Through functional screening of a miR-196b target network in the poor prognosis subgroup of MLL-rearranged AML, Meyer et al. reveal a therapeutically targetable Skp2/p27Kip1 regulatory axis that modulates leukemic stemness, differentiation, and survival.
Abstract
We have shown that antagomiR inhibition of miRNA miR-21 and miR-196b activity is sufficient to ablate MLL-AF9 leukemia stem cells (LSC) in vivo. Here, we used an shRNA screening approach to mimic miRNA activity on experimentally verified miR-196b targets to identify functionally important and therapeutically relevant pathways downstream of oncogenic miRNA in MLL-r AML. We found Cdkn1b (p27Kip1) is a direct miR-196b target whose repression enhanced an embryonic stem cell–like signature associated with decreased leukemia latency and increased numbers of leukemia stem cells in vivo. Conversely, elevation of p27Kip1 significantly reduced MLL-r leukemia self-renewal, promoted monocytic differentiation of leukemic blasts, and induced cell death. Antagonism of miR-196b activity or pharmacologic inhibition of the Cks1-Skp2–containing SCF E3-ubiquitin ligase complex increased p27Kip1 and inhibited human AML growth. This work illustrates that understanding oncogenic miRNA target pathways can identify actionable targets in leukemia.
Introduction
11q23 rearrangements account for ∼10% of chromosomal abnormalities in leukemia and result in fusion of the Mixed-Lineage Leukemia (MLL) gene with a multitude of partners (such as AF9). 11q23/MLL rearrangements (MLL-r) are associated with a poor prognosis in acute myeloid leukemia (AML). The overall survival is only 20–40% with current treatment, perhaps due to the high frequency of leukemia stem cells (LSCs; Huret et al., 2001; Cox et al., 2004; Somervaille and Cleary, 2006; Döhner et al., 2010; Grimwade et al., 2010). Thus, the identification of therapeutically targetable pathways maintaining LSC self-renewal or survival is a high priority.
Expression of MLL fusion proteins, such as MLL-AF9, is sufficient to transform normal bone marrow hematopoietic stem/progenitor cells (Corral et al., 1996; Krivtsov et al., 2006; Somervaille and Cleary, 2006; Chen et al., 2008b). Thus, new approaches for treating MLL-r leukemia use small molecules that specifically block the interaction of MLL fusion proteins with transcriptional complexes containing Menin (Grembecka et al., 2012), DOT1L (Daigle et al., 2011), and BET family proteins (Dawson et al., 2011) or inhibit key MLL target genes, such as CDK6 (Placke et al., 2014), which are known to be important for the transformation and maintenance of this leukemia subgroup. Although clinical trials are still underway for these newer classes of inhibitors in a variety of different cancers, reports of resistance mechanisms are beginning to emerge (Fong et al., 2015; O’Leary et al., 2016), suggesting that novel strategies that synergize with these agents to enhance their efficacy or restore drug sensitivity are needed.
MLL-fusion proteins direct expression of the HOXA cluster (HOXA9 and HOXA7) and MEIS1 oncogenes, but the role of specific HOX genes in transformation varies with the MLL-fusion partner (Armstrong et al., 2002; Ayton and Cleary, 2003; Kumar et al., 2004; So et al., 2004; Erfurth et al., 2008), suggesting that other factors may play a role in the oncogenesis of MLL/HOX signaling. The evolutionarily conserved miR-196 family MIR196B, MIR196A1, and MIR196A2 encoded within the HOX gene clusters are overexpressed in AML and share identical seed sequences. In particular, MLL-r leukemias overexpress miR-196b (Jongen-Lavrencic et al., 2008; Li et al., 2008; Marcucci et al., 2008; Schotte et al., 2010), and elevated miR-196b expression in AML is associated with reduced survival (Li et al., 2012), suggesting that miR-196b might play an important role in AML. In agreement with these observations, we previously showed that simultaneous inhibition of miR-21 and miR-196b reduced MLL-AF9 LSC and protected mice from established leukemia (Velu et al., 2014).
While miRNA target prediction algorithms can provide guidance about potential miRNA-regulated genes, they do not account for cellular context or functional relevance, and they penalize binding scores for noncanonical miRNA recognition elements. Hence, alternate approaches are needed that can identify functionally important direct miRNA–mRNA target interactions in specific cellular contexts, such as leukemia. We used a two-step unbiased experimental workflow combining a biotinylated miRNA mimic pulldown approach with pooled in vivo shRNA screening to identify miR-196b targets with important functional consequences in MLL-r leukemia. We identify Cdkn1b/p27Kip1 as a target of miR-196b, whose repression significantly accelerates MLL-AF9 leukemia. Follow-up analyses of signaling pathways revealed that when p27Kip1 expression was repressed in MLL-AF9 AML cells, an embryonic stem cell (ESC)–like self-renewal/proliferation program was activated, while myeloid differentiation programming was reduced. Conversely, either specific blockade of the miR-196b interaction with Cdkn1b mRNA or small molecule inhibition of SCFSKP2 E3-ubiquitin ligase complex increased p27Kip1 protein levels, promoted monocytic differentiation, significantly reduced leukemogenic potential, and increased cell death of AML cells. Finally, SCFSKP2 inhibition acted synergistically with Menin/MLL (Ml-1), CDK4/6 (Palbociclib), and BET (I-BET151) inhibitors to block the growth of human MLL-r–containing AML cells. Thus, our work provides global mechanistic insight into the function of an oncogenic miRNA and illustrates the utility of exploiting miRNA signaling to identify actionable targets in leukemia.
Results
Unbiased global identification of miR-196b functions in human 11q23 AM
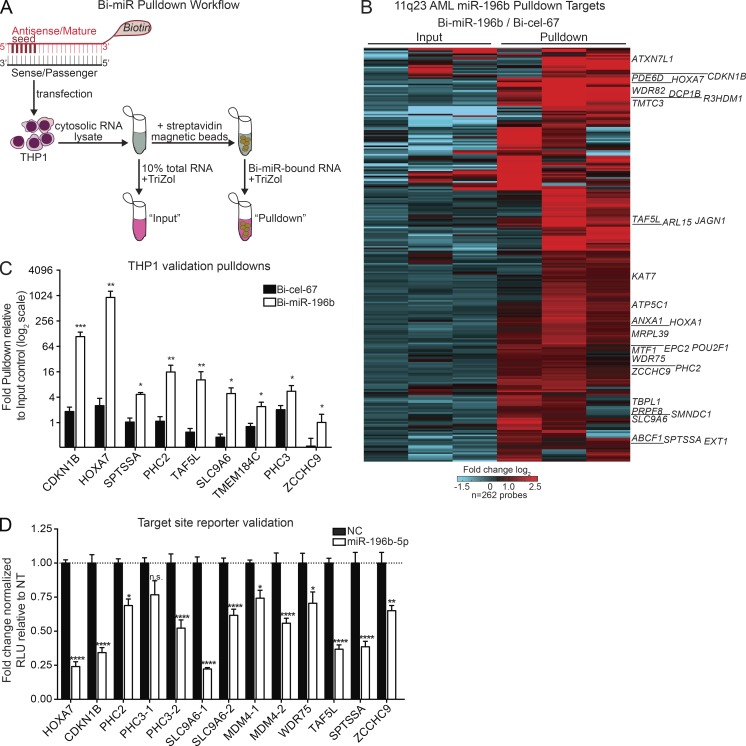
The miR-196b–directed signaling pathway in cancer, including 11q23 AML, is largely unknown. To globally define the miR-196b direct targets in AML, we adapted a biochemical approach to identify the mRNA transcripts bound by miR-196b–containing-RISC complexes in human AML cells harboring t(9;11), MLL-AF9 translocation (THP1 cells; Odero et al., 2000; Nonne et al., 2010; Lal et al., 2011). Using a miRNA target pulldown approach, transfection of biotinylated miRNA mimics (Bi-miR) permits purification of Bi-miR-196b target–containing RISC complexes using the biotin tag (Fig. 1 A). Identities of Bi-miR–associated transcripts were then revealed by microarray analyses, and validated by quantitative RT-PCR (RT-qPCR). We used two different human cell lines for pulldown studies: THP1 AML cells and HEK293 cells. Human embryonic kidney (HEK) cells express HOXA genes and miR-196b, permitting a comparative analysis of two miR-196b–relevant cellular contexts, one of which is leukemic. In both HEK293 and THP1 cells, Bi-miR-196b bound to previously known target HOXA7, which was enriched ∼500-fold compared with negative control Caenorhabditis elegans Bi-cel-67 pulldown (Fig. 1 C and Fig. S1 A; Yekta et al., 2004; Wong et al., 2015). Bi-miR-196b also repressed target protein expression (Fig. S1 B). Furthermore, microarray analyses identified 262 miR-196b target candidates with twofold or greater enrichment in Bi-miR-196b pulldown fractions compared with Bi-cel-67 control pulldowns in two or more independent experiments using THP1 AML cells (Fig. 1 B and Table S1). Analyses of each THP1 AML cell pulldown experiment independently identified an additional 1,708 candidate targets enriched at least twofold in THP1 cells (Table S1). These targets were reproducibly enriched greater than twofold in independent Bi-miR-196b pulldown experiments in THP1 AML cells, as well as HEK293 cells (Fig. 1 C and Fig. S1 A). Thus, the biotinylated miRNA pulldown approach reproducibly captures putative miR-196b targets from AML cells and also demonstrates context specificity for some miRNA target interactions.
Figure 1.
Unbiased identification of direct miR-196b targets in human 11q23 AML. (A) Schematic of biotinylated miRNA-mimic (Bi-miR) pulldown approach. (B) Heat map of putative miR-196b target gene pulldowns enrichments from three independent pulldowns in 11q23 mutant THP1 AML cells. Displayed genes were enriched at least twofold in Bi-miR-196b pulldowns relative to matched Bi-cel-67 pulldowns in two of three experiments. The corresponding fold change Bi-miR-196b versus Bi-cel-67 inputs are shown for each gene. (C) Independent RT-qPCR validation of miR target mRNA pulldown. The average fold pulldown (± SEM) relative to matched input controls for each Bi-cel-67 control (black bars) and Bi-miR-196b mimic (white bars) in at least three independent experiments in THP1 cells. Statistical significance by paired t tests for each gene versus Bi-cel-67 control. (D) Functional miR target validation. Average fold change ± SEM repression by miR-196b (miR-196b-5p) relative to negative control (NC) of indicated miR-196b binding sites in three independent Luciferase reporter assay experiments. Multiple binding sites in the same gene are distinguished by “1” and “2”. Statistical significance was determined using a two-way ANOVA versus NC. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. n.s., not significant.
To validate that the Bi-miR-196b pulldown assays identify novel targets, we first intersected pulldown targets with those predicated by computational algorithms PITA (Kertesz et al., 2007), miRanda (Enright et al., 2003; John et al., 2004; Betel et al., 2008), TargetScan7.0 (Lewis et al., 2003, 2005; Agarwal et al., 2015), and DIANA (Maragkakis et al., 2009a,b; Papadopoulos et al., 2009). Of the 162 pulldown targets mapped to known gene IDs from Fig. 1 B, 90 were not predicted by algorithms, suggesting that an experimental approach can identify novel miRNA targets (Fig. S1 C and Table S1). Next, to provide independent validation for miRNA binding to genes in our pulldown datasets, we manually located miR-196b binding sites with a minimum of six consecutive nucleotide-seed matches beginning at either α, β, or γ positions (Ellwanger et al., 2011). These sites were queried for binding by Argonaute (AGO) proteins using published high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP) datasets (Yang et al., 2015). CDKN1B is shown as a representative in Fig. S1 D. Of the 17 pulldown targets examined, 100% overlapped with at least one AGO-CLIP peak and 15/17 showed multiple AGO-CLIP datasets overlapping each miR binding site (Fig. S1 E). Using luciferase reporter assays, we validated 12/13 target sites located in 10 different pulldown target genes for miR-196b binding and luciferase repression (Fig. 1 D). Finally, we analyzed existing RNA-sequencing (RNA-seq) data from E9.5 embryos with limiting alleles of miR-196 (196a2+/−;b−/−, 196a2−/−;b+/−, or 196a2−/−;b−/− versus 196a2+/−;b+/+; Wong et al., 2015). Analysis of the RNA-seq data revealed increased expression of Bi-miR-196b pulldown targets in a miR-196 allele dose–dependent manner (Fig. S1 F and Table S1). In sum, these data demonstrate reproducibility of biotinylated miRNA target pulldowns, and confirm the direct binding of miR-196b and/or AGO complexes to pulldown target mRNAs.
An in vivo shRNA screen resolves oncogenic miR-196b signaling in MLL-AF9 leukemogenesis
To determine targets with functional importance in leukemogenesis, we devised an in vivo shRNA screen. Previously, similar screens have been used to identify novel therapeutic targets in MLL-AF9 leukemia or to phenocopy miRNA activity in vitro (Mavrakis et al., 2010; Zuber et al., 2011; Miller et al., 2013; Chen et al., 2015). Given that miRNA repress target mRNA stability and/or translation, we hypothesized that hairpins enriched in the shRNA screen would mimic miR-196b oncogenic activity. First, we performed an engraftment test on three mice transplanted with MLL-AF9 cells transduced with a randomly barcoded lentiviral library at constant multiplicity of infection (MOI) ∼0.2 to produce one unique barcode per cell after selection with puromycin. DNA from resultant AML in moribund mice was sequenced and compared with DNA from nontransplanted, transduced, and selected MLL-AF9 cells as input control, quantifying an engraftment efficiency of ∼4%. Based on these data, to ensure sufficient representation of each experimental hairpin in vivo, we estimated that shRNA should be divided into random pools of ∼100 hairpins per 1 million MLL-AF9 cells transplanted into 10 mice per pool. Thus, at a 4% engraftment rate, each hairpin was represented by 400 cells per mouse.
