Abstract
Pathogenic variants in BRCA1 or BRCA2 are identified in ∼20% of families with multiple individuals affected by early-onset breast and/or ovarian cancer. Extensive searches for additional highly penetrant genes or alternative mutational mechanisms altering BRCA1 or BRCA2 have not explained the missing heritability. Here, we report a dominantly inherited 5′ UTR variant associated with epigenetic BRCA1 silencing due to promoter hypermethylation in two families affected by breast and ovarian cancer. BRCA1 promoter methylation of ten CpG dinucleotides in families who are affected by breast and/or ovarian cancer but do not have germline BRCA1 or BRCA2 pathogenic variants was assessed by pyrosequencing and clonal bisulfite sequencing. RNA and DNA sequencing of BRCA1 from lymphocytes was undertaken to establish allelic expression and the presence of germline variants. BRCA1 promoter hypermethylation was identified in 2 of 49 families in which multiple women are affected by grade 3 breast cancer or high-grade serous ovarian cancer. Soma-wide BRCA1 promoter hypermethylation was confirmed in blood, buccal mucosa, and hair follicles. Pyrosequencing showed that DNA was ∼50% methylated, consistent with the silencing of one allele, which was confirmed by clonal bisulfite sequencing. RNA sequencing revealed the allelic loss of BRCA1 expression in both families and that this loss of expression segregated with the heterozygous variant c.−107A>T in the BRCA1 5′ UTR. Our results establish a mechanism whereby familial breast and ovarian cancer is caused by an in cis 5′ UTR variant associated with epigenetic silencing of the BRCA1 promoter in two independent families. We propose that methylation analyses be undertaken to establish the frequency of this mechanism in families affected by early-onset breast and/or ovarian cancer without a BRCA1 or BRCA2 pathogenic variant.
Keywords: BRCA1, promoter hypermethylation, familial breast and ovarian cancer, epigenetic, secondary epimutation
Introduction
Breast cancer (MIM: 114480) is the most common form of cancer in women.1 Germline heterozygous pathogenic variants in BRCA1 (MIM: 113705) and BRCA2 (MIM: 600185) account for 2%–3% of all cases2 and up to 15% of cases of epithelial ovarian cancer (MIM: 167000).3 In families with multiple individuals affected by early-onset disease, these percentages increase substantially: BRCA1 and BRCA2 variants explain approximately 20% of familial breast cancer and a higher proportion of familial ovarian cancer.4
Over the past 20 years, there have been exhaustive efforts to identify other breast and ovarian cancer susceptibility genes. This missing heritability has been postulated to be due to other highly penetrant genes, including TP53 (MIM: 191170); genes of modest effect, including PALB2 (MIM: 610355) and ATM (MIM: 607585); or polygenic risks due to the combination of multiple small-effect variants.5 However, no other genes that confer a high risk of both breast and ovarian cancer have been identified.
Genetic testing by DNA sequencing and copy-number analysis for pathogenic exonic variants in BRCA1 and BRCA2 is highly sensitive (it is estimated to detect over 90% of pathogenic variants)6, 7 and is now offered routinely to individuals at high familial risk of breast and/or ovarian cancer. Our previous studies using RNA sequencing in high-risk families have shown that deep intronic variants in BRCA1 or BRCA2 do not contribute significantly to this mutational spectrum.7 Detection of pathogenic variants is important for determining appropriate cancer surveillance for at-risk relatives, for reassuring relatives without the familial causative variant of their risk and removing the burden of unnecessary screening, and informing treatment choice, especially for poly ADP ribose polymerase (PARP) inhibitors.8
Gene promoter methylation has been proposed as an alternative mechanism for the transcriptional silencing of cancer-associated genes.9 Promoter hypermethylation has been associated with tumor-suppressor genes, both in the germline and as a somatic (acquired) event in tumor tissue,9 and results in transcriptional silencing.
