Abstract
The field of biomedical informatics experienced a productive 2015 in terms of research. In order to highlight the accomplishments of that research, elicit trends, and identify shortcomings at a macro level, a 19-person team conducted an extensive review of the literature in clinical and consumer informatics. The result of this process included a year-in-review presentation at the American Medical Informatics Association Annual Symposium and a written report (see supplemental data). Key findings are detailed in the report and summarized here. This article organizes the clinical and consumer health informatics research from 2015 under 3 themes: the electronic health record (EHR), the learning health system (LHS), and consumer engagement. Key findings include the following: (1) There are significant advances in establishing policies for EHR feature implementation, but increased interoperability is necessary for these to gain traction. (2) Decision support systems improve practice behaviors, but evidence of their impact on clinical outcomes is still lacking. (3) Progress in natural language processing (NLP) suggests that we are approaching but have not yet achieved truly interactive NLP systems. (4) Prediction models are becoming more robust but remain hampered by the lack of interoperable clinical data records. (5) Consumers can and will use mobile applications for improved engagement, yet EHR integration remains elusive.
Keywords: biomedical informatics, year in review, electronic health records, learning health system, consumer engagement
INTRODUCTION
The applications of informatics range from cutting-edge medical research in genomics to helping consumers find basic medical information. Due to its common core, however, informatics advances in one area often benefit other areas of the field. For example, a machine learning method for clinical research informatics might be usable in clinical decision support, a clinical decision support algorithm might be repurposed as a consumer mobile health app, a consumer engagement tool might provide insights into the development of improved interoperability standards, and adoption of effective standards might enable new forms of clinical research such as precision medicine. The different areas of informatics, therefore, can learn a lot from each other.
To accelerate the cross-pollination that can lead to further advances, several endeavors in the field have been proposed to expose researchers to the wider parts of informatics. These include year-in-review presentations at the AMIA Annual Symposium1,2 and in the annual International Medical Informatics Association (IMIA) Yearbook.3 Of necessity, a year-in-review benefits from examining a large number of articles and engaging in discussions among the review team.
For the 2015 AMIA Year-in-Review in Clinical and Consumer Health Informatics, we conducted a thorough literature review for an entire year within the scopes of clinical informatics, clinical research informatics, and consumer health informatics. Building on the AMIA Symposium presentation by Patricia Flatley Brennan, we produced a companion report (see online supplement) highlighting the most novel research and most interesting trends in 18 topics in informatics. While the presentation and report drew from the same base of reviewed articles, they differ in the articles each chose to highlight. The report details key findings, supported by over 250 citations, organized into 3 themes: methods of biomedical informatics, tasks supported by informatics, and trends in informatics. Table 1 lists all of the topics. In this perspective, we provide a summary of key observations and findings of the year, and argue for future directions informatics should take.
Table 1.
Topics covered in year-in-review
| Methods | Tasks | Trends |
|---|---|---|
|
|
|
METHODS
A review group of junior and senior scholars was empaneled to conduct the review. The group included 18 graduate students and postdoctoral trainees from the JAMIA Student Editorial Board and the 2014 AMIA Media Year-in-Review group. Of these, 11 contributed to the written report (the authors of this perspective). The group met monthly over 6 months during the process.