Given these parameters, to ensure adequate power and feasibility of the in vivo screen we restricted the number of miR-196b pulldown targets analyzed. Specifically, we selected 116 screen candidates that were enriched in at least one THP1 pulldown experiment, 60 of which were also identified in the HEK293 pulldown and 103 that were predicted targets by computational algorithms (PITA [Kertesz et al., 2007], miRanda [Enright et al., 2003; John et al., 2004; Betel et al., 2008], TargetScan7.0 [Lewis et al., 2003, 2005; Agarwal et al., 2015], and DIANA [Maragkakis et al., 2009a,b; Papadopoulos et al., 2009]; Table S1). Priority was given to genes with previously validated miR-196b target sites such as HoxA7 (Yekta et al., 2004), genes associated with oncogenic or tumor suppressive pathways such as Cdkn1b (Polyak et al., 1994; Zhang et al., 2013), Braf (Rajagopalan et al., 2002), Cdk12 (Blazek et al., 2011), Phc2 (Isono et al., 2005), and Mdm4 (Parant et al., 2001), and genes involved in stemness or differentiation pathways such as Anxa1 (Machado et al., 2016), Epha7 (Nguyen et al., 2017), Ikzf1 (Winandy et al., 1995; Wang et al., 1996), and Pou2f1 (Wang et al., 2004). In vivo examination of 116 candidates involved 632 individual hairpins (4–6 hairpins/gene) were divided into 8 pools (Fig. 2 A). Twelve hairpins against four control genes were also included in each pool.
Figure 2.
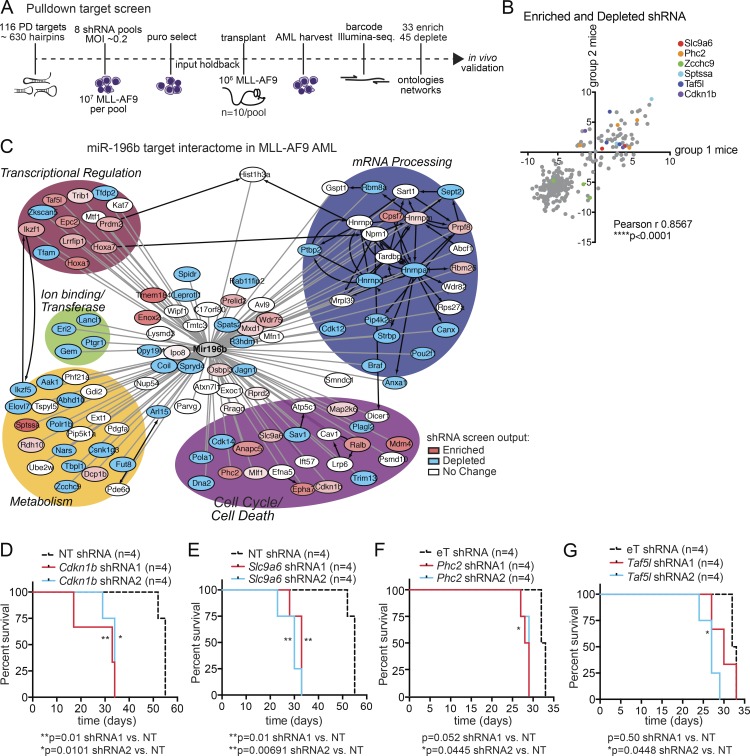
An in vivo shRNA screen functionally dissects miR-196b networks in MLL-AF9 leukemogenesis. (A) Schematic overview. In vivo shRNA-positive selection screen of miR-196b-pulldown targets in primary MLL-AF9 leukemia. Leukemic splenocytes were transduced with eight different lentiviral shRNA pools against 116 miR-196b target genes and transplanted into recipient mice (n = 10 mice/pool). Sequencing of pretransplant pools (input) versus leukemic splenocytes (from moribund mice) identified 33 enriched and 45 deleted genes of at least two hairpins/gene over input control. (B) Replicate assay correlation. For each enriched and depleted hairpin, recipient mice were divided into two groups/pool (n = 4–5 mice/group/pool) and plotted. The relatedness between these two groups of mice was evaluated by Pearson correlation demonstrating that in vivo selection of hairpin activity is not stochastic (r = 0.8567). Examples of genes with enriched (Scl9a6, Phc2, Sptssa, Taf5l, and Cdkn1b) or depleted (Zcchc9) shRNA are color coded. (C) Network analysis of screen targets by gene ontology biological processes. RNAi screen enriched gene nodes (red), depleted gene nodes (blue), and unchanged gene nodes (white). Protein–protein interactions are denoted by black edges and putative miR-196b target transcript interactions experimentally identified in miR target pulldown assays are denoted by gray edges. (D–G) In vivo validation of positive selection of individual targets. Kaplan-Meier survival curves of mice transplanted with MLL-AF9 leukemia expressing individual shRNA hairpins against the indicated genes (n = 4 mice/group; 2 hairpins/gene). Non-targeting (NT) or EV (eT) were used as controls. miR-196b pulldown targets Cdkn1b (D), Slc9a6 (E), Phc2 (F), and Taf5l (G) accelerated leukemia lethality. Significant p-values are reported by Log-rank (Mantel-Cox) test for each hairpin compared with control. *, P ≤ 0.05; **, P ≤ 0.01; ****, P < 0.0001.
As illustrated in Fig. 2 A, pooled shRNA-containing lentiviruses were transduced into MLL-AF9 leukemic splenocytes at a constant MOI of ∼0.2, selected with puromycin, and transplanted into sublethally irradiated mice (n = 10/pool). Of the 116 miR-196b target genes screened, 78 genes with at least two hairpins per gene showing reproducible effects in at least four mice per pool were subsequently scored. 33 genes showed enrichment, while 45 were depleted compared with control hairpins (Table S1). To test that hairpins were not stochastically selected in vivo, we randomly divided mice from each pool into two groups and found a significant correlation of hairpin enrichment and/or depletion across replicate mice in the two groups (Fig. 2 B). These data indicate a biological function of hairpin activity in vivo, and also demonstrate that the shRNA screen was adequately powered to resolve miRNA target function in vivo using shRNA.
To illustrate the biological processes impacted by the miR-196b target hairpins in our screen, we performed GO-Elite enrichment analyses of the 116 genes targeted by hairpins grouped by screen input versus screen output categories (enriched and depleted; Table S1). To illustrate these data, we created a miR-196b target interactome based on these ontologies (Fig. 2 C). Corresponding shRNA screen enrichment or depletion scores of genes involved in these different functional networks are graphed in Fig. S2 (A and B). We note examples in which both enriched and depleted hairpins target different genes involved in the same basic biological processes (e.g., gene expression; Fig. 2 C). To validate our pooled screen results, we assayed the activity of two hairpins per gene that were enriched in the MLL-AF9 screen against candidates from each biological process for in vivo effects of target repression on (1) disease latency as measured by overall animal survival (Fig. 2, D–G; and Fig. S2 C) and (2) knockdown of mRNA expression (Fig. S2 D). Compared with MLL-AF9 control (nontargeting [NT] or eT shRNA) transplanted mice, two independent enriched shRNA against Cdkn1b, Slc9a6, Phc2, and Taf5l (Fig. 2, D–G, respectively) significantly accelerated MLL-AF9 leukemogenesis, while shRNA against a depleted gene Zcchc9 had no effect on latency (Fig. S2 C). In contrast, independent validation studies using two independent shRNA that were enriched against HoxA7, Sptssa, Map2k6, Mdm4, Anapc5, Tmem184c, and Hnrnpm showed either incomplete (one out of two hairpins) or no (zero out of two hairpins) effect on disease latency in vivo as compared with MLL-AF9 control (data not shown). These data indicate that cyclin-dependent kinase Cdkn1b (p27Kip1), sodium-hydroxide exchanger Slc9a6 (Nhe6), Polycomb Group member Polyhomeotic homologue Phc2, and PCAF-associated factor Taf5l represent a select group of miR-196b targets with tumor suppressive activity in AML. In sum, repression of a subset of miR-196b targets in vivo is selectively enriched by cooperation with MLL-AF9 to promote leukemogenesis.
miR-196b direct-target Cdkn1b controls ESC-like programming to maintain MLL-AF9 leukemia
To gain deeper mechanistic insight into the contribution of the miR-196b targets in AML, we performed transcriptional profiling of Cdkn1b (p27Kip1) and Phc2 knockdown cells. Cdkn1b and Phc2 were robustly selected in Bi-miR-196b pulldown experiments, both are associated with cell cycle/cell death processes, independent shRNA targeting Cdkn1b or Phc2 were selected in the shRNA screen, and knockdown of both genes independently accelerated MLL-AF9 leukemogenesis in vivo. We identified 113 genes significantly differentially expressed in Cdkn1b-knockdown MLL-AF9 AML compared with NT control (Fig. 3 A; n = 2 per group). Gene set enrichment analyses (GSEA) were performed by exploring the MSigDB database, as well as other published gene sets previously enriched in MLL-AF9 leukemia (Table S2; Krivtsov et al., 2006; Somervaille et al., 2009). As expected, genes up-regulated in Cdkn1b-knockdown MLL-AF9 leukemia positively correlated to cell cycle (e.g., Cdk4 and Ccnd2). However, up-regulated genes also strongly correlated to an ESC core gene set (e.g., Hadh, Lmnb1, Msh2, and Prmt1; Fig. 3 B; Wong et al., 2008). This enrichment is not simply associated with accelerated leukemia pathogenesis, as Phc2 knockdown similarly accelerated leukemia, but showed reduced expression of the same ESC core gene set (Fig. S3, A and B; and Table S2). Instead, Phc2 knockdown leukemia showed increased expression of an adult stem cell gene set (Fig. S3 B; Wong et al., 2008), which was conversely down-regulated in Cdkn1b-knockdown MLL-AF9 leukemia (Fig. S3 C). Thus, there appear to be multiple miR-196b targets regulating distinct stem cell transcriptional programs important for the pathogenesis of MLL-AF9 leukemia. Importantly, genes down-regulated upon Cdkn1b knockdown in MLL-AF9 AML correspond with a poor prognosis gene signature in AML (Table S2; Yagi et al., 2003). Given the high frequency of MLL-r leukemia stem cells, which correlates with prognosis (Huret et al., 2001; Cox et al., 2004; Somervaille and Cleary, 2006; Döhner et al., 2010; Grimwade et al., 2010), and the stem cell signature activated by Cdkn1b knockdown, we focused subsequent studies on understanding the role and therapeutic potential of p27Kip1 in AML.
Figure 3.
