Abstract
Objective Clinical data warehouses have accelerated clinical research, but even with available open source tools, there is a high barrier to entry due to the complexity of normalizing and importing data. The Office of the National Coordinator for Health Information Technology's Meaningful Use Incentive Program now requires that electronic health record systems produce standardized consolidated clinical document architecture (C-CDA) documents. Here, we leverage this data source to create a low volume standards based import pipeline for the Informatics for Integrating Biology and the Bedside (i2b2) clinical research platform. We validate this approach by creating a small repository at Partners Healthcare automatically from C-CDA documents.
Materials and methods We designed an i2b2 extension to import C-CDAs into i2b2. It is extensible to other sites with variances in C-CDA format without requiring custom code. We also designed new ontology structures for querying the imported data.
Results We implemented our methodology at Partners Healthcare, where we developed an adapter to retrieve C-CDAs from Enterprise Services. Our current implementation supports demographics, encounters, problems, and medications. We imported approximately 17 000 clinical observations on 145 patients into i2b2 in about 24 min. We were able to perform i2b2 cohort finding queries and view patient information through SMART apps on the imported data.
Discussion This low volume import approach can serve small practices with local access to C-CDAs and will allow patient registries to import patient supplied C-CDAs. These components will soon be available open source on the i2b2 wiki.
Conclusions Our approach will lower barriers to entry in implementing i2b2 where informatics expertise or data access are limited.
Keywords: Medical Informatics, Information Storage and Retrieval, Meaningful Use, Systems Integration, Database Management Systems
Background and significance
The enterprise of clinical trials in the USA faces many challenges, according to the Institute of Medicine.1 The Patient Centered Outcomes Research Institute is investing over US$90 million in PCORnet, which seeks to reimagine the country's clinical research infrastructure and make progress to overcome these challenges.2 Clinical data analytics platforms will be used at over 100 sites in all 50 states to more easily match eligible patients to trials and to perform large scale comparative effectiveness research aided by retrospective clinical data. These goals agree with previous achievements associated with clinical data analytics platforms: increased local research funding, decreased cost to recruit patients to clinical trials,3 unprecedented access to local data to answer challenging questions,4 and distributed research across previously disconnected populations.5
Informatics for Integrating Biology and the Bedside (i2b2) is an open source freely available clinical data analytics platform funded by the National Institutes of Health. It is implemented at over 100 sites nationwide, including over one-third of PCORnet's sites.6–8 It has an active community of users and contributors, and a flexible data model that supports diverse implementations. Currently, six i2b2 networks share aggregate level data for population level research,9,10 and i2b2 components specifically designed for clinical trial recruitment are in development.11
However, even with open source solutions like i2b2, the barrier to entry for implementing a clinical data analytics platform is too high in some cases. Every implementation of i2b2 must define a custom approach to extract data from clinical systems, normalize it, and import it into i2b2. This process is called extract, transform, and load (ETL). This can be a complex expensive development process that is likely out of scope for small practices. (And in fact, most i2b2 installations are affiliated with large academic medical centers.12) Small practices could benefit from i2b2 in at least two significant ways. One, it would allow them to be part of clinical trial recruitment networks at low cost, which provides economic benefit to practices and empowers patients with the ability to contribute to research. There are multiple initiatives to build such networks of i2b2 repositories, including those of Patient Centered Outcomes Research Institute and the National Institutes of Health, which will be used to accrue for clinical trials. Two, i2b2 provides a low cost system for physicians to study their own patient population. This allows a wide variety of ‘on the fly’ research questions, such as pragmatic validation of complex treatment decisions4 or studying local trends for quality measurement or collective intelligence.13,14
Also, chronic disease registries typically do not have access to their patients’ medical record data and must rely on patients’ manual data entry. PCORnet includes 18 Patient Powered Research Networks (PPRNs) that represent patients with chronic disease, but less than half have an active registry,15 and a recent survey confirmed that most of the registries rely exclusively on patient reported data. PCORnet's long term goal is for PPRNs to import their patients’ electronic health record (EHR) data, which will enhance the networks’ research capabilities, but there is no plan to achieve this at present.
