Abstract
Objective: Phenotyping algorithms are capable of accurately identifying patients with specific phenotypes from within electronic medical records systems. However, developing phenotyping algorithms in a scalable way remains a challenge due to the extensive human resources required. This paper introduces a high-throughput unsupervised feature selection method, which improves the robustness and scalability of electronic medical record phenotyping without compromising its accuracy.
Methods: The proposed Surrogate-Assisted Feature Extraction (SAFE) method selects candidate features from a pool of comprehensive medical concepts found in publicly available knowledge sources. The target phenotype’s International Classification of Diseases, Ninth Revision and natural language processing counts, acting as noisy surrogates to the gold-standard labels, are used to create silver-standard labels. Candidate features highly predictive of the silver-standard labels are selected as the final features.
Results: Algorithms were trained to identify patients with coronary artery disease, rheumatoid arthritis, Crohn’s disease, and ulcerative colitis using various numbers of labels to compare the performance of features selected by SAFE, a previously published automated feature extraction for phenotyping procedure, and domain experts. The out-of-sample area under the receiver operating characteristic curve and F-score from SAFE algorithms were remarkably higher than those from the other two, especially at small label sizes.
Conclusion: SAFE advances high-throughput phenotyping methods by automatically selecting a succinct set of informative features for algorithm training, which in turn reduces overfitting and the needed number of gold-standard labels. SAFE also potentially identifies important features missed by automated feature extraction for phenotyping or experts.
Keywords: electronic medical records, phenotyping, data mining, machine learning
INTRODUCTION
Biorepository-linked electronic medical records (EMR) cohorts have become a valuable resource for research. These “virtual cohorts” have been utilized in a broad range of biomedical research, including genetic association studies as well as studies of comparative effectiveness and risk stratification.1–16 Performing comprehensive and statistically powerful studies of EMR cohorts involving multiple phenotypes is challenging, due to the difficulty in efficiently characterizing accurate phenotypes with EMR. Assigning phenotypes based on International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9) codes often results in misclassification and hampers the power of hypothesis tests in subsequent genomic or biomarker studies.17–20 Rules that combine ICD-9 codes, medication prescriptions, and laboratory and procedure codes can improve the accuracy of phenotyping,21,22 though designing these rules relies heavily on expert participation. Data-driven, machine learning–based phenotyping algorithms have become popular in recent years, in part due to their accuracy and portability and the promise of potential automation.23–31
Yet bottlenecks still exist that limit the ability to perform high-throughput phenotyping. To create a phenotyping algorithm, 2 important and rate-limiting steps are typically involved: collecting informative features that strongly characterize the phenotype, and developing a classification algorithm based on these features with a gold-standard training set. The most commonly used features include counts of a patient’s ICD-9 billing codes, codes of diagnostic and therapeutic procedures, medication prescriptions, and lab codes/values. In addition to the codified data, features can also be derived from the patient’s clinical narrative notes via natural language processing (NLP). One example of NLP features is the frequency of various medical concepts mentioned in each patient’s notes. Existing literature has proposed to use all possible unigrams, bigrams, and identified concepts of the Unified Medical Language System (UMLS)32 and other terminologies as candidate features for training with gold-standard labels.33–35 While such approaches avoid the need for feature curation and thus facilitate automation, they yield algorithms with poor out-of-sample classification accuracy due to overfitting induced by the huge number of irrelevant features.36 Less overfitted algorithms with higher generalizability can be built based on informative features manually curated by leveraging the knowledge of domain experts. However, such an approach is typically time-consuming and not ideal for the development of large biorepositories with many phenotypes. Automated feature extraction for phenotyping (AFEP)37 advances the capability for high-throughput phenotyping by curating features from 2 publicly available knowledge sources, Wikipedia and Medscape. AFEP subsequently selects informative features by only including features whose non-zero frequency and univariate rank correlation with the NLP count of the main concept exceed certain threshold values. Algorithms trained using features selected via AFEP and curated by experts were shown to achieve similar accuracies.
