Abstract
Internal initiation of translation can be mediated by specific internal ribosome entry site (IRES) elements that are located in certain mammalian and viral mRNA molecules. Thus far, these mammalian cellular and viral IRES elements have not been shown to function in the yeast Saccharomyces cerevisiae. We report here that a recently discovered IRES located in the genome of cricket paralysis virus can direct the efficient translation of a second URA3 cistron in dicistronic mRNAs in S. cerevisiae, thereby conferring uracil-independent growth. Curiously, the IRES functions poorly in wild-type yeast but functions efficiently either in the presence of constitutive expression of the eIF2 kinase GCN2 or in cells that have two initiator tRNAmet genes disrupted. Both of these conditions have been shown to lower the amounts of ternary eIF2-GTP/initiator tRNAmet complexes. Furthermore, tRNAmet-independent initiation was also observed in translation-competent extracts prepared from S. cerevisiae in the presence of edeine, a compound that has been shown to interfere with start codon recognition by ribosomal subunits carrying ternary complexes. Therefore, the cricket paralysis virus IRES is likely to recruit ribosomes by internal initiation in S. cerevisiae in the absence of eIF2 and initiator tRNAmet, by the same mechanism of factor-independent ribosome recruitment used in mammalian cells. These findings will allow the use of yeast genetics to determine the mechanism of internal ribosome entry.
Saccharomyces cerevisiae has been an invaluable tool in the study of mechanisms of cap-dependent translation initiation (1, 2). However, efforts to use yeast as a model system to study the mechanism of cap-independent internal initiation of translation have been hampered by the absence of functional internal ribosome entry site (IRES) elements that can direct the synthesis of selectable marker gene products. The well-studied viral IRES elements located in the RNA genomes of encephalomyocarditis virus, poliovirus, and hepatitis C virus do not function in living S. cerevisiae (3–5). The reasons for these IRES elements being inactive in yeast remain unclear. In the case of poliovirus and hepatitis C virus, a small inhibitor RNA has been detected, and it has been postulated that this inhibitor RNA sequesters factors that are needed for IRES-mediated translation (4, 5).
We have expressed approximately two million dicistronic mRNA species in yeast, containing unique nucleotide sequences in the intercistronic spacer. However, none of those sequences functioned as an IRES to mediate translation of the second cistron (unpublished observation). This finding was surprising because similar strategies have identified new synthetic IRES elements in mammalian cells (6, 7). The most likely reason why the yeast translation apparatus favors 5′ end-mediated translation over internal initiation is the synergy by which the 5′ terminal cap structure and the 3′ terminal polyadenosine sequences direct the binding of ribosomal subunits to the 5′ end of the mRNA (8). Thus, the translation machinery in S. cerevisiae does not seem to perform internal initiation in cells grown under normal laboratory conditions. Recently, Paz et al. discovered that a 140-nt RNA element from the lacI gene of Escherichia coli translated a second lacZ cistron in a dicistronic mRNA when yeast were in stationary phase (9). More recently, Zhou et al. reported that the leader regions in yeast YAP1 and TIF4631 could confer translation of a second luciferase cistron in logarithmically growing yeast (10). However, it is not clear whether these IRES elements can mediate translation of a second cistron encoding a selectable marker, allowing S. cerevisiae to grow under selectable conditions.
Very recently, we have discovered an IRES element in the intergenic region (IGR) of the cricket paralysis virus (CrPV) genome that can mediate internal initiation in the absence of any known eukaryotic initiation factors and without initiator tRNAmet (11, 12). Because the IGR-IRES element has such unusual properties, we wished to examine whether it could mediate internal initiation in S. cerevisiae.
Materials and Methods
Plasmids.