Here we present a straightforward reusable ETL approach that will allow both disease registries and small practices to more easily implement i2b2. It takes advantage of requirements mandated by stage 2 of the Meaningful Use incentive program (MU2): healthcare facilities are required to produce consolidated clinical document architecture (C-CDA) patient care summaries.16,17 All certified EHR systems will shortly be able to produce these machine readable documents with up to the minute patient information in standard terminologies. For example, Partners Healthcare (Boston, Massachusetts, USA) is building the capacity to generate at least 75 000 documents per day. Furthermore, patients must have access to visit summary information in MU2, and the popular Blue Button+ initiative is enabling individuals to access their own C-CDA documents.18–20 Meaningful Use stage 3 is expected to require support for Blue Button+ and might include a requirement for healthcare systems to submit C-CDAs to disease registries.19,21
Objective
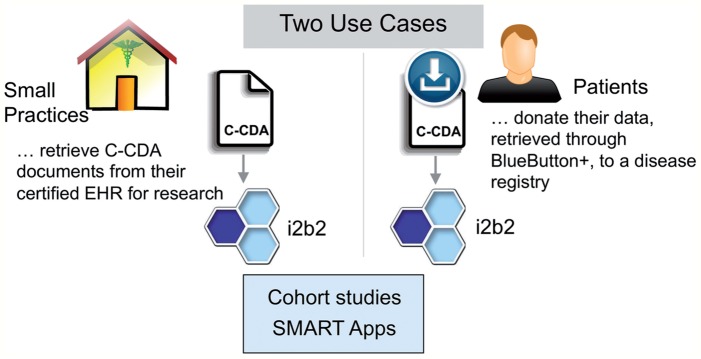
To demonstrate the ability to populate research databases from C-CDA documents, we created an i2b2 repository at Partners Healthcare using just C-CDA documents. We validated our ability to perform clinical research tasks with this repository using i2b2's graphical query tool6,22 and the SMART patient centric view.23 This proof of principle is readily extensible to other environments with C-CDAs, providing a straightforward approach for low volume data import for patient registries and small practices. An overview of this approach and its expected use cases is shown in figure 1. Reaching this objective involved developing a flexible i2b2 extension to import C-CDA documents and new ontology trees for the standard C-CDA terminologies. These components will soon be available as open source tools on the i2b2 wiki.24
Figure 1:
Anticipated uses for consolidated clinical document architecture (C-CDA) based Informatics for Integrating Biology and the Bedside (i2b2) import. Left: Small practices with low volume and certified electronic health records (EHRs) can use the C-CDA documents they already produce to populate an i2b2 data repository. Right: Patients will be able to export their C-CDA from healthcare systems through Blue Button+, which they can donate to disease registries for research. Bottom: i2b2 can then be used to study both cohorts and individual patients, through the i2b2 query tool and SMART apps, respectively (see figure 4).
Materials and methods
Design
i2b2 is a flexible relational data model and an extensible set of web services to interact with that data model.
i2b2 web services are logically organized into sets of independent software components, known as cells. These cells interact through web service calls, and together these cells make up the i2b2 ‘hive’. Many optional cells have been developed that extend the functionality of i2b2. i2b2 provides several standard user interaction motifs powered by these cells, including a graphical query builder used to find research cohorts and a patient ‘EHR view’ powered by SMART apps.23 These will be packaged together in the ‘i2b2 clinical trials’ platform.11
The underlying data model is a star schema design25 in which one large fact table stores most clinical observations. An observation can be any atomic fact, such as a diagnosis, procedure, medication, or even a computed value (eg, length of stay). Each observation may consist of several rows of information: the basic fact (eg, ‘diabetes mellitus’) and modifiers that provide additional context (eg, ‘admit diagnosis’). Each fact and modifier is associated with a code that is often used to define the code and coding system (eg, ‘ICD9:250.6’). To create the ‘star’, there are several ‘dimension’ tables that provide additional information about the facts, such as the patient, encounter, and provider dimensions.
Additionally, the set of known facts is navigable by ontology trees that are also stored in the data model. i2b2 provides several trees, others have been defined by individual i2b2 sites and projects that use i2b2, and they can be imported from the NCBO BioPortal.26
i2b2 can assign pseudoidentifiers to patients while keeping a mapping to the original medical record number (MRN) stored securely, separately from the rest of the patient data. This allows query results to be de-identified while allowing re-identification in appropriate circumstances. This is important in the present use case because i2b2 pseudoidentifiers are insufficient for C-CDA retrieval.
i2b2 exchanges clinical data among cells and applications using the i2b2 patient data object (PDO) XML format. This is a reflection of the underlying data model. PDO groups clinical data into groups of facts, with optional sections for dimension tables and patient mapping. i2b2's clinical research chart (CRC) cell includes a loader component that supports data import through the PDO.
