described above, but assay buffer plus 0.4 mg/ml casein. This concentration of casein blocking agent was maintained during subsequent experiments. The antibody used in Figure 2 was Anti Hklp2 Antibody, Catalogue Number AKIN13, from Cytoskeleton. In these experiments, polystyrene beads were incubated with motors (Carter and Cross, 2005). Beads were then added to assay buffer containing 1 mg/ml antibody and immediately introduced to the flow cell and imaged.
Flow cells for optical trapping
Flow cells were constructed using sonicator- and plasma-cleaned coverslips and Dow Corning High Vacuum Grease. Taxol-stabilized microtubules were diluted in assay buffer: 20 mM K.PIPES, 2 mM MgSO4, 1 mM ATP, 1 mM EGTA, 1 mM DTT, 3 mg/ml d-glucose, 4 μM taxol, and a glucose oxidase/catalase-based oxygen-scavenging system. Diluted microtubules were introduced into the flow cell and allowed to adsorb onto the surface for up to 5 min. The incubated bead–motor solutions were then diluted in an assay buffer and introduced into the flow cell, which was imaged immediately.
Force-ramp experiments
To gain insight into the behavior of the motor proteins at loads above stall, backward pulls were performed. In a backward pull the stage is moved, causing the motor to experience hindering loads that exceed its stall force. Stage movement was automatically triggered when the motor had walked to a set load of 3 pN. At this trigger point the stage was moved by piezoelectric positioning motors so that the trap center was shifted toward the premarked MT minus end. The imposed load was also preset, usually at 8 or 10 pN. At these levels of hindering load, motors take mostly backward steps. Unbinding load experiments require the motor to move past the MT at a constant rate. These experiments were performed by moving the stage with piezoelectric positioning motors (Physik Instrumente [PI] 50 μm × 50 μm XY piezoelectric translation stage). The stage was controlled by a 12 bit DAC (digital to analogue converter), allowing the 50-μm stage to be stepped in 12.2-nm movements in both the x and y directions. For data collection, the microtubule position and orientation were marked and the stage stepped so that the motor was dragged along the microtubule. Our microscope software collects data at 80–100 kHz, sampled down before storage to 45 kHz while performing calibrations and to 22 kHz while collecting motor stepping data. Laser trap stiffness calibration was performed daily using the average of stiffness determined using three methods; equipartition, Stokes, and drag force.
Data analysis
Steps were fitted to the raw data using a moving window t-test algorithm as described by Carter and Cross (2005). This algorithm requires the user to input a window size over which the t-test is to be performed and a t-value cutoff. Stepping data was binned into 1-pN bins for analysis of dwell time and F/B stepping ratio, and only bins where >5 steps were detected were used. The rate at which the load is applied can make a large difference to the distribution of the unbinding loads. When data sets were compared, the data were cropped so that for each set of data the loading rates were similar (5 ± 0.5 pN/s) and only a narrow range of loading rates was used (Figure 6O). In cases where the bead did not return to the trap center after a detachment (a backward slip greater than 16 nm), the second detachment was discounted.
Acknowledgments
R.A.C. is supported by a Wellcome Trust Senior Investigator Award (Grant 103895/Z/14/Z). A.D.M. is supported by a Wellcome Trust Senior Investigator Award (Grant 106151/Z/14/Z) and a Royal Society Wolfson Research Merit Award (Grant WM150020). T.M. was funded by an Engineering and Physical Sciences Research Council Doctoral Training Grant award (Grant 1090393). We thank Mathew Turner for insightful discussions.
Abbreviations used:
- Eg5
kinesin 5/KIF 11
- Kif15
kinesin-12
- MT
microtubule
- Tpx2
targeting protein for Xklp2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-03-0151) on May 17, 2018.
REFERENCES
- Balchand SK, Mann BJ, Titus J, Ross JL, Wadsworth P. (2015). TPX2 inhibits Eg5 by interactions with both motor and microtubule. J Biol Chem , 17367–17379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter NJ, Cross RA. (2005). Mechanics of the kinesin step. Nature , 308–312. [DOI] [PubMed] [Google Scholar]
- Cross RA, McAinsh A. (2014). Prime movers: the mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Biol , 257–271. [DOI] [PubMed] [Google Scholar]
- Drechsler H, McAinsh AD. (2016). Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci USA , E1635–E1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD. (2014). The Kinesin-12 Kif15 is a processive track-switching tetramer. Elife , e01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayek AS, Ohi R. (2014). Kinetochore-microtubule stability governs the metaphase requirement for Eg5. Mol Biol Cell , 2051–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Uemura S, Ishiwata S. (2003). Equilibrium and transition between single- and double-headed binding of kinesin as revealed by single-molecule mechanics. Biophys J , 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BJ, Balchand SK, Wadsworth P. (2017). Regulation of Kif15 localization and motility by the C-terminus of TPX2 and microtubule dynamics. Mol Biol Cell , 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milic B, Andreasson JOL, Hancock WO, Block SM. (2014). Kinesin processivity is gated by phosphate release. Proc Natl Acad Sci USA , 14136–14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muretta JM, et al (2018). A posttranslational modification of the mitotic kinesin Eg5 that enhances its mechanochemical coupling and alters its mitotic function. Proc Natl Acad Sci USA , E1779–E1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath OO, Kozielski FF. (2012). Kinesins and cancer. Nat Rev Cancer , 527–539. [DOI] [PubMed] [Google Scholar]
- Reinemann DN, Sturgill EG, Das DK, Degen MS, Vörös Z, Hwang W, Ohi R, Lang MJ. (2017). Collective force regulation in anti-parallel microtubule gliding by dimeric Kif15 Kinesin motors. Curr Biol , 2810–2820. e2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón SA, Lamson A, Blackwell R, Syrovatkina V, Fraisier V, Paoletti A, Betterton MD, Tran PT. (2017). Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast. Nat Commun , 15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM. (2007). Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol , R453–R454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Forth S, Kapoor TM. (2015). Measuring pushing and braking forces generated by ensembles of Kinesin-5 crosslinking two microtubules. Dev Cell , 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier SE, Ohi MD, Hwang W, Lang MJ, Ohi R. (2014). Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism. Curr Biol , 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Norris SR, Guo Y, Ohi R. (2016). Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol , 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R. (2013). Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol , 1280–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macu˚rek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema RH. (2009). Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol , 1703–1711. [DOI] [PubMed] [Google Scholar]
- Uemura S, Kawaguchi K, Yajima J, Edamatsu M, Toyoshima YY, Ishiwata S. (2002). Kinesin-microtubule binding depends on both nucleotide state and loading direction. Proc Natl Acad Sci USA , 5977–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesbeen RGHP, Tanenbaum ME, Medema RH. (2014). Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep , 948–956. [DOI] [PubMed] [Google Scholar]