The other important step (and bottleneck) in developing a phenotyping algorithm is creating gold-standard labels by domain experts via chart review. Labeling is both labor-intensive and time-consuming. The training sample sizes in previous studies typically ranged from 400 to 600,23–25,29,31,34,35,38,39 and have occasionally reached several thousand.28,33 Reviewing records for 100–200 gold-standard labels would increase the feasibility of developing algorithms for multiple phenotypes over the course of a year.
The 2 bottlenecks described above signify the need for automated feature selection methods that can choose a small set of informative features. Ideally, selection could be done without using any gold-standard labels to reduce overfitting, and only 100–200 gold-standard labels would be needed to train a generalizable algorithm with the selected features. The objective of this study is to develop such automated feature selection methods for high-throughput phenotyping through the use of easily available but noisy surrogates.
METHODS
The workflow of the proposed Surrogate-Assisted Feature Extraction (SAFE) procedure is illustrated in Figure 1.
Figure 1.
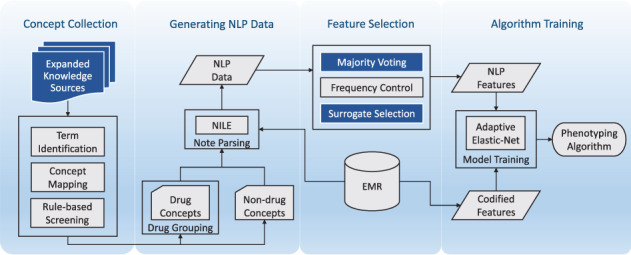
Flow chart of SAFE. Improvements over AFEP include expanded knowledge sources, majority voting selection, and surrogate-assisted selection.
The SAFE procedure
Concept collection
To form a set of candidate features, medical concepts are extracted using named entity recognition from a small number of topical articles from 5 publicly available knowledge sources: Wikipedia, Medscape, Merck Manuals Professional Edition, Mayo Clinic Diseases and Conditions, and MedlinePlus Medical Encyclopedia. URLs of the articles and detailed descriptions on the named entity recognition process are provided in sections 1–3 of the Supplementary Material. These 5 sources typically yield around 1000 UMLS concepts as candidate features for each phenotype.
Generating NLP data
We process the EMR clinical narratives with NLP (more details in section 4 of the Supplementary Material) to search for mentions of the candidate concepts extracted from the knowledge sources, using their UMLS terms as the dictionary. The patient-level counts of the concepts form a working dataset for feature selection. We only consider positive mentions that confirm presence of a condition, performance of a procedure, use of a medication, etc. Negated assertions, family history, and conditional problems such as drug allergies are not counted.
Feature selection
From the NLP output, we identify the concepts that appear in at least 3 of the 5 knowledge sources, a majority voting as a prescreening for importance. A second prescreen uses the frequency of the feature in the narrative notes. The concept must be mentioned in at least 5% of the notes where the target phenotype is mentioned, and it must be mentioned in the notes of no more than 50% of all patients. The ICD-9 and NLP counts of the target phenotype (referred to as the main ICD-9 count and the main NLP count hereafter), together with the other features that pass the voting and frequency control, denoted by , will serve as candidate features in the subsequent feature selection step. Here, the main ICD-9 count includes codes of all the subtypes of the phenotype, and the main NLP count corresponds to the UMLS concept of the phenotype.
The key idea behind the surrogate-assisted feature selection is that one can use the main ICD-9 and main NLP counts to create “silver-standard” labels, S, which can be viewed as a bespoke “probability” of having the phenotype. When S relates to a set of features F only through Y, it is statistically plausible to infer the predictiveness of F for Y based on the predictiveness of F for S. Since the main NLP and ICD-9 counts are the most predictive features for most phenotypes studied thus far,23–26 we use both of these variables to construct Ss. While it is known that the main ICD-9 and NLP counts by themselves are noisy surrogates of Y, they can accurately classify the phenotype status for a subset of patients who are “textbook cases” or clearly do not exhibit that phenotype. Specifically, patients with very high main ICD-9 or NLP counts generally have the phenotype, while patients with extremely low counts are unlikely to have the phenotype. That is, Y can be inferred accurately from the main NLP or ICD-9 counts within these extreme subsets.