System description
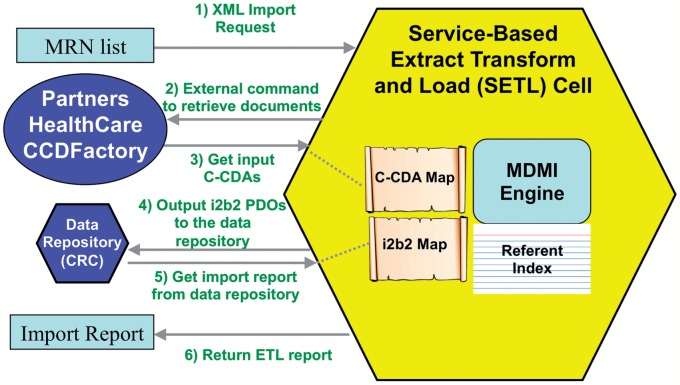
We designed an i2b2 cell, SETL (service based extract, transform, and load), to convert C-CDA documents into i2b2 format. This cell provides a web service that takes as input a list of patient MRNs. The cell calls a configurable command to retrieve a C-CDA for each patient MRN. This design allows each i2b2 site to define its own particular method for retrieving a C-CDA—they might exist in a database, in a folder on the local disk, or through an enterprise web service. On retrieving each C-CDA, SETL converts it to a PDO and sends it to the CRC loader for import into i2b2. The CRC loader automatically assigns a pseudoidentifier to the patient if one does not exist, and the actual MRN is securely stored separately. Finally, the SETL cell generates a report about the number of new patients, observations, and encounters added to the repository. For this first version, we convert demographics, encounters, diagnoses, and medications. Figure 2 (left) shows this data flow, with the external command configured for Partners Healthcare continuity of care document (CCD) factory (see Data source below).
Figure 2:
Technical design of the SETL (service based extract, transform, and load) cell import process. Left: The data flow among components in the architecture. (1) A request with a list of medical record numbers (MRNs) is sent to the SETL cell, which then (2) calls an external command to retrieve consolidated clinical document architecture (C-CDA) documents. For this study, this goes to our continuity of care document (CCD) factory connector at Partners Healthcare (Boston, Massachusetts, USA). (3) Retrieved documents are converted into Informatics for Integrating Biology and the Bedside (i2b2) patient data objects (PDOs) and (4) are sent to the data repository cell for storage, where MRNs are replaced by a unique pseudoidentifier. Finally (5, 6) a report on import errors and statistics is generated and returned to the user. ETL, extract, transform, and load. Right: The SETL cell embeds the Open Health Tools Model Driven Message Interoperability (MDMI) engine, which converts between healthcare data formats.
We are finalizing ontologies to support queries on the imported data. We use the following: for demographics, a subset of the existing i2b2 demographics ontology; for diagnoses, a SNOMED hierarchy of clinical findings converted from the NCBO BioPortal; and for medications, an RxNorm hierarchy organized by Veterans’ Administration drug class. We generated the RxNorm terminology using mappings present in the Unified Medical Language System that connect Veterans’ Administration drug classes with RxNorm generic and brand name medications.
The most problematic part of this ETL workflow is processing C-CDA documents. C-CDA is a set of restrictions on the extremely expressive clinical document architecture (CDA). The CDA is a specialization of the very broad HL7v3 Reference Implementation Model (RIM). The RIM is a general information model for expressing healthcare information through objects such as acts and the relationships between them. Although this design is very elegant theoretically, in practice an immense amount of work has gone into limiting expressivity to support computability and data exchange. Even at the level of the MU2 templates, there is room for differences of opinion on document structure, and implementations vary from site to site. C-CDA is also difficult to parse or understand because it is so complex and deeply nested. As an example, the first problem in a patient's problem list is eight levels deep in the XML hierarchy. The XPath expression is:
ClinicalDocument/component/structuredBody
/component[section/templateId
=′ 2.16.840.1.113883.10.20.22.2.5.1′]
/section/entry/act/entryRelationship/observation
[templateId= ′2.16.840.1.113883.10.20. 22. 4.4′]
We used Open Health Tools (OHT) to navigate the C-CDA XML hierarchy.27 OHT is a non-profit trade association responsible for growing and managing a diverse set of tools for healthcare interoperability. A particularly innovative emerging solution in OHT is the Model Driven Message Interoperability (MDMI) project,28 which utilizes an existing Object Management Group (OMG) standard for message exchange and applies it to healthcare. Leading experts in the banking industry developed MDMI for translation of data into a variety of proprietary formats. The innovation of MDMI over other message translation tools is that it is model based. MDMI defines a central ‘referent index’ that lists all of the data elements that a healthcare document could include. Maps can be written that define a message model. Then, particular message formats can be translated by these maps to and from the referent index, without concern about the ultimate inputs and outputs.
OHT-MDMI is an open source project; its first public release is scheduled to be available shortly.28,29 We programmatically integrated OHT-MDMI into the SETL cell to automatically translate input C-CDA documents into PDO. To accomplish this, the OHT-MDMI group provided us with a MU2 compliant C-CDA map that translates to and from the core referent index. Our message translation work was then to develop a map that translates the referent index into i2b2 PDO. We developed this using the included graphical map editor. This helped us navigate the complex structure of C-CDA. This integration is shown in figure 3 (right).
