Abstract

2-Deoxyribose-5-phosphate aldolase (DERA) catalyzes the reversible conversion of acetaldehyde and glyceraldehyde-3-phosphate into deoxyribose-5-phosphate. DERA is used as a biocatalyst for the synthesis of drugs such as statins and is a promising pharmaceutical target due to its involvement in nucleotide catabolism. Despite previous biochemical studies suggesting the catalytic importance of the C-terminal tyrosine residue found in several bacterial DERAs, the structural and functional basis of its participation in catalysis remains elusive because the electron density for the last eight to nine residues (i.e., the C-terminal tail) is absent in all available crystal structures. Using a combination of NMR spectroscopy and molecular dynamics simulations, we conclusively show that the rarely studied C-terminal tail of E. coli DERA (ecDERA) is intrinsically disordered and exists in equilibrium between open and catalytically relevant closed states, where the C-terminal tyrosine (Y259) enters the active site. Nuclear Overhauser effect distance restraints, obtained due to the presence of a substantial closed state population, were used to derive the solution-state structure of the ecDERA closed state. Real-time NMR hydrogen/deuterium exchange experiments reveal that Y259 is required for efficiency of the proton abstraction step of the catalytic reaction. Phosphate titration experiments show that, in addition to the phosphate-binding residues located near the active site, as observed in the available crystal structures, ecDERA contains previously unknown auxiliary phosphate-binding residues on the C-terminal tail which could facilitate in orienting Y259 in an optimal position for catalysis. Thus, we present significant insights into the structural and mechanistic importance of the ecDERA C-terminal tail and illustrate the role of conformational sampling in enzyme catalysis.
Keywords: aldolase, TIM-barrel fold, Hamiltonian replica exchange, NMR H/D exchange, NOE
Introduction
The fundamental principle of enzyme catalysis is the ability of an enzyme to decrease the transition-state energy (or destabilize the ground state) and thereby accelerate the chemical reactions which would otherwise not occur on biologically relevant time scales.1−3 Enzymes are intrinsically dynamic, and the emerging consensus is that conformational sampling plays a crucial role in enzyme catalysis.4−13 However, describing the direct link between protein motions and enzymatic function remains challenging. Under native conditions, enzymes sample various conformations separated by energy barriers that determine the rates of exchange between these substates.10,14,15 It has been shown that some of the substates sampled by enzymes in the absence of a substrate resemble the substrate-bound state,6,16−19 thus demonstrating the importance of conformational sampling in enzyme catalysis.9,20
2-Deoxyribose-5-phosphate aldolase (DERA, E.C. 4.1.2.4) is a class I aldolase with a triosephosphate isomerase (TIM) barrel fold and a highly conserved sequence in bacteria. In vivo, DERA catalyzes the reversible conversion of acetaldehyde and glyceraldehyde-3-phosphate (G3P) to generate deoxyribose-5-phosphate (dR5P)21,22 and provides key intermediates essential for different metabolic pathways, e.g., glycolysis, the pentose-phosphate pathway, and nucleotide catabolism.23 Therefore, DERA is considered as a promising pharmaceutical target against human bacterial pathogens.24,25 In addition to this, DERA is unique in catalyzing the cross-aldol condensation between two aldehydes and is utilized as an environmentally friendly biocatalyst for the synthesis of drugs such as statins.26−28 However, the absence of a phosphate group from the substrate severely curtails the catalytic efficiency of DERA,29 thereby restricting its use as an efficient biocatalyst to phosphorylated aldehydes. Additionally, mutations of residues interacting with the dR5P phosphate group have also been shown to drastically decrease the activity of ecDERA.30 Several efforts have been made to re-engineer DERAs for accelerated enzyme catalysis31 and acceptance of nonphosphorylated substrates.32
On the basis of Escherichia coli DERA (ecDERA) crystal structures with carbinolamine and Schiff base intermediates, as well as site-directed mutagenesis, Heine et al.33 have proposed a reaction mechanism of the reversible conversion of acetaldehyde and G3P to generate dR5P, which proceeds through a Schiff base formation with the active site K167 residue (Scheme 1). After the formation of an imine, a proton relay system composed of D102, K201, and a conserved water molecule facilitates the abstraction of a C2 proton (i.e., a proton attached to carbon adjacent to the aldehyde group), allowing the formation of an enamine. The enamine further performs a nucleophilic attack onto the carbonyl carbon of the acceptor aldehyde (G3P) to yield dR5P. The crystal structures also reveal that, within the ecDERA active site, the most important substrate coordinating residues are T18, D102, K167, and T170, whereas residues G205 and S238 directly coordinate the dR5P phosphate group, forming the major part of a phosphate-binding site.33
Scheme 1. Mechanism of the DERA-Catalyzed Aldol Reaction of Acetaldehyde (Magenta) and G3P (Green) Proceeding via a Schiff-Base Intermediate with K167, Giving dR5P.
In addition, D102, K201, and a water molecule were proposed to act as an acid (A)/base (B) in a proton relay system responsible for abstracting a C2 proton (highlighted in boldface).33
Despite several biochemical studies suggesting that the C-terminal tyrosine (i.e., Y259) is crucial for efficient catalysis of dR5P breakdown,33,34 the absence of electron density for the last eight C-terminal residues (residues D252–Y259 in ecDERA: i.e., the C-terminal tail) in all available crystal structures of DERAs containing a C-terminal tyrosine (including ecDERA)24,32,33,35,36 has precluded acquiring insights into the role of the tyrosine as well as the C-terminal tail. As observed in ecDERA, the C-terminal tyrosine residue of class I rabbit muscle fructose-1,6-disphosphate aldolase (FBPA) is important for the catalysis of dihydroxyacetone phosphate (DHAP) breakdown. Unlike ecDERA, however, crystal structures of FBPA reveal that, upon binding DHAP, the phosphate dianion recruits the disordered C-terminal region to the active site.37
To investigate the structural and functional basis of Y259 in the ecDERA catalytic mechanism, we have carried out a combined molecular dynamics (MD) and NMR spectroscopy study on its monomeric variant, DERAm (carrying K58E and Y96W mutations).32,36 Since the high molecular weight (56 kDa) of the dimeric wild-type ecDERA makes acquiring atomic-resolution NMR structural and dynamic information challenging, we chose DERAm, which has been shown to have a nearly identical tertiary structure and exhibits enzymatic activity similar to that of the dimeric ecDERA.32 In this study, we show that the C-terminal tail is flexible in solution and provide the first direct structural evidence that, in the absence of the substrate, it samples several substates including a catalytically relevant conformation where Y259 is located inside the active site (i.e., the closed state). We present the solution-state structure of the catalytically relevant closed state, derived using NOE distance restraints, in the substrate-free form of DERAm. We further reveal that, in the closed state conformation, the Y259 ηOH group is responsible for efficient substrate C2 proton exchange and, therefore, could enable a fast transition from the imine to enamine intermediates. Chemical shift perturbations with potassium phosphate reveal that ecDERA contains two distinct sets of phosphate-binding residues: the main phosphate-binding residues near the active site as observed from the reaction-intermediate bound crystal structures33 and previously unknown auxiliary phosphate-binding residues on the C-terminal tail, which could aid in stabilizing the catalytically active conformation of Y259. Thus, our data provide novel insights into DERA function and highlight the role of conformational sampling in the catalysis of dR5P breakdown.
Experimental Section
Protein Expression and Purification
Cloning of the DERAm construct was performed as described previously.38 Y259F, S257D/S258D, and the C-terminal tail deletion mutant of DERAm, D252stop (where the new C-terminal residue is G251), were generated with the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Düsseldorf-Ratingen, Germany) according to the manufacturer’s instructions using the primers given in Table S1. The recombinant protein production and purification were carried out as described previously.38
Activity Assay
Enzyme activity was determined at 20 °C in a retro-aldol reaction with 0.02–5.0 mM dR5P in 50 mM HEPES buffer (pH 6.8) using a coupled enzyme assay as described previously.32,39 In this assay, the G3P generated in the DERA-catalyzed retro-aldol reaction is isomerized to DHAP by triosephosphate isomerase. DHAP is further reduced to glycerol-3-phosphate by glycerol-3-phosphate dehydrogenase through the oxidation of NADH (Sigma-Aldrich Chemie GmbH, Munich, Germany). The decrease in NADH concentration was monitored at 340 nm.
NMR Experiments
[U-13C,15N] and [U-15N] DERAm and its mutants were prepared in 50 mM HEPES buffer (pH 6.8), unless stated otherwise, 0.03% (w/v) NaN3, and 1 mM 2,2,3,3-d(4)-3-(tetramethylsilyl)propionic acid sodium salt (TMSP, Thermo Fisher Scientific, Karlsruhe, Germany) in a 90%/10% (v/v) H2O/D2O mixture. The protein concentration was 800 μM. The NMR spectra were recorded on spectrometers operating at proton frequencies of 600, 800, and 900 MHz, equipped with cryogenically cooled z-gradient probes, and at 25 °C. The sample temperature was calibrated using a perdeuterated methanol sample as described by Findeisen et al.401H, 13C, and 15N backbone and side chain resonance assignments for DERAm were published previously (BMRB entry: 27048).38 The backbone assignments for DERAm mutants were confirmed using standard multidimensional heteronuclear NMR experiments.41 Spectra were processed using the NMRPipe package42 and analyzed with CcpNMR Analysis.43
{1H}-15N Heteronuclear NOE
{1H}15N NOE values for DERAm in 50 mM HEPES (pH 6.8) at 25 °C were derived from pairs of interleaved spectra recorded with (NOE) and without (reference) proton saturation during the recycle delay. A recycle delay of 14 s was used at 14.1 T. The {1H}15N NOE values were calculated from peak intensity ratios obtained from the NOE and reference spectra, with uncertainties estimated from background noise of the spectra.