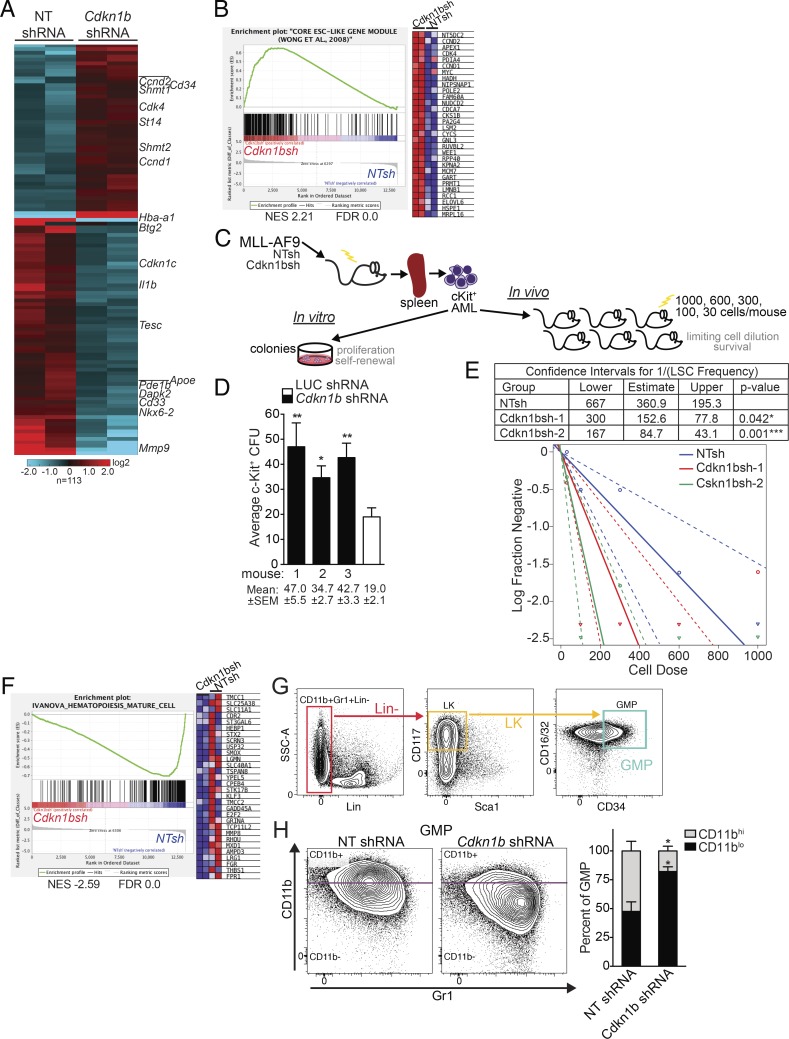
miR-196b targets Cdkn1b to regulate leukemia stemness and differentiation programs in MLL-AF9 leukemia. (A) Heatmap of gene expression in Cdkn1b shRNA-knockdown and NT-shRNA control leukemias. Hierarchical clustering of 113 differentially expressed genes showing greater than twofold change in expression by RNA-seq analysis of NT-shRNA control (n = 2) or Cdkn1b shRNA (n = 2) expressing MLL-AF9 leukemic splenocytes. (B) GSEA plot ranking ESC core gene set along descending fold change gene expression in Cdkn1b-knockdown (n = 2) versus NT control leukemias (n = 2) by RNA-seq. Expression of the top subset of leading edge genes is shown. NES, normalized enrichment score. (C) Schematic illustrating in vitro colony forming assay and in vivo limiting cell dilution transplantation assay to quantify functional stem cells from freshly isolated Cdkn1b shRNA and NT control MLL-AF9 leukemias. (D) In vitro leukemic stem cell analysis. Average CFU ± SEM from c-Kit+ leukemic splenocytes from individual Cdkn1b shRNA (n = 3) or control shRNA MLL-AF9 moribund mice. Mean numbers of CFU ± SEM indicated below. A representative experiment is shown. Statistical significance evaluated by t test. *, P ≤ 0.05; **, P ≤ 0.01. (E) In vivo quantitation of leukemic stem cells by limiting dilution. Sublethally irradiated mice transplanted with a cell dose range of 1,000, 600, 300, 100, and 30 cells (n = 6 mice/dose) were followed for 90 d. LSC frequencies and statistical comparisons for pairwise differences in active cell frequencies (table and log-fraction plot) between Cdkn1b-knockdown groups and NT control group were calculated by ELDA software (see Fig. S3 [E–G] for corresponding survival curves). Statistically significant differences in LSC frequencies of NTsh control (1/360.9) versus Cdkn1bsh1 (1/152.6); *, P = 0.042 and NTsh control (1/360.9) versus Cdkn1bsh2 (1/84.7) ***, P = 0.001. (F) GSEA plot ranking mature hematopoietic cells gene set along descending fold change gene expression in Cdkn1b-knockdown versus NT control leukemias by RNA-seq (n = 2/group). Expression of the top subset of leading edge genes is shown. (G) Leukemic GMP gating strategy: CD11b+Gr1+ lineage negative (Lin−, red), Lin−c-Kit+ (LK, orange), and LK CD34+CD16/32+ granulocyte monocyte progenitor gate (GMP, green). (H) Flow cytometric validation of differentiation status. Representative flow plots (left) and average percentage of CD11bhi and CD11blo leukemic GMP-gate populations ± SEM (right) from spleens of moribund Cdkn1b shRNA or control NT shRNA MLL-AF9 mice (n = 3/group). *, P < 0.05 for CD11bhi and CD11blo GMP in Cdkn1b shRNA versus NT shRNA by two-way ANOVA.
Based on enrichment for the ESC core gene set (Wong et al., 2008), we hypothesized that Cdkn1b knockdown would correspond to greater leukemia stem cell (LSC) activity, which may explain its accelerated phenotype. To this end, we used fresh c-Kit+ MLL-AF9 leukemic splenocytes from moribund animals to quantify colony-forming cells (CFCs) in vitro and the frequency of LSC by limiting cell dilution transplantation in vivo (Fig. 3 C). While MLL-AF9 cells freshly transduced with Cdkn1b or NT control shRNA exhibit no differences in clonogenicity or replating potential (Fig. S3 D), c-Kit+ Cdkn1b-knockdown MLL-AF9 leukemic splenocytes from moribund mice formed significantly greater numbers of colonies than shRNA control (Fig. 3 D). Sublethally irradiated mice transplanted with limiting numbers of c-Kit+ Cdkn1b or NT control shRNA expressing MLL-AF9 AML cells were monitored for survival over 60 d (Fig. S3 E-G), at which point LSC frequencies were estimated using the ELDA web tool (Hu and Smyth, 2009). Two independent Cdkn1b-knockdown tumors had LSC frequencies of 1/152.6 and 1/84.7, which were statistically significant from the frequency of LSCs (1/360.9) in NT control MLL-AF9 AML (Fig. 3 E). Thus, suppression of Cdkn1b increases the number of MLL-AF9 LSCs.
Cdkn1b-knockdown leukemia also showed down-regulation of a mature hematopoietic cell gene set (Fig. 3 F; Ivanova et al., 2002). To experimentally validate this observation, we performed flow cytometry to examine myeloid differentiation markers CD11b and Gr1 on Cdkn1b-knockdown compared with NT control MLL-AF9 leukemic splenocytes. As previously described, MLL-AF9 AML is characterized by immunophenotypically mature CD11b+Gr1+ granulocyte/monocyte progenitor (GMP)–like cells (Fig. 3 G; Krivtsov et al., 2006; Somervaille and Cleary, 2006). In agreement with the gene expression data, Cdkn1b-knockdown was associated with significantly reduced expression of CD11b on MLL-AF9 leukemic GMPs compared with NT control (Fig. 3 H and Fig. S3, H and I). Together, these data suggest lowering p27Kip1 levels is important for restricting myeloid differentiation and expanding LSCs within the MLL-r leukemic state.
In agreement with Cdkn1b-knockdown augmenting LSC activity, GSEA analyses showed increased expression of the ESC core gene set (Wong et al., 2008), and an independent gene ontology analysis revealed significant enrichment in cell proliferation and apoptosis processes (Fig. S3 J). It is possible that either similar proliferation rates of ESC and of MLL-AF9 LSC (Somervaille et al., 2009), or indirect effects on Myc (Kim et al., 2010), may explain the enrichment of ESC-like gene sets within cancer-derived gene expression signatures. To this end, we bioinformatically dissected Myc-regulated genes (Kim et al., 2008, 2010) from the ESC core gene set (Table S2; Wong et al., 2008). We also used a previously published ESC gene set with cell cycle annotations removed (Somervaille et al., 2009). Importantly, Cdkn1b-knockdown cells were significantly enriched for ESC core gene sets that were Myc independent or with cell cycle annotations removed (Fig. S3, K and L; and Table S2; Somervaille et al., 2009). Together, these data suggest that repression of the miR-196b target Cdkn1b enhances an ESC-like transcriptional program in leukemia cells, and that this gene expression program extends beyond known Myc and cell cycle effects. Altogether, these results suggest that p27Kip1 regulates (1) expression of an ESC core gene set (Wong et al., 2008), (2) LSC activity/clonogenicity, and (3) the differentiation of MLL-AF9 leukemic progenitors.
p27Kip1 interaction with cyclin–CDK complexes regulates ESC-like gene expression, MLL-r leukemia differentiation, and survival
Given the association of Cdkn1b-knockdown MLL-AF9 leukemia gene signature with that of AML with poor outcomes (Table S2; Somervaille et al., 2009), we posited that Cdkn1b may have clinical utility in MLL-r AML. As proof of concept, we force expressed Cdkn1b and a selectable marker (ZsGreen) in MLL-AF9 leukemic splenocytes. An empty vector (EV) served as control. ZsGreen+ cells were FACS-sorted and plated in methylcellulose colony assays or transplanted into recipient mice (Fig. 4 A, schematic). p27Kip1 overexpression significantly inhibited MLL-AF9 colony formation in vitro (Fig. 4 A). In vivo, p27Kip1 overexpression significantly delayed survival compared with EV control, but all mice succumbed to AML (Fig. 4 B). However, leukemic splenocytes harvested from mice transplanted with Cdkn1b-overexpressing MLL-AF9 cells were completely devoid of ZsGreen+ marker expression (black lines) as compared with EV control mice (Fig. 4 C, green lines). Moreover, immunoblot analyses revealed similar p27Kip1 protein levels in leukemic splenocytes from Cdkn1b or EV control MLL-AF9 transplanted mice (Fig. 4 C). In sum, p27Kip1 overexpression is not compatible with MLL-AF9 leukemogenesis in vitro and in vivo.
Figure 4.
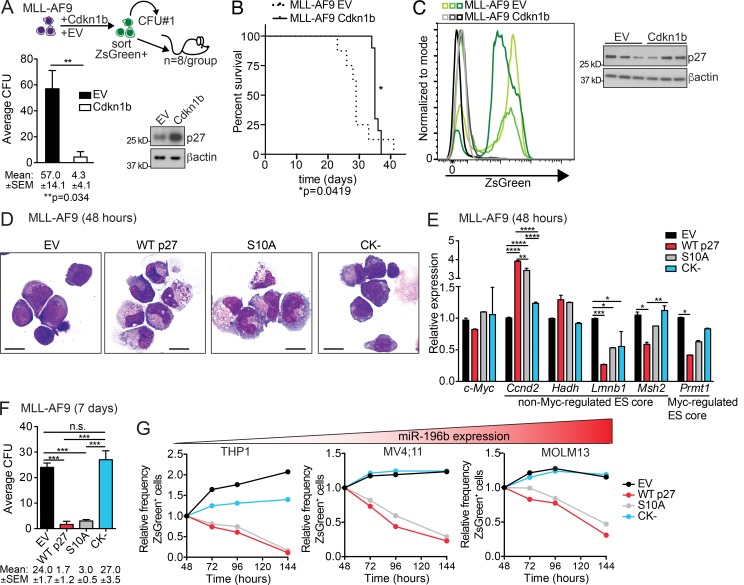
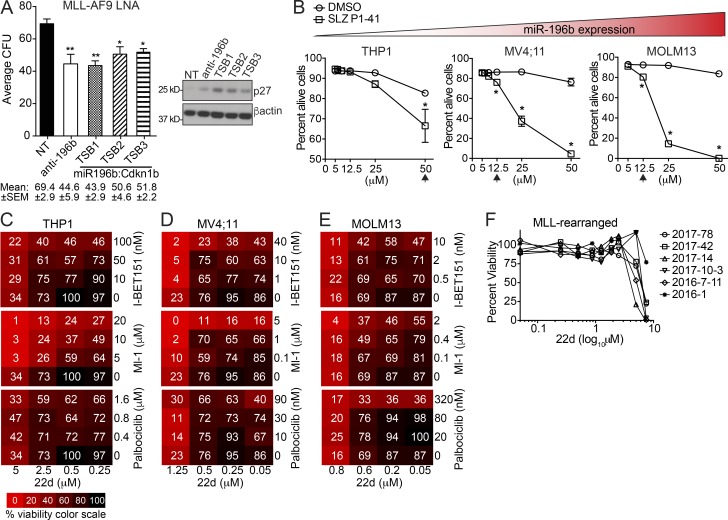
Cdkn1b/p27Kip1 suppresses MLL-r leukemia in a cyclin–CDK-dependent manner. (A) Top, schematic of CFU and transplantation of ZsGreen+ sorted Cdkn1b-ZsGreen (Cdkn1b) or EV-ZsGreen (EV) control expressing MLL-AF9 leukemia cells. Bottom, average number of colonies ± SEM (n = 3 replicates/group). Mean CFU numbers ± SEM indicated below bar graph. **, P ≤ 0.01 by t test. Immunoblot analyses of p27 performed on cells from CFU assays with β-actin as loading control. A representative of two independent experiments is shown with similar results. (B) Kaplan-Meier survival curve of mice transplanted with ZsGreen+ Cdkn1b overexpressing MLL-AF9 cells (n = 6/group). Significant differences in survival evaluated by Log-rank (Mantel-Cox) test; *, P ≤ 0.05. (C) Counter selection of p27 overexpression in vivo. Flow cytometric analyses for ZsGreen+ expression in EV (green lines) and Cdkn1b-overexpressing (black lines) MLL-AF9 leukemic splenocytes from moribund mice. Three representative mice/group are shown. Immunoblot analysis of p27 with β-actin as loading control (n = 3 mice/group). (D–F) Functional dissection of p27 activity. WT p27Kip1 (WT p27), nuclear localized p27Kip1 (S10A; increased cell cycle inhibition), or CDK-binding domain mutant p27Kip1 (CK−; no cell cycle inhibition) were expressed via lentiviral vectors in murine MLL-AF9 leukemic splenocytes. EV vector served as control. Cells were FACS sorted for ZsGreen+ 48 h after transduction and prepared for cytospin (D), RNA (E), or plated in CFU assay (F). Representatives of two independent experiments with similar results is shown. (D) Cytologies of ZsGreen+ EV, WT p27, S10A, and CK− expressing MLL-AF9 cells were visualized with Wright Giemsa stain and imaged at 60× magnification. Bars, 10 µm. (E) RNA extracted from ZsGreen+ EV, WT p27, S10A, and CK− MLL-AF9 cells was examined by RT-qPCR for changes in c-Myc expression as well as representative non–Myc-regulated (Ccnd2, Hadh, Lmnb1, and Msh2) and ESC core genes (Prmt1; n = 2 replicates/group; from Fig. S3, K and L). Expression was compared with Sdha as loading control and is graphed as average relative to EV ± SEM. Statistical significance was evaluated for each gene by two-way ANOVA Holm-Sidak multiple comparisons test. (F) Equal numbers of ZsGreen+ EV, WT p27, S10A, and CK− expressing MLL-AF9 leukemic splenocytes were plated in triplicate in methylcellulose. The average number colonies ± SEM enumerated after 7 d in methylcellulose is shown for three technical replicates with mean CFU ± SEM indicated below. A representative of two independent experiments is shown. Statistical significance was evaluated by one-way ANOVA Holm-Sidak multiple comparisons test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. n.s., not significant. (G) Human AML cell lines are organized according to their levels of miR-196b expression (lowest to highest, red gradient triangle; see also Fig. S4 B). The relative frequency of ZsGreen+ EV, WT p27, S10A, or CK− transduced THP1, MV4;11, and MOLM13 cells measured over time are shown as relative to the proportion of ZsGreen+ cells at the initial time point 48 h post-transduction. A representative experiment is shown.