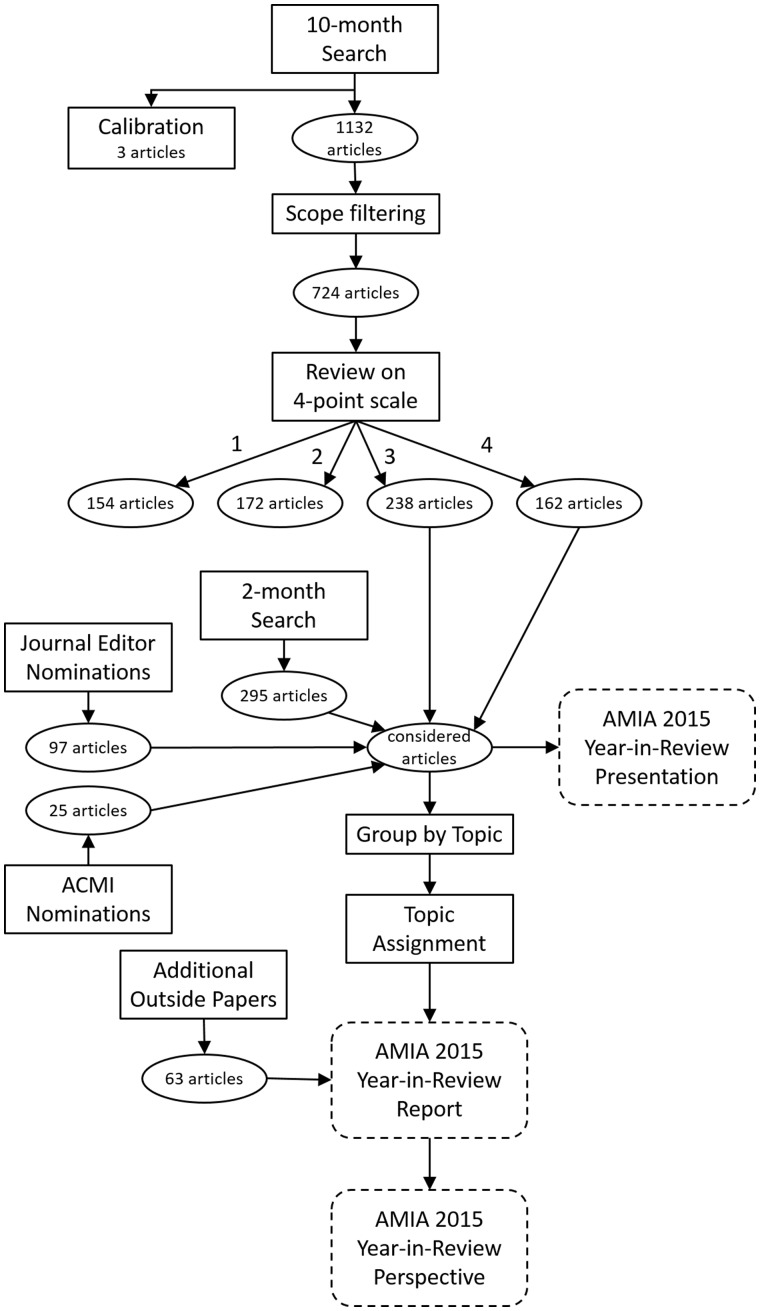
With the assistance of a health science librarian, a multistage review process of the published literature was conducted (see Figure 1). The scope for the search was English-language articles on topics in clinical or consumer informatics appearing online or in print between November 1, 2014, and October 31, 2015, in refereed journals indexed in PubMed, EBSCO, and Web of Science. The 3 search engines (see Table 2) returned an initial total of 1132 unique articles (after removal of 222 duplicates). This list was further filtered to eliminate articles that were, based on their citation information, clearly outside the scope of interest. This yielded 724 articles for review. The goal of this process was not to conduct a strictly thorough systematic review, but to gather a broad base of the literature. As such, important papers may have been missed, but other parts of the review process likely would have brought these back into consideration.
Figure 1.
Year-in-review process.
Table 2.
Search criteria
| PubMed |
| decision support systems [tiab] OR EHR [tiab] OR electronic health record*[tiab] OR M-health [tiab] OR consumer informatics [tiab] OR public health informatics [tiab] OR BD2K [tiab] OR PCORnet [tiab] OR precision medicine [tiab] OR meaningful use [tiab] OR CPCP [tiab] OR patient reported outcomes [tiab] OR clinical informatics [tiab] OR medical informatics [tiab] OR public health informatics [tiab] OR big data initiative* [tiab] OR health information exchange [tiab] OR telemedicine [tiab] OR evaluation of EHR [tiab] OR consumer health informatics [tiab] OR precision medicine [tiab] OR health informatics [tiab] |
| EBSCO |
| decision support systems [tia] OR EHR [tia] OR electronic health record*[tia] OR M-health [tia] OR consumer informatics [tia] OR public health informatics [tia] OR BD2K [tia] OR PCORnet [tia] OR precision medicine [tia] OR meaningful use [tia] OR CPCP [tia] OR patient reported outcomes [tia] OR clinical informatics [tia] OR medical informatics [tia] OR public health informatics [tia] OR big data initiative* [tia] OR health information exchange [tia] OR telemedicine [tia] OR evaluation of EHR [tia] OR consumer health informatics [tia] OR precision medicine [tia] OR health informatics [tia] |
| Web of Science |
| ((((((((((((((((((((“decision support systems” OR EHR) OR “electronic health record*”) OR “M-health”) OR “consumer informatics”) OR “public health informatics”) OR BD2K) OR cornet) OR “precision medicine”) OR “meaningful use”) OR CPCP) OR “patient reported outcomes”) OR “clinical informatics”) OR “medical informatics”) OR “public health informatics”) OR “big data initiative*”) OR “health information exchange”) OR telemedicine) OR “evaluation of EHR”) OR “consumer health informatics”) OR “health informatics”) |
All reviewers participated in a small calibration process with 3 papers to improve consistency on a 4-point scale (must include, maybe include, possibly include, and do not include). To be considered “must include,” an article had to provide a significant advance or novel application according to the reviewer’s (admittedly subjective) opinion. A survey system (Qualtrics™) was employed to distribute the initial 724 citations, collect assessments, and summarize results. Participants used the citation to obtain the full text. Papers scoring 3 (possibly include) or 4 (must include) by at least 2 reviewers were considered for inclusion in the presentation or report. The primary focus was on original informatics investigations, complemented by published review papers and policy perspectives according to the judgment of the team.