Wild-type and a mutated (IGRmut14) version of the IGR-IRES that disrupts a pseudoknot structure essential for IRES activity (11) were used in this study. The dicistronic reporter plasmids pLEU2 IGR URA3 and pLEU2 IGRmut14 URA3 plasmids were constructed as follows. The URA3 gene was amplified by PCR from wild-type yeast genomic DNA and C-terminally tagged with a FLAG epitope. The amplified DNA was digested with SacI and inserted into the SacI site of pRS313 (13). The LEU2 gene was cloned by PCR amplification; the DNA was digested with BglII and BamHI and inserted into BglII/BamHI sites of the URA3-containing plasmid, generating pLEU2-URA3. The pLEU2 IGR URA3 and pLEU2 IGRmut14 URA3 plasmids were generated by insertion of wild-type or mutated versions of the CrPV IGR-IRES, respectively, into the BamHI and XbaI sites located between the LEU2 and URA3 genes. The IGR-IRES begins eight nucleotides downstream of the LEU2 stop codon and initiates translation of a hybrid URA3 protein containing the first five amino acids of CrPV ORF2, followed by 10 amino acids encoded by vector sequences. Plasmids pCup1 LEU2 IGR URA3 and pCup1 LEU2 IGRmut14 URA3 were generated by inserting the LEU2-URA3 cassettes into the NcoI and SacI sites of the copper promoter-containing pSal I vector (14). The dicistronic dual luciferase plasmids have been previously described (15).
Yeast Strains and Genetic Methods.
Standard methods were used for culturing and transforming yeast strains (16). The yeast strains used in this study were: MBS (MATα, ade2–1, his3–11, his3–15, leu2–3,112, trp1–1, ura3–1, can1) (19), H1692 (MATα ura3–52, leu2–3,112, inol GCN2c-E1606G〈HIS4-lacZ ura3–52〉), H1613 (MATα ura3–52, leu2–3,112, inol GCN2c-E601K,-E1591K〈HIS4-lacZ ura3–52〉) (17), H1402 (MATα ura3–52, leu2–3,112, inol GCN2〈HIS4-lacZ ura3–52〉) (Alan Hinnebusch, personal communication), H2545 (MATa trp1-Δ1 ura3–52, IMT1 IMT2 imt3∷TRP1 imt4∷TRP1 leu2∷hisG GAL+), H2546 (MATa trp1-Δ1 ura3–52, IMT1 IMT2 imt3∷TRP1 imt4∷TRP1 leu2∷hisG GAL+ gcn2Δ) (18).
Northern Analysis.
Strains H1402, H1692, and H1613 were transformed with the indicated plasmids and grown in SD medium (16) supplemented with inositol and either with or without uracil at 30°C to an OD600 of 1. Cells were harvested from 10-ml cultures and washed once with H2O. Cells were disrupted by vortexing for 2 min with 200 μl of acid-washed glass beads/300 μl of RNA buffer (0.5 M NaCl/200 mM Tris⋅Cl, pH 7.5/10 mM EDTA)/300 μl of 5:1 phenol/chloroform, pH 5 (Amresco, Solon, OH). The aqueous phase was reextracted with 5:1 phenol/chloroform, pH 5, followed by ethanol precipitation with 3 volumes of 100% ethanol. Ten micrograms of total RNA was separated on a 0.8% formaldehyde agarose gel. The RNA was transferred to a nylon membrane and hybridized to a 32P-labeled anti-flag oligo (5′-GAGCTCTTACTTGTCGTCGTCGTCCTTGTAGTCAGCAGC-3′) or to a 32P-labeled StuI/XbaI fragment of URA3. Northern blots were analyzed by autoradiography.
Immunoblot Analysis.
Yeast strains H1402, H1692, and H1613 were grown in either SD medium supplemented with uracil and inositol (for cells transformed with plasmid) or in yeast extract/peptone/dextrose (for untransformed cells) at 30°C to an optical density at 600 nm of 1. Cells from 100-ml cultures were harvested by centrifugation for 5 min at 1,500 × g at 4°C and washed once with H2O. Cells were disrupted by vortexing four times with four volumes of glass beads for 1 min in three volumes of breaking buffer (20 mM NaHPO4, pH 7.2/50 mM NaCl/5 mM EDTA/2 mM PMSF/1 mM DTT/50 mM NaF/35 mM β-glycerolphosphate) at 4°C, and lysates were cleared by centrifugation at 10,000 × g. Fifty micrograms of protein was analyzed by SDS/PAGE, transferred to Immobilon-P (Millipore), and the FLAG-epitope-tagged Ura3p protein was visualized by using the anti-FLAG M2 monoclonal antibody (Sigma). Antibody CM-217 (gift from T. Dever, the National Institutes of Health) was used to detect both phosphorylated and nonphosphorylated forms of eIF2α; an antibody specific for the phosphorylated form of eIF2α was obtained from Research Genetics, Huntsville, AL. Immunoblots were developed by using an ECL kit (Amersham Pharmacia), as directed by the manufacturer.