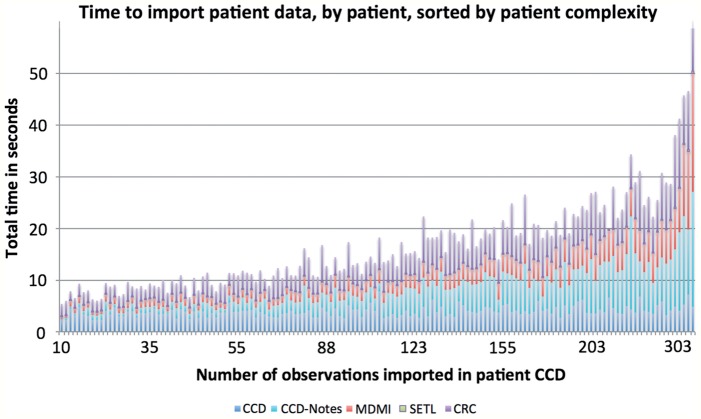
Figure 3:
Total time to import patient data, as number of imported observations increase. The time is split into color coded components. Continuity of care document (CCD) is Partners’ CCD factory time to generate a consolidated clinical document architecture (C-CDA). CCD-Notes is time to add encounter notes to the C-CDA from Partners’ outpatient notes service. Model Driven Message Interoperability (MDMI) is the time taken to convert the C-CDA into patient data object (PDO) by Open Health Tools-MDMI. SETL (service based extract, transform, and load) is all other processing of the PDO before sending to the clinical research chart (CRC) loader. CRC is the time taken by the CRC's loader service to import the PDO into the data repository.
For a site to implement our ETL process, the primary task would be to use the same graphical editor to modify the mapping of the source C-CDA for local variants. This does not require knowledge of the i2b2 PDO format. These changes are straightforward, and the map editor is intended for site administrators, not software developers. For an example of such a change, see the second paragraph of ‘Implementation’ below.
Data source
We used the Partners Healthcare CCD factory as our data source, which dynamically compiles patient information into MU2 compliant C-CDAs and is expected to handle capacities of over 75 000 documents generated daily.30 We wrote an adapter program to communicate with the CCD factory and retrieve C-CDAs. Because MU2 does not require encounter histories or encounter notes for the current C-CDA profiles, we retrieved those from an outpatient notes service and added these to the C-CDA document dynamically via our adapter. This study was approved by the institutional review board at Partners Healthcare.
Results
Implementation
For this initial release, we focused on importing patient information that was of high value for clinical trial recruitment: encounters, problems, medications, and demographics.31 We implemented the SETL cell in i2b2 1.7 as a JBoss 7 web service, like the other cells in the i2b2 hive. We also updated the SMART-i2b2 and CRC cells to fully support i2b2 1.7. We implemented the Partners Healthcare CCD adapter as a .NET program (that corresponds with the security model of the Partners Healthcare CCD factory) that can be called remotely over encrypted SSH within Partners’ firewall.
We were able to use OHT-MDMI's C-CDA map without modification for all the required entries in the aforementioned sections of the C-CDA. This map does not yet implement the optional sections of the C-CDA, and Partners Healthcare still uses the older C32 specification for some of these elements. Therefore, we used the MDMI map editor to add several Partners specific elements, such as provider information. To change the location of elements already in the map, we would simply navigate to that element in the editor's referent index, open the associated syntactic element, and change the XPath to reflect its location in the document. Because provider information is optional, it was not in the map, so it required an extra step. To add provider names for problems, we created a new syntax element and populated it with the appropriate XPath. Then we used an editor wizard to link it with a new semantic element that in turn links to ProblemPerformerName in the referent index. The total time to make this change was less than 5 min.
We then developed an i2b2 map with assistance from OHT-MDMI. MDMI maps contain two components: a set of semantic elements—the distinct blocks of information that have meaning in the data model; and one or more syntactic structures—descriptions of how the semantic elements precisely map to the output data format (eg, XML). MDMI provided an XML schema import tool that allowed us to create the syntax structure from our i2b2 PDO definition. We used their map editor to define semantic elements that are ‘transferred’ from the source data model. Table 1 summarizes the semantic elements in our i2b2 map. The key elements are the observations, which transfer clinical data from the C-CDA for storage in i2b2. All observations except encounters are mapped as a single core fact (eg, medication). We have not added modifiers to transfer additional detailed information (eg, medication route and frequency) because to do so efficiently will require a feature that will appear in the next release of MDMI (winter 2014).