Additional papers were included for consideration using other mechanisms: (1) a second search of the most recent papers (the final 2 months) was performed using the same search strategy; (2) 11 editors from well-known biomedical informatics journals were solicited to provide a “top 10” list for their respective journals; (3) American College of Medical Informatics members responded to a solicitation for top papers; and (4) members of the review team recommended papers. For the report, all papers included for consideration were organized according to the topics in Table 1 and re-reviewed by the respective section editor. Upon second review, the section editor might choose to drop the paper.
FINDINGS
A hallmark of a maturing field is having a complementary relationship between contributions to related fields while strengthening its core knowledge base. This year’s AMIA Year-in-Review of Clinical and Consumer Informatics reflects just that – growth in the application of informatics tools to new areas, consolidation of the base learning, and, importantly, revelation of areas yet to grow.
We organize this report in terms of contributions to 3 major policy objectives in informatics: (1) adopting, implementing, and improving use of electronic health records; (2) advancing the goals of the learning health system; and (3) engaging consumers in health and health care decision-making using information technology. Full details, including specific citations addressing the methods, tasks, and trends in biomedical informatics, are provided in the companion report, while a small number of citations are included here (when they directly support highlighted items).
Adopting, implementing, and improving use of electronic health records
With the Meaningful Use regulations shifting focus from adoption of electronic health records (EHRs) (which was widely seen as successful) to advancing specific EHR functions (which is more controversial), the informatics research community has shifted focus to how EHRs can improve patient care and outcomes. Thus the focus of papers this year is EHR implementation, not adoption; the results thus far have been mixed at best. The AMIA EHR-2020 Task Force4 issued a set of policy recommendations on how to improve the EHR. The state of the literature largely confirms the importance of these recommendations.
EHRs have an interesting impact on clinical documentation. Burke et al.5 found that EHRs improved clinical note quality for 12 elements (eg, chief complaint, problem list, family history) in outpatient visit notes. This should be contrasted with the EHR-2020 Task Force’s concern about documentation burden and bloat (eg, from copy/paste). This suggests the trade-offs related to increased documentation should be more fully explored, and it remains important to address the various contributors to EHR note quality, including provider behaviors, technical features, and local culture.
EHRs are very good at improving additional specific elements of clinical processes, such as decision support6 and risk prediction.7 EHRs can increase rates of needed counseling8 and even reduce medical malpractice.9 In spite of specific improvements, there remains some burden arising from documentation time and mixed effects on patient interaction.10 To date, the lack of improvement in outcomes may not justify the risks: not only financial costs and security concerns, but lack of data availability during downtimes.11 To improve outcomes, we must reconsider what aspects of the clinical process really do affect patient health as opposed to what aspects are simple to automate.