Solvent Paramagnetic Relaxation Enhancement (sPRE)
2D 1H–15N HSQC spectra of DERAm and the Y259F mutant (300 μM each) in 50 mM HEPES (pH 6.8) at 25 °C were recorded in the presence of 0.5 and 1.0 mM [Gd(DTPA-BMA)]44 and in the absence of the paramagnetic agent. The HSQC spectra were recorded with 1024* × 140* complex data in the 1H and 15N dimensions, respectively, with acquisition times of 106.5 ms (tHN) and 67.7 ms (tN). The recycle delay was 5 s, and 16 scans were collected over 6.5 h per experiment. The experiment was performed in duplicate. The ratio of peak intensities in the presence and absence of the paramagnetic agent (Ipara/Idia) was used to qualitatively assess the solvent accessibility for DERAm and the Y259F mutant.
NMR Structure Calculation
All NOESY-HSQC experiments were recorded at 800 MHz and 25 °C. Distance restraints were obtained from 13C- and 15N-edited NOESY spectra recorded with mixing times of 100–110 ms.
The NOE distance restraint derived structural ensemble of DERAm closed state conformation was calculated starting from the crystal structure (PDB ID: 5EKY).32 Residues missing from the crystal structure (i.e., N-terminal M1–T2 and C-terminal D252–Y259) were added with Swiss-PdbViewer 4.1.0,45 and hydrogen atoms were added with NIH version 1.2.1 of X-PLOR 3.851 (XPLOR-NIH 1.2.1).46,47 Structure preparation was followed by 1000 steps of Powell minimization48 based on the bond geometry parameter file parallhdg.pro modified as described previously49 with harmonic restraints to keep the heavy atoms as close to the crystal structure as possible (atomic RMSDs of 0.28 Å for the backbone and 0.37 Å for all heavy atoms) while reducing the deviation from the ideal covalent geometry of the force field used for the subsequent structure calculation. The resulting heavy-atom positions of residues D3–H250 were kept fixed, except for the side chains of L245, K246, and H250 and the carbonyl group of H250. The closed conformation of the C-terminal region was determined from 48 unambiguous medium- to long-range distance restraints derived from the three-dimensional NOESY spectra in an iterative procedure. The NOEs were manually classified into medium (<3.8 Å) and long (<5.3 Å) distance restraints on the basis of their corresponding cross peak volumes, i.e., medium and weak. Intra-residual, sequential, and possibly also medium-range NOE cross-peaks in the highly mobile C-terminal region are expected to contain significant contributions from conformations other than the closed state conformation and were therefore not included in the structure calculation. These experimental restraints served as an input for the calculation of 120 structures using restrained molecular dynamics according to a three-stage simulated annealing protocol50 using floating assignment of prochiral groups51 with XPLOR-NIH 1.2.1,46,47 as described previously.52 The Gaussian conformational database potential53 with a cutoff of 10.0 standard deviations54 was included in the target function in order to improve the stereochemical properties. The 26 structures showing the lowest energy values (excluding conformational database potential) and fewest distance restraint violations were selected for further characterization using XPLOR-NIH 1.2.146,47 and PROCHECK-NMR 3.4.55
Chemical Shift Perturbation Analysis
Potassium phosphate (KPi) at concentrations ranging from 0 to 150 mM was titrated against 800 μM DERAm and the Y259F, D252stop, and S257D/S258D mutants in 50 mM HEPES buffer (pH 6.8) to obtain the affinity toward inorganic phosphate. 2D 1H–15N HSQC spectra for each titration point were recorded at 25 °C and 600 MHz (DERAm and the S257D/S258D and D252stop mutants) and 800 MHz (Y259F mutant). The data matrix of the 2D 1H–15N HSQC at 600 MHz consisted of 160* × 1024* complex data points with acquisition times of 77.5 ms (tN) and 107 ms (tHN) and 160* × 832* data points and 59 ms (tN) and 64.5 ms (tHN) at 800 MHz. Sixteen scans per titration point were collected with a recycle delay of 1.3 s. The total measuring time was 2 h per 2D 1H–15N HSQC experiment. The equilibrium dissociation constant, KD, was obtained from changes in the weighted average chemical shift differences Δδav = [(Δδ2HN + Δδ2N/25)/2]1/2 assuming a two-state model56
 |
1 |
where [P0] is the total protein concentration and X is the molar ratio of ligand to protein. In total, chemical shift changes for 21 residues were used to report the average KD values for DERAm and the Y259F mutant and 16 residues for the D252stop mutant, whereas 23 residues could be used for the S257D/S258D mutant to describe the respective affinities toward Pi.
1D 1H Hydrogen/Deuterium Exchange Experiments
A 200 mM solution of propanal (Sigma-Aldrich Chemie GmbH, Munich, Germany) was incubated in 50 mM potassium phosphate in degassed D2O (pD 6.8) with 0.5 mM TMSP. In aqueous solution propanal exists in equilibrium with its hydrate, propane-1,1-diol. The chemical shift assignments of the 1D 1H spectrum of propanal were confirmed using a 2D 1H–1H COSY spectrum. A reference sample without DERA was recorded as time point t = 0. After addition of 5 μM DERAm to the reaction mixture, 1D 1H spectra were recorded with a relaxation delay of 20 s, 16384 data points, a spectral width of 9579 Hz, and four scans resulting in 128 s per experiment. A series of 1D spectra was recorded for 3 h. In another experiment, upon the addition of 5 μM Y259F mutant, 1D 1H spectra were recorded using the same acquisition parameters with 32 scans and resulting in 13 min 57 s per 1D experiment, over a period of 4 days. Peak heights were extracted using NMRDraw.42 To quantify the rate of hydrogen/deuterium (H/D) exchange of the C2 proton, the collapse of the triplet representing the methyl group of propanal at 1.06 ppm (t, 3JH–H = 7.3 Hz) and propane-1,1-diol at 0.91 ppm (t, 3JH–H = 7.5) to a doublet (with coupling constants of d, 3JH–H = 7.4 Hz for propanal and d, 3JH–H = 7.6 Hz for propane-1,1-diol) was monitored and the ratio of the upfield peak and the downfield peak of each triplicate was plotted as a function of time. The intensity ratios could be fit to the monoexponential equation
| 2 |
with R0 = 1/τ where y is the intensity ratio, y0 is the intensity ratio of the reference (without DERA) representing t = 0, R0 is the rate and τ is the time constant describing the H/D exchange kinetics. The fitting was performed using OriginPro 8.5G (OriginLab Corporation, Friedrichsdorf, Germany).
1D 31P NMR Experiments
1D 31P NMR spectra of 50 mM KPi (pH 6.8) with 800 μM DERAm and its mutants were recorded on a Bruker Avance III HD with a Prodigy cryoprobe at 600 MHz and 25 °C with 16384* complex data points and an acquisition time of 672.8 ms and 86161 scans with an experimental time of 27.5 h per experiment.
MD Simulations
System Setup
The crystal structure of DERAm (PDB ID: 5EKY),32 resolved at 1.1 Å resolution, was used as the starting structure for MD simulations. As this structure contains several unassigned residues due to the absence of electron density (i.e., N-terminal M1–T2 and C-terminal D252–Y259), we used the MODELER 9.14 tool57 to prepare the full-length protein structure. A model where the C-terminus is positioned laterally to the TIM barrel was selected (K167–Y259 distance of ∼26 Å).
For MD simulations, GROMACS 5.1 suite58,59 was used. The protein was represented with the Amber 99SB*-ILDN force field60−62 in combination with the explicit TIP3P water model.63 For titratable residues, the protonation states were assigned to a pH of 6.8, on the basis of the PROPKA 3 estimate.64 The protein was centered in a cubic box, at least 15 Å away from any edge. A larger than usual box size was used so that the flexible C-terminus does not come in contact with the periodic image even in the extended state. The simulation box was filled with ∼22000 water molecules, and the charge was neutralized with Na+ ions. The steepest descent algorithm was used to minimize the system (maximal force of 500 kJ mol–1 nm–1) before equilibrations. In NVT equilibration, the system was heated to 298 K (v-rescale thermostat65) and equilibrated for 200 ps. At this stage, positional restraints were applied to all protein atoms (force constant of 1000 kJ mol–1 nm–2). During 2.5 ns NPT equilibration, the restraints were gradually reduced from 1000 to 5 kJ mol–1 nm–2, while the pressure was kept at 1 bar (Berendsen barostat66).
The system was modeled under periodic boundary conditions, and the particle mesh Ewald method67 was used to treat electrostatic interactions. The short-range nonbonded interactions were calculated under the cutoff of 10 Å. All bonds were constrained using the LINCS algorithm.68 An integration step of 2.0 fs was used.
HREX-MD Simulation
The Hamiltonian replica exchange (HREX)-MD was performed in GROMACS patched with the Plumed 2.3 plugin.69,70 Twelve replicas were simulated, where the Hamiltonian affecting the C-terminus and several loops over the active site were scaled (i.e., residues 19–25, 75–82, 168–178, 202–208, 249–259). The Hamiltonian scaling factors were 1.00, 0.95, 0.91, 0.87, 0.83, 0.79, 0.76, 0.72, 0.69, 0.66, 0.63, and 0.60, which corresponds to the temperature range of 298–497 K. The exchanges between replicas were attempted every 4 ps, leading to an exchange acceptance rate of ∼30%. Production HREX-MD simulation was carried out with the NPT ensemble (v-rescale thermostat and the Parrinello–Rahman barostat71). Each replica was simulated for 220 ns (accumulated sampling time of 2.64 μs), with coordinates of the system collected every 5 ps. The replica with the unperturbed Hamiltonian (i.e., scaling factor of 1.00) was used for the analysis.