Table 1:
Semantic elements in i2b2 and their corresponding representation in C-CDA, in this initial version of the SETL cell
| i2b2 semantic element | C-CDA semantic elements | Description | i2b2 database storage |
|---|---|---|---|
| i2b2 patient map | PatientID | MRN |
Stored in the patient mapping table
Mapped to a pseudoid by the CRC loader |
| i2b2 encounter map |
EncounterEffectiveTime.low
Global ID (computed) |
An i2b2 encounter is identified by the date of each C-CDA encounter. One global encounter is used for all non-encounter-based observations |
Stored in the encounter mapping table
Used by the query tool to search within or across encounters |
| Concepts | displayName and code for each code in the observations listed below. | Not stored (duplicates information in the ontology trees) but used by SMART apps | |
| Patient |
PatientDateofBirth
PatientLanguage PatientAdministration GenderCode PatientRace Age (computed) |
Stored in the patient dimension table | |
| Observers | PerformerPersonName of each provider in the observations listed below. | Stored in the provider dimension table | |
| Demographic observations | Same set as those in ‘patient’, but as observations | Stored in the observation fact table | |
| Problem observations |
ProblemObservationType.value
ProblemConcernEffectiveTime |
SNOMED code Start/end date |
|
| Medication observations |
MedicationProductCode.value
MedicationEffectiveTime |
RxNorm code Start/end date |
|
| Encounter observations |
EncounterFreeText
EncounterEffectiveTime EncounterPerformerPersonName EncounterPlayingEntityName EncounterType.displayName |
Patient note Start/end date Provider name Practice name Encounter subject |
C-CDA, consolidated clinical document architecture; CRC, clinical research chart; i2b2, Informatics for Integrating Biology and the Bedside; MRN, medical record number; SETL, service based extract, transform, and load.
All of the i2b2 components and ontologies described here are available separately. The updated CRC cell will be part of future i2b2 1.7 service packs. The remaining i2b2 components (SETL cell, upgraded SMART-i2b2 cell, and ontologies) will be released on the i2b2 wiki.32,33 OHT-MDMI's map editor will be available on the OHT website.29 We expect site specific changes will only require modifying the C-CDA and i2b2 maps using the OHT-MDMI map editor.
Evaluation
We deployed i2b2 1.7 and our additions on a 3 GHz Intel Xeon virtual machine (VM) inside the secure Partners environment. The VM had 2 GB of RAM and 15 GB of disk space. We created an i2b2 project on the physical SQL Server database (8 processors, 64 GB RAM) used for many of our internal research projects, which we directed our VM to use. To evaluate the import process, we loaded 150 patients into i2b2 from C-CDA documents. We measured the speed and information loaded, and we verified the ability to perform both cohort queries and individual patient review via SMART apps. Because we used our standard production environment, we imagine this is a good average case test of our components.
We used the Partners data repository to select patients between 18 and 65 years seen at a Massachusetts General Hospital or Brigham and Women's Hospital outpatient facility between January 1, 2013 and March 1, 2014. This resulted in 661 420 patients. The average number of observations recorded on each of those patients since January 1, 2013 was 186 (SD 415). Therefore, to choose ‘average’ patients, we selected all of these patients between the average and 1 SD greater than the average (between 186 and 601 observations), resulting in 80 501 patients. We randomly chose 150 of these patients. We removed five of these patients from consideration: in two cases, Partners’ CCD factory could not generate a C-CDA and in the remaining three the supported sections of the C-CDA were empty or contained only invalid dates and unmapped codes.
We imported problems, demographics, medications, and encounter notes for the remaining 145 patients, and recorded both number of observations imported and the time to import them. We also re-ran the import without notes to determine the impact of importing notes. This is instructional for implementers, because notes are not required in the MU2. Server and service load impacted import speed, but we ran our import after 17:00 to minimize this effect. Results are shown in table 2 and figure 3. Each C-CDA was imported in, on average, 3 s without notes and 10 s with notes. Partners’ CCD factory added an additional overhead of 9 ± 5 s/patient. Problem and medication lists were on average five items long and the majority of items were added in the past 3 years (57% for problems, 94% for medications).
Table 2:
Imported elements, number of patients with those elements, and average number of elements per patient, from the cohort of 145
| No of patients | Mean count per patient | Total | |
|---|---|---|---|
| Unmapped | 117 | 2 | 303 |
| Demographics | 145 | 4 | 546 |
| RxNorm | 133 | 5 | 739 |
| SNOMED | 135 | 5 | 741 |
| Subtotal | 14 | 2026 | |
| Notes | 145 | 105 | 15 175 |
| Total | 119 | 17 201 |
Each C-CDA took, on average, 3 s to import without notes and 10 s with notes, not including the Partners specific CCD factory generation time.
CCD, continuity of care document; C-CDA, consolidated clinical document architecture.
We analyzed the completeness of our ontologies against the imported data by comparing the set of imported SNOMED and RxNorm codes to those in our ontologies. We found that 95.9% of the problem list codes are listed in our SNOMED ontology and 97.3% of the medication list codes are listed in our RxNorm ontology. For RxNorm, six of the missing codes were drug combination packs such as ethinyl estradiol/etonogestrel that we are currently adding. The remainder were obsolete or invalid codes. In SNOMED, the missing codes were not diagnoses but procedures or clinical statements (like ‘family history’). Our current SNOMED ontology includes only clinical findings, but we have developed ontologies for other parts of SNOMED that we are considering including. At present, the i2b2 workbench provides an ‘edit terms’ view that is available for adding additional codes to existing ontology trees as necessary.