It seems clear that instead of altering clinical workflows to gather process information, the focus should be on utilizing available technology and advancing research to enable seamless data capture. An important component for this is natural language processing (NLP), which the EHR-2020 Task Force advocates as a mechanism to perform structured data capture and improve data entry. Clinical NLP research continues to focus on general-purpose processing (eg, extracting and mapping concepts), secondary-use applications such as the i2b2 challenge on identifying risk factors for heart disease,12 mining social media data for adverse drug reactions,13 and supporting systematic reviews.14 However, few advances have been made in the dynamic use of NLP in clinical care processes to improve data capture and decision support while reducing burden. On the other hand, computerized innovations should be permitted to alter clinical workflow when doing so would improve health care. For example, data visualization holds great potential to improve clinical cognition about patients, but workflows will have be changed to ensure that visualizations are integrated into practice. Visualizations have the potential to provide more intuitive data representations, saving time while reducing cognitive load. There is an increasing number of such visualization tools, for both clinicians and patients.15,16
Beyond clinical use of EHRs, an important recent trend is patient access to health data. Initiatives such as OpenNotes and Blue Button are enabling patients to access data17–19 that they have long had a legal right to obtain. Roadblocks, including patients’ ability to understand their own data and the impact on clinical practice workflows,17,20 remain and represent emerging areas of informatics research.
Patient control of data is another emerging area, including access control21,22 and even the ability to add data to the EHR from sources such as wearable technology and mobile health (mHealth) apps.4 While many view the integration of patient-generated data into the EHR with skepticism, it has the potential to provide a patient’s social, environmental, and functional contexts. As we enter an era of precision medicine, these contexts should provide invaluable information sources that are well understood to have important health implications.
The information infrastructure for the learning health system
The concept of the learning health system (LHS)23 influences much of the research performed in biomedical informatics. From the long understood need to standardize data, to large-scale analysis (“big data”), to applications that analyze evidence at ever-finer levels (eg, precision medicine), research reports are beginning to address the informatics challenges inherent in meeting LHS goals.
As access to clinical data increases, research demonstrating methods for extracting knowledge from clinical data keeps pace. A core feature of much of this research is the ability to automatically identify a cohort of patients according to some predefined phenotype. Frequently, artificial intelligence methods are utilized, such as NLP, information retrieval, and machine learning.12,24–26 Such methods have been used for clinical trial eligibility assessment,27 readmission reduction,28,29 and quality assessment.7
Prediction models are also popular. These include: (1) disease risk stratification for baseline risk prediction, disease progression, or mortality;30–34 (2) clinical effectiveness research, such as treatment for patients at risk for a side effect;35–37 and (3) hospital admission prediction, whether for readmission29,38 or estimating no-shows.39 There have even been several visualization methods to aid in this type of analysis that may help spot trends that are difficult to define before they arise.40
There are several barriers, however, that can limit the utility of clinical data in the LHS. Since this is observational data, new forms of bias are constantly being studied, such as biases toward sicker patients41 and observation frequency.42 Many confounding factors have been identified, including social risks affecting outcomes,43,44 geographic effects on rehabilitation,35 and even how birth month impacts lifetime disease risk.45 Finally, clinical data is often siloed, limiting the ability to study rare diseases and geographic effects. Several initiatives, some general purpose and some condition-specific, are under way to enable inter-institution analysis.46–48
The movement toward the LHS is thus very promising, if slower than ideal. One trend toward speeding up the LHS cycle is the use of social media data, especially for pharmacovigilance. Using NLP, adverse effects can be detected long before they appear in traditional clinical sources.13,49,50 Possibly what we need most, however, are approaches to close the LHS loop. Given the constant stream of new factors that affect the diagnosis and treatment of disease, it is clear that every patient truly is unique. General-purpose tools to find “patients like mine” and compare their characteristics, treatments, and outcomes would allow doctors to decide which factors are important on a case-by-case basis. Turning physicians into statisticians and data scientists is not without its perils, but this would create an organic LHS.
Engaging patients in health and health care through information technology
Information technologies, particularly mHealth and telemedicine, hold promise for engaging patients outside the clinic, especially in improving patient-provider communication for patients with chronic diseases. Several papers this year demonstrate how many of the barriers to patient engagement with health information technology can be overcome, including incorporating foundational informatics features such as data standards or decision support.
