Abstract
Our aim was to uncover unrecognized phenomic relationships using force-based network visualization methods, based on observed electronic medical record data.
A primary phenotype was defined from actual patient profiles in the Multiparameter Intelligent Monitoring in Intensive Care II database. Network visualizations depicting primary relationships were compared to those incorporating secondary adjacencies. Interactivity was enabled through a phenotype visualization software concept: the Phenomics Advisor.
Subendocardial infarction with cardiac arrest was demonstrated as a sample phenotype; there were 332 primarily adjacent diagnoses, with 5423 relationships. Primary network visualization suggested a treatment-related complication phenotype and several rare diagnoses; re-clustering by secondary relationships revealed an emergent cluster of smokers with the metabolic syndrome.
Network visualization reveals phenotypic patterns that may have remained occult in pairwise correlation analysis. Visualization of complex data, potentially offered as point-of-care tools on mobile devices, may allow clinicians and researchers to quickly generate hypotheses and gain deeper understanding of patient subpopulations.
Keywords: Medical informatics applications, Data display, Phenotype, Data mining, Decision making; computer-assisted
Background and significance
Clinical phenomics is the measurement of the diversity of disease states across human subjects. The massive accumulation of clinical data accrued automatically inside electronic medical records (EMRs) with each episode of patient care through clinical, laboratory, and billing systems has enabled a new type of phenomic research using clinical data.1–3 When such phenotype data are extracted, these large data sets, called phenomes, can provide useful snapshots of disease prevalence, distribution, and correlation. Correlation, especially through the employment of phenome-wide association study (PheWAS), may yield valuable insights, including the linking of genome to phenome, as has been successfully demonstrated by our group and others.4–7
Although tabular reports may convey adequate analytic information for limited exercises in phenomic association, the phenotype space is dauntingly large. For example, the International Classification of Diseases, Clinical Modification (ICD-9-CM) diagnosis code set has roughly 14 000 codes; ICD-10-CM has about 68 000 codes, a scale that begins to approach the lower end of ‘-omics’ studies. Not surprisingly, therefore, the Manhattan Plot, the visualization tool widely adopted in genome-wide association studies, has emerged as the best-known visualization tool for phenome exploration. These plots can be generated using the R PheWAS package8 or via tools such as PheWAS-View9; the latter also allows for construction of pairwise correlation heat maps. We have also introduced a two-dimensional variant of the Manhattan Plot that presents a ‘view from above’ for visual analytics of clinical features with continuous values, for example, most laboratory tests and time intervals.10,11 This approach allows for the identification of ‘microphenotypes’ that may only apply within certain contexts and over specific intervals; for example, the microphenotype of hospital-acquired complication is most evident for the longest decile of hospitalization in a critically ill cohort.11
However, none of the aforementioned approaches except for pairwise correlation takes into account the interaction of phenotype features. As in the underlying biologic systems, disparate phenotypes can be directly related, induced, or inhibited by other phenotypes. For example, type II diabetes mellitus (T2DM) may simultaneously induce a phenotype of neuropathy and inhibit a phenotype of foot pain (since the neuropathy can mask the pain due to numbness). As another example, the seemingly unrelated phenotypes of rash, arthralgia, and abnormal blood counts may be the manifestation of underlying autoimmunity (e.g., systemic lupus erythematosus). Furthermore, the action of one phenotype upon another may be through one or more intermediaries. For these reasons, we propose that network visualization of a primary phenotype and its immediate neighborhood may present patterns, upon inspection, that yield clinical insight and hypothesis generation.
Objective
To assist clinical phenomics research, we propose a clinical software package for networked phenotype visualization, a ‘Phenomics Advisor.’12 This software would provide a patient-centered view into a phenomic database, with interactive visualization tools for clinician use. This paper presents the concept and illustrates it with a real-world example from EMR data.
Materials and methods
For the purposes of this pilot, we have used the Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC II) database as a source of phenotype information.13 MIMIC II contains extensive information on more than 25 000 hospitalized patients with critical illness, including laboratory values, medication exposures, and demographics. For simplicity, we use ICD-9-CM codes to represent phenotype. All investigators completed appropriate human subjects training prior to accessing MIMIC II, which is completely de-identified and classified as Institutional Review Board exempt.