To understand how p27Kip1 exerts anti-leukemic effects, we expressed either WT p27Kip1 (WT p27) or two p27Kip1 mutants in primary murine MLL-AF9 leukemia. Mutation of p27Kip1 at Ser10 to Ala (S10A) mimics the dephosphorylation of p27Kip1 at this residue inhibiting its nuclear export and promoting assembly into cyclin–CDK complexes (Besson et al., 2006). Thus, the S10A mutation increases p27Kip1-mediated cell cycle inhibition. Conversely, mutation of the amino acids required for p27Kip1 interaction with cyclins (R30A and L32A) and CDKs (F62A and F64A) generates the p27Kip1 (CK−) mutant. The CK− mutant abolishes p27Kip1-mediated cell cycle inhibition (Besson et al., 2006). Expression of WT p27 or the S10A mutant induced G1 cell cycle arrest (Fig. S4 A) and monocytic differentiation (Fig. 4 D) of MLL-AF9 leukemic blasts as compared with EV. The CK− mutant expressing cells were similar to EV cells (Fig. 4 D), suggesting that cell cycle and differentiation effects are linked. Additionally, ESC-like genes Lmnb1, Msh2, and Prmt1 (from Fig. 3 B and Fig. S3, F and G) were significantly repressed by WT p27 expression, whereas expression of c-Myc and Hadh was unchanged (Fig. 4 E). The CK− mutant abrogated repression of Msh2 and Prmt1, while repression of Lmnb1 was observed in all groups (Fig. 4 E, compare CK− with EV). This suggests that p27Kip1 interaction with cyclin–CDK complexes is important for the expression of a subset of ESC-like genes regulated in response to p27Kip1 signaling in leukemia cells. Moreover, WT p27 and S10A, but not CK− expression, significantly reduced MLL-AF9 clonogenicity (Fig. 4 F). Next, we extended these analyses into three human MLL-r AML cell lines with differing levels of miR-196b expression (Fig. S4 B). Similar to murine AML, human AML cells expressing WT p27 and the S10A mutant, but not the CK− mutant, underwent G1 arrest (Fig. S4 C) and were counter-selected over time (Fig. 4 G). Altogether, these data demonstrate that elevating p27Kip1 has tumor suppressive activity in both human and murine MLL-r AML at least in part through coordinated induction of growth arrest and differentiation, and repression of ESC-like gene expression.
p27Kip1 as a therapeutic target in MLL-r leukemia
Given that Cdkn1b overexpression had anti-leukemic effects (Fig. 4), we next tested whether disruption of mechanisms regulating endogenous p27Kip1 protein expression and turnover would also negatively affect MLL-AF9 leukemogenicity. First, we disrupted the miR-196b direct interaction with Cdkn1b mRNA using sequence-specific miRNA target site blockers (TSB) constructed with locked nucleic acid (LNA) chemistry (Petersen and Wengel, 2003; Fazi et al., 2005; Ceppi et al., 2009; Wynendaele et al., 2010; Dajas-Bailador et al., 2012). Primary murine MLL-AF9 leukemia cells were treated with TSB sequences homologous to the validated miR-196b binding site in the Cdkn1b 3′UTR (Fig. 1 D), as well as anti-miR-196b-5p and scrambled nontargeting control LNAs. Anti–miR-196b (positive control) or any of the miR-196b-Cdkn1b TSB treatments significantly reduced colony formation (Fig. 5 A) accompanied by derepression of p27Kip1 protein expression compared with NT control LNA (Fig. 5 A, immunoblot). These data indicate that the miR-196b/Cdkn1b signaling axis is important to maintain MLL-AF9 clonogenic capacity.
Figure 5.
RNAi disruption of miR-196b activity or pharmacologic inhibition of SCFSKP2 elevate p27Kip1 and inhibit human AML growth. (A) MLL-AF9 leukemic splenocytes were treated with indicated LNA and plated in triplicate for CFU assays. The sequences of the LNAs coded for a scramble nontargeting control (NT), homologous sequence to miR-196b-5p (anti-196b), or three variant target site–blocking (TSB1-3) LNAs designed with sequence homology to the Cdkn1b 3′UTR miR-196b target binding site (miR196b:Cdkn1b). CFU results are shown as the average number of colonies ± SEM for each LNA in two independent experiments (left), with mean CFU ± SEM indicated below. Statistical significance was evaluated by t test for anti-196b and TSB LNAs compared with NT control. Immunoblots for p27 (right) were performed on LNA-treated CFU to detect target engagement with β-actin as loading control. (B) Average ± SEM percentage of alive (AnnexinV−PI−) THP1, MV4;11, and MOLM13 human AML cells treated for 3 d in duplicate with the indicated amounts (μM) of SLZ P1-41 (squares) or equivalent volume of DMSO control (circles). Statistical significance was evaluated for each cell line by multiple unpaired t tests Holm-Sidak multiple testing correction. *, P ≤ 0.05; **, P < 0.01. Relative miR-196b expression indicated by triangular color scale (see also Fig. S4 B). (C–E) Cell viability heat maps of THP1 (C), MV4;11 (D), and MOLM13 (E) cells treated for 3 d with single drugs or combinations of 22d, IBET-151, Ml-1, or Palbociclib. Color scale is denoted in C. A representative of at least two experiments with similar results is shown for each. See Table S3 for analyses of drug synergies. (F) Primary human MLL-r AML patient samples (n = 6) were treated with increasing concentrations of 22d. Cell viability was measured by MTS for all doses after 3-d 22d treatment.
In addition to miR-196b regulation, the ubiquitin-proteasome pathway regulates p27Kip1 protein expression. The F-box protein S-phase kinase-associated protein 2 (Skp2) is the substrate recognition subunit of the SCF E3 ubiquitin ligase complex (SCFSkp2) that targets proteins including p27Kip1 for proteasome degradation (Carrano et al., 1999; Sutterlüty et al., 1999; Tsvetkov et al., 1999; Kossatz et al., 2004). Thus, an orthogonal way to elevate p27Kip1 therapeutically would be to block SCFSkp2-mediated protein turnover. SLZ P1-41 is a small molecule inhibitor of the SCFSkp2 E3 ubiquitin ligase complex that blocks Skp2–Skp1 binding. Previously, SLZ P1-41 was shown to decrease ubiquitination of SCFSkp2 targets (including stabilization of p27Kip1) and delay prostate tumor growth in vivo (Chan et al., 2013). Treatment of human MLL-r AML cell lines THP1, MOLM13, and MV4;11 with SLZ P1-41 resulted in significant AML cell death in a dose-dependent manner (Fig. 5 B). In contrast, the Cdc34 E2 ubiquitin enzyme inhibitor CC0651 (Ceccarelli et al., 2011) had no effect (data not shown). Despite differing sensitivities of AML cells to SLZ P1-41, the lowest dose that elicited a cell-killing effect was associated with increased p27Kip1 protein levels in all cases (Fig. S5 A). While there are likely multiple Skp2-regulated proteins that may affect AML cell death, these results suggest that accumulation of p27Kip1, through disruption of miR-196b and/or Skp2 activity, represents a potential new therapeutic opportunity for AML.
Inhibitors of Menin/MLL (Ml-1), BET family (IBET-151), and CDK4/6 (Palbociclib) are currently being tested for MLL-r AML in the clinic. Given encouraging results with SLZ P1-41, we sought to identify a more potent SCFSkp2 inhibitor with a lower relative IC50 that could be used to compare the effects of chemically perturbing p27Kip1 protein levels to the effects of existing small-molecule therapeutics. To that end, we evaluated N-(5-(pyridin-3-yl)quinolin-8-yl)-3-(trifluoromethyl)benzenesulfonamide, referred to as 22d (Singh et al., 2015). 22d is a potent (0.17 µM IC50) inhibitor of the Skp2-Cks1 interaction that is required for SCFSkp2 ubiquitination of p27Kip1 (Ganoth et al., 2001; Spruck et al., 2001). First, we performed dose titrations of 22d measuring cell viability after 3 d of treatment to determine the relative IC50 doses of 3, 0.9, and 0.6 µM 22d in THP1, MV4;11, and MOLM13, respectively (data not shown). Notably, p27Kip1 protein levels were increased in a dose-dependent manner upon 22d treatment in all three AML cell lines (Fig. S5 B). Next, we independently titrated inhibitors of Menin/MLL (Ml-1), BET family (IBET-151), and CDK4/6 (Palbociclib) to identify the relative IC50 for each drug on THP1, MOLM13, and MV4;11 human AML cells (data not shown). Third, AML cells were treated with three to four doses of Ml-1, IBET-151, or Palbociclib with and without 22d using a dose range that spanned the relative IC50 dose determined for each drug in each AML cell line. After 3 d, cell viability was measured relative to vehicle control treated cells (Fig. 5, C–E; and Table S3). Synergy with 22d plus IBET-151 was observed in THP1 and MV4;11, but not MOLM13 cells. Conversely, 22d plus Palbociclib showed synergy in THP1, but not MV4;11 or MOLM13 cells. Overall, 22d plus the Menin/MLL inhibitor Ml-1 showed the most consistent synergistic effects across all cell lines tested (Table S3). Thus, inhibiting Skp2-Cks1 interaction may be a complementary target to those currently under investigation. Finally, we determined whether primary human MLL-r AML are sensitive to 22d. After 3 d in vitro treatment with 22d, we observed growth inhibition of MLL-r AML patient samples (Fig. 5 F and Table S3), at 22d concentrations similar to those required to inhibit MLL-r AML cell lines.
Discussion
Target predication algorithms can be useful, cost-effective tools to identify miRNA target interactions. However, predication algorithms alone are biased for canonical miRNA seed pairing, predict thousands of targets for a given miRNA, and have no cellular context. These shortfalls often lead to a high error rate when attempting to validate miRNA binding and functional importance in a biological system. With the use of experimental endogenous miRNA–mRNA target discovery methods, including biotinylated miR-mimic pulldown, HITS-CLIP, PAR-CLIP, and TAP-Tar, it is now believed that noncanonical target recognition (i.e., seedless) may play a large role in miRNA-mediated regulation, binding can occur outside of the 3′UTR, and target regulation is often context specific. We identified miR-196b targets in human MLL-r AML cells using an experimental biotinylated miR-mimic pulldown approach, which identified candidate miRNA target genes that are not predicted by one or even any prediction algorithm. Thus, the use of an experimental approach can identify novel, disease-relevant miRNA targets.
miRNA are universally down-regulated in cancers compared with normal tissues (Lu et al., 2005), perhaps because miRNA can function as tumor suppressors, or miRNA biogenesis may be tumor suppressive (Kumar et al., 2007; Chang et al., 2008). However, mutations in DICER1 have been identified in human cancers, and Dicer1 haploinsufficiency is more permissive to tumorigenesis than either WT or Dicer1 knockout in mice (Kumar et al., 2009). Thus, some degree of miRNA signaling might be required for oncogenesis. Indeed, miR-196b provides an example of a miRNA signaling network important for the stemness and survival of MLL-AF9 leukemia. Functional screening of a subset of direct miR-196b targets during leukemia expansion in vivo revealed shRNA-mediated repression of targets were largely counter-selected or unchanged, yet repression of some targets (∼25%) exhibited significant pro-leukemic effects. Thus, miR-196b signaling balances both tumor growth and tumor-suppressive effects that can be functionally dissected in vivo to identify which are important for oncogenic miRNA signaling.
We find Cdkn1b (p27Kip1) is one of the top miR-196b targets consistently bound by miR-196b in several different target-validation methods. While miRNA buffer normal stem cell maintenance and myeloid differentiation (Emmrich et al., 2014; Lechman et al., 2016), our studies extend the functional requirement for miRNA buffering to malignant stem cell biology. Cdkn1b-knockdown by shRNA, which mimics miR-196b activity, cooperates with MLL-AF9 to accelerate leukemogenesis, increases LSC output, and signals a poor prognosis gene signature. Conversely, specific disruption of the miR-196b-Cdkn1b target interaction increases p27Kip1 levels, significantly reducing MLL-AF9 clonogenicity. We note that the effect of Cdkn1b-knockdown on MLL-AF9 leukemia maintenance is dissimilar to germline deletion of Cdkn1b (Zhang et al., 2013). Germline deletion of Cdkn1b impacts proliferation of normal hematopoietic progenitors (Cheng et al., 2000), a population capable of transformation by MLL-AF9 (Krivtsov et al., 2006). Thus, our data provide an important distinction between the effects of (1) complete deletion of Cdkn1b on MLL-AF9 leukemia initiation (Zhang et al., 2013), (2) sustaining low-level p27Kip1 expression in leukemia stem cell maintenance, and (3) derepressing p27Kip1 levels triggering anti-leukemic effects. Altogether, our studies support the conclusion that miR-196b activity functions to regulate the levels of its targets, such as tumor suppressor p27Kip1, for leukemia stem cell maintenance and survival.