To proceed, we consider 3 choices of S:
where , , , and are lower and upper thresholds that can be determined via domain knowledge or percentiles of the observed data. For each with , we define extreme subsets as patients whose take the value 0 or 1. We randomly sample patients from those with , and from those with , to form a working dataset. Then we fit an adaptive elastic-net40,41 penalized logistic regression model to the working dataset, with being the response variable and transformed excluding the variables involved in being the predictors. Thus, when is used as the response, the main ICD-9 count is not included as a feature; when is used as the response, the main NLP count is not included; and when is used as the response, neither the main ICD-9 nor the main NLP counts are included. The penalized logistic regression penalizes model complexity (ie, the number of features included in the model) and shrinks the coefficients of uninformative features to zero. The tuning parameters controlling the amount of penalty for model complexity are selected based on the Bayesian information criterion.36 To determine which of the features are uninformative in the presence of uncertainty and randomness of the data, we repeatedly sample patients from the extreme subsets and train the models many times for each . A feature is selected only if its coefficient is not zero at least 50% of the time, averaged over the above repeated fittings and 3 choices of . These selected features, along with the main ICD-9 and NLP counts, the total number of notes, and the age and gender of the patient, denoted by , are included as features for the algorithm training with gold-standard labels.
Training phenotyping algorithm with gold standard labels
The final algorithm is then trained by fitting an adaptive elastic-net penalized logistic regression, with gold-standard labels being the response and being the predictors. All count variables are again transformed by. The tuning parameter is selected via the Bayesian information criterion.
Data and metrics for evaluation
We applied various feature selection methods to generate phenotyping algorithms for coronary artery disease (CAD), rheumatoid arthritis (RA), Crohn’s disease (CD), and ulcerative colitis (UC). We utilized 2 Partners HealthCare EMR datamarts, 1 for RA and 1 for inflammatory bowel diseases (IBDs), originally used to create machine learning algorithms for classifying these 4 phenotypes.23,24,29 The RA datamart, created in June 2010, includes 46 568 patients who had at least 1 ICD-9 code of 714.x (Rheumatoid arthritis and other inflammatory polyarthropathies) or had been tested for anticyclic citrullinated peptide, a diagnostic marker for RA. The IBD datamart, created in November 2010, included 34 033 patients who had at least 1 ICD-9 code of 555.x (Regional enteritis) or 556.x (Ulcerative enterocolitis). Gold-standard labels were available for 435 patients randomly selected from the RA datamart. Among the 4446 patients predicted to have RA by Liao et al.,23 758 patients who had at least 1 ICD-9 code or a free-text mention of CAD were reviewed to create a training set for CAD. For IBD, UC labels were obtained for 600 patients selected randomly from those with at least 1 ICD-9 code of UC, and CD labels were obtained for 600 patients selected from those with at least 1 ICD-9 code for CD. In the original studies, features for these algorithms were manually curated via multiple iterations between clinical domain and NLP experts. The prevalence of CAD, RA, CD, and UC was estimated as 40.1%, 22.5%, 67.5%, and 63.0%, respectively.
Since one main goal of automated feature selection is to reduce the number of labels needed for creating phenotype algorithms with low generalization errors, we trained algorithms based on features selected via different approaches with n = 100, 150, 200, 250, and 300 labels. For each n, we randomly selected n labeled samples for algorithm training and used the remaining labels to assess the out-of-sample accuracy, quantified by the area under the receiver operating characteristic curve (AUC) and the F-score at the 95% specificity level.42 To obtain stable estimates, we repeatedly sampled the labeled data randomly 200 times, and the results reported are based on the average.