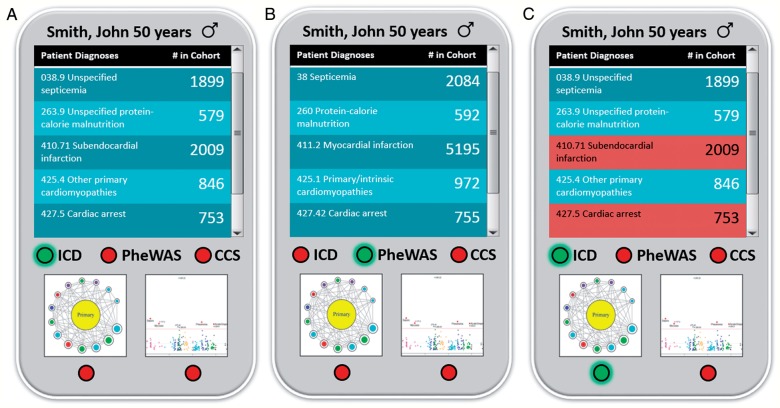
A simple interface is provided to the user for access the Phenomics Advisor (figure 1). Because the application is intended for use at the point-of-care, the initial view is of an individual patient (‘John Smith’). At the top, a table lists the patient's diagnostic codes and the number of patients in the cohort with one or more identical codes. Underneath the table is a radio button to switch association to one of the following: (1) un-aggregated ICD-9-CM codes; (2) minor aggregation using PheWAS codes (http://phewas.mc.vanderbilt.edu/); or (3) major aggregation using the single-level Agency for Healthcare Research and Quality (AHRQ) Clinical Classifications Software (CCS) codes (http://www.hcup-us.ahrq.gov/).14 The bottom radio button set switches between network visualization of the ‘Phenotype Neighborhood’ and conventional Manhattan Plot visualization.
Figure 1:
Initial view of the Phenomic Advisor. In panel A, the default view shows International Classification of Diseases, Clinical Modification (ICD-9-CM) codes by chapter, along with their counts in the cohort (Multiparameter Intelligent Monitoring in Intensive Care II in this example). In the middle panel B, the user has selected to display phenome-wide association study (PheWAS) codes and the aggregate counts are recalculated accordingly. On the right, the user has selected a combined phenotype as well as the Phenotype Neighborhood view to drill further into phenotypic relationships.
In Phenotype Neighborhood visualization mode, the focus of this paper, a user-selected phenotype is displayed as the Primary Phenotype. For custom phenotype aggregations, the software allows the user to select one or more diagnosis codes (heretofore, ICD-9-CM) to define the Primary Phenotype. The default view is a polar plot visualization15 with first-degree adjacencies only, where the Primary Phenotype is the central vertex and the distance between the center and the first-degree vertices is determined by a weighted Fruchterman-Reingold model.16 Edge weight is defined as the ratio of two components: (1) the number of co-occurrences of the Primary Phenotype and an adjacent vertex, and (2) a ‘counterweight’ of the number of co-occurrences of the adjacent vertex with any out-of-neighborhood vertex. By definition, out-of-neighborhood vertices will only occur in patients not having the Primary Phenotype. Alternatively, the user may choose to display both primary and secondary adjacencies using a layout determined by the edge weights as described above, as well as additional edge weights applied to the secondary adjacencies, here defined as the ratio of co-occurrences in cases to the total number of co-occurrences in the database.
For clarity, by default only edges representing two or more co-occurrences in the database are applied to the layout model; singly connected vertices are hidden. Users can choose to reveal these hidden vertices or to alter the co-occurrence threshold parameter. Primary edges are colored faintly and can be hidden by the user, if their presence introduces unwanted visual clutter; secondary edges (when displayed) are colored darkly so as to emphasize their presence. Vertices are colored by their respective ICD-9-CM chapter and sized proportionate to the distinct number of patients in the database with at least one occurrence of the ICD-9-CM code. In order to reduce labeling clutter, only vertices representing more than 2500 patients are labeled numerically, in descending order of size.
R V.3.0.2 (R Foundation for Statistical Computing) was used for the calculations.17 Networks were displayed using the igraph R package,18 with coloration based on RColorBrewer qualitative palettes.19 Preliminary R code is available upon request.
The Phenomics Advisor could be readily implemented as a SMART (Substitutable Medical Applications & Reusable Technology) app20 for use on a variety of EMR and data warehouse architectures, such as i2b2.21 The Phenomics Advisor requires a patient identifier and linked access to a patient's diagnosis codes (ICD-9-CM or others) to represent phenotype.