MD Simulations
A set of 120 frames, where Y259 was observed in the active site (distance between K167 ζN and Y259 ηO < 6 Å) was selected from HREX-MD. From each frame, an unrestrained MD simulation was set up. The water box was reduced to at least 10 Å away from any edge, while preserving the cubic shape. The system was minimized and equilibrated as initially described. Production MD simulations were 50 ns long, providing a total of 6 μs of sampling, and coordinates of the system were saved every 20 ps.
Structural Analysis
The analysis of MD trajectories was performed using GROMACS tools and the MDTraj library.72 Hydrogen bonds were identified according to the Baker–Hubbard criteria,73 i.e., where the H···acceptor distance was shorter than 2.5 Å and the donor–H···acceptor angle was greater than 120°, in at least 30% of frames. For the identification of the hydrophobic contacts between residues, all side chain C atoms not directly bound to a heteroatom were considered. Given a residue pair, a hydrophobic contact was defined if the shortest distance between the two residues was lower than 4.5 Å, in at least 30% of the frames.
Results
Efficient Catalysis by ecDERA Requires Y259 ηOH
The ecDERA reaction mechanism, proposed by Heine et al.,33 describes the reversible conversion of acetaldehyde and G3P to dR5P, through a Schiff-base intermediate with the catalytic K167. Several studies have indicated that the side chain hydroxyl group of the C-terminal tyrosine is crucial for DERA catalysis33,34 and the Y259F mutation results in a reduction in the catalytic activity of 2 orders of magnitude.33 Similarly, the DERAm variant, used in our study, shows an ∼100-fold decrease in the kinetic rate constant due to the Y259F mutation (kcat, Table 1), further highlighting the importance of this residue in the catalytic mechanism.
Table 1. Catalytic Activities of DERAm and Its Y259F Mutant Using dR5P as a Substrate.
| DERA variant | kcat (s–1) | KM (mM) | kcat/KM (s–1 M–1) |
|---|---|---|---|
| DERAm | 19.0 ± 1.0 | 0.31 ± 0.01 | (6.1 ± 0.1) × 104 |
| DERAm Y259F | 0.20 ± 0.05 | 0.16 ± 0.01 | (1.3 ± 0.3) × 103 |
C-Terminal Tail of DERAm Is Flexible in Solution and Transiently Occupies the Active Site
In all the available crystal structures of DERAs containing a C-terminal tyrosine,24,32,33,35,36,74 the electron density for the C-terminal tail is poorly defined, which prevents understanding the structural and functional basis of the participation of Y259 in catalysis. To characterize the dynamics of the C-terminal tail in solution, we acquired {1H}15N heteronuclear NOE (hetNOE) values for DERAm. The hetNOE values, obtained for 204 out of total 251 nonproline residues, suggest that the TIM-barrel fold of DERAm (residues 2–250) is rigid on the picosecond to nanosecond time scale with only G205 and D147 showing hetNOE values lower than 0.65 (Figure 1A). The C-terminal tail shows hetNOE values consistently lower than 0.65, indicating high flexibility in solution, which explains the lack of well-defined electron density in the crystal structures.
Figure 1.
Flexibility of DERAm in solution. (A) Steady-state heteronuclear {1H}15N NOE values recorded at 25 °C and 14.1 T. The dashed black line is drawn at an NOE value of 0.65. (B) Intensity ratio (Ipara/Idia) of the backbone amide resonances of DERAm at two concentrations of the paramagnetic agent [Gd(DTPA-DMA)]: 0.5 mM (blue) and 1.0 mM (red). A schematic representation of the secondary structure elements is shown at the top. Residues in the C-terminal tail are highlighted in yellow.
Many recent experimental and computational studies have shown that, under native conditions, enzymes have an intrinsic ability to sample open and closed forms in the absence of a substrate.9,20,75,76 Upon substrate binding one of these conformations gets stabilized through local rearrangements.20 For the possible participation of DERAm Y259 in the enzymatic reaction, it is plausible that the C-terminal tail would need to sample conformations which allow Y259 to enter the active site (closed state). However, for the substrate binding and product release, the enzyme should preferentially be in a conformation that does not block the active site (i.e., open state). A highly flexible C-terminal tail of DERA leads potentially to many open states which would have higher solvent accessibility in comparison to the closed conformation. Paramagnetic relaxation enhancements, induced using soluble paramagnetic compounds (sPREs), provide a straightforward approach toward structural and dynamic analysis of biomolecules using solvent accessibilities.77−79 sPREs have been used to detect transiently populated conformers and changes in the solvent accessibilities of intrinsically disordered regions of proteins.80,81
In our study, we used sPREs of the backbone amide group, with [Gd(DTPA-DMA)] as a paramagnetic agent, to assess the solvent accessibility of DERAm. Intensity ratios (Ipara/Idia) of resonances in 2D 1H–15N HSQC spectra recorded with (0.5 mM and 1.0 mM [Gd(DTPA-DMA)]) and without the paramagnetic solvent indicate that several loop regions (N21–E26, A93–A98, E115–V117, G224, A225, A230, R231) and the N-terminus of helix 3 (residues Y49–I53) are the most surface exposed regions of the protein (Figure 1B). In contrast, the flexible C-terminal tail shows significantly higher Ipara/Idia values in comparison with the aforementioned surface-exposed residues. These observations indicate that the C-terminal tail may sample conformations with low solvent-accessible surface area: e.g., a closed state.
To investigate the possibility of the C-terminal tail adopting a closed state, thereby allowing Y259 to enter the active site, we recorded 2D 1H–15N HSQC spectra of DERAm Y259F mutant. A quantitative analysis of the backbone amide chemical shift differences between DERAm and the Y259F mutant revealed that most of the significant chemical shift changes (i.e., Δδav > 0.015 ppm) occurred to residues located within the DERAm active site (i.e., T18, L20, T168, T170, G171, A203, G204, G205, and R207) (Figure 2). These residues have been shown to be crucial for DERA–substrate interaction.33 The observed perturbations demonstrate that the flexible C-terminal tail of DERAm can sample the closed conformation, thereby bringing Y259 into the active site, in the absence of the substrate. Additionally, the Ipara/Idia ratios between DERAm and the Y259F mutant in the presence of 1 mM [Gd(DTPA-DMA)] are nearly identical with each other (Figure S2). This indicates that the absence of the ηOH group of Y259 does not result in large scale conformational changes and the aforementioned chemical shift perturbations arise primarily due to the absence of the Y259 side chain hydroxyl group.
Figure 2.
Weighted 1H–15N chemical shift differences (Δδav) between DERAm and the Y259F mutant. (A) The black dotted line represents the average Δδav value, whereas the red dotted line represents the average Δδav plus one standard deviation (SD). A schematic representation of the secondary structure elements is shown at the top. (B) The Δδav values are mapped onto the ecDERA crystal structure (PDB ID: 1JCL) using the coloring scheme shown. The catalytic K167 side chain is shown in a ball and stick representation.
MD Simulations Suggest That the Closed State Is Stabilized by a Weak-Interaction Network
The inability to resolve the conformation of the C-terminal tail of ecDERA from X-ray crystallography32,33,35 was at the origin of an idea that the tail is intrinsically disordered and samples many states.33,35 However, the early kinetic investigation pointed out the crucial role of the C-terminus for catalysis by DERA (Table 1). Although the C-terminus is long enough to extend and protrude into the active site,33 no structure of such a conformation is available. To understand the closed conformation of DERA, where Y259 is in the active site, we explored the conformational ensemble of DERAm using enhanced sampling HREX-MD simulations. We recently demonstrated how HREX-MD can be used to study conformational ensembles of enzyme active sites10 and here present the sampling on a larger scale. To that extent, we enhanced the sampling of the C-terminal tail together with the loops on the catalytic face. While the secondary structure elements of the TIM (α/β)8 barrel fold remain stable during 220 ns of HREX-MD (RMSD value of 2 Å from the initial structure), the complete protein backbone indicates much more dynamics (RMSD of up to 6 Å, Figure S3) which comes from the highly flexible C-terminal tail.
To analyze if the C-terminus protrudes into the active site, we measured the distance between the catalytic K167 and C-terminal Y259 residue. Figure S4 shows that the C-terminal tail of DERAm explores a wide range of conformations. While there are multiple open states which present the majority of the complete conformational ensemble, a low closed-state population was identified where the K167–Y259 distance was shorter than 6 Å (Figure S4A). In the open states, the C-terminal tail samples a wide range of conformations, as indicated by structures representing maxima on the distance distribution plot (Figure S4B). The high relative free energy of the closed state, in comparison to the open states, leads to a low occurrence of this state in the HREX-MD simulations. To further sample the closed state, we performed an additional 6 μs of MD (120 simulations starting from different closed state frames identified in the HREX-MD, each 50 ns long). Figure S5A shows that a much higher number of the closed state conformations were sampled, which allowed us to investigate the interactions that stabilize this state.
We used the complete closed state ensemble, identified with MD (around 80000 structures), to investigate the noncovalent interactions in the closed state, which we roughly divide into polar (Figure 3A) and hydrophobic (Figure 3B). However, as the C-terminal tail was found to open in each of the 120 closed state simulations (Figure S5B), these stabilizing interactions are only transient.