To evaluate SAFE and further understand the effects of the various building blocks of the procedure (ie, the 2 main features of ICD-9 and NLP counts, concept collection from the original 2 and expanded 5 knowledge sources, the majority voting, and the surrogate-assisted feature selection), we trained multiple algorithms using features selected from the following procedures: expert curation; M2, the 2 main features; AFEP; A5, expanded 5 sources + AFEP selection; A5V, expanded 5 sources + majority voting + AFEP selection; S2, original 2 sources + surrogate-assisted selection, and SAFE. For S2 and SAFE, the thresholds and were set to 0, while and were set to 10, and was set to 400.
RESULTS
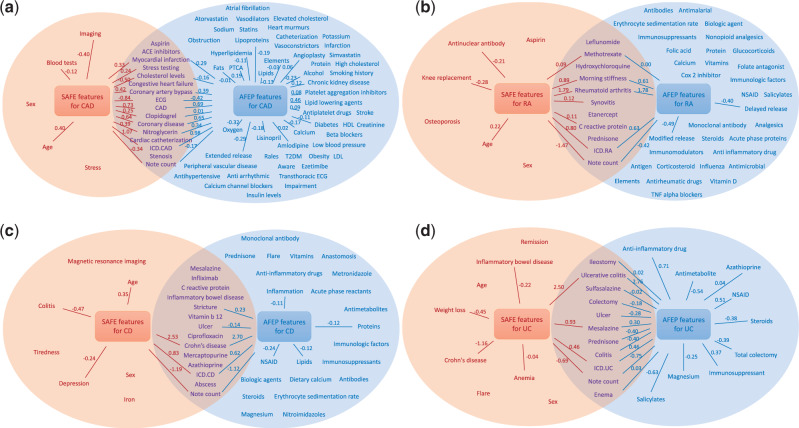
The 5 source articles were accessed on April 28, 2015, for RA, on May 20, 2015, for CD and UC, and on April 4, 2016, for CAD. Table 1 shows the numbers of concepts/features selected from each procedure. SAFE generally selects fewer features than AFEP and domain experts. Figure 2 compares the selected features from SAFE and AFEP and their coefficients in the fitted models. While the features selected by AFEP and SAFE had overlaps, SAFE tended to select more clinically meaningful features, with the majority of noninformative features removed during the feature selection process. Section 5 of the Supplementary Material gives a comparison of the SAFE and expert-curated features.
Table 1.
Comparison of feature numbers across the methods
| Phenotype |
||||
|---|---|---|---|---|
| CAD | RA | CD | UC | |
| Number of concepts extracted from source articles | 805 | 1067 | 1057 | 700 |
| Number of expert-curated features (after frequency control) | 36 | 23 | 49 | 50 |
| Number of features from AFEP | 68 | 42 | 35 | 20 |
| Number of features from A5 | 75 | 43 | 37 | 23 |
| Number of features from A5V | 30 | 22 | 23 | 15 |
| Number of features from S2 | 19 | 16 | 10 | 16 |
| Number of features from SAFE | 21 | 17 | 18 | 19 |
Numbers in bold are the numbers of features used for the final training with the gold-standard labels
Figure 2.
Comparison of features selected by SAFE and AFEP for (A) CAD, (B) RA, (C) CD, and (D) UC. Left and right circles include features from SAFE and AFEP methods, respectively. Edges indicate features with non-zero beta coefficients in the final SAFE- or AFEP-based algorithms, trained with the entire training set, where coefficients are shown as weights.