Results
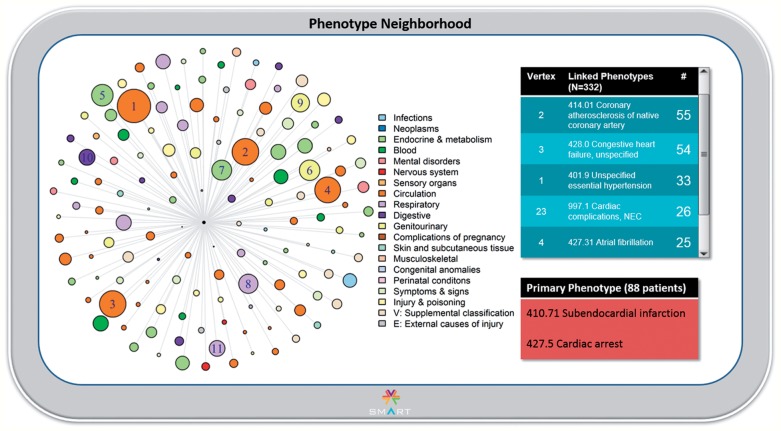
For the pilot, an adult subject ‘John Smith’ was randomly chosen from the MIMIC II database. As shown in figure 1, Mr. Smith has been coded as having had a relatively mild form of myocardial infarction (subendocardial MI, 410.7122) but also cardiac arrest (427.5). This phenotype pair is designated the Primary Phenotype, to further explore this somewhat unusual combination. Indeed, as shown in figure 2, only 88 patients (4.4% of the 2009 patients with subendocardial MI) have the co-occurrence of cardiac arrest. In this view, there are 138 primary edges in the network (excluding 194 singly connected nodes). We observe that ICD-9-CM codes in the Circulatory system chapter are generally enriched, although many are not in close proximity to the Primary Phenotype, suggesting that these are commonly observed ICD-9-CM codes with high counterweights; an exception is coronary atherosclerosis, both primary (414.01) and of grafts (414.02). Through interaction with the network visualization (not shown), we find that several rare phenotypes are in very close proximity to the Primary Phenotype, including Moyamoya disease (437.5), iliac artery dissection (443.22), and, quite interestingly, complications of cardiac catheterization (E879.0). This visualization therefore suggests that the Primary Phenotype may actually be a complication related to treatment for MI.
Figure 2:
The Phenotype Neighborhood view. On the left, a polar plot is displayed with the Primary Phenotype in the center, since it is by definition the most connected vertex. Phenotypes linked by first-degree adjacency are displayed. The ‘largest’ phenotypes are labeled by descending order of frequency; the underlying International Classification of Diseases, Clinical Modification (ICD-9-CM) code is available as a pop-up when the user scrolls over the vertex of interest (not shown). Vertex color is by ICD-9-CM chapter, and size is proportionate to the number of occurrences of the particular phenotype in the overall database. On the upper right, vertices are listed in tabular format in descending order by number of co-occurrences. The Primary Phenotype, with the number of patients represented, is shown in the lower right.
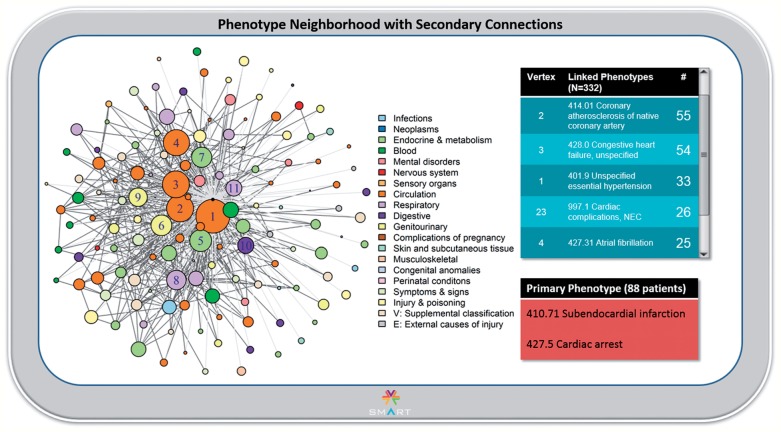
When secondary adjacencies are introduced, there are 616 edges (excluding 4807 singly occurring co-occurrences) and the network configuration changes significantly (figure 3). The network collapses towards a centroid to the left of the Primary Phenotype, and the common Circulation codes are now in close proximity. This suggests that the Primary Phenotype may occur in the context of a particular pattern of underlying disease. Inspection of loose clustering close to the Primary Phenotype vertex and investigation into some of the underlying clustered codes suggested apparent coordination of the vertices representing unspecified essential hypertension (#1), coronary atherosclerosis (#2), T2DM (#5), and tobacco use disorder (the medium-sized pink vertex between #3 and #11). This coordination, which raised the suspicion of a particularly unhealthy underlying phenotype, was not at all evident until the secondary connections were introduced. Several other interesting effects also surfaced. Of note, the rare phenotypes that were in close proximity to the Primary Phenotype do not move much, even after the introduction of the secondary connections, suggesting a true relationship not subject to confounding. Also notably, there are vertices that remain peripheral, indicating a looser connection with the phenotype cluster as a whole. One example is atrial fibrillation (#4). Indeed, atrial fibrillation is a very uncommon cause of MI or cardiac arrest.