Figure 3.
Transient noncovalent interactions stabilize the closed state conformation. (A) Polar and (B) hydrophobic interactions stabilize the C-terminal tail in the closed state conformation. The side chain of the catalytic K167 is highlighted in dark gray. (a1) The hydrophobic interaction between the K172 side chain and the aromatic ring of Y259 as well as the salt bridge between the K172 ζN-group and the C-terminal COO– group. (a2) Hydrogen bonds between A256 and G253 backbone as well as S255 N to Q35 εO. (b1) The C-terminal tyrosine Y259 is further stabilized through hydrophobic interactions with T18 and L20. (b2) Residues A256 and D252 have hydrophobic interactions with residues L241 and A242 located at the C-terminal helix 10.
The hydrophobic contacts are defined here based on the proximity of nonpolar C atoms in any residue pair, i.e., below the cutoff of 4.5 Å. The distance distributions for hydrophobic contacts are shown in Figure S6. These interactions, stabilizing the closed conformation of the C-terminal tail of DERAm, could be split into two groups: those stabilizing N-terminal half of the tail and those stabilizing Y259 in the active site. The former group involves a particularly complex network of contacts through the aliphatic side chain of L248, K5, L9, L12, and V40, which seems to tether the C-terminal helix 10 (S239–L248 in PDB ID: 5EKY) with the rest of the protein. L245, located on the helix 10, forms contacts with residues H250 and D252 on the tail (Figure S6L,M). D252 also forms contacts with A242, and A256 is further in contact with L241 and A242, both located on the helix 10 (Figure 3b2). Y259 forms an extensive network of hydrophobic contacts with aliphatic side chains of residues located in the active site: i.e., T18, L20, and K172 (Figure 3a1,b1).
Polar interactions were further split into salt bridges and hydrogen bonds (Figure S7). Only one salt bridge was identified in the closed state, between K172, located on the loop over the active site, and the Y259 C-terminal COO– group (Figure 3a1 and Figure S7A). Furthermore, several hydrogen bonds were identified between the C-terminal tail and the protein core (i.e., G249–K246, H250–L245, and S255–Q35, Figure S7B–D) and also within the tail itself (A256–G253, S257–K254, and S258–S255, Figure S7E–G). However, the last three hydrogen bonds observed in the C-terminal tail (i.e., residues 253–258) are rarely present simultaneously, appearing together only in 5% of the total closed state ensemble, whereas at least one of the three is present in more than 70% of the sampled conformations.
NOE-Derived NMR Structure of the Closed State
The classical MD simulations indicate that the closed state of the DERAm C-terminal tail gets stabilized through a range of hydrophobic interactions with residues in the C-terminal helix 10 (Figure 3A,b1,b2). To verify the closed state structural representation and confirm the MD-observed interactions that stabilize the closed state in solution, we recorded a 2D 1H–15N HSQC spectrum of the D252stop mutant. A quantitative analysis of the backbone amide chemical shift differences between DERAm and the D252stop mutant revealed that most of the significant chemical shift changes (i.e., Δδav > 0.038 ppm) occurred not only to residues located around the active site but also to residues located in the helix 10 (A242, S243, L244, L245, and K246) (Figure S8). These chemical shift changes coincide with the hydrophobic contacts observed from the MD simulations. Therefore, we recorded NOESY spectra in an attempt to obtain distance restraints between the C-terminal tail and rest of the protein.
Analyses of the NOESY spectra revealed the presence of an unambiguous NOE network between residues in the C-terminal tail and the rest of the protein: in particular, the active site (Figure 4). Although the hetNOE values and TALOS-N secondary structure prediction, using NMR chemical shifts,38 show conclusively that the C-terminal tail is disordered in solution (Figure 1A), the presence of NOEs indicates that the closed conformations of the C-terminal tail could nevertheless be populated in the conformational equilibrium for a substantial fraction of the time. Owing to the steep r–6 dependence of the NOE intensity on the interproton distance, r, long-range contacts are expected to give rise to detectable NOE cross-peaks even if these contacts are only transiently populated for a fraction of the NOESY mixing time used.82
Figure 4.
NOE distance restraint derived structural ensemble of DERAm C-terminal tail closed state. (A) A set of 48 NOEs satisfies the closed state ensemble. The inset shows that the presence of Y259 inside the active site is supported by NOEs (dashed lines) to nearby residues. Strips from 13C-edited NOESY spectra showing inter-residue NOE interactions for 259 Hδ* (B), 259 Hε* (C), and 18 Hγ2* (D) protons. Weak NOE between 259 Hδ* and 20 Hδ1* is highlighted by a black circle in (B). 13C chemical shifts for 259 Cδ*, 259 Cε*, and 18 Cγ2* are displayed at the top of the NOESY strips in (B)–(D), respectively.
We used a total of 48 unambiguous NOE distance restraints (Table S2), between the residues in the C-terminal tail and the rest of the protein (i.e., helix 10 and active site), for the calculation of an ensemble of 26 structures representing the closed state of DERAm (Figure 4A), which is provided as a PDB file in the Supporting Information. Each of these 26 structures individually satisfies the full set of 48 distance restraints without seriously violating the stereochemical quality. None of the C-terminal tail residues lie in the disallowed region of the Ramachandran plot, and only S257 and S258 are in the generously allowed region.
The contribution of the distance restraints to the target function was 0.43 ± 0.29 kcal mol–1, the root-mean-square deviation from the distance restraints was 0.013 ± 0.004 Å, and no distance restraint was violated by more than 0.12 Å. The transient presence of Y259 in the active site is conclusively supported by an extensive network of NOEs between the Y259 side chain and residues in the active site (Figure 4A inset). A comparison of inter-residue cross-peak intensities of NOEs between Y259 and other active site residues (Figure 4B–D) indicates that the ε atoms of Y259 protrude more deeply in the active site than the δ groups. The distance between the side chain amino group of the active site K167 and the ηOH group of Y259 in the NMR ensemble was 4.3 ± 0.5 Å (n = 26), in support of the observations made by the MD simulations. The population of the closed state represented by the calculated NMR structure was estimated to be ∼13% from intensities of NOEs between L20 Hδ1* and Y259 Hε*. To estimate the population, we assumed that in open states the contribution to the NOE intensity is zero due to larger interproton distances. The interproton distances were averaged with r–6 weighting, and intra-residue NOE between L20 Hδ1* and L20 Hα was used as an internal distance reference.
It is important to note that although the NMR ensemble satisfies all 48 experimentally derived distance restraints and exhibits structural features very similar to those observed in the MD simulations, this closed state represents only a small—albeit significant—fraction of the full conformational ensemble sampled by the intrinsically disordered C-terminal tail. Moreover, our MD simulations point out that not all stabilizing interactions are simultaneously required to maintain the DERAm closed state: i.e., to keep Y259 inside the active site. This is also illustrated by fluctuations of the distance between the side chain amino group of the active site K167 and the hydroxyl group of Y259 in the apoenzyme (Figure S5). Therefore, due to the flexible nature of the C-terminal tail, we cannot rule out the existence of a more diverse closed state conformational ensemble in comparison to that calculated using our limited unambiguous NOE data.
Y259 Facilitates C2 Proton Exchange in Propanal
The importance of the Y259 ηOH group in the ecDERA catalytic reaction is highlighted by an ∼100-fold reduction in the kcat value reported herein (Table 1) and in previous studies.33,34 To understand the role of the Y259 ηOH group, we monitored the real-time H/D exchange of the C2 proton of propanal in the presence of DERAm and the Y259F mutant using 1D 1H NMR in D2O and KPi buffer (Figure 5A). Propanal was chosen over acetaldehyde (natural substrate) because the latter can react in a sequential manner to form side products which covalently attach to and inactivate ecDERA.32 For monitoring of the C2 deprotonation, the transformation of the methyl group of both propanal and its hydrate, propane-1,1-diol, from a triplet into a doublet was monitored over time. The addition of DERAm leads to an immediate decrease in the intensity of the downfield resonance of the methyl group triplet and a simultaneous increase in the upfield resonance (Figure 5B), signifying exchange of a C2 proton with a deuterium. Fitting the change in the intensity ratio of the upfield and downfield peaks to a monoexponential equation (eq 2) yielded a time constant (τ) of 19.7 ± 2.5 min for DERAm-catalyzed C2 deprotonation (Figure 5C). However, the change in the aforementioned intensity ratio in the presence of the Y259F mutant was observed to be significantly slower, with the fits yielding a time constant of 62.7 ± 5.9 h (Figure 5D), ∼190-fold higher than τDERAm. This drastic increase in τ indicates that the C2 proton abstraction is significantly more efficient in the presence of the Y259 side chain hydroxyl group than in its absence.
Figure 5.
Substitution of the C2-proton of propanal with deuterium in the presence of DERAm and the Y259F mutant studied with 1D proton NMR spectroscopy highlights the role of Y259. (A) Schematic of the DERA-catalyzed H/D exchange reaction. The proton undergoing exchange is highlighted in red. (B) Transformation of the propanal methyl group triplet into a doublet over time in the presence of 5 μM DERAm. The ratio of the peaks at 1.068 and 1.043 ppm (marked with asterisks in (B)) was fit to a single-exponential equation in the presence of 5 μM DERAm (C) and 5 μM Y259F mutant (D), respectively. The errors in (C) and (D) were estimated from triplicate measurements.