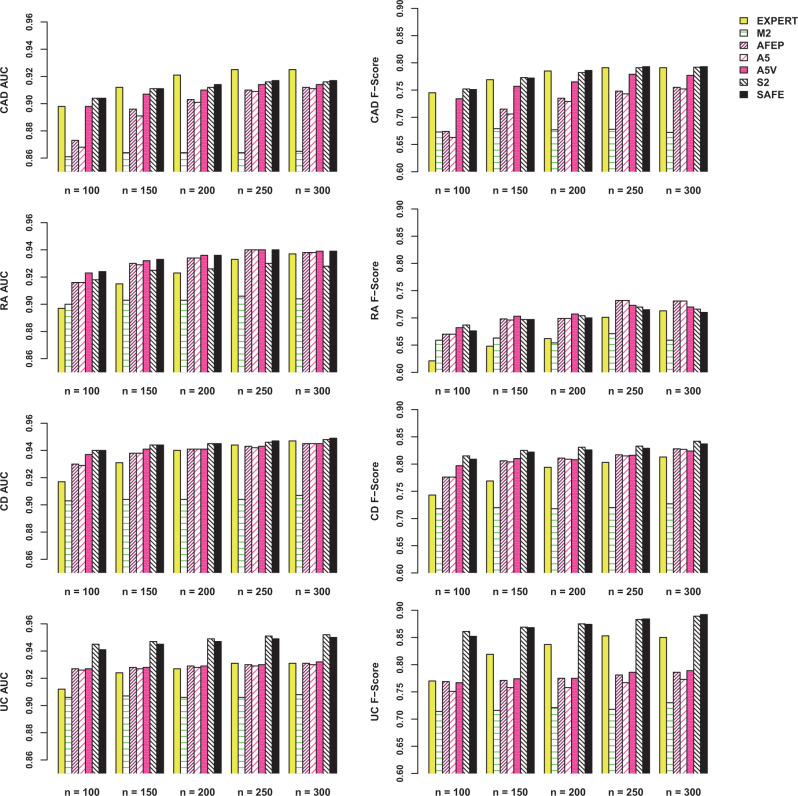
Figure 3 shows the out-of-sample AUC and the F-scores for the algorithms trained using various features with n = 100, 150, 200, 250, and 300 labels (a tabular presentation is shown in section 6 of the Supplementary Material). The algorithms trained with features selected by SAFE outperformed those using features selected by AFEP or experts by varying degrees, and the improvements are most significant when n is small. The out-of-sample performance of SAFE-based algorithms tended to stabilize with 150 labels, while 200–300 labels were typically required for algorithms based on AFEP to achieve similar performance. For CAD, the algorithm using expert-curated features attained a slightly higher AUC than the SAFE-based algorithm for larger n, but their F-scores are nearly identical. This is due in part to the inclusion of a feature covering several CAD-specific procedures manually created by domain experts, while automated procedures such as SAFE and AFEP do not have such a feature. For UC, SAFE resulted in consistently more accurate algorithms compared to those based on AFEP or expert-curated features. The algorithms trained using all gold-standard labels are shown in section 7 of the Supplementary Material.
Figure 3.
AUC and F-scores (when specificity = 0.95) of algorithms trained with n gold-standard labels, using features selected by EXPERT, expert-curated features; M2, the main ICD and NLP features only; AFEP, the original AFEP procedure; A5, expanded 5 sources + AFEP selection (frequency control + rank correlation selection); A5V, expanded 5 sources + majority voting + AFEP selection; S2, original 2 sources + surrogate-assisted selection; and SAFE, the proposed procedure with 5 sources and majority voting, plus surrogate-assisted selection.