Patients consider self-tracking to be work,51 the solution to which is not only more usable apps, but involving the patient’s social network (which itself is quite complex).52 Research has also shown that when providers utilize self-tracking data, patient engagement improves.53 But researchers also found that providers generally lack the time and expertise to review this data,53 suggesting that new care models are needed to enable providers to better engage patients with chronic diseases. One compromise solution is telemedicine, which can improve engagement while staying close to traditional patient-provider communication. Telemedicine, however, faces several legal and regulatory (as opposed to technical) barriers. To help overcome these, new frameworks have been proposed, including one based on the Meaningful Use regulations.54 These all suggest that informatics solutions to patient engagement must consider the wider technological, social, economic, and political contexts.
CONCLUSION
We have described high-level advances and trends in biomedical informatics discovered during our review process across 3 broad areas: electronic health records, the learning health system, and consumer/patient engagement. These advances and trends indicate that informatics is a growing and thriving field, integrating advances in artificial intelligence and consumer technology while helping to advance medical understanding and improve health.
ACKNOWLEDGMENTS
The authors would like to thank Stephanie Kaitlyn Hendren and Mary Hitchcock, librarians at the Ebling Health Science Library, University of Wisconsin-Madison, as well as the additional members of the review group: Dean F Gawum, Regina Wysocki, Jacqueline Brixey, Brittany Partridge, Edward Krause, Stacey Slager, and Zhe He. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the US government.
FUNDING
K.R. was supported by US National Library of Medicine (NLM) grant 1K99LM012104, as well as the intramural research program at NLM. M.R.B. is supported by NLM training grant T15LM00707 at Columbia University. U.B. is supported in part by NLM training grant T15LM007442 at the University of Washington. K.H.Y. is a Winston Chen Stanford Graduate Fellow and Howard Hughes Medical Institute International Student Research Fellow.
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
K.R. and P.F.B. drafted the original manuscript. K.R., M.R.B., L.P., J.D., A.B., M.G., R.H., R.F.S., U.B., K.H.Y., and Y.J. all contributed sections to the supplemental report and suggested important items to include in the perspective. All authors read and approved the manuscript.
REFERENCES
- 1. Masys D. AMIA Informatics Years in Review. http://faculty.washington.edu/dmasys/YearInReview/. Accessed 13 July 2016. [Google Scholar]
- 2. Hersh W, Ash J. AMIA Annual Symposium 2014 Year in Review. http://skynet.ohsu.edu/∼hersh/yearinreview2014.html. Accessed 13 July 2016. [Google Scholar]
- 3. IMIA. Yearb Med Inform. 2015;(1):1–233. [Google Scholar]
- 4. Payne TH, Corley S, Cullen TA, et al. Report of the AMIA EHR 2020 Task Force on the Status and Future Direction of EHRs. J Am Med Inform Assoc. 2015;22(5):1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burke HB, Sessums LL, Hoang A, et al. Electronic health records improve clinical note quality. J Am Med Inform Assoc. 2015;22(1):199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ben-Assuli O, Shabtai I, Leshno M. Using electronic health record systems to optimize admission decisions: the Creatinine case study. Health Inform J. 2015;21(1):73–88. [DOI] [PubMed] [Google Scholar]
- 7. Anderson JE, Chang DC. Using electronic health records for surgical quality improvement in the era of big data. JAMA Surgery. 2015;150(1):24–29. [DOI] [PubMed] [Google Scholar]
- 8. Sharifi M, Adams WG, Winickoff JP, Guo J, Reid M, Boynton-Jarrett R. Enhancing the electronic health record to increase counseling and quit-line referral for parents who smoke. Acad Pediatrics. 2014;14(5):478–484. [DOI] [PubMed] [Google Scholar]
- 9. Wright A, Maloney FL, Wien M, et al. Assessing information system readiness for mitigating malpractice risk through simulation: results of a multi-site study. J Am Med Inform Assoc. 2015;22(5):1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carayon P, Wetterneck TB, Alyousef B, et al. Impact of electronic health record technology on the work and workflow of physicians in the intensive care unit. Int J Med Inform. 2015;84(8):578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sittig DF, Gonzalez D, Singh H. Contingency planning for electronic health record–based care continuity: a survey of recommended practices. Int J Med Inform. 2014;83(11):797–804. [DOI] [PubMed] [Google Scholar]