Promoter hypermethylation of BRCA1 is present in the tumor tissue of approximately 10% of sporadic breast cancers10, 11 and in breast tumors of women with BRCA1 germline pathogenic variants12 and is more common in triple-negative (estrogen receptor, progesterone receptor, and HER2) breast cancer.13 Constitutional methylation of the BRCA1 promoter has been reported in individuals with breast cancer,14 but this has always been at low “mosaic” levels (maximum 20%), and there has been no convincing evidence that this is inherited from one generation to the next. In contrast, inherited variants associated with promoter hypermethylation of MLH1 (MIM: 120436)15 and MSH2 (MIM: 609309)16 have been reported in familial colorectal cancer (MIM: 114500). In this study, we describe two families who are affected by breast and ovarian cancer and carry an inherited germline variant that results in transcriptional silencing of BRCA1 through promoter hypermethylation (secondary epimutation). This mutational mechanism for BRCA1 has important implications for diagnostic testing of individuals at high risk of breast and/or ovarian cancer and for optimum treatment selection.17
Material and Methods
Subjects and Family Members
Screening for BRCA1 promoter methylation was undertaken in the lymphocyte-derived DNA of 49 unrelated individuals from families affected by breast and/or ovarian cancer and with a Manchester score > 34 without a germline BRCA1 or BRCA2 pathogenic variant. A Manchester score represents the likelihood of detecting a pathogenic variant in BRCA1 or BRCA2.7, 18, 19 In our local population, 158 of 220 (71.8%) families with a Manchester score > 34 have had pathogenic variants in BRCA1 or BRCA2 identified by conventional genetic testing of DNA sequencing and multiplex ligation-dependent probe analysis (MLPA).
Blood, buccal mucosa, tumor, and hair samples were collected (where possible) from affected and unaffected family members with breast or ovarian cancer when BRCA1 promoter methylation was detected. Cancer diagnoses were confirmed from hospital records or through the North West (England) Cancer Intelligence Service, which has data on all individuals with any malignancy from 1960 onward. DNA was extracted from blood by Chemagen (Perkin Elmer), from hair with the QIAamp DNA Investigator Kit (QIAGEN), from buccal mucosa with the QIAGEN EZ1 system, and from tumor cells with the Cobas DNA Sample Preparation Kit (Roche). The study was approved by the Central Manchester Research Ethics Committee (10/H1008/24 and 11/H1003/3), and written informed consent was obtained from each participant.
BRCA1 Promoter Methylation Assays
Genomic DNA was bisulfite converted with the EZ DNA Methylation Kit (Zymo Research) for distinguishing between methylated and unmethylated DNA. BRCA1 promoter methylation was determined by pyrosequencing (QIAGEN) across 10 CpG dinucleotides within the BRCA1 promoter. The core promoter of BRCA1 encompasses the non-coding exon 1 and part of intron 1 of BRCA1 and exon 1 and part of intron 1 of the neighboring gene NBR2, as annotated by Ensembl (chr17: 43,168,800–43,172,601). The 10 CpG dinucleotides fall within the non-coding exon 1 of BRCA1. The methylation status was quantified in DNA derived from hair follicles, buccal mucosal cells, peripheral-blood lymphocytes, and tumor cells (Supplemental Material and Methods).
Clonal bisulfite sequencing on a minimum of 37 clones was performed for determining whether the methylation pattern was allele specific (Supplemental Material and Methods).
RNA and DNA Analysis
To measure BRCA1 expression, we collected whole blood in PAXgene Blood RNA tubes (PreAnalytiX) and extracted RNA. RNA was converted to cDNA by RT-PCR with the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). We genotyped five SNPs in BRCA1 exon 11 (rs1799949, rs16940, rs799917, rs16941, and rs16942) by Sanger sequencing to determine whether there was a difference in allelic ratios between the RNA and DNA genotypes and thus silencing of one allele (Supplemental Material and Methods and Table S1).
Haplotype Analysis
To determine relatedness between families identified with BRCA1 promoter methylation, we genotyped 12 BRCA1 intragenic SNPs by Sanger sequencing to determine ancestral haplotypes (Supplemental Material and Methods).20 In addition, genotyping using Affymetrix Genome-Wide SNP6.0 arrays was undertaken according to the manufacturer’s protocol. Genotypes and copy-number data were generated within the Affymetrix Genotyping Console (v.4.1.3.840) via the Birdseed V2 algorithm and SNP 6.0 CN/LOH algorithm, respectively.
Whole-Genome Sequencing
We performed whole-genome sequencing in order to identify any potential unique variants present in individuals with promoter methylation and not in unaffected individuals. PCR-free paired-end whole-genome sequencing (TruSeq DNA PCR-Free, Illumina) was undertaken on a HiSeqX platform. Reads were aligned against the human assembly GRCh38 (UCSC Genome Browser) via the Burrows-Wheeler Aligner (v.0.6.2), and variants were called with the Genome Analysis Toolkit (3.4-0-g7e26428). Annotation was performed with Ensembl v.89 and compared with variation identified in the Genome Aggregation Database (gnomAD)21 (Supplemental Material and Methods).