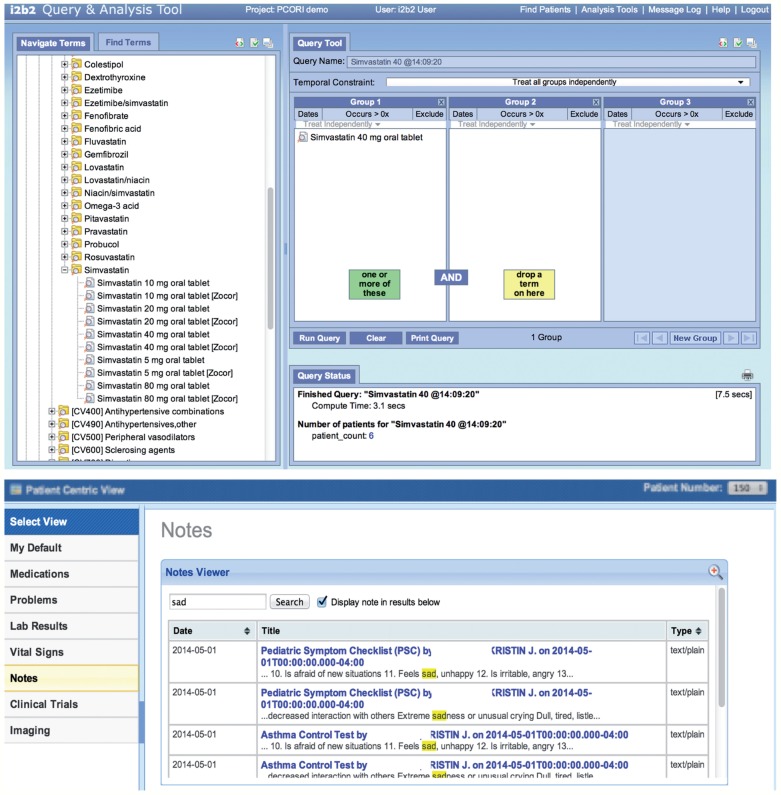
We also selected three random test patients generated by Partners Healthcare. Using the same methodology described above, we imported these three test patients for indepth testing. We created several cohort queries using the ontologies and verified that the counts returned were the same as the same queries run against the imported database. For example, we performed a query for all patients using a simvastatin 40 mg tablet and found that this number was equivalent to the number of patients with a fact entry for ‘RXNORM:198211’. Finally, we ran SMART apps to view information about individual imported patients to verify that notes, medications, and problems displayed correctly. Figure 4 shows a patient count query on the imported data and the note list of a test patient viewed in a SMART app.
Figure 4:
Imported data can be studied in aggregate or by patient. (Top) The Informatics for Integrating Biology and the Bedside (i2b2) query tool shows that six imported patients are using simvastatin 40 mg. (Bottom) The i2b2-SMART note viewer app shows the list of patient notes containing the word ‘sad’ for a test patient.
Discussion
We successfully generated an i2b2 data repository from 145 C-CDA documents representing ‘average’ patients at Partners Healthcare, importing approximately 17 000 facts in about 24 min. These patients had an average medication list and problem list of five entries each, with the majority of data coming from notes. We were able to explore this repository with the graphical query tool and view patient notes, medications, and diagnoses with the SMART apps we previously developed for i2b2.23
This experiment demonstrates that this standards based import approach can successfully populate an i2b2 repository using only C-CDA documents. Our approach combines existing open source tools with new tools and ontologies that we are also in the process of releasing into open source. Our tools are readily extensible to other sites with minimal technical work: only the MDMI maps need to be edited to match local C-CDAs.
Our evaluation represents a reasonable test case for target users: small practices and disease registries such as the PCORnet PPRNs. While not a replacement for the custom direct to database ETL scripts written for site specific high volume import, we expect our approach is more than sufficient for these low volume environments. Small practices can leverage their investment in MU2 compliant EHR technologies that must already produce these C-CDA documents. Disease registries can set up a research repository from patients’ C-CDAs, and patients are increasingly gaining access to these documents due to MU2 requirements.
Limitations and future directions
The primary limitation of our approach is that implementations of C-CDAs are variable, and the documents might be incomplete or inaccurate snapshots of patient care. For disease registries in particular, C-CDAs required by MU2 might not be granular enough to perform meaningful research. The utility of C-CDA data for research and trial recruitment is largely untested, but concerns exist. A recent MITRE report found that the majority of codes referenced by Meaningful Use Clinical Quality Measures are not present in the clinical data in C-CDAs collected by the Massachusetts eHealth Collaborative.34
Practically, implementation of our approach in other C-CDA environments could prove challenging. At Partners, OHT-MDMI's map worked without modification for all required sections of the C-CDA. A recent study showed that across 17 MU2 certified vendor systems, C-CDA documents were generally structurally compatible. This is a reasonable finding, because all certified systems must produce C-CDA documents that pass a structural validation test. However, this study found minor structural differences and a range of semantic issues (eg, improper dates, codes, or coding systems).35 We expect a reduction in these incompatibilities as C-CDA becomes more widely implemented, but the number of map changes required at sites is presently unknown.