- 12. Stubbs A, Kotfila C, Xu H, Uzuner O. Identifying risk factors for heart disease over time: overview of 2014 i2b2/uthealth shared task track 2. J Biomed Inform. 2015;58Supplement:S67–S77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015;22(3):671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marshall IJ, Kuiper J, Wallace BC. RobotReviewer: evaluation of a system for automatically assessing bias in clinical trials. J Am Med Inform Assoc. 2015;23(1):193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kopanitsa G, Veseli H, Yampolsky V. Development, implementation and evaluation of an information model for archetype based user responsive medical data visualization. J Biomed Inform. 2015;55:196–205. [DOI] [PubMed] [Google Scholar]
- 16. Warner JL, Denny JC, Kreda DA, Alterovitz G. Seeing the forest through the trees: uncovering phenomic complexity through interactive network visualization. J Am Med Inform Assoc. 2015;22(2):324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker J, Meltsner M, Delbanco T. US experience with doctors and patients sharing clinical notes. BMJ,. 2015;350:g7785. [DOI] [PubMed] [Google Scholar]
- 18. Nazi KM, Turvey CL, Klein DM, Hogan TP, Woods SS. VA OpenNotes: exploring the experiences of early patient adopters with access to clinical notes. J Am Med Inform Assoc. 2015;22(2):308–389. [DOI] [PubMed] [Google Scholar]
- 19. Bell SK, Folcarelli PH, Anselmo MK, Crotty BH, Flier LA, Walker J. Connecting patients and clinicians: the anticipated effects of open notes on patient safety and quality of care. Jt Comm J Qual Patient Saf. 2015;41(8):378–384. [DOI] [PubMed] [Google Scholar]
- 20. Pell JM, Mancuso M, Limon S, Oman K, Lin CT. Patient access to electronic health records during hospitalization. JAMA Intern Med. 2015;175(5):856–858. [DOI] [PubMed] [Google Scholar]
- 21. Caine K, Kohn S, Lawrence C, Hanania R, Meslin EM, Tierney WM. Designing a patient-centered user interface for access decisions about EHR data: implications from patient interviews. J Gen Intern Med. 2015;30:S7–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leventhal JC, Cummins JA, Schwartz PH, Martin DK, Tierney WM. Designing a system for patients controlling providers’ access to their electronic health records: organizational and technical challenges. J Gen Intern Med. 2015;30:S17–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Institute of Medicine. Best Care at Lower Cost: The Path to Continuously Learning Health Care in America. Washington, DC: The National Academies Press; 2013. [PubMed] [Google Scholar]
- 24. Hanauer DA, Mei Q, Law J, Khanna R, Zheng K. Supporting information retrieval from electronic health records: a report of University of Michigan’s nine-year experience in developing and using the Electronic Medical Record Search Engine (EMERSE). J Biomed Inform. 2014;55:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu S, Liao KP, Shaw ST, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015;22:933–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Joffe E, Pettigrew EJ, Herskovic JR, Bearden CF, Bernstam EV. Expert guided natural language processing using one-class classification. J Am Med Inform Assoc. 2015;22:962–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miotto R, Weng C. Case-based reasoning using electronic health records efficiently identifies eligible patients for clinical trials. J Am Med Inform Assoc. 2015;22(e1):e141–e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hao S, Wang Y, Jin B, et al. Development, Validation and Deployment of a Real Time 30 Day Hospital Readmission Risk Assessment Tool in the Maine Healthcare Information Exchange. PLoS ONE. 2015;10(10):e0140271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shadmi E, Flaks-Manov N, Hoshen M, Goldman O, Bitterman H, Balicer RD. Predicting 30-day readmissions with preadmission electronic health record data. Medical Care. 2015;53(3):283–289. [DOI] [PubMed] [Google Scholar]
- 30. Perotte A, Ranganath R, Hirsch JS, Blei D, Noemie E. Risk prediction for chronic kidney disease progression using heterogeneous electronic health record data and time series analysis. J Am Med Inform Assoc. 2015;22(4):872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh A, Nadkarni G, Gottesman O, Ellis SB, Bottinger EP, Guttag JV. Incorporating temporal EHR data in predictive models for risk stratification of renal function deterioration. J Biomed Inform. 2015;53:220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Z, Dong W, Duan H. A probabilistic topic model for clinical risk stratification from electronic health records. J Biomed Inform. 2015;58:28–36. [DOI] [PubMed] [Google Scholar]