Figure 3:
The Phenotype Neighborhood view with secondary adjacencies. In this view, vertex layout has been recalculated after the introduction of secondary edges between the vertices immediately adjacent to the Primary Phenotype vertex. Collapse and re-clustering are evident, with previously distant vertices now in close proximity to the Primary Phenotype vertex. The Primary Phenotype is no longer central to the network, although it clearly remains the most connected. Primary edges appear in light gray with secondary edges in dark gray to emphasize these connections.
Discussion
The human clinical phenome is highly complex, as evidenced by the fact that even master clinicians can have trouble synthesizing an unusual constellation of signs and symptoms into a pathophysiologically robust diagnosis.23 As we have demonstrated, a user of the Phenomics Advisor can quickly appreciate that the combined phenotype of subendocardial MI and cardiac arrest is unusual, occurring in <5% of patients in the MIMIC II database who had subendocardial MI, and in ∼12% of those having cardiac arrest. Inspection of primary adjacencies suggests that this phenotype may be a treatment-related complication, or possibly associated with a rare diagnosis, Moyamoya disease.24 When secondary adjacencies are introduced, two further ‘findings’ surface: (1) the ‘Primary’ phenotype is probably not primary at all, since the mass of the secondarily clustered graph appears to be well to the left of center; and (2) a cluster of smokers with the metabolic syndrome appears to emerge. While it is not surprising that this particularly unhealthy population will tend to experience grave cardiac outcomes,25,26 this relationship was not clearly evident (after the application of some specific clinical domain knowledge) until the secondary adjacencies were introduced and the graph layout was recalculated. Less evident but essential for hypothesis generation are the movements of individual vertices with the change in network configuration, which could trigger further, but still rapid, investigation.
Irrespective of the value of broader phenotype definitions than ICD-9-CM-based claims data (which will only get more challenging with ICD-10-CM), our study shows that claims data can yield considerable insights, especially when approached as a network of relationships. Within the context of genetic association data, we have previously shown that billing codes can replicate 66% of known associations with an area under the receiver operator curve of 0.83.6 It may be possible that a networked phenotype approach could improve upon this accuracy, for example through the use of imputation.
To be accessible to clinicians, we chose force-based network layout visualization, as it does not require expertise in the interpretation of data structures.27 Other visualization methods, such as chord diagrams,28 adjacency matrices,29 and hive plots,30 may however offer other insights, so we intend to explore them in the future. We also plan to enhance the Phenomics Advisor with: (1) Bayesian weighting; (2) visualization of higher order (tertiary or greater) vertices with dimensionality reduction and filtering; (3) visual and algorithmic ‘knock out’ of the Primary Phenotype to reveal adjacencies that otherwise are concealed or confounded; and (4) temporal elements. Finally, we plan to conduct usability evaluations of the planned interactive software product, so as to ascertain usefulness and improve usability in actual clinical settings.
Conclusion
We have introduced the Phenomics Advisor, a set of visual analytic techniques, software, and phenomics database, which together permit detection and study of the complex relationships that characterize the human phenome. The example we used in our pilot demonstrates the potential for rapid research investigation or even point-of-care usage (i.e., within clinical workflow) to allow a physician to rapidly explore for ‘patients like this one.’ Such a data-driven approach, built into clinical or research systems, for example, i2b231 or the Vanderbilt University Synthetic Derivative,32 could enable prompt considerations of alternative diagnoses. A means to visually explore a patient's diagnosis against the backdrop of a large population of patient phenotype data could aid clinicians facing difficult or rare diagnostic situations. The same tool could also help researchers characterize the human phenome further, which will be necessary to achieve the ‘human phenome project.’33,34
Contributors
JLW had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. JLW, DK, and GA conceived the study design; JCD provided thoughts on direction; JLW performed the experiments and analyzed the data; all authors contributed to the manuscript writing and approved the final manuscript.
Funding
This work was supported in part by grants NLM R01LM010685, NHGRI 1R01HG004836-01, NLM 4R00LM009826-03, and ONC 90TR0001/01.
Competing interests
None.
Provenance and peer review
Not commissioned; externally peer reviewed.
Data sharing
Data was all from MIMIC II, a publicly available de-identified EMR database.