DISCUSSION
SAFE is built upon the framework of AFEP, but with major innovations in how feature selection is performed. The key idea behind SAFE is that the noisy surrogates, the main ICD-9 and NLP counts, can be used to create silver-standard labels, Ss, to well approximate the gold-standard labels for patients in extreme subsets. Using data from the extreme subsets, we can effectively select additional features that are predictive of Y by fitting penalized regression models using S as the response. The results showed that SAFE was highly effective in removing noninformative or redundant features, resulting in feature sets typically smaller in size when compared to those from AFEP. In contrast, AFEP uses univariate rank correlation analysis, which limits its ability to remove redundant features. The smaller size of the feature set reduces overfitting and hence the number of gold-standard labels required to train a generalizable algorithm. The advantage of using a small candidate feature set was most evident when the training sample size n was small and became less apparent as n increased. This is expected, since overfitting is generally less concerning for larger n. SAFE also potentially selects important features not identified by AFEP or domain experts. This was evident in the UC example, where “Crohn’s disease” and “weight loss” were both missing in AFEP, and the latter was also missed by the expert. Crohn’s disease is a differential diagnosis of UC, and weight loss is a common symptom in patients with CD but rare in those with UC. Including these 2 important features enabled SAFE to consistently outperform algorithms using AFEP or expert selected features. Moreover, SAFE is not very sensitive to the choice of parameters like and in defining the silver-standard labels (see section 8 of the Supplementary Material). Interestingly, features curated by domain experts generally led to worse or comparable algorithm performance compared to SAFE. This further highlights the advantage of the automated feature extraction procedure.
The use of silver-standard labels to select additional features plays an important role in the SAFE procedure. Both the main ICD-9 and NLP counts are obvious choices for creating the silver labels. Adding the combined label to the process is important for increasing the robustness of the selection. Since the NLP and ICD-9 counts are highly correlated with each other, regression modeling with response derived from one count and then the other count as a feature tends to underestimate the importance of the remaining features. Including and removing both of these 2 main features from the candidate set enables SAFE to evaluate the informativeness of the remaining features more robustly. As an example, morning stiffness, an important feature for classifying RA, would not be selected if was not used as part of the feature selection process. In addition, other choices of may be available for specific phenotypes if there are additional strong predictors. For example, HgA1c laboratory results can be used for diabetes mellitus, the count of disease-modifying antirheumatic drug prescriptions can be used for RA, and percutaneous coronary intervention or coronary artery bypass graft procedure codes can be used for CAD. However, not all phenotypes have such strong predictors, and the selection may require input from domain experts.
Using 5 knowledge sources followed by majority voting to create candidate features instead of 2, as was done for AFEP, improved the robustness of the procedure. Extracting from a larger number of knowledge sources allows SAFE to include more candidate concepts, thus reducing the risk of missing important features. On the other hand, the majority voting effectively removes a large number of noninformative concepts based on the assumption that more important concepts are more frequently mentioned in multiple articles. Although the S2 procedure based on the original 2 sources performed similarly to the SAFE procedure, SAFE has the advantage of being more robust than the choice of the knowledge sources. In the 4 examples, the algorithms based on S2 and SAFE achieved similar accuracy except for RA, where SAFE had a slightly higher AUC than S2. For RA, SAFE also selected more clinically meaningful features. For example, “morning stiffness” was selected as a feature by SAFE but omitted by S2. This also explains the slightly worse prediction performance of S2.
Comparing SAFE to M2, A5, and A5V revealed more details regarding the impact of the various building blocks of SAFE in improving the algorithm’s performance. Among all these procedures and across all 4 phenotypes, M2, with only the 2 main features, generally had the worst performance, highlighting the importance of including additional informative features. On the other hand, both AFEP and A5 generally did not perform well either, due to the inclusion of too many features, resulting in a significant overfitting issue, particularly when the training size n was small. This further confirms the value of feature selection. The A5V procedure with majority voting performed better than AFEP and A5, but still performed generally worse than SAFE. For UC, AFEP-based procedures performed substantially worse than SAFE, due to the ability of SAFE to identify 2 important features missed by AFEP, Crohn’s disease and weight loss. For example, with 150 labels, the AUC was 0.95 (F-score 0.87) based on SAFE but only 0.93 (F-score 0.77) based on A5V. Thus, the use of multiple knowledge sources and selections with both majority voting and silver-standard labels together make SAFE the overall best performing and most robust method.