In Vitro Translation Assays.
Yeast translation extracts were prepared as described previously (19) with omission of micrococcal nuclease treatment. Briefly, late logarithmically growing yeast cells were harvested from yeast extract/peptone/dextrose medium. The cells were lysed by agitation with 0.5-mm glass beads in ice-cold buffer (30 mM Hepes–KOH, pH 7.4/100 mM KOAc/2 mM MgOAc/8.5% mannitol/2 mM DTT/0.5 mM PMSF). Cellular debris was removed by centrifugation at 4°C, 38,700 × g for 5 min. Extracts were quick frozen on dry ice and stored at −80°C.
Extracts were treated with micrococcal nuclease (19) just before using, and in vitro translation was performed in 50% yeast extract programmed with 0.5 μg of in vitro transcribed capped dicistronic mRNA. Reactions had a final concentration of 37 mM Hepes–KOH, pH 7.4/170 mM KOAc/3 mM MgOAc/0.75 mM ATP/0.1 mM GTP/25 mM creatine phosphate/0.04 mM each amino acid/2.7 mM DTT/0.25 mM PMSF/0.24 mM CaCl2/1 mM EGTA/90 units/ml of micrococcal nuclease/4 μg of creatine phosphokinase/4 units RNasin. Reactions were incubated at 25°C for 90 min. In the edeine study, the extract was preincubated with the indicated concentrations of edeine for 5 min at room temperature before addition of RNA.
Results
The CrPV IGR-IRES Functions Poorly in Wild-Type Yeast Cells.
Wild-type yeast strain H1402 (17) was transformed with plasmid pCup1 LEU2 IGR URA3, which contains the tightly regulated CUP1 promoter (14, 20) linked to a dicistronic LEU2-IGR-URA3 gene (Fig. 1A) containing the wild-type IGR-IRES from CrPV (11). The growth of the transformed strain was monitored on plates that contained 100 μM Cu2+ and lacked uracil. As can be seen in Fig. 1B, neither the wild-type IGR-IRES nor the mutated IGRmut14 IRES mediated the synthesis of sufficient Ura3p to allow uracil-independent growth.
Figure 1.
A growth assay for the cricket IGR-IRES function in vivo. (A) A diagram of the dicistronic reporter construct that contains a CrPV IGR-IRES element preceding the second cistron. The expression of the reporter mRNA is directed by the copper promoter (Cup1). (B) Growth assay for yeast strains H1402 (isogenic wild type), H1692 (GCN2c), and H1613 (GCN2c) transformed with either pCup1 LEU2 IGR URA3 (IGR) or pCup1 LEU2 IGRmut14 URA3 (mut14) plasmids containing the dicistronic reporter. Transformants were streaked on minimal SD medium supplemented with 100 μM Cu2+, inositol, with or without uracil, as indicated.
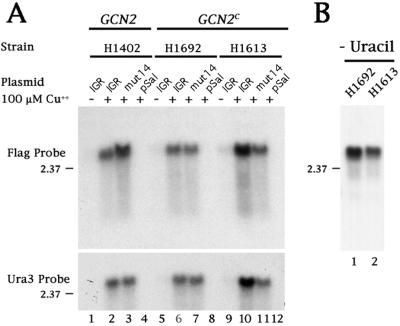
To determine whether full-length mRNAs were produced from the CUP1 promoter in the presence of added Cu2+, Northern analysis was performed. Fig. 2A shows that full length mRNAs that contained URA3 sequences were produced in transformed yeast cells in the presence (lane 2) but not in the absence (lane 1) of Cu2+. Importantly, the IRES-encoding DNA sequences did not display cryptic promoter activities that would have led to the synthesis of smaller functionally monocistronic URA3-encoding mRNAs. These experiments showed that both wild-type and mutated IGR-IRES-containing dicistronic mRNAs could be expressed in H1402 yeast cells, but these IGR-IRES elements did not mediate efficient internal initiation of the second cistron.