Results
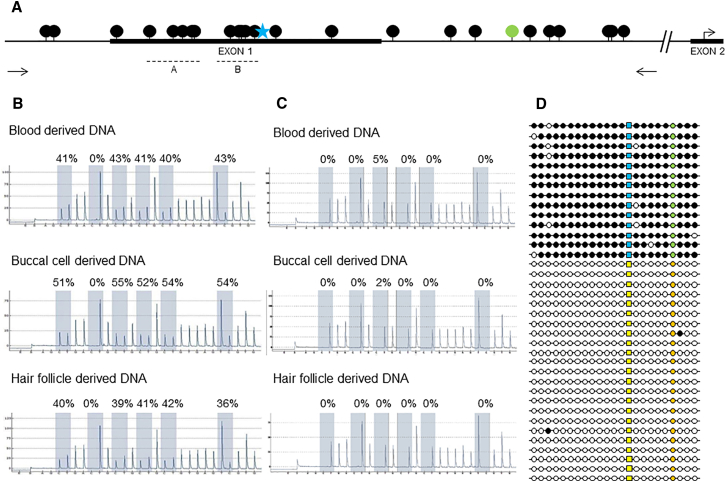
To determine whether promoter hypermethylation of BRCA1 could result in familial breast and/or ovarian cancer, we undertook methylation assays. BRCA1 promoter hypermethylation was identified in two women from a screen of 49 unrelated individuals with familial breast and/or ovarian cancer (and a Manchester score > 34) in whom previous Sanger sequencing and MLPA of BRCA1 and BRCA2 coding exons had not identified a pathogenic single-nucleotide or copy-number variant. In individuals with a Manchester score > 34, there is a >70% likelihood of detecting a BRCA1 or BRCA2 germline pathogenic variant.7, 18, 19 Promoter hypermethylation was detected in a woman (II-4 in family 1; Figure 1A) with a strong family history of breast cancer and in whom breast cancer was diagnosed at 39 years of age and a poorly differentiated serous ovarian cancer was diagnosed at 48 years (Manchester score 43) and in a woman (III-2 in family 2; Figure 1B) in whom bilateral grade 3 triple-negative breast cancer was detected at 38 and 46 years of age (Manchester score 35). In the two women, pyrosequencing assays on lymphocyte-derived DNA were consistent with BRCA1 promoter hypermethylation across 10 CpG dinucleotides (Figure 2A) (averages 43% and 41%), indicating that one allele was fully methylated (Table 1, Figures 2B and 2C, and Figure S1). This hypermethylation pattern was consistent in DNA extracted from buccal mucosa (54% and 69%) and hair follicles (38% and 43%) (Table S2), representing endoderm and ectoderm derived tissues, respectively. Clonal bisulfite sequencing orthogonally confirmed the BRCA1 promoter hypermethylation pattern in the two affected women (Figure 2D).
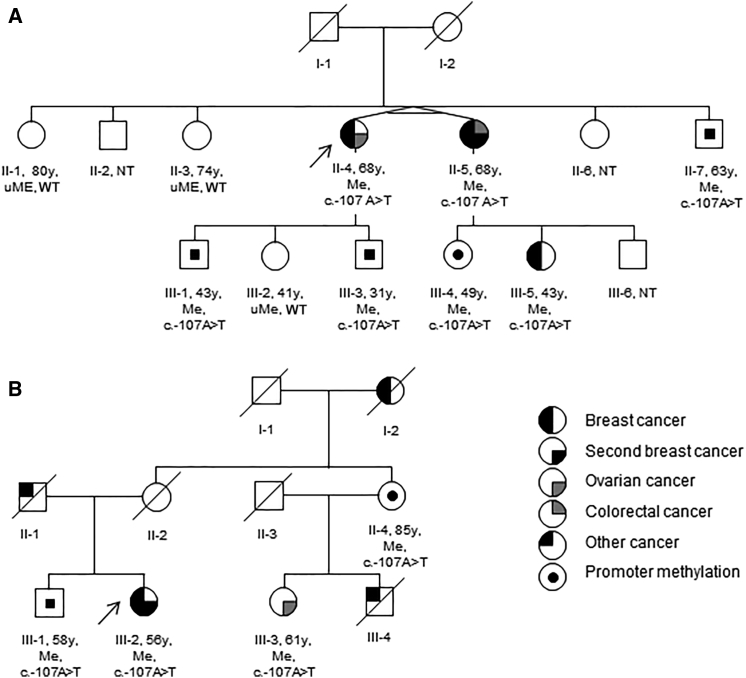
Figure 1.