One limitation of the study is that the NLP data were extracted using HITEx43 (Health Information Text Extraction) for the domain expert curated features, but NILE44 (Narrative Information Linear Extraction system) for AFEP and SAFE. Although the SAFE procedure is not expected to be overly sensitive to the choice of NLP software, generalizability to other NLP software as well as to additional phenotypes and other EMR systems warrants further research.
CONCLUSION
In this paper, we introduced SAFE as a high-throughput method to efficiently curate features for EMR phenotyping algorithms, leveraging publicly available knowledge sources and a large set of unlabeled data. SAFE advances current methods by expanding the knowledge sources for collection of medical concepts and employs a majority voting step to coarsely estimate the importance of a concept. The novel data-driven feature selection process employed by SAFE removes noninformative and redundant features via penalized regression modeling using easily available silver-standard labels to approximate the gold-standard labels for patients in extreme subsets. The results show that the SAFE method can identify fewer but more informative features than the AFEP method. The automation in feature curation eliminates the need for domain experts to manually create a list of relevant clinical terms and for NLP experts to identify optimal concept mapping. The reduction in the size of candidate features achieved by SAFE also leads to a substantial reduction in the number of gold-standard labels needed for algorithm training. Thus, employing SAFE in the training of EMR-based phenotyping algorithms can greatly improve efficiency, allowing for high-throughput phenotyping. These types of methods will play a key role in effectively leveraging big data for precision medicine studies.45
Supplementary Material
FUNDING
This work was supported by National Institutes of Health grants U54-HG007963, U54-LM008748, R01-GM079330, K08-AR060257, and K23- DK097142, as well as the Harold and Duval Bowen Fund and internal funds from Partners HealthCare.
COMPETING INTERESTS
None.
CONTRIBUTORS
All authors made substantial contributions to conception and design; acquisition, analysis, and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
REFERENCES
- 1. Ryan PB, Madigan D, Stang PE, et al. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Stat Med. 2012;31:4401–15. [DOI] [PubMed] [Google Scholar]
- 2. Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther. 2011;90:133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castro VM, Clements CC, Murphy SN, et al. QT interval and antidepressant use: a cross sectional study of electronic health records. BMJ. 2013;346:f288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. L. Masica A, Ewen E, A. Daoud Y, et al. Comparative effectiveness research using electronic health records: impacts of oral antidiabetic drugs on the development of chronic kidney disease. Pharmacoepidemiol Drug Saf. 2013;22:413–22. [DOI] [PubMed] [Google Scholar]
- 5. Pantalone KM, Kattan MW, Yu C, et al. The risk of developing coronary artery disease or congestive heart failure, and overall mortality, in type 2 diabetic patients receiving rosiglitazone, pioglitazone, metformin, or sulfonylureas: a retrospective analysis. Acta Diabetol. 2009;46:145–54. [DOI] [PubMed] [Google Scholar]
- 6. Pantalone KM, Kattan MW, Yu C, et al. The risk of overall mortality in patients with Type 2 diabetes receiving different combinations of sulfonylureas and metformin: a retrospective analysis. Diabet Med. 2012;29:1029–35. [DOI] [PubMed] [Google Scholar]
- 7. Douglas I, Evans S, Smeeth L. Effect of statin treatment on short term mortality after pneumonia episode: cohort study. BMJ. 2011;342:d1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stakic SB, Tasic S. Secondary use of EHR data for correlated comorbidity prevalence estimate. In: 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). 2010;3907–10. [DOI] [PubMed] [Google Scholar]
- 9. Wu L-T, Gersing K, Burchett B, et al. Substance use disorders and comorbid Axis I and II psychiatric disorders among young psychiatric patients: findings from a large electronic health records database. J Psychiatr Res. 2011;45:1453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–28. [DOI] [PubMed] [Google Scholar]