Supplementary Material
REFERENCES
- 1. Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–428. [DOI] [PubMed] [Google Scholar]
- 2. Blair DR, Lyttle CS, Mortensen JM, et al. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology. 2014;141:157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pendergrass SA, Brown-Gentry K, Dudek SM, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol. 2011;35:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pendergrass SA, Dudek SM, Crawford DC, et al. Visually integrating and exploring high throughput Phenome-Wide Association Study (PheWAS) results using PheWAS-View. BioData Min. 2012;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Warner JL, Alterovitz G. Phenome-based analysis as a means for discovering context-dependent clinical reference ranges. AMIA Annu Symp Proc. 2012;2012:1441–1449. [PMC free article] [PubMed] [Google Scholar]
- 11. Warner JL, Zollanvari A, Ding Q, et al. Temporal phenome analysis of a large electronic health record cohort enables identification of hospital-acquired complications. J Am Med Inform Assoc. 2013;20:e281–e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Warner J, Denny J, Kreda D, et al. Analytic approaches to phenotypic complexity. Stud Health Technol Inform. 2013;192:1267. [Google Scholar]
- 13. Saeed M, Villarroel M, Reisner AT, et al. Multiparameter Intelligent Monitoring in Intensive Care II: a public-access intensive care unit database. Crit Care Med. 2011;39:952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cusack CM, Shah S. Web-based tools from AHRQ's National Resource Center. AMIA Annu Symp Proc. 2008:1221. [PubMed] [Google Scholar]
- 15. Draper GM, Livnat Y, Riesenfeld RF. A survey of radial methods for information visualization. IEEE Trans Vis Comput Graph. 2009;15:759–776. [DOI] [PubMed] [Google Scholar]
- 16. Fruchterman TM, Reingold EM. Graph drawing by force-directed placement. Softw Pract Exper. 1991;21:1129–1164. [Google Scholar]
- 17. Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2013. ISBN 3-00051-7-0. [Google Scholar]
- 18. Csardi G, Nepusz T. The igraph software package for complex network research. Inter Journal Complex Systems. 2006;1695. [Google Scholar]
- 19. Neuwirth E. RColorBrewer: ColorBrewer palettes, 2011. R package version 1.0-5.
- 20. Mandl KD, Mandel JC, Murphy SN, et al. The SMART Platform: early experience enabling substitutable applications for electronic health records. J Am Med Inform Assoc. 2012;19:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wattanasin N, Porter A, Ubaha S, et al. Apps to display patient data, making SMART available in the i2b2 platform. AMIA Annu Symp Proc. 2012;2012:960–969. [PMC free article] [PubMed] [Google Scholar]
- 22. Madigan NP, Rutherford BD, Frye RL. The clinical course, early prognosis and coronary anatomy of subendocardial infarction. Am J Med. 1976;60:634–641. [DOI] [PubMed] [Google Scholar]
- 23. Szolovits P. Uncertainty and decisions in medical informatics. Methods Inf Med. 1995;34:111–121. [PubMed] [Google Scholar]
- 24. Ahn YK, Jeong MH, Bom HS, et al. Myocardial infarction with Moyamoya disease and pituitary gigantism in a young female patient. Jpn Circ J. 1999;63:644–648. [DOI] [PubMed] [Google Scholar]
- 25. Park YW, Zhu S, Palaniappan L, et al. The metabolic syndrome: prevalence and associated risk factor findings in the US population from the Third National Health and Nutrition Examination Survey, 1988–1994. Arch Intern Med. 2003;163:427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Teo KK, Ounpuu S, Hawken S, et al. Tobacco use and risk of myocardial infarction in 52 countries in the INTERHEART study: a case-control study. Lancet. 2006;368:647–658. [DOI] [PubMed] [Google Scholar]
- 27. Chittaro L. Information visualization and its application to medicine. Artif Intell Med. 2001;22:81–88. [DOI] [PubMed] [Google Scholar]
- 28. Vassiliev V. Cohomology of knot spaces. Adv Sov Math. 1990:23–69. [Google Scholar]
- 29. Abello J, van Ham F. Matrix zoom: A visual interface to semi-external graphs. 2004 INFOVIS 2004 IEEE Symposium on Information Visualization; IEEE, 2004:183–190. [Google Scholar]
- 30. Krzywinski M, Birol I, Jones SJ, et al. Hive plots—rational approach to visualizing networks. Brief Bioinform. 2012;13:627–644. [DOI] [PubMed] [Google Scholar]
- 31. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc. 2010;17:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Danciu I, Cowan JD, Basford M, et al. Secondary use of clinical data: the Vanderbilt approach. J Biomed Inform. 2014. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008;24:103–106. [DOI] [PubMed] [Google Scholar]
- 34. Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.