At present, our work supports only demographics, encounter notes, medications, and problems, because these are most important in screening for clinical trial eligibility. Other sections of the C-CDA, such as laboratory results and procedures, can be very important for other research questions. Supporting the remainder of the C-CDA will involve expanding the i2b2 map and developing new ontologies for the remaining sections. To import the full richness of C-CDA, the OHT-MDMI map will need to support the optional elements in the standard, and the i2b2 map will need to utilize a C-CDA specific ontology like the one previously developed for Query Health.36
Further ontology development is needed. A national collaboration to coordinate standard ontology development is now underway as part of PCORnet. This collaboration is presently developing common ontologies to enable interoperability and implementation of the PCORnet Common Data Model, with an eye to expanding to Meaningful Use standards.
Conclusion
This work successfully paves a standards-based pathway to reuse Meaningful Use required CCD for clinical research. This could give PCORnet's PPRNs a method to import their patients’ EHR data, and it lowers the barrier to entry for small practices to participate in clinical research initiatives and to study their own patient populations. The tools we have developed and validated are open source extensions to the i2b2 clinical data analytics platform that will be available shortly, and are designed to be easily extensible to other environments.
Supplementary Material
Acknowledgments
Thanks to the other members of the i2b2 team who have made this work possible: Martin Rees and Vivian Gainer. Thanks also to the OHT-MDMI team, with whom we have closely collaborated on this endeavor: Ken Lord, Sean Muir, Gabriel Oancea, and Sally Conway.
Contributors
JGK led the project and the collaboration with OHT-MDMI, wrote the majority of the manuscript, and developed and tested the SETL cell. MM is the lead developer for i2b2, and he upgraded the CRC cell. LCP developed the terminology trees to support C-CDA. APG performed proof of concept work. BHR and HSG developed and maintain the Partners CCD factory, and assisted with design of the CCD factory connector. NW is lead developer for SMART-i2b2 and upgraded the SMART-i2b2 cell. These authors each contributed to the manuscript as it related to their individual contribution. SNM is Director of Research Computing and Information Systems at Partners Healthcare and is the chief architect of i2b2. He provided guidance on the preparation of this manuscript and the design of the SETL cell. All authors gave final approval of the version to be published.
Funding
This work was supported in part by ONC 90TR0001/01, NIH National Library of Medicine U54LM00874, and NIH NIGMS 5R01GM104303-02.
Competing interests
None.
Ethics approval
The study was approved by the institutional review board at Partners Healthcare.
Provenance and peer review
Not commissioned; externally peer reviewed.
Data sharing
The software components developed herein, with the exception of Partners Healthcare specific software, will be available open source on the i2b2 wiki (https://community.i2b2.org/wiki/dis play/SETL/SETL) and the OHT-MDMI website (https://www.projects.openhealthtools.org/sf/projects/mdmi).
REFERENCES
- 1. Envisioning a Transformed Clinical Trials Enterprise in the United States: Establishing an Agenda for 2020: Workshop Summary. http://books.nap.edu/openbook.php?record_id= 13345&page=74. Accessed April 21, 2014.
- 2. Collins FS, Hudson KL, Briggs JP, et al. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21:576–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nalichowski R, Keogh D, Chueh HC, et al. Calculating the benefits of a research patient data repository. AMIA Annu Symp Proc. 2006;2006:1044. [PMC free article] [PubMed] [Google Scholar]
- 4. Frankovich J, Longhurst CA, Sutherland SM. Evidence-based medicine in the EMR era. N Engl J Med. 2011;365:1758–1759. [DOI] [PubMed] [Google Scholar]
- 5. Natter MD, Quan J, Ortiz DM, et al. An i2b2-based, generalizable, open source, self-scaling chronic disease registry. J Am Med Inform Assoc. 2013;20:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc. 2010;17:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. i2b2. Informatics for Integrating Biology and the Bedside. https://www.i2b2.org/. Accessed January 7, 2014.
- 8. Mandl KD, Kohane IS, McFadden D, et al. Scalable Collaborative Infrastructure for a Learning Healthcare System (SCILHS): architecture. J Am Med Inform Assoc. 2014;21:615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc. 2009;16:624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McMurry AJ, Murphy SN, MacFadden D, et al. SHRINE: enabling nationally scalable multi-site disease studies. PLoS ONE. 2013;8:e55811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wattanasin N, Mendis M, Porter A, et al. Components and Workflow for Patient Identification Using i2b2 for Cllinical Trials (i2b2-CT). In: AMIA Joint Summits 2014;2014. http://knowledge.amia.org/amia-56636-cri2014-1.977698/t-003-1.978301/a-060-1.978326/a-060-1.978327/ap-056-1.978328. [Google Scholar]
- 12. i2b2 installations. https://www.i2b2.org/work/i2b2_instal lations.html. Accessed May 16, 2014.