Pedigrees of Families Carrying the 5′ UTR BRCA1 Variant
Pedigrees of family 1 (A) and 2 (B). Abbreviations are as follows: Y, age (in years) tested; uMe, unmethylated BRCA1 promoter; Me, methylated BRCA1 promoter; WT, wild-type; and NT, not tested. Arrows indicate probands.
Figure 2.
Methylation Analysis of BRCA1 Promoter Region
(A) Schematic overview of BRCA1 promoter region (black dots, CpG sites; blue star, c.−107; green dot, rs799905; arrows, primer locations for clonal bisulfite sequencing; dotted lines, pyrosequencing regions [A and B]).
(B and C) Representative pyrograms (region B) show the level of BRCA1 promoter methylation in lymphocytes, buccal mucosa, and hair-derived DNA of an affected and unaffected individual. Five CpGs and a control site (0%) (to ensure complete bisulfite conversion) are shaded, and the level of methylation as a ratio of C:T peak heights is calculated at each site (representing methylated versus unmethylated cytosine). (B) Affected individual II-4 from family 1. (C) Unaffected individual II-1 from family 1. Further pyrogram data (region A) indicating methylation across the BRCA1 promoter are available in Figure S1.
(D) Schematic overview of clonal bisulfite sequencing results. Allelic discrimination is made on the basis of rs799905 C>G (orange, C; green, G). The variant c.−107A>T is present on the methylated allele (yellow, A; blue, T; black, methylated; white, unmethylated).
Table 1.
Summary of BRCA1 Promoter Methylation Status in Lymphocyte-Derived DNA and Clinical Phenotypes and Genotypes for the c.−107A>T Variant in All Tested Individuals
| Individual | BRCA1 Promoter Methylation (Mean %) | c.−107A>T | Clinical Status (Age at Diagnosis in Years) | Sex | Age Tested (Years) |
|---|---|---|---|---|---|
| Family 1 | |||||
| II-1 | 1 | AA | unaffected | female | 80 |
| II-3 | 0 | AA | unaffected | female | 74 |
| II-4 | 43 | AT | breast (39) and ovarian (48) cancer | female | 68 |
| II-5 | 37 | AT | bilateral breast cancer (30 and 32), colorectal cancer (64) | female | 68 |
| II-7 | 41 | AT | unaffected | male | 63 |
| III-1 | 38 | AT | unaffected | male | 43 |
| III-2 | 1 | AA | unaffected | female | 41 |
| III-3 | 44 | AT | unaffected | male | 31 |
| III-4 | 41 | AT | unaffected | female | 49 |
| III-5 | 32 | AT | breast cancer (39) | female | 43 |
| Family 2 | |||||
| II-4 | 44 | AT | unaffected | female | 85 |
| III-1 | 44 | AT | unaffected | male | 58 |
| III-2 | 41 | AT | bilateral breast cancer (38 and 46) | female | 56 |
| III-3 | 43 | AT | ovarian cancer (48) | female | 61 |
Segregation analysis for BRCA1 promoter hypermethylation was undertaken in the two families. In family 1, the proband’s identical twin (II-5), affected by bilateral grade 3 breast cancer at age 30 and 32 years (no receptor status available) and colorectal cancer at 64 years, and II-5’s daughter (III-5), who had been affected by high-grade triple-negative breast cancer at 39 years (Figure 1A), both had hypermethylation of the BRCA1 promoter at allele frequencies similar to that of the proband (Table 1 and Table S2). Samples from the parents of the affected twins were not available, but both were deceased and neither had a history of cancer.
Samples from seven other family members (II-1, II-2, II-7, III-1, III-2, III-3 and III-4) were available. Of these, four showed a soma-wide hypermethylated BRCA1 promoter in blood, buccal mucosa, and hair follicles, and three showed a normal methylation pattern (Table 1, Table S2, Figures 2B and 2C, and Figure S1). In family 2, the maternal first cousin (III-3) of the proband (III-2) had been diagnosed with high-grade serous ovarian cancer at 48 years and also had soma-wide hypermethylation of the BRCA1 promoter. The mother of III-2, who had no history of cancer, was deceased (as a result of myocardial infarction at 76 years of age). Her sister and the mother of III-3 (II-4), who also had no history of cancer, was alive at 85 years and had a similar level of hypermethylation (43%) as her affected daughter and niece. The healthy brother (III-1) of the proband also showed hypermethylation of the BRCA1 promoter.