- 11. Liao KP, Kurreeman F, Li G, et al. Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non–rheumatoid arthritis controls. Arthritis Rheum. 2013;65:571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ritchie MD, Denny JC, Zuvich RL, et al. Genome- and phenome-wide analysis of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;12713:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pathak J, Wang J, Kashyap S, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc 2011;18:376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benesch C, Witter DM, Wilder AL, et al. Inaccuracy of the International Classification of Diseases (ICD-9-CM) in identifying the diagnosis of ischemic cerebrovascular disease. Neurology. 1997;49:660–4. [DOI] [PubMed] [Google Scholar]
- 18. Birman-Deych E, Waterman AD, Yan Y, et al. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care. 2005;43:480–85. [DOI] [PubMed] [Google Scholar]
- 19. White RH, Garcia M, Sadeghi B, et al. Evaluation of the predictive value of ICD-9-CM coded administrative data for venous thromboembolism in the United States. Thromb Res. 2010;126:61–67. [DOI] [PubMed] [Google Scholar]
- 20. Zhan C, Battles J, Chiang Y-P, et al. The validity of ICD-9-CM codes in identifying postoperative deep vein thrombosis and pulmonary embolism. Jt Comm J Qual Patient Saf. 2007;33:326–31. [DOI] [PubMed] [Google Scholar]
- 21. McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of electronic health record oriented phenotyping algorithms. AMIA Annu Symp Proc. 2011;2011:274. [PMC free article] [PubMed] [Google Scholar]
- 23. Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res. 2010;62:1120–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of Crohn's disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis. 2013;19:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xia Z, Secor E, Chibnik LB, et al. Modeling disease severity in multiple sclerosis using electronic health records. PLoS ONE. 2013;8:e78927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castro V, Shen Y, Yu S, et al. Identification of subjects with polycystic ovary syndrome using electronic health records. Reprod Biol Endocrinol. 2015;13:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castro VM, Minnier J, Murphy SN, et al. Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am J Psychiatry. 2014;172:363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu S, Kumamaru KK, George E, et al. Classification of CT pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing. J Biomed Inform. 2014;52:386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS ONE. 2015;10:e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ. 2015;350:h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012;19:e162–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Libr Assoc. 1993;81:170. [PMC free article] [PubMed] [Google Scholar]
- 33. Pakhomov SV, Buntrock J, Chute CG. Identification of patients with congestive heart failure using a binary classifier: a case study. In: Proceedings of the ACL 2003 Workshop on Natural Language Processing in Biomedicine, Volume 13. Stroudsburg, PA: Association for Computational Linguistics; 2003:89–96. [Google Scholar]
- 34. Bejan CA, Xia F, Vanderwende L, et al. Pneumonia identification using statistical feature selection. J Am Med Inform Assoc. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carroll RJ, Eyler AE, Denny JC. Naïve electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc 2011;2011:189. [PMC free article] [PubMed] [Google Scholar]
- 36. Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer; 2009. [Google Scholar]
- 37. Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015;225:993-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar V, Liao K, Cheng S-C, et al. Natural language processing improves phenotypic accuracy in an electronic medical record cohort of type 2 diabetes and cardiovascular disease. J Am Coll Cardio. 2014;63(12_S). [Google Scholar]
- 39. Love TJ, Cai T, Karlson EW. Validation of psoriatic arthritis diagnoses in electronic medical records using natural language processing. Semin Arthritis Rheum. 2011;40:413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B. 2005;67:301–20. [Google Scholar]
- 41. Zou H, Zhang HH. On the adaptive elastic-net with a diverging number of parameters. Ann Stat. 2009;37:1733–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett 2006;27:861–74. [Google Scholar]
- 43. HITEx Manual. https://www.i2b2.org/software/projects/hitex/hitex_manual.html. Accessed January 14, 2014. [Google Scholar]
- 44. Yu S, Cai T. A short introduction to NILE. ArXiv13116063 Cs Published online first: November 23, 2013. http://arxiv.org/abs/1311.6063. Accessed August 15, 2014. [Google Scholar]
- 45. Delude CM. Deep phenotyping: The details of disease. Nature. 2015;527:S14–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.