C-Terminal Tail Contains Auxiliary Phosphate-Binding Residues
The catalytic efficiency of ecDERA is severely reduced for nonphosphorylated substrates. The kcat value is 68 ± 1 s–1 with dR5P as the acceptor, whereas it decreases by a factor of ∼600 to 0.11 ± 0.01 s–1 when deoxyribose (dR) is used as the acceptor aldehyde.29 A strong increase in KM from 0.64 mM (dR5P) to 57.7 mM (dR) suggests that the phosphate group is vital for the enzyme–substrate interaction. Additionally, under our reaction conditions, we could not detect any catalytic activity for DERAm toward dR.
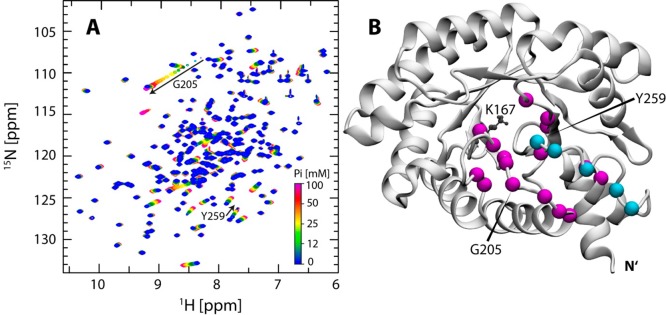
Considering that the phosphate group is crucial for enzyme–substrate interaction, we investigated the interaction between DERAm and phosphate using 2D 1H–15N HSQC spectra recorded at different phosphate concentrations ranging from 0 mM to 100 mM KPi (Figure 6A). As expected, the main phosphate-binding residues in the vicinity of the active site identified by X-ray crystallography (i.e., T18, L20, T170, G171, G204, G205, and A237)33 show significant chemical shift changes upon titration of KPi. Residues undergoing fast to intermediate exchange on the NMR time scale were used to extract KD values (eq 1). The chemical shift changes for 16 residues in DERAm (Table S3) were used to calculate an average KD value of 18.8 ± 3.6 mM for the main phosphate-binding residues. Additionally, several residues on the C-terminal tail showed chemical shift changes upon interaction with phosphate. The average KD value for these auxiliary phosphate-binding residues was 19.0 ± 0.7 mM (n = 5, Table S3), nearly identical with that for the main set of residues. The 31P NMR spectrum, recorded on DERAm variant in 50 mM KPi buffer (pH 6.8), shows the presence of one additional phosphate resonance in addition to the buffer resonance. This observation indicates that both binding sites may simultaneously coordinate the same molecule of inorganic phosphate (Figure S9).
Figure 6.
Phosphate-binding sites in DERAm. (A) Overlay of 2D 1H–15N HSQC spectra of DERAm recorded in the presence of KPi ranging from 0 mM to 100 mM. The main and auxiliary phosphate-binding residues show significant changes in backbone amide chemical shift. In addition to chemical shift changes for various residues, extensive changes in line widths are observed. (B) Residues showing significant chemical shift changes upon addition of KPi are highlighted (magenta, main; cyan, auxiliary) on a model of the DERAm closed state. The catalytic K167 side chain is shown in a ball and stick representation.
To identify the key C-terminal tail auxiliary phosphate-binding residues, we carried out 1H–15N HSQC titration of KPi with 15N-labeled DERAm mutants: Y259F and S257D/S258D. The affinity toward Pi for the main and auxiliary binding residues in the Y259F mutant were 17.9 ± 2.4 mM (n = 16) and 19.3 ± 0.4 mM (n = 5), respectively. The near-identical affinities for both sets of binding residues, as well as those to the corresponding binding sites in DERAm, indicate that the Y259 ηOH group does not play any role in phosphate coordination. This is supported by the presence of a single protein-bound phosphate resonance in the 31P NMR spectrum, which has a chemical shift identical with that of the bound phosphate resonance in the 31P spectrum of DERAm variant (Figure S9).
For the S257D/S258D mutant, 2D 1H–15N HSQC spectra were recorded with KPi concentrations ranging from 0 to 150 mM, in anticipation of the decreased phosphate affinity due to the presence of negatively charged aspartic acid residues. Residues Y259 and A256, flanking the mutation site, show a decrease in Pi affinity by more than 200%, whereas residues D258, G253, L241, A237, and T208 show an affinity decrease of more than 100% in comparison to their respective affinities in DERAm and the Y259F mutant (Table S3). The remaining residues used for KD determination located around the active site (i.e., D16, T18, L20, T170, N176, A177, A203, G204, G205, and R207) show a decrease in affinity between 47 and 87% (Table S3). Additionally, a 31P NMR spectrum recorded on the S257D/S258D mutant shows a single protein-bound phosphate resonance which has a distinct chemical shift in comparison with the bound phosphate resonance observed in 31P NMR spectra of DERAm and Y259F mutant (Figure S9). This observation indicates that S257 and S258 might be the key phosphate interacting residues on the C-terminal tail and demonstrates that the perturbation of the phosphate affinity of the auxiliary phosphate-binding residues has a direct effect on the affinity of the main phosphate-binding residues.
Discussion
In this report, we show that mutation of the C-terminal Y259 into a phenylalanine leads to a drastic drop in the catalytic activity of the monomer variant of ecDERA (i.e., DERAm) (Table 1), similar to the effect observed in wild-type ecDERA.33 Such an observation indicates that the ηOH group of Y259 plays a crucial role in the function of ecDERA. Despite the presence of significant structural and biochemical work on ecDERA,29,30,32,33,35,36 precise structural information on how its intrinsically disordered C-terminal tail facilitates the catalytic process has been largely unaddressed. Using NMR chemical shift perturbations, we provide the first evidence demonstrating that the intrinsically disordered C-terminal tail samples conformations which allow Y259 to enter the active site (i.e., closed state), in the absence of the substrate (Figure 2). Such presampling of functionally relevant conformations in the absence of substrates has been observed for several enzymes, highlighting the importance of conformational flexibility for the catalytic process. For example, loops within the structural core of enzymes such as cyclophilin A,6 adenylate kinase,18,83 and RNase A16 have the ability to sample catalytically relevant “closed” state conformations in the absence of a substrate, and the frequency of the sampling of such conformations correlates with the catalytic turnover rates. We also attempted to quantify the exchange rate between the open and closed states of the DERAm C-terminal tail. However, even at 5 °C, the exchange process was too fast to obtain reliable quantitative data from NMR CPMG relaxation dispersion experiments (i.e., kex > 10000 s–1). Since DERAm-catalyzed dR5P breakdown proceeds with a kcat value of 19 s–1, fast sampling of the open and closed states, in the substrate-free form, is unlikely to be the rate-limiting step of DERAm catalysis. Experiments to probe the effect of the natural substrate (dR5P) on the C-terminal tail conformational sampling were hampered by the instability of the system. Upon addition of dR5P, chemical shift as well as line width changes in a series of 2D [1H–15N] HSQC spectra were observed over time (Figure S10). These time-dependent changes could be caused by acetaldehyde molecules, formed during the reversible aldol reaction, which can react in a sequential manner to form side products32 which may covalently attach to the enzyme.
Despite the conformational plasticity of the C-terminal tail, the population of the closed state was sufficiently large to give rise to NOEs between the C-terminal tail and DERAm active site as well as helix 10 (Figure 4B–D). Using the unambiguous NOE distance restraints, we were able to derive a structural ensemble of the catalytically relevant closed state. The NMR ensemble concurs with a closed state ensemble generated through the MD simulations. Medium- to weak-intensity NOEs were observed between residues showing hydrophobic interactions in the MD simulations: i.e., Y259–L20, A256–A242, A246–L241, and others (Table S2). From a structural point of view, it appears that the closed state is not ideal for the substrate entry and/or product exit, as Y259 blocks the entrance to the active site pocket (Figure S11). Therefore, a transition between open and closed states may be necessary for substrate and product diffusion, as observed for rabbit muscle FBPA.37,84 Additionally, deletion of the C-terminal tail (D252stop mutant) leads to a significant decrease in DERAm activity in comparison to full-length DERAm. Interestingly, the activity of D252stop mutant (kcat = 0.48 s–1 and KM = 0.12 mM) was moderately higher (2.4-fold increase in kcat) than that of the Y259F mutant. This increase could be attributed to efficient substrate entry and/or product release in the absence of the C-terminal tail (i.e., the open conformation) and indicates that Y259 could be the only C-terminal tail residue participating in the catalytic process.
In spite of the abundance of biochemical data stressing the importance of Y259 in DERA catalysis, the role of this crucial residue remained inconclusive. Studies with Salmonella typhimurium DERA, which is 96.5% sequence identical with ecDERA, have shown that Y259 does not play a role in facilitating active site Schiff base formation.34 In FBPA, the C-terminal tyrosine residue is proposed to be a general base in the reaction with DHAP, enabling the transformation of the imine to enamine intermediate.37,85 However, Heine et al.33 used a combination of X-ray crystallography and 1D 1H NMR to rule out the role of Y259 as a general base in the catalytic mechanism of ecDERA. Instead, it was proposed that a structurally conserved water molecule participates in a proton relay, together with D102 and K201, responsible for the C2 proton abstraction from the substrate.