DNA extracted from formalin-fixed paraffin-embedded breast tumor was available from individual III-5 (family 1). Genotyping showed loss of the wild-type allele across five informative intragenic SNPs (Table S3) (i.e., only the alleles of the variants not expressed in the cDNA were present), consistent with loss of BRCA1 as the second hit in the tumor tissue.
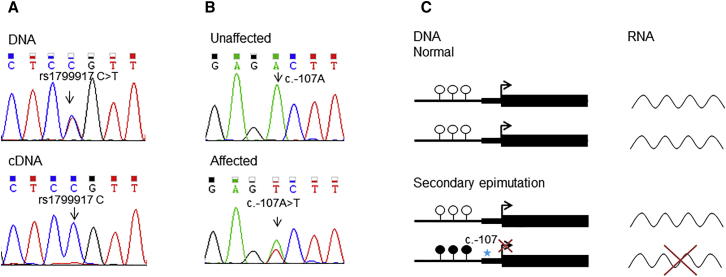
Expression analysis of BRCA1 in RNA extracted from lymphocytes was undertaken in individuals with promoter hypermethylation. Absence of heterozygosity across five SNPs with high minor allele frequencies within the BRCA1 cDNA suggested allelic imbalance (Figure 3A) secondary to the loss of expression of one allele as a result of hypermethylation of the BRCA1 promoter (Figure 3C). Sanger sequencing upstream of the BRCA1 translation start site identified the heterozygous variant c.−107A>T (g.43125358A>T [GenBank: NM_007294.3]) in a woman affected by BRCA1 promoter hypermethylation in each family (Figure 3B). This variant segregated with the hypermethylated BRCA1 allele in all tested individuals in both families and was absent in individuals lacking the hypermethylated allele, confirming that it was in cis (Table 1 and Table S2). None of the other 47 families carried this variant. This variant was absent in gnomAD,21 a database that includes whole-exome and whole-genome sequencing data on 123,136 and 15,496 individuals, respectively. The variant has not been reported in any individual with breast or ovarian cancer in disease-specific databases, including the BRCA Exchange.
Figure 3.
DNA and RNA Analysis of BRCA1
(A) Representative Sanger sequencing traces demonstrating allelic loss of expression of rs1799917 C>T in exon 11 of BRCA1. In the DNA trace, both the C and T nucleotides are present, whereas in the cDNA trace only the C nucleotide is present.
(B) Representative Sanger sequencing traces for the heterozygous c.−107A>T variant, which is present in the individual with a methylated BRCA1 promoter and absent in an individual with an unmethylated promoter.
(C) Schematic representation of the normal pattern of gene expression and transcription and abnormal gene expression and transcription caused by a germline variant (c.−107), the latter of which results in hypermethylation of the promoter (secondary epimutation) and silencing of one allele.
The two families (both non-consanguineous white British families from North West England) were not knowingly related to each other. All individuals in the two families with promoter hypermethylation and the c.−107A>T variant carried the previously described B1 haplotype (Tables S4 and S5).20 To identify any additional germline variants that could result in promoter hypermethylation, we undertook SNP arrays and whole-genome sequencing. SNP array analysis of II-5, III-2, and III-5 (family 1) and III-2 and III-3 (family 2) did not identify any other rare or unreported copy-number variants. Whole-genome sequencing analysis was restricted to a candidate region (chr17: 42,044,295–44,215,483, UCSC Genome Browser hg38) 1 Mb upstream and downstream of BRCA1. We performed segregation analyses to identify variants in a heterozygous state in the two unrelated affected individuals (III-5 in family 1 and III-2 in family 2) and absent in the unaffected individual (II-2 in family 1). This restricted analysis identified 14 variants that were absent from both the gnomAD dataset and dbSNP. Two variants (one in intron 2, c.80+661_80+667delAAAAAAA [g.43123349–43123356delAAAAAAA (GenBank: NM_007294.3)] [Supplemental Material and Methods and Figure S3], and the previously identified c.−107A>T) were determined to be within the genomic region for BRCA1. Three variants within the candidate interval were present within DNase I hypersensitivity sites characterized across 125 cell types. In combination, these analyses identified c.−107A>T as single candidate variant linked to hypermethylation of the promoter (Figure S2).