Figure 2.
Northern blot analysis of total RNA isolated from yeast strains transformed with various dicistronic reporter plasmids. (A) Yeast strains H1402, H1692, or H1613 were transformed with the plasmids described in Fig. 1 or with the pSal I (pSal) parental vector and grown in SD medium supplemented with (+) or without (−) 100 μM Cu2+, inositol, and uracil. Total RNA was isolated, separated on denaturing gels, transferred to nitrocellulose, and hybridized with a probe complementary to either the C-terminal FLAG epitope or the N-terminal coding region of URA3. The slight differences observed in mobility are also observed by ethidium bromide staining of the ribosomal RNAs and were not consistently observed, so it is unlikely that they reflect a true difference in mobility. (B) Yeast strains H1692 and H1613 transformed with the pCup1 LEU2 IGR URA3 plasmid were grown in SD medium supplemented with 100 μM Cu2+ and inositol. Northern analysis was performed as above by using the probe complementary to the N-terminal coding region of URA3.
The CrPV IGR-IRES Functions Efficiently in Yeast Cells That Express GCN2 Mutations That Result in Enhanced Phosphorylation of eIF2α.
In mammalian cells, the IGR-IRES functions poorly when ternary eIF2-GTP/initiator tRNAmet complexes are abundant. However, ribosome recruitment is efficient when the amount of intracellular ternary complex is lowered, for example by induction of kinases that phosphorylate eIF2α (12).
To test whether this dependence on ternary complex concentration can also be observed in yeast, we examined IGR-IRES activity in two different yeast strains that express dominant constitutive mutants of the eIF2 kinase GCN2 (GCN2c) (17). These GCN2c alleles have been shown to lower the abundance of ternary complex by increasing the phosphorylation of eIF2α (17, 21); as a result, phosphorylated eIF2 cannot be recycled to eIF2-GTP (21). Strain H1692 contains a E1606G change in the ribosome binding and dimerization domain of GCN2p, and strain H1613 contains double E601K-E1591K mutations mapping to the protein kinase and ribosome-binding domains of GCN2p, respectively (17, 22). Strains H1692 and H1613 were transformed with the dicistronic plasmids containing wild-type (IGR) or mutated (mut14) IGR-IRES elements. The CUP1 promoter was induced, and the growth of the strains was examined on plates lacking uracil. Fig. 1B shows that both GCN2c allele-expressing yeast strains could grow as colonies if the dicistronic mRNA contained a wild-type IGR-IRES (IGR); in contrast, yeast cells that expressed dicistronic mRNAs harboring the mutated IRES (mut14) failed to grow.
To examine whether full length dicistronic mRNAs were produced in the GCN2c-expressing strains, the size of dicistronic mRNAs was examined by Northern analysis by using radiolabeled probes that hybridize to the Flag epitope-encoding 3′ end of URA3 or to the URA3 coding region. Fig. 2A (lanes 5–12) shows that dicistronic RNAs of similar size were produced in the strains when the CUP1 promoter was active. As shown in Fig. 1, the majority of these strains will not grow in the absence of uracil in the media; therefore, Northern analysis (Fig. 2A) was performed on RNA isolated from cells grown in the presence of uracil. To address the formal possibility that H1692 and H1613 strains express monocistronic messages when grown in the absence of uracil, a Northern analysis was performed. As shown in Fig. 2B (lanes 1 and 2), only full length dicistronic mRNAs were detected, suggesting that Ura3p was produced from the dicistronic mRNA and not from smaller RNAs generated by degradation or activation of a cryptic promoter.