Our study shows that the C2 proton of propanal is abstracted by DERAm and the Y259F mutant, similarly to the observations by Heine et al.33 However, analysis of the real-time propanal H/D exchange performed herein, using a 14-fold lower concentration of the enzyme (relative to Heine et al.), reveals an ∼190-fold increase in the time constant for the Y259F mutant catalyzed C2 proton abstraction (Figure 5), making it the rate-limiting step for this mutant. A similar effect was observed for FBPA after hydrolysis of the carboxy–terminal peptide bond through carboxypeptidase treatment.85 Therefore, participation of Y259 in the ecDERA catalytic mechanism, together with residues D102 and K201, cannot be ruled out. Two putative mechanisms could explain the role that the Y259 ηOH group plays in lowering the activation energy (ΔΔG⧧Y259F-DERAm value of ∼3 kcal mol–1, obtained from kcat in Table 1 using the Eyring equation) for the proton-abstraction step. (1) Y259 ηOH acts as a general base to directly abstract the C2 proton, as suggested previously for other class I aldolases.37,86 In its absence, an alternative mechanism such as the participation of a conserved water molecule could drive the substrate proton abstraction, albeit inefficiently. (2) Y259 ηOH participates in the proton-relay system, originally proposed for ecDERA,33 by maintaining a desirable electrostatic environment for fast proton abstraction. The absence of the Y259 ηOH group (e.g., in the Y259F and D252stop mutants) perturbs this conducive environment, leading to a much slower proton abstraction.
Overlay of the NMR structure of the DERAm closed state with the crystal structure of ecDERA bound to the carbinolamine intermediate (PDB: 1JCL) reveals the presence of a steric clash between Y259 and the substrate (Figure 7). Therefore, after the substrate enters the active site, a reorientation of the side chains of Y259 and other active site residues might be necessary.
Figure 7.

Y259 side chain clashed with the carbinolamine intermediate. An overlay of the NMR DERAm closed state structure (yellow) with the crystal structure of ecDERA in complex with the carbinolamine reaction intermediate (PDB: 1JCL) reveals the presence of a steric clash between Y259 and the carbinolamine intermediate.
Using a combination of chemical shift perturbation analysis and 31P NMR with different DERAm mutants, we have revealed that the C-terminal tail coordinates the active site phosphate moiety through previously unknown auxiliary phosphate-binding residues (Figure 6 and Figure S9) involving amino acids 256–259. For FBPA, a water-mediated coordination of the C-terminal Y363 with inorganic phosphate and DHAP phosphate was proposed to generate a reactive phenolate ion which carries out stereospecific proton abstraction to generate the enamine intermediate.37 However, for DERAm we observed identical phosphate-binding affinities regardless of the C-terminal residue: i.e., Y259 or F259. Furthermore, the deprotonation of Y259 through a phosphate coordinating water molecule, to generate a phenolate ion, can also be ruled out for DERAm due to the near-identical chemical shifts of the Y259 epsilon carbon (Cε) in 50 mM HEPES and 50 mM KPi buffers (Y259 εC, 118.00 ppm in HEPES buffer;38 and Y259 εC, 118.04 ppm in KPi buffer). Therefore, the auxiliary phosphate-binding residues might have an alternative function.
The phosphate-binding region of ecDERA active site shows the presence of A237, S238, and S239, forming a noncanonical phosphate-binding site.30 Interestingly, the auxiliary phosphate-binding residues form a noncanonical motif (residues A256, S257, and S258) as well. Mutation of S257 and S258 into aspartic acids decreased Pi affinity not only at the C-terminal tail but also in the active site. This demonstrates that the residues in the two noncanonical phosphate-binding regions coordinate the same phosphate molecule located at the active site. Since deletion of the C-terminal tail results in a slight increase (1.9-fold, Table S3) in phosphate affinity in the vicinity of the active site, the auxiliary phosphate-binding residues are not required for the phosphate interaction of the main phosphate-binding residues. Therefore, we hypothesize that these auxiliary residues allow the C-terminal tail to dock onto the phosphate group of the substrate, thereby stabilizing the closed state conformation. Such docking may allow reorientation Y259 in an optimal position, avoiding a steric clash with the substrate and enabling participation in the C2 proton exchange step of the DERA catalytic reaction.
Conclusions
We have uncovered novel insights into the role of the rarely studied intrinsically disordered C-terminal tail on a structural level and provided a more detailed picture of the ecDERA catalytic reaction mechanism. Although a comparison between the static crystal structures of ecDERA in the apo- and substrate-bound states has provided insights into understanding the DERA–substrate interactions,32,33,35 our data demonstrate that conformational sampling of the C-terminal tail is a key feature for efficient catalysis. The dynamic aspects related to enzyme catalysis are becoming more evident with the development of both experimental (e.g., NMR87 and multitemperature and X-ray-free electron laser crystallography14) and computational (e.g., MD88 and Monte Carlo simulations89) methods. The TIM-barrel fold is one of the most common among enzymes, and many recent attempts to design de novo enzymes showed an incredible evolvability of the TIM-barrel scaffolds.90−92 The importance of loop dynamics within the TIM-barrel fold family of enzymes, for the catalytic activity, has been highlighted previously.93−97 However, to our knowledge, the results provided herein represent the first structural evidence in the TIM-barrel fold family where an intrinsically disordered C-terminal tail, located outside the fold, not only samples catalytically relevant conformations in the absence of a substrate but also participates in the catalytic reaction. Therefore, our current contribution not only provides significant insights related to DERAs specifically but also deepens the understanding of the involvement of the intrinsic motions in catalysis of the TIM-barrel fold enzymes.
Furthermore, DERAs found in several human pathogens, e.g. Salmonella typhimurium, Klebsiella pneumoniae, Vibrio cholerae, and others (Figure S12), show a high sequence identity (81% for V. cholerae and >94% identity for the others presented) with ecDERA, unlike human DERA with only 38% identity.98 All of these pathogens (except V. cholerae) have an identical C-terminal tail sequence. Since DERA plays a crucial role in microbial survival by participating in nucleotide catabolism and providing key metabolic intermediates such as acetaldehyde and G3P, it is considered to be a promising drug target against human pathogens.24,25 Considering the high sequence identity between the aforementioned bacterial DERAs, their structural and dynamic properties should be identical with those of ecDERA. The insights uncovered herein, together with the closed state structure, should therefore aid in not only the development of better DERA variants for efficient biocatalysis but also small-molecule inhibitors targeting DERAs from human bacterial pathogens.
Acknowledgments
We thank Professors Tobias Madl, Lynn Kamerlin, and Gunnar Schröder for fruitful discussions and critical reading of the manuscript. The authors acknowledge access to the Jülich-Düsseldorf Biomolecular NMR Center that is jointly run by the Forschungszentrum Jülich and Heinrich-Heine-Universität Düsseldorf and the computing time granted by JARA-HPC Vergabegremium and VSR commission on the supercomputer JURECA (project JICS69 to D.P. and Prof. Birgit Strodel) at Forschungszentrum Jülich. We thank Dr. Thomas Classen for providing the plasmid encoding DERAm. M.S. acknowledges the Ministry of Innovation, Science, and Research of the German federal state of North Rhine-Westphalia and the Heinrich-Heine-Universität Düsseldorf (scholarship within the CLIB-Graduate Cluster Industrial Biotechnology) for financial support. D.P. was financially supported by the Jürgen Manchot Foundation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscatal.7b04408.
Author Present Address
# D.P.: Department of Cell and Molecular Biology, Uppsala University, BMC Box 596, S-751 24 Uppsala, Sweden.
The authors declare no competing financial interest.