Discussion
Here, over 20 years after the initial report that pathogenic variants in BRCA1 result in familial breast cancer,22 we demonstrate a previously undescribed dominantly inherited 5′ UTR variant associated with epigenetic silencing of BRCA1 in two families affected by early-onset breast and ovarian cancer. A constitutional epimutation describes an epigenetic change (e.g., promoter hypermethylation) that results in the transcriptional silencing of a gene that is normally active across a range of normal tissues and predisposes to disease. Sloane et al.23 set out four criteria for establishing the presence of a constitutional epimutation, and these criteria were met in our two families in that promoter hypermethylation is confined to one allele in normal tissues derived from the mesoderm (blood), hair follicles (ectoderm), and buccal mucosa (endoderm); the level (∼50%) and presence of hypermethylation are demonstrated by at least two independent methods (pyrosequencing and clonal bisulfite sequencing); and the methylated allele is transcriptionally silent and co-segregates with the phenotype.23
Inherited variants resulting in epigenetic silencing have rarely been described in familial cancer, notably in Lynch syndrome, which is due to hypermethylation of the MLH1 promoter15 or MSH2 promoter.16 MLH1 promoter hypermethylation has been reported both in the context of a cis-acting germline variant, c.−27C>A, and more recently c.−63−delins18 (secondary epimutations) and in the absence of any detectable genetic alteration (primary epimutation).24, 25 In contrast, MSH2 promoter hypermethylation has always been associated with a cis-acting deletion encompassing the 3′ end of the adjacent EPCAM.16, 26 Here, we identified a BRCA1 exon 1 variant, c.−107A>T, in cis with the hypermethylated promoter and confirmed that it segregates with the phenotype in both families.
In these families, we found no evidence to determine whether male-to-female vertical transmission of BRCA1 promoter methylation results in a breast or ovarian cancer phenotype in the next generation. Future predictive testing of the at-risk daughters of male carriers will be able to establish this. However, given that there is a linked upstream variant (c.−107A>T), it is likely that transmission will result in promoter methylation and a phenotype.
The c.−107A>T BRCA1 variant is found on an ancestral B1 haplotype20 in both families. Although the families are not known to be related to each other, this indicates that the two families could share a common ancestry. It will be important to determine whether this variant occurs in other affected individuals to establish whether this variant has arisen more than once and whether other non-coding variants can result in BRCA1 promoter hypermethylation. The c.−107 nucleotide is not highly conserved through mammalian species, and in silico tools are not informative when predicting its pathogenicity. Notably, exon 1 is not normally sequenced in clinical BRCA1 testing, and so the c.−107A>T variant would not have been detected by routine testing. Even if it had been identified by sequence analysis, without the methylation studies it would be classified as a variant of unknown significance. Therefore, studies of promoter methylation should clarify the functional effect of all rare or previously unreported 5′ variants in BRCA1. The specific mechanism by which the 5′ variant results in promoter hypermethylation remains unknown.
Importantly, the clinical presentation of the affected individuals in the two families is consistent with the phenotype in other families affected by BRCA1 pathogenic variants and does not indicate any specific clinical features that would prioritize individuals with familial breast or ovarian cancer without coding BRCA1 pathogenic variants for methylation analysis. Although based on two families, the penetrance of the variant causing a hypermethylated BRCA1 promoter is 71.4% in informative women. This is consistent with estimates of cumulative risks by age 80 years for females with pathogenic BRCA1 variants of 75% for breast cancer.5 The two unaffected female variant carriers were born before 1940, when the penetrance of BRCA1 pathogenic variants was much lower.27 For the male relatives, as expected, there is no evidence of an elevated cancer risk.28 Variable (mosaic) levels of BRCA1 promoter methylation were detected in normal somatic tissues from individuals carrying the 5′ variant and ranged from 24% in hair in individual II-5 in family 1 to 69% in buccal mucosa in individual III-2 in family 2; both women have bilateral breast cancer. There is no correlation between these levels of promoter methylation and the clinical phenotype; for example, the variant carrier (II-4) in family 2 has >40% methylation but does not have cancer at 85 years of age.