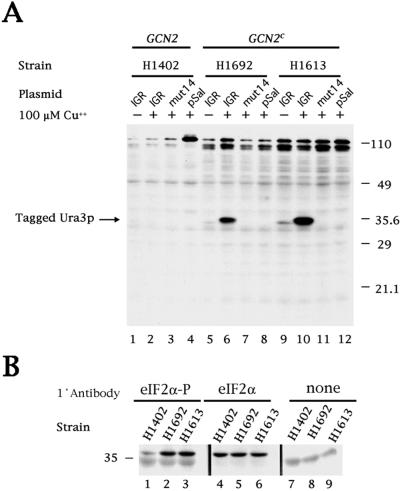
To test whether the growth observed in the absence of uracil correlated with the amount of Flag-tagged Ura3p protein produced, the amount of intracellular Ura3p was measured in immunoblots (Fig. 3A). Large amounts of Ura3p were produced only in GCN2c strains expressing the wild-type IGR-IRES (lanes 6 and 10), whereas little or no Ura3p was detected in the H1402 GCN2 strain (lanes 1–4) or in strains transfected with the mutated IGR-IRES (mut14) (lanes 3, 7, and 11) or the plasmid pSal, which lacks the dicistronic cassette (lanes 4, 8, and 12). The low amount of Ura3p produced in the GCN2c strain that expressed the wild-type IGR-IRES in the absence of Cu2+ (lanes 5 and 9) is likely to reflect a low level of uninduced transcription from the CUP1 promoter. As is supported by overexposure of the Northern shown in Fig. 2A, a very small amount of only full length dicistronic mRNA is detected when the promoter is not induced (in the absence of added Cu2+).
Figure 3.
Immunoblot analysis of CrPV IGR-dependent translation of tagged Ura3p and levels of eIF2α phosphorylation. (A) The transformed yeast strains are described in the legend to Fig. 1. An immunoblot was prepared and incubated with the anti-FLAG M2 monoclonal antibody to detect expression of Ura3p with a C-terminal FLAG tag. An image of the developed blot is shown. (B) Immunoblots from yeast strains H1402, H1692, and H1613 were prepared and incubated, as indicated, with anti-eIF2α polyclonal antibody, which detects only the phosphorylated form of eIF2α (eIF2α-P), with CM-217 polyclonal antibody, which detects both phosphorylated and nonphosphorylated forms of eIF2α (eIF2α) or without primary antibody (none). Images of the developed blots are shown. The 36-kDa eIF2α subunit migrates slightly above the 35-kDa marker protein.
To test whether the ability of the IGR-IRES to direct translation of Ura3p correlated with the predicted phosphorylation of eIF2α in the GCN2c strains, the abundance of eIF2α was examined in the various yeast strains. The immunoblot in Fig. 3B shows that the overall abundance of eIF2α was similar in all strains tested (lanes 4–6); however, the abundance of phosphorylated eIF2α was clearly higher in the GCN2c-expressing strains (lanes 2 and 3) than in the GCN2-expressing H1402 strain (lane 1).
IGR-IRES Functions in Yeast Cells That Contain Two Disrupted Initiator tRNAmet Genes.
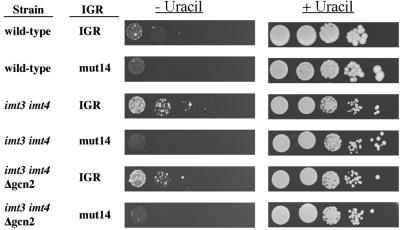
Although it is most likely that the diminished amounts of ternary complexes resulting from phosphorylation of eIF2α caused enhanced IGR-IRES activity, it was possible that constitutively active GCN2 could have stimulated the IGR-IRES by other means. Thus, IGR-IRES activity was examined in yeast strains in which the intracellular pool of functional ternary complex was diminished by lowering the abundance of initiator tRNAmet (18). To this end, strain H2545, which contains disrupted IMT3 and IMT4 initiator tRNAmet genes (18), was transformed with the various IGR-IRES plasmids, and growth was examined on plates lacking uracil. Fig. 4 shows that the wild-type IGR-IRES, but not the mutated IGR-IRES (mut14), conferred uracil-independent growth to the H2545 strain (imt3 imt4). However, the wild-type H1402 strain, which contains a full set of initiator tRNAmet genes, displayed much less uracil-independent growth on induction of the CUP1 promoter, demonstrating the IGR-IRES was not active at higher intracellular initiator tRNAmet concentrations.