Repression of Cdkn1b (p27Kip1) was anticipated to influence cell cycle and apoptosis since these p27Kip1 functions are well described (Polyak et al., 1994; Toyoshima and Hunter, 1994; Katayose et al., 1997; Wang et al., 1997); however, our studies also uncovered increased expression of genes associated with an ESC-like transcriptional program. Notably, Cdkn1b-knockdown leukemia gene expression was enriched for the ESC gene set with cell cycle annotations removed (Somervaille et al., 2009). This ESC core signature is distinct from an adult stem cell signature (Wong et al., 2008) and is enriched in a variety of cancers with high frequencies of cancer stem cells, including functionally validated MLL-AF9 LSCs (Somervaille et al., 2009). Whether or not an ESC-like program is responsible for cancer stem cell self-renewal remains controversial. Oncogenic c-Myc activity might specify ESC-like signatures in cancer (Kim et al., 2010); however, we find that Cdkn1b-knockdown leukemia was still enriched for a c-Myc–independent ESC-core gene set. Thus, at least part of p27Kip1 ESC programming is c-Myc independent, in agreement with our finding that forced expression of p27Kip1 did not affect c-Myc transcript levels. Instead, forced expression of p27Kip1 significantly repressed the expression of a subset of these ESC-like genes. In sum, these data support an MLL-AF9-miR-196-Cdkn1b pathway that controls leukemia stem cell activity through the regulation of ESC-like core genes (independent of c-Myc or cell cycle).
Failure to eradicate leukemia stem cells within minimal residual disease leads to relapse. Consequently, AML subtypes with high frequencies of LSC are associated with poorer outcome (van Rhenen et al., 2005; Ghiaur et al., 2012; Terwijn et al., 2014). Thus, pathways that maintain LSC self-renewal, survival, or suppress differentiation represent important therapeutic opportunities for the treatment of AML. Our work illustrates how unbiased global dissection of oncogenic miRNA signaling can be exploited to identify actionable LSC-relevant targets. MLL-AF9 and its downstream target HoxA9 transactivate miR-196b expression (Popovic et al., 2009; Velu et al., 2014). Here we define a unique subset of miR-196b direct targets involved in transcriptional and cell cycle processes that help to explain oncogenic miR-196b activity. Our work uncovered p27Kip1 as a potential therapeutic target for the MLL-r AML subtype. We find that inhibiting the E3 ubiquitin ligase activity of SCFSkp2 complexes results in elevation of p27Kip1 protein levels (Chen et al., 2008a; Ceccarelli et al., 2011; Chan et al., 2013), and that MLL-r AML are sensitive to Skp2 inhibitors. There should be a significant therapeutic window for this approach, as genetic deletion of Skp2 in mice decreases HSC quiescence and increases HSC proliferation without adversely affecting HSC long-term repopulating potential (Wang et al., 2011). Finally, we identify that Skp2 inhibition acts synergistically with Menin/MLL, BET, or CDK4/6-targeted therapeutics in AML contexts where treatment as single agents are less effective. For example, we observe THP1 cells are relatively resistant to Palbociclib as compared with MV4;11 or MOLM13 cells. However, cotreatment with Palbociclib and the Skp2-Cks1 inhibitor 22d synergize to induce growth arrest and/or cell death of THP1 AML cells, but not MV4;11 or MOLM13 cells. In conclusion, we have elucidated a critical miRNA pathway that can be exploited therapeutically to tip the balance from tumor growth to tumor suppression. Our study establishes an experimental approach for future studies that combines in vitro miRNA target identification and in vivo shRNA screening to identify clinically important miRNA targets to better understand miRNA signaling pathways in cancer.
Materials and methods
Transgenic animals, mouse maintenance, and murine studies
All animal studies were approved by and performed in compliance with the Institutional Review Board Committee on Animal Research at Cincinnati Children’s Hospital Medical Center (CCHMC). All animal procedures were conducted in accord with the Institutional Animal Care and Use Committee at CCHMC. Mouse strains used in viral transduction and transplantation studies included CD45.2+ C57BL/6-Ly5.2 (JAX and Charles River) and CD45.1+ C57BL/6-Ly5.1 (Charles River and CCHMC Comprehensive Mouse Core). All transplant recipient mice were male between 1–3 mo of age. Murine CD45.1+ MLL-AF9 leukemia was a gift from N. Zeleznik-Le (Loyola University Chicago, Chicago, IL). MLL-AF9 leukemic splenocytes were expanded in house by transplantation into eight recipient mice. Single cell suspensions from all moribund mouse spleens were pooled, red blood cells (RBCs) lysed by ammonium-chloride-potassium (ACK) buffer (Gibco) and archived as ∼15 × 106 cell aliquots in 10% DMSO liquid nitrogen frozen to create a homogeneous stock from which all shRNA and gene overexpression studies were performed. For shRNA and gene overexpression studies, the replicate hairpins per gene and matched controls were performed simultaneously, and no blinding was used when recording animal survivals. For independent shRNA hairpin validations and gene overexpression studies, no statistical method was used to predetermine sample size.
Human AML cell line validation
For cell line validation, DNA was harvested from our THP1, MV4;11, and MOLM13 cells actively growing in culture using the Blood and Tissue kit (Qiagen). DNA was sent to Genetica DNA Laboratories, who performed the SNP analyses. ATCC database search for the SNPs reported in our human AML cell lines match with 100% identity to the SNPs in the correctly corresponding human AML cells at ATCC (Table S3).
Biotinylated miR mimic pulldown
The biotinylated miRNA mimic–mRNA target pulldown approach was adapted from Lal et al. (Lal et al., 2011) with modifications. miRIDIAN miRNA mimics hsa-miR-196b and C. elegans cel-67 miRNA control were purchased from Dharmacon (ThermoFisher Scientific) with a biotin modification at the 3′ end of the mature/anti-sense strand (Bi-miR-196b and Bi-cel-67). THP1 cells were a gift from J. Parentesis (CCHMC), and were SNP verified by Genetica DNA Laboratories (see Supplemental material). HEK293T cells were cultured in DMEM (ThermoFisher Scientific) containing 10% heat-inactivated FBS (Atlanta Biologics), Pen-Strep (ThermoFisher Scientific), and l-Glutamine (ThermoFisher Scientific). For Bi-miR pulldowns, 0.2 × 106 HEK 293 cells were plated overnight then cotransfected the following day with 100 nM biotinylated miRNA mimics and 0.5 µg/ml GFP-Max (Lonza) with CaCl2 BBS. THP1 cells were cultured in RPMI 1640 (ThermoFisher Scientific) containing 20% heat-inactivated FBS (Atlanta Biologics), Pen-Strep (ThermoFisher Scientific), and l-Glutamine (ThermoFisher Scientific). Cells were maintained in liquid culture at subconfluency at low passage at 37°C in 5% CO2. For Bi-miR pulldowns, 106 THP1 cells were cotransfected with 100 nM biotinylated miR mimics and 0.5 µg/ml GFP-Max by electroporation using Amaxa Nucleofector 4D and cell line SG kit (Lonza) according to manufacturer instructions.
24 h after transfection, cells were lysed on ice in a cytosolic lysis buffer RLN (Qiagen) with 1× HALT protease inhibitor tab (Roche), RNaseI (Ambion), and 1 mM DTT (Sigma). Nuclei were pelleted at 10,000 g for 10 min at 4°C and discarded. 10% of cytosolic lysates were mixed with TriZol LS (Ambion) as “Input” control then frozen at −80°C, while the remaining lysates were subjected to “Pulldown” by mixing with M-280 Dynabeads (Invitrogen) for 4 h rotating at 4°C. Dynabeads were prepared in advance for RNA use according to manufacturer instructions, then rinsed in cold lysis buffer and blocked with rotation at 4°C for 2 h with yeast tRNA (Invitrogen) and BSA (Invitrogen). After the pulldown incubation, beads were washed on a magnetic stand three times with ice-cold wash buffer (Nonne et al., 2010) for 3 min rotating at 4°C. After the last wash beads were resuspended in 0.75 ml TriZol LS and brought to 1 ml with nuclease free water (ThermoFisher Scientific) then frozen at −80°C. Isolated RNAs of both input and pulldown fractions were examined for target enrichment by TaqMan RT-qRCR using the delta Ct method as follows in Eq. 1 and Eq. 2 to calculate pulldown enrichment ratios. Enrichment = 2-ΔΔCt; Eq. 1 = ΔCt; Eq. 2 = ΔΔCt.
| (1) |
| (2) |
RNA extraction, TaqMan quantitative PCR gene, and miRNA expression analyses
RNA was extracted using TriZol or TriZol LS (Ambion) as previously described (Meyer et al., 2009). Total RNA was converted into cDNA using the High Capacity cDNA Archive kit (Life Technologies) for standard gene expression analyses and or with miRNA TaqMan RT assays for hsa-miR-196b-5p, mmu-miR-196b-5p, hsa-miR-196a-5p, and mmu-miR-196a-5p (Life Technologies) according to manufacturer instructions. All gene and miRNA expression were quantified using TaqMan assays (Life Technologies) with the 2× Universal TaqMan Master Mix (Life Technologies) according to the manufacturer instructions on the StepOne Plus Instrument (Applied Biosystems, Inc.) using equal amounts of starting cDNA. Standard inventoried gene expression TaqMan assays recommended by the manufacturer (Life Technologies) for best coverage were purchased when available. For human gene expression assays: CDKN1B (NM_004064.3), SPTSSA (NM_138288.3), HOXA7 (NM_006896.3), SLC9A6 (NM_001042537.1, NM_001177651.1, NM_006359.2), MDM4 (NM_001204171.1, NM_001204172.1, NM_001278516.1, NM_001278517.1, NM_001278518.1, NM_001278519.1, NM_002393.4), TAF5L (NM_001025247.1, NM_014409.3), ZCCHC9 (NM_001131035.1, NM_001131036.1, NM_032280.2), PHC2 (NM_004427.3, NM_198040.2), PHC3 (NM_024947.3), TMEM184C (NM_018241.2), HOXA9 (NM_001277238.1, NM_010456.3), SDHA (NM_004168.2). For mouse gene expression assays: HoxA7 (NM_010455.2), HoxB9 (NM_008270.2), and Meis1 (NM_001193271.1, NM_010789.3), Phc2 (NM_001195083.1, NM_001195130.1, NM_018774.4), Taf5l (NM_133966.2), Zcchc9 (NM_145453.2), Cdkn1b (NM_009875.4), Sptssa (NM_134054.2), Slc9a6 (NM_172780.3), Mdm4 (Mm.426531), c-Myc (NM_001177352.1, NM_001177353.1, NM_001177354.1, NM_010849.4), Ccnd2 (NM_009829.3), Hadh (NM_008212.4), Lmnb1 (NM_010721.2), Msh2 (NM_008628.2), Prmt1 (NM_001252476.1, NM_001252477.1, NM_019830.3, NR_045521.1), and Sdha (NM_023281.1). For human mature miRNA expression assays (Life Technologies): hsa-miR-196b-5p, hsa-miR-196a-5p, and RNU6B (NR_002752). For mouse mature miRNA expression assays (Life Technologies): mmu-miR-196b-5p, mmu-miR-196a-5p, and U6 snRNA (NR_004394). Gene expression data were analyzed using the comparative Ct method (2-ΔΔCt). SDHA and RNU6B served as loading controls. For Bi-miR pulldown validation assays the fold change pulldown and inputs (Eq. 1) was calculated (see Biotinylated miR mimic pulldown method).
Affymetrix microarray and data analyses
Microarrays were performed that the CCHMC Microarray Core Facility. In brief, miR mimic pulldown and input control RNA samples were analyzed on an Agilent Bioanalyzer with a RNA integrity number >8.0 were selected for microarray processing. 550–1,000 pg of matched input and pulldown RNA samples from three independent THP1 experiments and one HEK293 experiment were processed using the Ovation Pico WTA System V.2 (Nugen) and hybridized to Human Gene 1.0 ST arrays (Affymetrix). For HEK293 and THP1 pulldowns, gene expression was obtained using the RMA algorithm output from the GCOS Affymetrix software with a raw value cut-off of 5 in pulldown samples. The Bi-miR-196b input and pulldown samples were normalized separately to show change relative to the Bi-cel-67 control samples. Results were visualized in GeneSpring and AltAnalyze version 2.0.9 for any Ensembl version 72 gene aligning probe sets transcript clusters (Salomonis et al., 2009, 2010; Emig et al., 2010). miR-196b target genes are reported for independent pulldowns with a twofold increase in the miR-196b pulldown fractions compared with the cel-67 pulldown controls. (Fig. 1 B and Table S1). Venny 2.0 (Oliveros, 2015) or InteractiVenn (Heberle et al., 2015) was used to overlap pulldown target lists with target site computational prediction algorithms PITA (Kertesz et al., 2007), miRanda (Enright et al., 2003; John et al., 2004; Betel et al., 2008), TargetScan7.0 (Lewis et al., 2003, 2005; Agarwal et al., 2015), and DIANA (Maragkakis et al., 2009a,b; Papadopoulos et al., 2009) to aide identification of pulldown targets for the shRNA screen (Table S1).