We detected the secondary epimutation in 2 of 49 families ascertained in North West England with a Manchester score > 34. Therefore, this mechanism accounts for at least 1.25% of BRCA1 pathogenic variants in our very high-risk familial breast and ovarian cancer cohort and increases sensitivity from 71.8% to at least 72.7% in families with a high likelihood of a BRCA1 or BRCA2 pathogenic variant. Therefore, this mechanism is more common in our population than deep intronic mutations.7 Further, in our familial breast and ovarian cancer cohort, next-generation sequencing of a panel of genes associated with an increased risk of breast cancer increased the diagnostic yield for familial breast cancer by a similar amount but revealed variants in genes (ATM and CHEK2 [MIM: 604373]) with less clear actionability.5, 7 The uplift achieved by methylation testing would argue that testing for BRCA1 promoter methylation is a valuable adjunct to sequence and copy-number analysis for individuals with a strong family history of breast and/or ovarian cancer.
In summary, we have identified two families carrying the dominantly inherited 5′ UTR variant c.−107T>A linked to allele-specific promoter methylation of BRCA1; it is present in all three germ layers and results in transcriptional silencing of one allele. This mechanism could explain some of the missing heritability in families affected by familial breast and/or ovarian cancer.
Declaration of Interests
T.A. declares an honorarium from Illumina for speaking.
Acknowledgments
We thank the family members for their participation and co-operation. This work was funded by Prevent Breast Cancer (GA 12-006 and GA 15-002) and the Manchester NIHR Biomedical Research Centre (IS-BRC-1215-20007). D.G.R.E. is an NIHR Senior Investigator (NF-SI-0513-10076). J.M.E. is funded by a postdoctoral research fellowship from the Health Education England Genomics Education Programme (HEE GEP). The views expressed in this publication are those of the authors and not necessarily those of the HEE GEP. No funding bodies had a role in the design or conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. D.G.R.E. and E.M.v.V. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All tested individuals gave consent to present their personal information in this manuscript. We thank Jeanette Rothwell for sample collection and Natasha Leo for technical assistance, as well as the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about.
Published: August 2, 2018
Footnotes
Supplemental Data include three figures, five tables, and Supplemental Material and Methods, including BRCA1 promoter methylation assays, DNA and RNA analysis, haplotype analysis, whole-genome sequencing, and high-resolution capillary electrophoresis and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.07.002.
Contributor Information
D.Gareth R. Evans, Email: gareth.evans@mft.nhs.uk.
William G. Newman, Email: william.newman@manchester.ac.uk.
Accession Numbers
The accession number for the c.−107A>T BRCA1 variant reported in this paper is LOVD: BRCA1_005077.
Web Resources
BRCA Exchange, http://brcaexchange.org
Ensembl, https://www.ensembl.org
Genome Aggregation Database, http://gnomad.broadinstitute.org
OMIM, http://omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Anglian Breast Cancer Study Group Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br. J. Cancer. 2000;83:1301–1308. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsop K., Fereday S., Meldrum C., deFazio A., Emmanuel C., George J., Dobrovic A., Birrer M.J., Webb P.M., Stewart C. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: A report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012;30:2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eccles S.A., Aboagye E.O., Ali S., Anderson A.S., Armes J., Berditchevski F., Blaydes J.P., Brennan K., Brown N.J., Bryant H.E. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15:R92. doi: 10.1186/bcr3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Easton D.F., Pharoah P.D., Antoniou A.C., Tischkowitz M., Tavtigian S.V., Nathanson K.L., Devilee P., Meindl A., Couch F.J., Southey M. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palma M.D., Domchek S.M., Stopfer J., Erlichman J., Siegfried J.D., Tigges-Cardwell J., Mason B.A., Rebbeck T.R., Nathanson K.L. The relative contribution of point mutations and genomic rearrangements in BRCA1 and BRCA2 in high-risk breast cancer families. Cancer Res. 2008;68:7006–7014. doi: 10.1158/0008-5472.CAN-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byers H., Wallis Y., van Veen E.M., Lalloo F., Reay K., Smith P., Wallace A.J., Bowers N., Newman W.G., Evans D.G. Sensitivity of BRCA1/2 testing in high-risk breast/ovarian/male breast cancer families: Little contribution of comprehensive RNA/NGS panel testing. Eur. J. Hum. Genet. 