Figure 4.
Growth assays for strains H1402 (wild-type), H2545 (imt3 imt4), and H2546 (imt3 imt4 Δgcn2) transformed with the plasmids described in Fig.1B. The figure shows the growth of serially diluted yeast cells on plates containing minimal SD medium supplemented with 100 μM Cu2+, inositol, and with or without uracil, as indicated.
Finally, strain H2546, which lacks the GCN2 gene in addition to functional IMT3 and IMT4 genes (imt3 imt4 Δgcn2) (18), allowed uracil-independent growth (Fig. 4), arguing that IGR-IRES activity did not depend on the presence of either wild-type or constitutively activated GCN2. Enhanced IGR-IRES activity suggests that the yeast translation apparatus is capable of performing internal initiation on this IGR-IRES during low abundance of eIF2-GTP or initiator tRNAmet.
The IGR-IRES Functions in Translation-Competent Yeast Extracts.
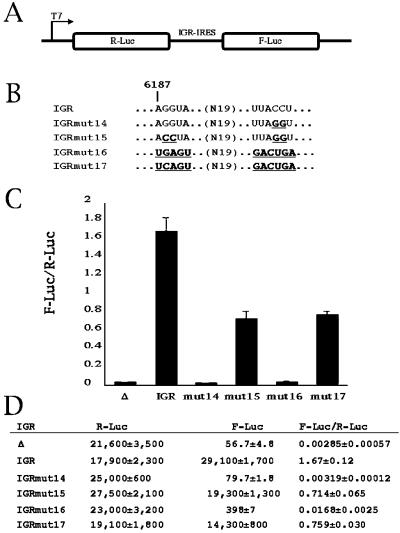
We have reported recently that the IGR-IRES can assemble mammalian 80S ribosomes without initiation factors or initiator tRNAmet, with the IGR-IRES occupying the ribosomal P site (12). To examine whether yeast ribosomes can be recruited to the IGR-IRES by a similar mechanism, we tested first whether the IGR-IRES can mediate internal initiation in translation-competent yeast extracts. Dicistronic luciferase reporter mRNAs (Fig. 5A) were synthesized by T7 RNA polymerase (11) and translated in translation extracts prepared from yeast (Fig. 5 C and D) (19). Only the first cistron was translated from transcripts that lacked the IGR-IRES in the intercistronic spacer (see Δ). However, inclusion of the IGR-IRES stimulated the translation of the second cistron by ≈500-fold (see IGR). As has been observed in cultured cells, IGRmut14 did not mediate internal initiation in the yeast lysate; however, IGRmut15, which contains compensatory mutations to restore the pseudoknot disrupted in IGRmut14 (Fig. 5B) (11), partially restored translation of the second cistron (Fig. 5 C and D).
Figure 5.
IGR-IRES activity in yeast extracts. (A) Diagram of dicistronic mRNAs containing the Renilla luciferase (R-Luc) as the first cistron and the firefly luciferase (F-Luc) as the second cistron. (B) Genotype of the wild-type (IGR) and various mutant IGR-IRES elements. Nucleotide sequences that differ from the wild-type IGR are shown in bold and underlined. (C) Translation-competent yeast extracts were programmed with various dicistronic RNAs, and the ratios of F-Luc to R-Luc production are indicated. (D) The primary data are shown. Δ represents the dicistronic mRNA without any IGR-IRES sequences inserted. Standard error of three experiments is indicated.
By using mutagenesis and toeprinting assays, we have recently reported that 80S ribosomes can initiate translation on IGRmut17, which contains a functional pseudoknot and a stop codon in the ribosomal P site (Fig. 5B) (12). Fig. 5 shows that yeast ribosomes initiated translation on IGRmut17 but not on IGRmut16 (Fig. 5 C and D) that lacks a functional pseudoknot. These findings indicate that yeast ribosomes can perform internal initiation on IGR-IRES elements as observed in living yeast cells.