Supplementary Material
References
- Andrews L. D.; Fenn T. D.; Herschlag D. Ground state destabilization by anionic nucleophiles contributes to the activity of phosphoryl transfer enzymes. PLoS Biol. 2013, 11, e1001599. 10.1371/journal.pbio.1001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Schramm V. L. Ground-state destabilization in orotate phosphoribosyltransferases by binding isotope effects. Biochemistry 2011, 50, 4813–4818. 10.1021/bi200638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold W. R.; Castro J. N.; Fisher A. J.; Toney M. D. Ground-state electronic destabilization via hyperconjugation in aspartate aminotransferase. J. Am. Chem. Soc. 2012, 134, 8436–8438. 10.1021/ja302809e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes-Schiffer S. Catalytic efficiency of enzymes: A theoretical analysis. Biochemistry 2013, 52, 2012–2020. 10.1021/bi301515j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovic S. J.; Hammes-Schiffer S. A perspective on enzyme catalysis. Science 2003, 301, 1196–1202. 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- Eisenmesser E. Z.; Millet O.; Labeikovsky W.; Korzhnev D. M.; Wolf-Watz M.; Bosco D. A.; Skalicky J. J.; Kay L. E.; Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature 2005, 438, 117–121. 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- Campbell E.; Kaltenbach M.; Correy G. J.; Carr P. D.; Porebski B. T.; Livingstone E. K.; Afriat-Jurnou L.; Buckle A. M.; Weik M.; Hollfelder F.; Tokuriki N.; Jackson C. J. The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 2016, 12, 944–950. 10.1038/nchembio.2175. [DOI] [PubMed] [Google Scholar]
- Luk L. Y.; Loveridge E. J.; Allemann R. K. Protein motions and dynamic effects in enzyme catalysis. Phys. Chem. Chem. Phys. 2015, 17, 30817–30827. 10.1039/C5CP00794A. [DOI] [PubMed] [Google Scholar]
- Boehr D. D.; Nussinov R.; Wright P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796. 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrović D.; Frank D.; Kamerlin S. C. L.; Hoffmann K.; Strodel B. Shuffling active site substate populations affects catalytic activity: The case of glucose oxidase. ACS Catal. 2017, 7, 6188–6197. 10.1021/acscatal.7b01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis A.; Risso V. A.; Sanchez-Ruiz J. M.; Kamerlin S. C. L. Cooperativity and flexibility in enzyme evolution. Curr. Opin. Struct. Biol. 2018, 48, 83–92. 10.1016/j.sbi.2017.10.020. [DOI] [PubMed] [Google Scholar]
- Wolfenden R. Massive thermal acceleration of the emergence of primordial chemistry, the incidence of spontaneous mutation, and the evolution of enzymes. J. Biol. Chem. 2014, 289, 30198–30204. 10.1074/jbc.R114.567081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm V. L. Transition states and transition state analogue interactions with enzymes. Acc. Chem. Res. 2015, 48, 1032–1039. 10.1021/acs.accounts.5b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keedy D. A.; Kenner L. R.; Warkentin M.; Woldeyes R. A.; Hopkins J. B.; Thompson M. C.; Brewster A. S.; Van Benschoten A. H.; Baxter E. L.; Uervirojnangkoorn M.; McPhillips S. E.; Song J.; Alonso-Mori R.; Holton J. M.; Weis W. I.; Brunger A. T.; Soltis S. M.; Lemke H.; Gonzalez A.; Sauter N. K.; Cohen A. E.; van den Bedem H.; Thorne R. E.; Fraser J. S. Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography. eLife 2015, 4, e07574. 10.7554/eLife.07574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten R.; Liu L.; Kenner L. R.; Clarkson M. W.; Mavor D.; Tawfik D. S.; Kern D.; Fraser J. S. Rescue of conformational dynamics in enzyme catalysis by directed evolution. Nat. Commun. 2018, 9, 1314. 10.1038/s41467-018-03562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach H.; Cole R.; Gill M. L.; Loria J. P. Conservation of μs-ms enzyme motions in the apo- and substrate-mimicked state. J. Am. Chem. Soc. 2005, 127, 9167–9176. 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- Boehr D. D.; McElheny D.; Dyson H. J.; Wright P. E. The dynamic energy landscape of dihydrofolate reductase catalysis. Science 2006, 313, 1638–1642. 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K. A.; Thai V.; Lei M.; Ott M.; Wolf-Watz M.; Fenn T.; Pozharski E.; Wilson M. A.; Petsko G. A.; Karplus M.; Hubner C. G.; Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature 2007, 450, 838–844. 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Jackson C. J.; Foo J. L.; Tokuriki N.; Afriat L.; Carr P. D.; Kim H. K.; Schenk G.; Tawfik D. S.; Ollis D. L. Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 21631–21636. 10.1073/pnas.0907548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar I.; Chennubhotla C.; Tobi D. Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Curr. Opin. Struct. Biol. 2007, 17, 633–640. 10.1016/j.sbi.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machajewski T. D.; Wong C. H. The catalytic asymmetric aldol reaction. Angew. Chem., Int. Ed. 2000, 39, 1352–1375. . [DOI] [PubMed] [Google Scholar]
- Hoffee P. Deoxyribose-5-phosphate aldolase from Salmonella typhimurium. Methods Enzymol. 1975, 42, 276–279. 10.1016/0076-6879(75)42127-9. [DOI] [PubMed] [Google Scholar]
- Tozzi M. G.; Camici M.; Mascia L.; Sgarrella F.; Ipata P. L. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 2006, 273, 1089–1101. 10.1111/j.1742-4658.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- Cao T. P.; Kim J. S.; Woo M. H.; Choi J. M.; Jun Y.; Lee K. H.; Lee S. H. Structural insight for substrate tolerance to 2-deoxyribose-5-phosphate aldolase from the pathogen Streptococcus suis. J. Microbiol. 2016, 54, 311–321. 10.1007/s12275-016-6029-4. [DOI] [PubMed] [Google Scholar]
- Tonkin M. L.; Halavaty A. S.; Ramaswamy R.; Ruan J.; Igarashi M.; Ngo H. M.; Boulanger M. J. Structural and functional divergence of the aldolase fold in Toxoplasma gondii. J. Mol. Biol. 2015, 427, 840–852. 10.1016/j.jmb.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ošlaj M.; Cluzeau J.; Orkić D.; Kopitar G.; Mrak P.; Časar Z. A highly productive, whole-cell DERA chemoenzymatic process for production of key lactonized side-chain intermediates in statin synthesis. PLoS One 2013, 8, e62250. 10.1371/journal.pone.0062250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein S.; Schürmann M.; Wolberg M.; Hilker I.; Luiten R.; Wubbolts M.; Mink D. Directed evolution of an industrial biocatalyst: 2-deoxy-D-ribose-5-phosphate aldolase. Biotechnol. J. 2006, 1, 537–548. 10.1002/biot.200600020. [DOI] [PubMed] [Google Scholar]
- Greenberg W. A.; Varvak A.; Hanson S. R.; Wong K.; Huang H.; Chen P.; Burk M. J. Development of an efficient, scalable, aldolase-catalyzed process for enantioselective synthesis of statin intermediates. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 5788–5793. 10.1073/pnas.0307563101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis G.; Liu J.; Clark D. P.; Heine A.; Wilson I. A.; Wong C. H. Structure-based mutagenesis approaches toward expanding the substrate specificity of D-2-deoxyribose-5-phosphate aldolase. Bioorg. Med. Chem. 2003, 11, 43–52. 10.1016/S0968-0896(02)00429-7. [DOI] [PubMed] [Google Scholar]
- Ma H.; Szeler K.; Kamerlin S. C.; Widersten M. Linking coupled motions and entropic effects to the catalytic activity of 2-deoxyribose-5-phosphate aldolase (DERA). Chem. Sci. 2016, 7, 1415–1421. 10.1039/C5SC03666F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J.; Meneghini L. M.; Raston C. L.; Weiss G. A. Accelerating enzyme catalysis using vortex fluidics. Angew. Chem., Int. Ed. 2016, 55, 11387–11391. 10.1002/anie.201604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick M.; Hartmann R.; Weiergräber O. H.; Bisterfeld C.; Classen T.; Schwarten M.; Neudecker P.; Willbold D.; Pietruszka J. Mechanism-based inhibition of an aldolase at high concentrations of its natural substrate acetaldehyde: structural insights and protective strategies. Chem. Sci. 2016, 7, 4492–4502. 10.1039/C5SC04574F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine A.; DeSantis G.; Luz J. G.; Mitchell M.; Wong C.-H.; Wilson I. A. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science 2001, 294, 369–374. 10.1126/science.1063601. [DOI] [PubMed] [Google Scholar]
- Hoffee P.; Snyder P.; Sushak C.; Jargiello P. Deoxyribose-5-P aldolase: Subunit structure and composition of active site lysine region. Arch. Biochem. Biophys. 1974, 164, 736–742. 10.1016/0003-9861(74)90087-3. [DOI] [PubMed] [Google Scholar]
- Heine A.; Luz J. G.; Wong C. H.; Wilson I. A. Analysis of the class I aldolase binding site architecture based on the crystal structure of 2-deoxyribose-5-phosphate aldolase at 0.99A resolution. J. Mol. Biol. 2004, 343, 1019–1034. 10.1016/j.jmb.2004.08.066. [DOI] [PubMed] [Google Scholar]
- Dick M.; Weiergräber O. H.; Classen T.; Bisterfeld C.; Bramski J.; Gohlke H.; Pietruszka J. Trading off stability against activity in extremophilic aldolases. Sci. Rep. 2016, 6, 17908. 10.1038/srep17908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean M.; Sygusch J. Stereospecific proton transfer by a mobile catalyst in mammalian fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 2007, 282, 31028–31037. 10.1074/jbc.M704968200. [DOI] [PubMed] [Google Scholar]
- Schulte M.; Stoldt M.; Neudecker P.; Pietruszka J.; Willbold D.; Panwalkar V. 1H, 13C, and 15N backbone and sidechain resonance assignments of a monomeric variant of E. coli deoxyribose-5-phosphate aldolase. Biomol. NMR Assignments 2017, 11, 197–201. 10.1007/s12104-017-9747-6. [DOI] [PubMed] [Google Scholar]
- Nicholas P. C. Effects of buffer concentrations, pH, triose phosphate isomerase and glycerol-3-phosphate dehydrogenase on the measurement of the activity of aldolase and glyceraldehyde-3-phosphate dehydrogenase. Biochem. Soc. Trans. 1988, 16, 752–753. 10.1042/bst0160752. [DOI] [Google Scholar]
- Findeisen M.; Brand T.; Berger S. A 1H-NMR thermometer suitable for cryoprobes. Magn. Reson. Chem. 2007, 45, 175–178. 10.1002/mrc.1941. [DOI] [PubMed] [Google Scholar]