2016;24:1591–1597. doi: 10.1038/ejhg.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., Friedlander M., Arun B., Loman N., Schmutzler R.K. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 9.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 10.Rice J.C., Massey-Brown K.S., Futscher B.W. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M., Silva J.M., Dominguez G., Bonilla F., Matias-Guiu X., Lerma E., Bussaglia E., Prat J., Harkes I.C., Repasky E.A. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 12.Tapia T., Smalley S.V., Kohen P., Muñoz A., Solis L.M., Corvalan A., Faundez P., Devoto L., Camus M., Alvarez M., Carvallo P. Promoter hypermethylation of BRCA1 correlates with absence of expression in hereditary breast cancer tumors. Epigenetics. 2008;3:157–163. doi: 10.4161/epi.3.3.6387. [DOI] [PubMed] [Google Scholar]
- 13.Yamashita N., Tokunaga E., Kitao H., Hitchins M., Inoue Y., Tanaka K., Hisamatsu Y., Taketani K., Akiyoshi S., Okada S. Epigenetic inactivation of BRCA1 through promoter hypermethylation and its clinical importance in triple-negative breast cancer. Clin. Breast Cancer. 2015;15:498–504. doi: 10.1016/j.clbc.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Hansmann T., Pliushch G., Leubner M., Kroll P., Endt D., Gehrig A., Preisler-Adams S., Wieacker P., Haaf T. Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Hum. Mol. Genet. 2012;21:4669–4679. doi: 10.1093/hmg/dds308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hitchins M.P., Wong J.J., Suthers G., Suter C.M., Martin D.I., Hawkins N.J., Ward R.L. Inheritance of a cancer-associated MLH1 germ-line epimutation. N. Engl. J. Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 16.Ligtenberg M.J., Kuiper R.P., Chan T.L., Goossens M., Hebeda K.M., Voorendt M., Lee T.Y., Bodmer D., Hoenselaar E., Hendriks-Cornelissen S.J. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 17.Stefansson O.A., Villanueva A., Vidal A., Martí L., Esteller M. BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy in breast and ovarian cancer. Epigenetics. 2012;7:1225–1229. doi: 10.4161/epi.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans D.G., Eccles D.M., Rahman N., Young K., Bulman M., Amir E., Shenton A., Howell A., Lalloo F. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J. Med. Genet. 2004;41:474–480. doi: 10.1136/jmg.2003.017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans D.G., Harkness E.F., Plaskocinska I., Wallace A.J., Clancy T., Woodward E.R., Howell T.A., Tischkowitz M., Lalloo F. Pathology update to the Manchester Scoring System based on testing in over 4000 families. J. Med. Genet. 2017;54:674–681. doi: 10.1136/jmedgenet-2017-104584. [DOI] [PubMed] [Google Scholar]
- 20.Frosk P., Burgess S., Dyck T., Jobse R., Spriggs E.L. The use of ancestral haplotypes in the molecular diagnosis of familial breast cancer. Genet. Test. 2007;11:208–215. doi: 10.1089/gte.2006.0518. [DOI] [PubMed] [Google Scholar]
- 21.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 23.Sloane M.A., Ward R.L., Hesson L.B. Defining the criteria for identifying constitutional epimutations. Clin. Epigenetics. 2016;8:39. doi: 10.1186/s13148-016-0207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitchins M.P., Rapkins R.W., Kwok C.T., Srivastava S., Wong J.J., Khachigian L.M., Polly P., Goldblatt J., Ward R.L. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5’UTR. Cancer Cell. 2011;20:200–213. doi: 10.1016/j.ccr.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Morak M., Ibisler A., Keller G., Jessen E., Laner A., Gonzales-Fassrainer D., Locher M., Massdorf T., Nissen A.M., Benet-Pagès A., Holinski-Feder E. Comprehensive analysis of the MLH1 promoter region in 480 patients with colorectal cancer and 1150 controls reveals new variants including one with a heritable constitutional MLH1 epimutation. J. Med. Genet. 2018;55:240–248. doi: 10.1136/jmedgenet-2017-104744. [DOI] [PubMed] [Google Scholar]
- 26.Hitchins M.P. Constitutional epimutation as a mechanism for cancer causality and heritability? Nat. Rev. Cancer. 2015;15:625–634. doi: 10.1038/nrc4001. [DOI] [PubMed] [Google Scholar]
- 27.Evans D.G., Shenton A., Woodward E., Lalloo F., Howell A., Maher E.R. Penetrance estimates for BRCA1 and BRCA2 based on genetic testing in a Clinical Cancer Genetics service setting: Risks of breast/ovarian cancer quoted should reflect the cancer burden in the family. BMC Cancer. 2008;8:155. doi: 10.1186/1471-2407-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran A., O’Hara C., Khan S., Shack L., Woodward E., Maher E.R., Lalloo F., Evans D.G. Risk of cancer other than breast or ovarian in individuals with BRCA1 and BRCA2 mutations. Fam. Cancer. 2012;11:235–242. doi: 10.1007/s10689-011-9506-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.