Target site sequences, cloning, and luciferase reporter assays
Following the miRNA target interaction principles demonstrated by Kertesz et al. and Ellwanger et al. (Kertesz et al., 2007; Ellwanger et al., 2011), a 22-nucleotide (nt) target site was identified beginning with minimal seed pairing (6 consecutive nt) and extending the natural length of the targeting miRNA region of homology in the mRNA target. An additional 3 nt flanking at the 5′ end and 15 nt flanking the 3′ end of the full-length (22 nt) target site for a total of 40 nt. We used two different approaches to clone target sites into the psiCHECK2 Dual Luciferase reporter vector (Promega). All target site inserts were sequence verified at the CCHMC sequencing core.
Either miR-196b target sites sequences (listed below) were ordered as double stranded DNA (dsDNA) gblocks from IDT (Integrated DNA Technologies) with 24 nucleotide overhangs on both 5′ (5′-CGAGCAGTAATTCTAGGCGATCGC-3′) and 3′ (5′-TCGAGCCCGGGAATTCGGTTAAAC-3′) ends homologous to the 5′ and 3′ ends of the psiCHECK2 vector (Promega) cut with the Xho1 restriction enzyme (New England Biolabs). The target site gblocks were cloned into the Xho1 site at the 3′ end of Renilla Luciferase in psiCHECK2 dual luciferase reporter vector using Gibson Assembly (New England Biolabs).
CDKN1B: 5′-ATTATGCAATTAGGTTTTTCCTTATTTGCTTCATTGTACTACCTGTGTATATAGTTTTTACCTTTTATGTAGCACATAAA-3′.
PHC2: 5′-ACTTCTTTGCCTATAAATCTTCCTAGCAGCAATTTGTTGAGCTACCTGAGGAGGAGGCAGGGCAGAAAGGGCGAGGGCCTGCC-3′.
SLC9A6-1: 5′-TTGTAAAATTTGCACATGTGATTGTGAAGAAATTTGTACTACCTAAAAGTCCCAGTGCATGTCTCTGAATGTGTAAGCTA-3′.
SLC9A6-2; 5′-TTATTCAAGTTGTAAAAGGTTATACAATAATTTAACAACTACCTTTTTTATTCTGTCGGGTTACTGACCTCACTTTATGT-3′.
WDR75: 5′-AAAAACTAAACGAAACTTTAGAGAATGAGCTGGTACAACTACCCTTAACAGAAAACATACCCGCAATTAGTGAGCTTCTT-3′.
TAF5L: 5′-TAACAAGTACGTGGTCCGTCTCCAAGAAGACAGCTACAACTACCTTATCCGCTACCTCCAAAGTGACAACAATACTGCCCT-3′.
SPTSSA: 5′-TTGTATTGTTTTTTTTAAGGTTTTTATTCCTTAAATGTAAAATGACTACCTAATTTTTTGATGTAAATACATTAAATTCAA-3′.
ZCCHC9: 5′-TGTTAATTTTTGATAACAGCTAGCACTATCATGAGTTACTACCTCATTGTTACTTTCTAAACCAGGCCCGCTTCACGAGT-3′.
Elsewise, miR-196b target sites sequences (listed below) were synthesized as single stranded complimentary DNA strands (Upper and Lower) with 5′ XhoI and 3′ NotI restriction sites on the ends (Upper, XhoI/NotI 5′-TCGAG…GC-3′ and Lower, NotI/XhoI 5′-GGCCGC…C-3′; ThermoFisher Scientific). Equimolar amounts of the two strands were annealed before T4 DNA ligase (New England Biolabs) mediated ligation to XhoI/NotI cut psiCHECK2 dual luciferase reporter vector (Promega).
HOXA7: 5′-ACTACCTACTCCCCTAAAACTACCTATTTTGTGCTGGCTG-3′.
MDM4-1: 5′-GACGATATCCCCACACTGCCTACCTCAGAGCATAAATGCA-3′.
MDM4-2: 5′-CATGTTTTGATCCTGATGCTACCTTTGCTAAAAATGGCCA-3′.
PHC3-1: 5′-TGAAAGGGCCTCATCTCACTACCTCTCTAAGGCCTCTAGC-3′.
PHC3-2: 5′-AGGTTCTTCCTACTTTTGCTACCTAAGTTTGCATTTTCTG-3′.
CDKN1B: 5′-CTTATTTGCTTCATTGTACTACCTGTGTATATAGTTTTTA-3′.
The number of cells, transfection reagents, reporters, miR mimics, and luciferase assay reagents were all titrated and optimized for use in 96-well format with the PerkinElmer EnVision plate reader with injectors. HEK293T cells were seeded in regular growth medium, as described above, at 8 × 104 c/ml in 96-well plates. The following day cells were cotransfected with miR mimics and luciferase reporter plasmids using DharmaFect Duo (Dharmacon) according to manufacturer’s instructions with the following final concentrations per well: 50 nM miR mimic (nontargeting or miR-196b-5p purchased from ThermoFisher Scientific), 50 ng of the indicated psiCHECK2 plasmid, 50 ng pBlueScript plasmid (to bring total DNA to optimal recommended range), and 0.4 µl DharmaFect Duo in a total volume of 100 µl growth medium. Cells were incubated with transfection mixture for 24 h, and then luciferase activity was measured using the Dual Luciferase Assay System (Promega) according to manufacturer instructions with the following modifications. Cells were lysed in 40 µl 1× Passive Lysis Buffer (Promega) for 15 min to 1 h at room temperature, and then 30 µl lysate was transferred to a white flat-bottom plate. The EnVision injectors were filled with LARII and Stop+Glo (Promega) and the Dual Luciferase Assay protocol dispensed 25 µl LARII, shake plate 1 s at 300 rpm, incubated 10 s, read Firefly luminescence (400–700 nM), dispensed 25 µl Stop+Glo, shake plate 1 s at 300 rpm, incubated 10 s, and read Renilla luminescence. Each miR mimic/reporter construct combination was performed in triplicate per assay, and at least three independent assays were performed.
Data analyses were performed according to Promega manufacturer instructions. In brief, the Firefly luminescence values (RLU) measured in each well was normalized for transfection efficiency to the Firefly luminescence in psiCHECK2 EV control transfected wells for each plate. Then Renilla luminescence (RLU) was corrected for transfection efficiency (Renilla RLU/normalized Firefly RLU) within each well. The replicates for each psiCHECK2 construct cotransfected with nontargeting (NT) control mimics were averaged and used to calculate the fold change in miR-196b-5p transfected wells for each corresponding psiCHECK2 target reporter construct. Data shown are an average or three independent assays.
Pulldown target site comparison with argonaute CLIP databases
To further validate the Bi-miR-196b pulldown target binding sites, all human cell line CLIP datasets for AGO1-4 CLIP-seq studies were downloaded from CLIPdb (Yang et al., 2015) as PARalayzer processed .bed files (Table S1). Each dataset was individually uploaded as a unique track in USCS genome browser (Kent, 2002; Kent et al., 2002; Raney et al., 2014) mapped to GRCh37/hg19. Since the binding site of the miRNA of interest must be known for AGO-CLIP data interrogation, we manually searched for each miR1-96b binding site from a filtered selection of pulldown targets. All miR-196a-5p and miR-196b-5p miRNA–mRNA interactions were downloaded from starBase v2.0 (Yang et al., 2011) using human assembly hg19, with medium stringency of two or more supporting experiments. This resulted in 874 miR-196 target genes. starBase predictions were overlapped with THP1 pulldown, HEK293 pulldown, and finally the 116 shRNA screen candidates using Venny 2.0 (Oliveros, 2015). Based on the mature miR-196-5p seed nucleotide sequence target sites were manually searched and identified using BLAT feature in the Genome Browser, the corresponding genomic coordinates are indicated in Fig. S1 (C and D). All target-binding sites were counted within 22 nucleotides in either direction of the exact target site coordinates.
MOI determination
To determine the amount of lentivirus required to transduce murine MLL-AF9 leukemic splenocytes with an MOI of ∼0.2, an indirect surrogate measurement of transduction efficiency was experimentally determined. Titration of a random barcoded lentivirus pool of similar 107 transducing units/ml titer not containing any hairpins was used. MLL-AF9 leukemic splenocytes were thawed from liquid nitrogen and allowed to recover for 48 h in standard growth medium RPMI 1640 (Gibco) supplemented with 20% FBS (Hyclone), 60 ng/ml stem cell factor (SCF), 20 ng/ml IL6, and 20 ng/ml IL3 (Miltenyi Biotech). 106 MLL-AF9 leukemia cells in 100 µl RPMI 1640 supplemented with 20% FBS were mixed with 0, 25, 50, 100, 200, 300, or 400 µl lentiviral supernatant plus 60 ng/ml SCF, 20 ng/ml IL6, 20 ng/ml IL3, and 10 µg/ml polybrene. Cells were incubated with virus for 6 h at 37°C with 5% CO2. After 6 h, the virus was removed from cells and replaced with standard growth medium. Cells were allowed to rest of 48 h in standard growth medium, at which point they were divided into two replicate wells, one receiving 1 µg/ml puromycin and the other treated with vehicle. After 48 h, cells were mixed with trypan blue and the number of live cells in each well was enumerated. Transduction efficiency was calculated using the equation: (no. infected + puro / no. infected − puro) – (no. uninfected + puro / no. uninfected − puro) = transduction efficiency. Based on prior studies at the Broad Institute, a transduction efficiency between 30–50% yields ∼80% of cells getting a single shRNA/cell. We selected the amount of virus/106 MLL-AF9 cells that achieved 35% transduction efficiency for our shRNA screen.
MLL-AF9 leukemia engraftment assay, pool size, and animal number determination
To determine the size of each shRNA pool to obtain sufficient representation of each shRNA hairpin from the resultant leukemias in the in vivo RNAi screen, murine MLL-AF9 leukemic splenocytes were prestimulated in growth medium containing RPMI 1640 supplemented with 20% FBS (Hyclone), 60 ng/ml mouse SCF, 20 ng/ml mouse IL6, and 20 ng/ml mouse IL3 (Miltenyi Biotech). 15 × 106 MLL-AF9 cells were transduced in growth medium plus 10 µg/ml polybrene with a lentiviral pool containing over 199,000 unique barcodes (Broad Institute of MIT Harvard) at an MOI of ∼0.1–0.2. After transduction, cells were washed then incubated in growth medium for 48 h before puromycin selection. Transduced cells and 106 untransduced control cells were treated with 1 µg/ml puromycin for 48 h. Three sublethally irradiated mice (500 Rad) were each transplanted with 106 puromycin-selected MLL-AF9 cells. In addition, 106 puromycin selected transduced cells were frozen at −80°C in PBS as pretransplant control. Moribund mice were sacrificed, spleens were harvested, and single-cell suspensions were treated with ACK to lyse RBCs. Genomic DNA was prepared from purified MLL-AF9 leukemic cells and pretransplant control cells using the QIAamp DNA Blood kit (Qiagen). The Broad Institute performed DNA sequencing and read alignment. Unique barcode aligned reads for each mouse tumor sample were summed and compared with the aligned reads for each barcode in the pretransplant control. Random barcoded lentiviruses exert no selective pressure on MLL-AF9 cells in vivo as they do not specifically target any murine coding sequences. By comparison of the number of barcodes in the pretransplant hold-back control, we calculated an engraftment of ∼4% of bulk MLL-AF9 leukemic splenocytes and thus we estimated that shRNA should be divided into random pools of ∼100 hairpins each, with each pool containing all control gene (EV, Luciferase, RFP, GFP, and LacZ) targeting hairpins, and 10 mice/pool to give sufficient representation of each hairpin in vivo.
Selection of gene-targeting hairpins for RNAi screen
Candidate miR-196b pulldown targets were identified from three independent experiments performed in THP1 AML cells (Table S1). Targets also identified in HEK293 pulldown and/or in the cumulative predicted miR-196b target list from four computational prediction algorithms PITA (Kertesz et al., 2007), miRanda (Enright et al., 2003; John et al., 2004; Betel et al., 2008), TargetScan7.0 (Lewis et al., 2003, 2005; Agarwal et al., 2015), and DIANA (Maragkakis et al., 2009a,b; Papadopoulos et al., 2009) were determined (Table S1). Targets were also examined by gene ontology analyses using the ToppGene Suite (Chen et al., 2007, 2009a,b). Genes with cancer-related biological pathway and biological processes gene ontologies, and previously validated miR-196b target binding sites were given priority. The final candidate targets included in the screen were enriched in at least one THP1 pulldown experiment (n = 116; 38/116 were enriched in ≥2/3 THP1 pulldown experiments represented in Fig. 1 B) and either (1) present in HEK293 pulldown experiments (n = 60), (2) predicted by computational algorithms (n = 103), or (3) in HEK293 pulldowns and predicted (n = 48; Table S1).
RNAi screen transduction
MLL-AF9 leukemic splenocytes were thawed from liquid nitrogen and allowed to recover for 48 h in standard growth medium RPMI 1640 (ThermoFisher Scientific) containing 20% FBS (HyClone) supplemented with mouse recombinant SCF (60 ng/ml), IL6 (20 ng/ml), and IL3 (20 ng/ml; Miltenyi Biotech). 106 MLL-AF9 leukemia cells in 400 µl standard growth medium containing FBS were mixed with 100 µl lentiviral supernatant. Cytokines (60 ng/ml SCF, 20 ng/ml IL6 and IL3) and polybrene (10 µg/ml, Sigma) were added to the final volume. Cells were incubated with virus for 6 h at 37°C with 5% CO2, then virus supernatant was replaced with standard growth medium containing FBS and cytokines. Cells were incubated for 48 h, then for an additional 48 h with 1 µg/ml puromycin in fresh growth medium. 106 puromycin-selected cells were transplanted into 10 each sublethally irradiated (500 Rads) recipient mice, and 5 × 106 cells were held back in 100 µl PBS at −80°C as pretransplant input control. The transduction transplantation procedure was repeated for each of the eight shRNA lentivirus pools. Mice were monitored for illness and were sacrificed in accord with Institutional Animal Cancer and Use Committee–approved procedures. Leukemic splenocytes were harvested and DNA isolated as described.