IGR-IRES Is Not Inhibited by Edeine at Concentrations That Inhibit Translation of Capped mRNAs.
The compound edeine interferes with the recognition of the AUG start codon by the preinitiation complex (23). Therefore, inhibition by edeine is a good indication that translational initiation requires a functional ternary complex. We have previously shown that the IGR-IRES directs initiation of protein synthesis from a non-AUG codon and is insensitive to edeine at concentrations that inhibit tRNA delivery to the ribosomal P site (12). Addition of this compound to yeast translation extracts programmed with dicistronic, IGR-IRES-containing mRNAs inhibited translation of the first Rluc cistron in a dose-dependent manner (Fig. 6), whereas translation of the second cistron was insensitive at the edeine concentrations that inhibited translation of the first cistron, i.e., 0.05–0.5 μM. However, at higher edeine concentrations known to affect both the initiation and the elongation phases of protein synthesis (24, 25), translation mediated by the IGR-IRES was inhibited as well. Therefore, both the in vivo and in vitro results support the hypothesis that the IGR-IRES is translated by mammalian and yeast ribosomes by an unusual mechanism of internal initiation at a non-AUG codon that does not involve an eIF2-GTP-initiator tRNAmet ternary complex.
Figure 6.
The in vitro translation activity of the wild-type CrPV IGR-IRES in the presence of edeine. Translation-competent yeast extract was preincubated for 5 min with the indicated concentrations of edeine and programmed with the IGR dicistronic mRNA diagramed in Fig. 5. Translation efficiencies of F-Luc (shaded) and R-Luc (unshaded) are shown. Error bars represent the standard error of three experiments.
Discussion
Our results show that the IGR-IRES from CrPV functions in yeast when the intracellular concentration of ternary eIF2-GTP/initiator tRNAmet complex is lowered. Although it is known that diminishing the amount of ternary complexes in yeast enhances the translation of certain mRNAs, such as GCN4, by a reinitiation mechanism (21), it is unlikely that the IGR-IRES element mediated translation by the reinitiation mechanism used by GCN4. First, reinitiation operates on 5′ leader sequences that contain short ORFs at times when abundance of ternary complex is low. For the URA3 reporter used in this study, the 5′ sequences in the dicistronic mRNA are ≈1,000 nt long, consisting of the 5′ leader, LEU2 coding region, and the CrPV IGR-IRES; it is difficult to imagine how ribosomes would traverse these sequences to translate the URA3 coding region by a reinitiation mechanism. Even if some 40S subunits failed to disengage from the mRNA after reaching the stop codon of the first cistron, LEU2, such scanning 40S subunits would have to traverse several ORFs in the IGR-IRES before 80S complexes could be reassembled at the start codon of the second URA3 cistron in the dicistronic mRNA. Secondly, the 2-nt mutation in IGRmut14 completely abolished translation of the second cistron. Such a mutation should not affect the scanning of a 40S subunit unless this mutation would fortuitously stabilize higher ordered RNA structural motifs in the preceding IRES. However, this scenario is unlikely because it was shown that the IGRmut14 destabilizes a predicted pseudoknot structure that is essential for IRES activity (11). Taken together, the in vivo and in vitro experiments indicate that the IGR element functions as an IRES.
Although it has been known for some time that translation-competent extracts prepared from S. cerevisiae can perform internal initiation mediated by some IRES elements (19, 26), only recently have IRES elements been reported to function in living yeast (9, 10). As shown here, the IGR-IRES allowed the expression of a selectable marker gene in the GNC2c strains. This finding will allow use of yeast genetics to analyze the process of internal initiation of this unusual IRES element. Genes that regulate the efficiency of the IRES can now be identified, and interactions of the IRES with components of the translation apparatus can be examined in genetic screens. For example, we have shown that IGR-IRES function is favored by conditions that reduce intracellular concentrations of eIF2-GTP/initiator tRNAmet, arguing that components of the translation apparatus that direct the initiator tRNA to the ribosomal P site are not needed for IGR-IRES-mediated translation initiation. Thus, mutations in genes that affect the positioning of the mRNA in the ribosomal P site might be identified in such screens.