- 33. Miller GW, Mugler JP, 3rd, Sá RC, Altes TA, Prisk GK, Hopkins SR. Advances in functional and structural imaging of the human lung using proton MRI. NMR Biomed. 2014;27(12):1542–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J, Maslove DM, Dubin JA. Personalized mortality prediction driven by electronic medical data and a patient similarity metric. PLoS ONE. 2015;10(5):e0127428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reistetter TA, Kuo YF, Karmarkar AM, et al. Geographic and facility variation in inpatient stroke rehabilitation: multilevel analysis of functional status. Arch Phys Med Rehabil. 2015;96(7):1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pell JM, Cheung D, Jones MA, Cumbler E. Don’t fuel the fire: decreasing intravenous haloperidol use in high risk patients via a customized electronic alert. J Am Med Inform Assoc. 2014;21(6):1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kirkendall ES, Spires WL, Mottes TA, Schaffzin JK, Barclay C, Goldstein SL. Development and performance of electronic acute kidney injury triggers to identify pediatric patients at risk for nephrotoxic medication-associated harm. Appl Clin Inform. 2014;5(2):13–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olson CH, Dierich M, Adam T, Westra BL. Optimization of decision support tool using medication regimens to assess rehospitalization risks. Appl Clin Inform. 2014;5(3):773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Y, Hanauer DA. Patient no-show predictive model development using multiple data sources for an effective overbooking approach. Appl Clin Inform. 2014;5(3):836–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. West VL, Borland D, Hammond WE. Innovative information visualization of electronic health record data: a systematic review. J Am Med Inform Assoc. 2015;22(2):330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rusanov A, Weiskopf NG, Wang S, Weng C. Hidden in plain sight: bias towards sick patients when sampling patients with sufficient electronic health record data for research. BMC Med Inform Decis Mak. 2014;14(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hripcsak G, Albers DJ, Perotte A. Parameterizing time in electronic health record studies. J Am Med Inform Assoc. 2015;22(4):794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hripcsak G, Forrest CB, Brennan PF, Stead WW. Informatics to support the IOM Social and Behavioral Domains and Measures. J Am Med Inform Assoc. 2015;22(4):921–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie ZH, He Z. The study on the development of decision support systems in response to catastrophic social risks. Proceedings of the 3rd International Conference on Computer Science and Service System 2014;109:461–464. [Google Scholar]
- 45. Boland MR, Shahn Z, Madigan D, Hripcsak G, Tatonetti NP. Birth month affects lifetime disease risk: a phenome-wide method. J Am Med Inform Assoc. 2015;22(5):1042–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deserno TM, Haak D, Brandenburg V, Deserno V, Classen C, Specht P. Integrated image data and medical record management for rare disease registries. A general framework and its instantiation to the German Calciphylaxis Registry. J Digit Imaging. 2014;27(6):702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walji MF, Kalenderian E, Stark PC, et al. BigMouth: a multi-institutional dental data repository. J Am Med Inform Assoc. 2014;21(6):1136–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roelofs E, Dekker A, Meldolesi E, et al. International data-sharing for radiotherapy research: an open-source based infrastructure for multicentric clinical data mining. Radiotherapy Oncol. 2014;110(2):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adrover C, Bodnar T, Huang Z, Telenti A, Salathé M. Identifying adverse effects of HIV drug treatment and associated sentiments using Twitter. JMIR Public Health Surveill. 2015;1(2):e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sarker A, Ginn R, Nikfarjam A, et al. Utilizing social media data for pharmacovigilance: a review. J Biomed Inform. 2015;54:202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ancker JS, Witteman HO, Hafeez B, Provencher T, Van de Graaf M, Wei E. “You Get Reminded You’re a Sick Person”: personal data tracking and patients with multiple chronic conditions. J Med Internet Res. 2015;17(8):e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Valdez RS, Brennan PF. Exploring patients’ health information communication practices with social network members as a foundation for consumer health IT design. Int J Med Inform. 2015;84(5):363–374. [DOI] [PubMed] [Google Scholar]
- 53. Chung CF, Cook J, Bales E, Zia J, Munson SA. More than telemonitoring: health provider use and nonuse of life-log data in irritable bowel syndrome and weight management. J Med Internet Res. 2015;17(8):e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vo A, Shore J, Waugh M, et al. Meaningful use: a national framework for integrated telemedicine. Telemed J E Health. 2015;21(5):355–363. [DOI] [PubMed] [Google Scholar]