RNAi screening and data analyses
Leukemic splenocytes were harvested from moribund mice transplanted with MLL-AF9 cells transduced with shRNA pools, and single-cell suspensions were treated with ACK to lyse RBCs. 5 × 106 cells/mouse were frozen in PBS at −80°C and 2 × 106 cells were examined by FACS analysis for expression of known cell surface markers on MLL-AF9 AML cells CD117, CD11b, and Gr1 expression. The remaining cells were frozen in liquid nitrogen. Genomic DNA was harvested from all mouse tumor and holdback pretransplant input control samples using the QIAamp DNA Blood kit (Qiagen). DNA sequencing and read counts were aligned to the unique barcodes for each individual hairpin shRNA were performed at the Broad Institute of MIT and Harvard.
To calculate the final representation score for each hairpin, the following data analysis pipeline was used. Read counts (in counts per million) for each hairpin were normalized to the sum of all counts per sample then log2 transformed. Any mouse leukemic splenocyte sample with a sum <10,000 counts per million was excluded from further analyses. Note that no control samples were excluded. The average log2 normalized value for control hairpins GFP, RFP, and Luc was calculated for each sample. Individual experimental hairpins were then normalized to the control average for each sample, and finally the fold change enrichment or depletion scores of each hairpin in the tumor samples were expressed as relative to pretransplant input control sample. Enriched shRNA was scored with a fold change >0 in at least 4/10 mice and depleted with a fold change less than −8 in at least 4/10 mice. To account for potential off-target effects, final shRNA representation lists included only genes with at least two hairpins achieving the same directionality in fold change. Genes with hairpins scoring on both enrichment and depletion lists were considered “no change” and removed from final representations. To confirm that our scoring method and selection criteria were not randomly selecting for enriched or depleted hairpins but that these represent reproducible shRNA activity in vivo, replicate mice for each hairpin were divided into two groups per pool and plotted against each other. The relatedness between these two groups of mice was compared by Pearson correlation analysis using Prism 6.0.
RNAi screen gene ontology and network analyses
Gene ontology analyses were performed on the datasets: (1) 116 genes shRNA screen Input shRNA, (2) 33 genes shRNA screen Enriched, and (3) 45 genes shRNA screen Depleted. GO-Elite analyses were performed on all three datasets as Input in AltAnalyze with a z-score cut-off of 1.96, requirement of at least five changed genes per category, and the remaining default parameters. Network analyses were performed using AltAnalyze's NetPerspective. The Cytoscape Sets plugin was used to layout the network according to Gene Ontology terms assigned by GO-Elite (labels and colored circles added manually).
Lentivirus production, transduction, and gene overexpression
For gene overexpression lentivirus constructs, the pLVX-EIF1α-IRES-ZsGreen (Clonetech) vector was digested with EcoRI. The coding sequence (CDS) for Cdkn1b (NM_009875.4) was downloaded from National Center for Biotechnology Infromation. Mouse Cdkn1b coding region does not contain any miR-196b target sites, thus was used directly for overexpression. cDNA sequences of Cdkn1b were ordered as gblocks from IDT with 5′ (5′-GATCTATTTCCGGTGAATTC-3′) and 3′ (5′-AGAATTCCTCGAGACTAGTTC-3′) ends homologous to the 5′ and 3′ ends of the EcoRI site in pLVX. The Kozak sequence 5′-GCCACC-3′ was also added just before the ATG start site. Gibson Assembly (New England Biolabs) was used to recombine the Cdkn1b gblock with the pLVX-ZsGreen vector.
Cdkn1b CDS: 5′-ATGTCAAACGTGAGAGTGTCTAACGGGAGCCCGAGCCTGGAGCGGATGGACGCCAGACAAGCGGAGCACCCCAAGCCTTCCGCCTGCAGAAATCTCTTCGGCCCGGTCAATCATGAAGAACTAACCCGGGACTTGGAGAAGCACTGCCGGGATATGGAAGAAGCGAGTCAGCGCAAGTGGAATTTCGACTTTCAGAATCATAAGCCCCTGGAGGGCAGATACGAGTGGCAGGAGGTGGAGAGGGGCAGCTTGCCCGAGTTCTACTACAGGCCCCCGCGCCCCCCCAAGAGCGCCTGCAAGGTGCTGGCGCAGGAGAGCCAGGATGTCAGCGGGAGCCGCCAGGCGGTGCCTTTAATTGGGTCTCAGGCAAACTCTGAGGACCGGCATTTGGTGGACCAAATGCCTGACTCGTCAGACAATCCGGCTGGGTTAGCGGAGCAGTGTCCAGGGATGAGGAAGCGACCTGCTGCAGAAGATTCTTCTTCGCAAAACAAAAGGGCCAACAGAACAGAAGAAAATGTTTCAGACGGTTCCCCGAACGCTGGCACTGTGGAGCAGACGCCCAAGAAGCCCGGCCTTCGACGCCAGACGTAA-3′.
The Cdkn1b/p27 Ser10 to Ala mutant (S10A) and p27 cyclin/CDK-binding mutant Arg30 to Ala, Leu32 to Ala, Phe62 to Ala and Phe64 to Ala (CK−) were described previously (Vlach et al., 1997; Besson et al., 2006). The point mutations were introduced into the murine Cdkn1b CDS (NM_009875.4). The final sequences were codon optimized for murine expression using publically available software from Integrated DNA Technologies. Sequences were ordered as gblocks from IDT with 5′ (5′-GATCTATTTCCGGTGAATTC-3′) and 3′ (5′-AGAATTCCTCGAGACTAGTTC-3′) ends homologous to the 5′ and 3′ ends of the EcoRI site in pLVX. The Kozak sequence 5′-GCCACC-3′ was also added just before the ATG start site. Gibson Assembly (New England Biolabs) was used to recombine the Cdkn1b gblock with the pLVX-ZsGreen vector.
Cdkn1b S10A CDS: 5′-ATGTCAAACGTCAGGGTATCTAATGGAGCACCTAGTTTGGAACGCATGGACGCTCGGCAAGCCGAGCACCCCAAACCCTCTGCCTGTCGGAACTTGTTCGGCCCTGTTAATCACGAGGAGCTCACCAGGGACCTGGAGAAGCACTGCAGAGACATGGAGGAGGCAAGCCAGCGAAAATGGAATTTCGACTTTCAGAACCACAAACCACTGGAGGGCAGGTACGAGTGGCAGGAGGTGGAGAGGGGGTCTCTGCCAGAATTTTATTACCGGCCCCCTCGCCCACCTAAAAGCGCTTGTAAGGTACTGGCCCAAGAAAGCCAAGACGTAAGTGGCAGCAGACAGGCTGTTCCCCTCATCGGGTCACAAGCCAACTCTGAGGACAGGCACCTTGTCGACCAGATGCCCGATTCCAGCGATAATCCTGCAGGACTCGCCGAGCAGTGTCCTGGGATGCGAAAAAGGCCAGCCGCTGAAGACTCTAGCTCTCAGAACAAGAGAGCCAACCGCACTGAAGAGAACGTGTCTGACGGCAGTCCCAATGCCGGGACAGTCGAGCAGACACCGAAAAAGCCTGGACTCCGACGGCAGACGTAA-3′.
Cdkn1b CK− CDS: 5′-ATGAGTAATGTTAGGGTGTCTAACGGGTCTCCTTCTCTGGAGAGAATGGACGCTCGGCAGGCCGAGCATCCGAAGCCATCCGCCTGCGCCAATGCCTTCGGTCCTGTGAATCACGAGGAGCTCACAAGAGATCTGGAGAAACATTGCCGGGACATGGAGGAGGCCTCTCAAAGAAAATGGAACGCCGATGCCCAGAACCACAAGCCTCTTGAGGGTCGGTATGAATGGCAGGAAGTCGAGAGGGGATCTCTCCCCGAGTTCTATTATCGGCCTCCTCGCCCACCAAAGTCTGCATGCAAAGTGCTGGCCCAGGAGTCCCAGGATGTGAGTGGAAGTAGGCAGGCCGTCCCTCTGATAGGTTCTCAGGCTAACTCAGAAGACAGGCACCTTGTTGATCAAATGCCTGACTCTAGCGACAACCCAGCCGGGCTGGCAGAACAGTGTCCGGGGATGCGCAAGAGGCCAGCCGCCGAGGACTATCATCTCAGAACAAACGCGCTAATAGAACTGAGGAGAATGTTAGCGACGGGTCTCCTAACGCAGGCACAGTGGAGCAGACCCCCAAAAAGCCGGGACTGCGACGCCAGACCTAA-3′.
For lentivirus production, lentiviral vectors Cdkn1b-ZsGreen (WT p27), S10A, CK− or EV control were cotransfected with lentiviral packaging plasmid Δ8.9 (Naldini et al., 1996) and envelope plasmid VsVg (Burns et al., 1993) using TransIT-LT1 (Mirus Bio) into Lenti-X 293T cells (Clonetech). Lenti-X 293T (Clonetech) cells were cultured in DMEM containing 10% heat-inactivated FBS (Atlanta Biologics), Pen-Strep (Gibco), and l-Glutamine (Gibco). Virus-containing supernatant was collected 48 and 72 h post-transfection, and either used immediately or frozen at −80°C in aliquots.
MLL-AF9 leukemic splenocytes were transduced as described for the RNAi screen, but with a second transduction repeated the following day. Cells were allowed to rest of 48 h in standard growth medium then sorted for ZsGreen+ expression using MoFloXDP (BD Biosciences) at the CCHMC flow cytometry core. 104 sorted cells were divided evenly into methylcellulose M3434 (Stem Cell Technologies) colony forming assays in triplicate, or 5 × 104 cells were transplanted into sublethally irradiated recipient mice (n = 6 per group). For p27 mutant experiments, RNA and cytospins were also performed using ZsGreen+ sorted cells 48 h post-transduction, and 103 cells were plated in methylcellulose M3434 (Stem Cell Technologies) colony forming assays in triplicate. Human AML cell lines were transduced with EV, WT p27, S10A, and CK− lentivirus expressing the ZsGreen fluorescent marker. AML cell lines were transduced as described for the RNAi screen in one round of transduction and allowed to rest for 48 h. Beginning at 48 h post-transduction, and every 24 h thereafter for a total of 5 d, an aliquot of each group of AML cells were examined for ZsGreen+ expression by flow cytometric analyses using an LSR flow cytometer (BD Biosciences). The percentage of ZsGreen-expressing cells overtime was measured for each group and expressed as the frequency of ZsGreen+ cells relative to the initial time point at 48 h.
Flow cytometric analyses and cell sorting
For the GMP myeloid differentiation analyses, 5 × 106 MLL-AF9 leukemia cells were incubated for 1 h at room temperature in 50 µl FACS buffer (1% FBS (HyClone), 0.1% NaAzide, PBS) with a biotinylated lineage antibody cocktail 0.5 µl/antibody CD3 (clone 145-2C11, BioLegend), CD4 (clone RM4-5, eBiosciences), CD8 (clone 53- 6.7, BD Biosciences), CD19 (clone 6D5, BioLegend), CD45R (clone RA3-6B2, BioLegend), CD127 (clone B12-1, BD Biosciences), and Ter119 (clone Ter-119, BioLegend) plus 0.5 µl of CD16/32-PerCP-eF710 (clone 93, eBiosciences), CD11b-BV605 (clone M1/70, BD Biosciences), Gr1-PE-Cy5 (clone RB6-8C5, eBiosciences), CD117-BV650 (clone RAM34, BD Biosciences), Sca1-PE-Cy7 (clone D7, BD Biosciences), and 1.0 µl CD34-BV421 (clone RAM34, BD Biosciences). Cells were washed with FACS buffer then resuspended in 50 µl FACS buffer plus 0.5 µl Streptavidin-APC-Cy7 (BD Biosciences) and incubated at 4°C until just before analyzing, cells were washed once with FACS buffer, then analyzed on Fortessa 2 flow cytometer (BD Biosciences). For bone marrow chimerism, 106 ACK treated bone marrow cells were stained in 100 µl FACS buffer with 1 µl FcγRIII/FcγRII block (clone 2.4G2, BD Biosciences), 1 µl CD45.1-BV605 (clone A20, BioLegend), and 1 µl CD45.2-FITC (Clone 104, BioLegend) for 1 h at RT. Cells were washed and resuspended in FACS buffer plus PI and analyzed on the Fortessa 2 flow cytometer. For all analyses, cells were gated on the live populations with doublets excluded. Cells analyzed on Fortessa 2 flow cytometer (BD Biosciences). All biotin-lineage (Lin) positive cells were excluded leaving Cd11b+Gr1+Lin− cells from which the c-Kit+ population was further defined as GMP by CD34+ and CD16/32+. Leukemic GMP cells were divided for CD11bhi and CD11blo or Gr1hi and Gr1lo expression by setting the gates for all samples based on the median CD11b or Gr1 expression in WT MLL-AF9 control cells. For all analyses cells were gated on the live populations with doublets excluded.