Acknowledgments
We thank Karla Kirkegaard for stimulating discussions and critical reading of the manuscript. We are grateful to Alan Hinnebusch for the suggestion to use GCN2c-expressing yeast strains in this study. We also thank Tom Dever [the National Institutes of Health (NIH)] for suggesting use of strains with disrupted initiator tRNA genes and for providing yeast strains and antibodies. This work was supported by grants from NIH (AI10485 to S.R.T., GM17766 to K.D.G., and GM55979 to P.S.).
Abbreviations
- IRES
internal ribosome entry site
- IGR
intergenic region
- CrPV
cricket paralysis virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 12866.
References
- 1.Donahue T F. In: Translational Control of Gene Expression. Sonenberg N, Hershey J W B, Mathews M B, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 487–502. [Google Scholar]
- 2.McCarthy J E. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evstafieva A G, Beletsky A V, Borovjagin A V, Bogdanov A A. FEBS Lett. 1993;335:273–276. doi: 10.1016/0014-5793(93)80745-g. [DOI] [PubMed] [Google Scholar]
- 4.Coward P, Dasgupta A. J Virol. 1992;66:286–295. doi: 10.1128/jvi.66.1.286-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S, Ott M, Yamane A, Venkatesan A, Gupta S, Dasgupta A. Front Biosci. 1998;3:D1241–D1252. doi: 10.2741/a359. [DOI] [PubMed] [Google Scholar]
- 6.Owens G C, Chappell S A, Mauro V P, Edelman G M. Proc Natl Acad Sci USA. 2001;98:1471–1476. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkatesan A, Dasgupta A. Mol Cell Biol. 2001;21:2826–2837. doi: 10.1128/MCB.21.8.2826-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preiss T, Hentze M W. Nature (London) 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 9.Paz I, Abramovitz L, Choder M. J Biol Chem. 1999;274:21741–21745. doi: 10.1074/jbc.274.31.21741. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Edelman G M, Mauro V P. Proc Natl Acad Sci USA. 2001;98:1531–1536. doi: 10.1073/pnas.98.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson J E, Powell M J, Hoover S E, Sarnow P. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson J E, Pestova T V, Hellen C U, Sarnow P. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 13.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mascorro-Gallardo J O, Covarrubias A A, Gaxiola R. Gene. 1996;172:169–170. doi: 10.1016/0378-1119(96)00059-5. [DOI] [PubMed] [Google Scholar]
- 15.Johannes G, Carter M S, Eisen M B, Brown P O, Sarnow P. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Greene and Wiley; 1989. [Google Scholar]
- 17.Ramirez M, Wek R C, Vazquez de Aldana C R, Jackson B M, Freeman B, Hinnebusch A G. Mol Cell Biol. 1992;12:5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dever T E, Yang W, Astrom S, Bystrom A S, Hinnebusch A G. Mol Cell Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka N, Najita L, Franzusoff A, Sarnow P. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butt T R, Sternberg E J, Gorman J A, Clark P, Hamer D, Rosenberg M, Crooke S T. Proc Natl Acad Sci USA. 1984;81:3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinnebusch A G. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 22.Hinnebusch A G. In: Translational Control of Gene Expression. Sonenberg N, Hershey J W B, Mathews M B, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 185–243. [Google Scholar]
- 23.Kozak M, Shatkin A J. J Biol Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 24.Carrasco L, Battaner E, Vazquez D. Methods Enzymol. 1974;30:282–289. doi: 10.1016/0076-6879(74)30031-6. [DOI] [PubMed] [Google Scholar]
- 25.Szer W, Kurylo-Borowska Z. Biochim Biophys Acta. 1970;224:477–486. doi: 10.1016/0005-2787(70)90580-0. [DOI] [PubMed] [Google Scholar]
- 26.Altmann M S, Blum J, Pelletier N, Wilson T M A, Trachsel H. Biochim Biophys Acta. 1990;1050:155–159. doi: 10.1016/0167-4781(90)90158-x. [DOI] [PubMed] [Google Scholar]