- 13. Weber GM, Kohane IS. Extracting physician group intelligence from electronic health records to support evidence based medicine. PLoS ONE. 2013;8:e64933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klann JG, Szolovits P, Downs S, et al. Decision support from local data: creating adaptive order menus from past clinician behavior. J Biomed Inform. 2014;48:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daugherty SE, Wahba S, Fleurence R, et al. Patient-powered research networks: building capacity for conducting patient-centered clinical outcomes research. J Am Med Inform Assoc. 2014; 21:583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Health Level Seven (HL7). Implementation Guide for CDA® Release 2: IHE Health Story Consolidation, DSTU Release 1.1.Ann Arbor, MI, July2012; Report No: CDAR2_ IG_IHE_CONSOL_DSTU_R1.1_2012JUL. [Google Scholar]
- 17. Meaningful Use Stage 2 Rules Finalized—InformationWeek. Informationweek. http://www.informationweek.com/health care/policy/meaningful-use-stage-2-rules-finalized/24 000 6128. Accessed August 23, 2012.
- 18. Conn J. Blue Button gains fans, apps. Simple tech from VA puts interoperability to work. Mod Healthc. 2012;42:14. [PubMed] [Google Scholar]
- 19. Blue Button Implementation Guide. 2013. http://bluebuttonplus.org/. Accessed February 20, 2014.
- 20. Turvey C, Klein D, Fix G, et al. Blue Button use by patients to access and share health record information using the Department of Veterans Affairs’ online patient portal. J Am Med Inform Assoc. 2014;21:657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Health IT Policy Committee. Request for Comment Regarding the Stage 3 Definition of Meaningful Use of Electronic Health Records. January2013.
- 22. Murphy SN, Gainer V, Chueh HC. A visual interface designed for novice users to find research patient cohorts in a large biomedical database. AMIA Annu Symp Proc. 2003;2003:489–493. [PMC free article] [PubMed] [Google Scholar]
- 23. Wattanasin N, Porter A, Ubaha S, et al. Apps to display patient data, making SMART available in the i2b2 platform. Proceedings of the AMIA Symposium; 2012. [PMC free article] [PubMed] [Google Scholar]
- 24. The i2b2 Community Wiki. I2b2 Wiki 2014. https://com munity.i2b2.org/wiki/. Accessed March 13, 2014.
- 25. Giovinazzo WA. Object-oriented data warehouse design: building a star schema. Upper Saddle River, NJ, USA: Prentice Hall PTR, 2000. [Google Scholar]
- 26. i2b2. NCBO Ontology Tools—i2b2 Sponsored Project. https://community.i2b2.org/wiki/display/NCBO/NCBO+Ontology+Tools. Accessed February 19, 2013.
- 27. OpenHealthTools. http://www.openhealthtools.org/. Accessed Junuary 6, 2014.
- 28. OHT/MDMI Project. SemantX Inc 2013. http://www.semant xinc.com/ohtmdmi-project.html. Accessed February 20, 2014.
- 29. Open Health Tools’ Model-Driven Message Interoperability. 2014. https://www.projects.openhealthtools.org/sf/projec ts/mdmi/. Accessed April 23, 2014.
- 30. Goldberg HS, Paterno MD, Rocha BH, et al. A highly scalable, interoperable clinical decision support service. J Am Med Inform Assoc. 2014;21:e55–e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murphy SN, Morgan MM, Barnett GO, et al. Optimizing healthcare research data warehouse design through past COSTAR query analysis. Proc AMIA Symp. 1999;1999:892–896. [PMC free article] [PubMed] [Google Scholar]
- 32. Query Health Reference Implementation. Standards and Interoperability Framework http://code.google.com/p/query-health/source/browse
- 33. SETL—Standards-based Extract, Transform, and Load. I2b2 Wiki 2014. https://community.i2b2.org/wiki/display/SETL/Home. Accessed April 23, 2014. [Google Scholar]
- 34. Hadley M, McCready R, Delano D, et al. Assessing the Feasibility of Meaningful Use Stage 2 Clinical Quality Measure Data Elements at the Massachusetts eHealth Collaborative. September2013; Report No: MITRE 13-3360 http://projectkamira.org/docs/kamira_cqm_data_feasibility_maehc.pdf
- 35. D'Amore JD, Mandel JC, Kreda DA, et al. Are meaningful use stage 2 certified EHRs ready for interoperability? Findings from the SMART C-CDA Collaborative. J Am Med Inform Assoc. 2014;21:1060–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klann JG, Buck MD, Brown J, et al. Query Health: standards-based, cross-platform population health surveillance. J Am Med Inform Assoc. 2014;21:650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.