- Sattler M.; Schleucher J.; Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999, 34, 93–158. 10.1016/S0079-6565(98)00025-9. [DOI] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct., Funct., Genet. 2005, 59, 687–696. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Pintacuda G.; Otting G. Identification of protein surfaces by NMR measurements with a paramagnetic Gd (III) chelate. J. Am. Chem. Soc. 2002, 124, 372–373. 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- Guex N.; Peitsch M. C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Schwieters C. D.; Kuszewski J. J.; Tjandra N.; Marius Clore G. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Brünger A. T.X-PLOR: version 3.1: A system for X-ray crystallography and NMR; Yale University Press: New Haven, CT, USA, 1992. [Google Scholar]
- Powell M. J. D. Restart procedures for the conjugate gradient method. Math. Program. 1977, 12, 241–254. 10.1007/BF01593790. [DOI] [Google Scholar]
- Neudecker P.; Nerkamp J.; Eisenmann A.; Nourse A.; Lauber T.; Schweimer K.; Lehmann K.; Schwarzinger S.; Ferreira F.; Rosch P. Solution structure, dynamics, and hydrodynamics of the calcium-bound cross-reactive birch pollen allergen Bet v 4 reveal a canonical monomeric two EF-hand assembly with a regulatory function. J. Mol. Biol. 2004, 336, 1141–1157. 10.1016/j.jmb.2003.12.070. [DOI] [PubMed] [Google Scholar]
- Kharrat A.; Macias M. J.; Gibson T. J.; Nilges M.; Pastore A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995, 14, 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holak T. A.; Nilges M.; Oschkinat H. Improved strategies for the determination of protein structures from NMR data: The solution structure of acyl carrier protein. FEBS Lett. 1989, 242, 218–224. 10.1016/0014-5793(89)80473-9. [DOI] [PubMed] [Google Scholar]
- Neudecker P.; Schweimer K.; Nerkamp J.; Scheurer S.; Vieths S.; Sticht H.; Rosch P. Allergic cross-reactivity made visible: Solution structure of the major cherry allergen Pru av 1. J. Biol. Chem. 2001, 276, 22756–22763. 10.1074/jbc.M101657200. [DOI] [PubMed] [Google Scholar]
- Kuszewski J.; Clore G. M. Sources of and solutions to problems in the refinement of protein NMR structures against torsion angle potentials of mean force. J. Magn. Reson. 2000, 146, 249–254. 10.1006/jmre.2000.2142. [DOI] [PubMed] [Google Scholar]
- Neudecker P.; Sticht H.; Rosch P. Improving the efficiency of the Gaussian conformational database potential for the refinement of protein and nucleic acid structures. J. Biomol. NMR 2001, 21, 373. 10.1023/A:1013369107271. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; MacArthur M. W.; Moss D. S.; Thornton J. M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Grzesiek S.; Stahl S. J.; Wingfield P. T.; Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Ned. Mapping of the Nef binding surface by NMR. Biochemistry 1996, 35, 10256–10261. 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- Šali A.; Blundell T. L. Comparitive protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- van der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Pronk S.; Páll S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; Hess B.; Lindahl E. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K.; Piana S.; Palmo K.; Maragakis P.; Klepeis J. L.; Dror R. O.; Shaw D. E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct., Funct., Genet. 2010, 78, 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R. B.; Hummer G. Optimised molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Phys. Chem. B 2009, 113, 9004–9015. 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct., Funct., Genet. 2006, 65, 712–725. 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Olsson M. H. M.; Søndergaard C. R.; Rostkowski M.; Jensen J. H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F.; DiNola A.; Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Hess B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008, 4, 116–122. 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- Bonomi M.; Branduardi D.; Bussi G.; Camilloni C.; Provasi D.; Raiteri P.; Donadio D.; Marinelli F.; Pietrucci F.; Broglia R. A.; Parrinello M. PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 2009, 180, 1961–1972. 10.1016/j.cpc.2009.05.011. [DOI] [Google Scholar]
- Bussi G. Hamiltonian replica exchange in GROMACS: A flexible implementation. Mol. Phys. 2014, 112, 379–384. 10.1080/00268976.2013.824126. [DOI] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- McGibbon R. T.; Beauchamp K. A.; Harrigan M. P.; Klein C.; Swails J. M.; Hernandez C. X.; Schwantes C. R.; Wang L. P.; Lane T. J.; Pande V. S. MDTraj: A modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015, 109, 1528–1532. 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. N.; Hubbard R. E. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984, 44, 97–179. 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Jiao X.-C.; Pan J.; Xu G.-C.; Kong X.-D.; Chen Q.; Zhang Z.-J.; Xu J.-H. Efficient synthesis of a statin precursor in high space-time yield by a new aldehyde-tolerant aldolase identified from Lactobacillus brevis. Catal. Sci. Technol. 2015, 5, 4048. 10.1039/C5CY00537J. [DOI] [Google Scholar]
- Wurm J. P.; Holdermann I.; Overbeck J. H.; Mayer P. H.; Sprangers R. Changes in conformational equilibria regulate the activity of the Dcp2 decapping enzyme. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 6034–6039. 10.1073/pnas.1704496114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo E.; Saladino G.; Lambrughi M.; Lindorff-Larsen K.; Gervasio F. L.; Nussinov R. The role of protein loops and linkers in conformational dynamics and allostery. Chem. Rev. 2016, 116, 6391–6423. 10.1021/acs.chemrev.5b00623. [DOI] [PubMed] [Google Scholar]
- Hocking H. G.; Zangger K.; Madl T. Studying the structure and dynamics of biomolecules by using soluble paramagnetic probes. ChemPhysChem 2013, 14, 3082–3094. 10.1002/cphc.201300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl T.; Güttler T.; Görlich D.; Sattler M. Structural analysis of large protein complexes using solvent paramagnetic relaxation enhancements. Angew. Chem., Int. Ed. 2011, 50, 3993–3997. 10.1002/anie.201007168. [DOI] [PubMed] [Google Scholar]
- Hartlmüller C.; Günther J. C.; Wolter A. C.; Wöhnert J.; Sattler M.; Madl T. RNA structure refinement using NMR solvent accessibility data. Sci. Rep. 2017, 7, 5393. 10.1038/s41598-017-05821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidis L.; Schütz U.; Tripsianes K.; Madl T.; Radke J.; Rucktäschel R.; Wilmanns M.; Schliebs W.; Erdmann R.; Sattler M. Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. Nat. Commun. 2017, 8, 14635. 10.1038/ncomms14635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Friedman J. I.; Stivers J. T. Cosolute paramagnetic relaxation enhancements detect transient conformations of human uracil DNA glycosylase (hUNG). Biochemistry 2011, 50, 10724–10731. 10.1021/bi201572g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J.; Wright P. E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004, 104, 3607–3622. 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- Wolf-Watz M.; Thai V.; Henzler-Wildman K.; Hadjipavlou G.; Eisenmesser E. Z.; Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat. Struct. Mol. Biol. 2004, 11, 945–949. 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- Rago F.; Saltzberg D.; Allen K. N.; Tolan D. R. Enzyme substrate specificity conferred by distinct conformational pathways. J. Am. Chem. Soc. 2015, 137, 13876–13886. 10.1021/jacs.5b08149. [DOI] [PubMed] [Google Scholar]
- Rose I. A.; O’Connell E. L.; Mehler A. H. Mechanismof the aldolase reaction. J. Biol. Chem. 1965, 240, 1758–1765. [PubMed] [Google Scholar]
- Zhu X.; Tanaka F.; Lerner R. A.; Barbas C. F. III; Wilson I. A. Direct observation of an enamine intermediate in amine catalysis. J. Am. Chem. Soc. 2009, 131, 18206–18207. 10.1021/ja907271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. G. III Enzyme dynamics from NMR spectroscopy. Acc. Chem. Res. 2015, 48, 457–465. 10.1021/ar500340a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss G.; Çelebi-Ölçüm N.; Moretti R.; Baker D.; Houk K. Computational enzyme design. Angew. Chem., Int. Ed. 2013, 52, 5700–5725. 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- Mandell D. J.; Kortemme T. Backbone flexibility in computational protein design. Curr. Opin. Biotechnol. 2009, 20, 420–428. 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Sterner R.; Höcker B. Catalytic versatility, stability, and evolution of the (βα) 8-barrel enzyme fold. Chem. Rev. 2005, 105, 4038–4055. 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]
- Röthlisberger D.; Khersonsky O.; Wollacott A. M.; Jiang L.; DeChancie J.; Betker J.; Gallaher J. L.; Althoff E. A.; Zanghellini A.; Dym O.; Albeck S.; Houk K. N.; Tawfik D. S.; Baker D. Kemp elimination catalysts by computational enzyme design. Nature 2008, 453, 190–195. 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- Siegel J. B.; Zanghellini A.; Lovick H. M.; Kiss G.; Lambert A. R.; Clair J. L. S.; Gallaher J. L.; Hilvert D.; Gelb M. H.; Stoddard B. L.; Houk K. N.; Michael F. E.; Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Adler reaction. Science 2010, 329, 309–313. 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi F.; Wang C.; Palmer A. G. Solution NMR and computer simulation studies of active site loop motion in triosephosphate isomerase. Biochemistry 2006, 45, 10787–10794. 10.1021/bi060764c. [DOI] [PubMed] [Google Scholar]
- Williams J. C.; McDermott A. E. Dynamics of the flexible loop of triose-phosphate isomerase: The loop motion is not ligand gated. Biochemistry 1995, 34, 8309–8319. 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- Guallar V.; Jacobson M.; McDermott A.; Friesner R. A. Computational modeling of the catalytic reaction in triosephosphate isomerase. J. Mol. Biol. 2004, 337, 227–239. 10.1016/j.jmb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Katebi A. R.; Jernigan R. L. The critical role of the loops of triosephosphate isomerase for its oligomerization, dynamics, and functionality. Protein Sci. 2014, 23, 213–228. 10.1002/pro.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph D.; Petsko G.; Karplus M. Anatomy of a conformational change: Hinged “lid” motion of the triosephosphate isomerase loop. Science 1990, 249, 1425–1428. 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- Salleron L.; Magistrelli G.; Mary C.; Fischer N.; Bairoch A.; Lane L. DERA is the human deoxyribose phosphate aldolase and is involved in stress response. Biochim. Biophys. Acta, Mol. Cell Res. 2014, 1843, 2913. 10.1016/j.bbamcr.2014.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.