For isolation of c-Kit+ MLL-AF9 leukemic splenocytes fresh or frozen leukemia cells were incubated with mouse CD117 Microbeads (130-091-224, Miltenyi) according to the manufacturer’s instructions. The c-Kit+ and c-Kit− fractions were sorted on the AutoMACs Pro (Miltenyi).
Cell cycle analyses
ZsGreen+ EV, WT p27, S10A, or CK− transduced murine MLL-AF9 leukemic splenocytes and human AML cell lines THP1, MV4;11, and MOLM13 were treated with 1 µl Vybrant DyeCycle violet stain (ThermoFisher Scientific) at 106 cells in respective growth medium and incubated for at least 30 min at 37°C at 5% CO2 in accordance with manufacturer instructions. Cells were analyzed for violet dye using the violet laser and acquired at <1,000 events/second on the LSR flow cytometer (BD Biosciences) in linear scale. Cell cycle analyses were performed on ZsGreen+ gated alive single cells using the Cell Cycle platform in FlowJo v10.0.7 (FlowJo, LLC). Cell cycle analyses were confirmed for each p27 construct or EV control in all three cell lines in at least two independent experiments.
RNA-seq analyses
Leukemic splenocytes were harvested from mice transplanted with MLL-AF9 leukemia transduced with either Cdkn1b shRNA (n = 2 mice), Phc2 shRNA (n = 3 mice), or NT control shRNA (n = 2 matched experimental control mice in Cdkn1b-knockdown study group and n = 3 mice matched experimental controls in Phc2-knockdown study group). RBCs were lysed with ACK, and 5 × 106 leukemia cells were lysed with Trizol. RNA quality and quantity were measured by Agilent Bioanalyzer at CCHMC Microarray Core. Then samples with a RNA integrity number >8.0 were processed by poly-A selection, stranded True-seq, paired-end 75 bp 20 million reads by the CCHMC Sequencing Core. The fastq files were aligned to mouse genome Mm9 using BowTie 2 and TopHat 2 (Trapnell et al., 2009; Kim et al., 2013). Gene level fragments per kilobase of transcript per million mapped reads were obtained using Cufflinks 2 (Trapnell et al., 2010) and further processed in AltAnalyze 2.0.8 to identify differentially expressed genes using default parameters moderated t test with Benjamini-Hochberg multiple testing correction, raw p-value cut-off of 0.05, and an expression cut-off of twofold.
GSEA analyses
Log2 RNA-seq expression derived from total leukemic splenocytes from two independent Cdkn1b shRNA (“Cdkn1bsh”) with two independent NT shRNA controls (“NTsh”), and three independent Phc2 shRNA (“Phc2sh”) with three independent NT shRNA controls (“NTsh”) were imported into GSEA v.2.2.0 (Mootha et al., 2003; Subramanian et al., 2005). A fold change ranked gene list was created comparing the Cdkn1bsh or Phc2sh to NTsh control samples using the Diff_of_Classes ranking metric and genes were sorted by descending order real mode. GSEA analyses were run using 1,000 gene_set permutations of gene sets in the website gene matrix database with the weighted enrichment statistic. Results are reported for the most highly positively correlated and negatively correlated hematopoietic-related gene sets with leading edge analyses.
To dissect the Myc-regulated, from non-Myc regulated genes within with Wong ESC core gene set (Wong et al., 2008), all of the Myc ChIP-Chip identified direct target genes in ESC (Kim et al., 2008) and the Myc core module genes (Kim et al., 2010) were subtracted from the Wong ESC core gene set. Then GSEA analysis was performed as described above for enrichment of the “Myc-regulated ES core” (n = 168 genes) and “non-Myc regulated ES core” gene sets (n = 124 genes) in Cdkn1bsh MLL-AF9 cells compared with NTsh control. See Table S2 for gene sets.
Gene ontology analyses
Gene ontology analyses were preformed using ToppFun in the ToppGene Suite (Chen et al., 2007, 2009a,b).
In vitro colony forming assays
Equal numbers of cells were mixed with methylcellulose M3434 complete with cytokines SCF, IL6, IL3, and EPO (Stem Cell Technologies) according to manufacturer’s instructions in triplicate. 104 cells/plate were seeded in all studies using transduced bone marrow cells, 103 cells/triplicate were divided evenly/plate for studies using purified c-Kit+ leukemic splenocytes, and 103 ZsGreen+ leukemic splenocytes were seeded per plate in triplicate for p27 overexpression studies. On day 7 immortalized/transformed colonies >1,000 cells were enumerated. Colonies were dissociated with PBS, single cell suspensions were counted, and either equal numbers of cells were replated or used in other indicated downstream applications. Each group was performed in triplicate and results were repeated in independent experiments at least twice.
Limiting dilution transplantation assay
Leukemic splenocytes were isolated from moribund CD45.1 C57BL/6J mice (BoyJ, Jackson Labs) transplanted with MLL-AF9 leukemia cells expressing Cdkn1b-targeting or nontargeting control shRNA. c-Kit+ cells were enriched using the CD117 microbead kit and AutoMACS Pro (Miltenyi) according to the manufacturer’s specifications and then diluted to 1000, 600, 300, 100, or 30 cells/200 µl in PBS. BoyJ transplant recipients (n = 6 mice per cell dose) were conditioned with a split dose radiation of 500 cGy and 200 cGy 3 h apart, then cells were injected via tail vein. Recipients were sacrificed when moribund or at 90 d after transplant. Leukemia stem cell frequency was calculated using ELDA software (Hu and Smyth, 2009).
Immunoblot analyses
Cells were lysed in radioimmunoprecipitation (RIPA) buffer (50 mM Tris, pH 7.4, 1% TX-100, 0.2% Na deoxycholic acid, and 0.2% SDS, HALT complete tab [Roche]). Proteins were quantified using the Pierce BCA Protein Assay kit (ThermoFisher Scientific) and equal amounts of protein were run on 4–12% Bis-Tris gradient gels (Invitrogen) according to manufacturer’s instructions. Proteins were transferred by semi-dry electrophoresis (BioRad) onto Immobilon-P PVDF (EMD Millipore). Membranes were incubated overnight at 4°C with anti-p27 3688 (Cell Signaling Technologies) or HoxA7 ab70027 (Abcam), washed then incubated with anti-mouse or anti-rabbit HRP conjugated secondary antibodies (GE Healthcare). Membranes were developed on film with WesternSure Premium Chemiluminescent Substrate (LI-COR). The same Immunoblots were reprobed with β-actin clone AC-15 (Sigma) as loading control. Biotinylated miRNA target immunoblots and p27Kip1 immunoblots were repeated on lysates generated from at least two independent experiments each.
In vitro RNAi therapeutic treatments
Standard commercially available NT LNA 5′-ACGTCTATACGCCCA-3′ and anti–mmu-miR-196b-5p LNA 5′-CAACAGGAAACTACCT-3′ were purchased from Exiqon. Custom design LNAs (Exiqon) to specifically block miR-196b binding to the validated target sites in human and mouse Cdkn1b include TSB1 5′-ATACACAGGTAGTAC-3′, TSB2 5′-ACACAGGTAGTACAA-3′, and TSB3 5′-ATATACACAGGTAGT-3′. For MLL-AF9 LNA treatments, nontargeting control, anti-miR-196b, and custom miR-196b/Cdkn1b target site blocker LNAs were purchased from Exiqon. MLL-AF9 leukemic splenocytes were plated at 5 × 104 c/ml in 96-well plates and treated with 500 nM of LNA. Treated cells were incubated overnight. 0.21 × 104 cells from each group were retreated and plated in triplicate in methylcellulose M3434 in 24-well format. Colonies were enumerated after 7 d, and then single cell suspensions were generated and lysed in RIPA buffer for Western analyses. Experiments were repeated three times.
Skp2 inhibitors, cell death, and drug synergy assays
THP1 cells were cultured in RPMI 1640 (ThermoFisher Scientific) containing 20% heat-inactivated FBS (Atlanta Biologics), Pen-Strep (ThermoFisher Scientific), and l-Glutamine (ThermoFisher Scientific). Cells were maintained at subconfluency and low passage at 37°C in 5% CO2. MV411 and MOLM13 cells were cultured similarly except in supplemented with 10% heat-inactivated FBS. For cell death analyses, cells were plated in duplicate at 0.3 × 106 c/ml and treated with 0, 12.5, 25, or 50 µM SLZ P1-41 (Tocris) or equivalent DMSO vehicle control and incubated for 3 d. Cells were then stained for AnnexinV–Pacific Blue (BioLegend) and propidium iodide (PI; BD Biosciences) in AnnexinV Binding Buffer (BioLegend) according to manufacturer instructions. Cells were analyzed on LSRII or Fortessa 2 flow cytometers (BD Biosciences). Results are graphed as average percentage alive cells as defined by AnnexinV− and PI−. The amount of inhibitor chosen for p27 immunoblots corresponded with the lowest dose of inhibitor to have a significant effect on cell viability in the 3-d treatment time course studies. Cells were treated for 24 h with indicated amounts of inhibitor then lysed in RIPA buffer and frozen at −80°C followed by immunoblot analyses to detect p27. Experiments were repeated at least twice.
For synergy assays, we synthesized the previously published Skp2-Cks1 inhibitor N-(5-(pyridin-3-yl)quinolin-8-yl)-3-(trifluoromethyl)benzenesulfonamide, referred to as compound 22d (Singh et al., 2015), resuspended in DMSO. 22d was titrated on THP1, MOLM13, and MV;411 cell lines with and without Ml-1 (Sigma), IBET-151 (GSK1210151A, Sigma), or Palbociclib (PD0332991, Selleckchem). Three doses of ML1, I-BET151, or Palbociclib were determined by dose titration studies by CellTiter Glo assay, and three concentrations surrounding the relative IC50 concentrations for each drug in each cell line were used for synergy studies (data not shown). AML cell lines were plated in triplicate at 0.106 cells/ml. Cell lines were treated with inhibitors 22d, IBET-151, Ml-1, or Palbociclib, or vehicle and cultured for 72 h. CellTiter Glo (Promega) was used according to manufacturer instructions to measure cell viability. The relative cell viabilities were calculated for each drug or drug combination compared with vehicle treated control cells. Drug synergy was calculated using CompuSyn software by ComboSyn, Inc. (Chou, 2006).
Primary human AML
De-identified viably frozen purified mononuclear cells from peripheral blood and bone marrow of AML patients were obtained from patients at CCHMC. Informed consent was obtained before sample banking. Institutional Review Board (IRB) approval for this study was obtained at CCHMC. This study was performed in accordance with the Declaration of Helsinki. Six different AML patient samples were used with MLL-rearrangements (see Table S3 for complete genetic information). 50,000 cells were plated in duplicate in a 96-well plate in IMDM containing 20% heat-inactivated FBS plus 10 ng/ml of each SCF, IL3, IL6, TPO, and FLT3. 22d was added at the indicated concentrations and cells were incubated with drug for 3 d, at which point cell viability was measured by MTS assay (Promega) according to manufacturer’s instructions.
Statistical analyses
Except where described for Microarray, RNA-Seq, and RNAi screening, all statistical analyses were performed using Prism 6 (GraphPad Software). For RT-qPCR differences in gene expression or pulldown, enrichments were evaluated by ratio paired parametric t test assuming Gaussian distribution. For ESC core gene expression RT-qPCR, differences were evaluated by two-way ANOVA Holm-Sidak multiple comparisons test. For flow cytometric analyses differences in populations were compared by two-way ANOVA Bonferroni’s multiple comparisons test. Differences in average colony numbers (CFU assays) were evaluated statistically using two-way ANOVA Bonferroni’s multiple comparisons test or one-way ANOVA Holm-Sidak multiple comparisons test as indicated. Differences in target site dual luciferase reporter assay repressions were evaluated by two-way ANOVA Sidak’s multiple comparisons test. Differences in Skp2 inhibitor SLZ P1-41 dosage response were evaluated by multiple unpaired t tests with Holm-Sidak multiple testing correction.
Data availability
Microarray and RNA-seq data can be accessed in the Gene Expression Omnibus accession no. GSE75843.
Online supplemental material
Supplementary information includes five figures and three tables. Fig. S1 shows the optimization, characterization, and validation of biotinylated miR-196b mimic target pulldowns. Derepression of miR-196b pulldown targets in miR196 knockout mice is shown. Fig. S2 shows enrichment and depletion scores of shRNA performance in the in vivo shRNA screen and examples of shRNA-mediated knockdown of miR-196b pulldown target RNA levels in MLL-AF9 AML. Fig. S3 shows heat map of Phc2 knockdown MLL-AF9 gene expression and Cdkn1b knockdown effects on MLL-AF9 transcriptional programming and immunophenotype. Fig. S4 shows p27Kip1 induces cyclin/cdk-dependent cell cycle arrest in MLL-r AML. Fig. S5 shows p27Kip1 levels in human AML treated with SCFSkp2 inhibitors. Table S1 contains 10 tabs of biotinylated miR mimic pulldown microarray enrichment results, accession nos. of AGO-CLIP data, miR-196 target predictions, details of each hairpin in the shRNA screen, average enrichment and depletion scores for shRNA screen hairpins, shRNA screen gene ontologies, and RNA-seq on E9.5 embryos with limiting miR-196 alleles. Table S2 contains seven tabs of RNA-seq and GSEA results from Cdkn1b and Phc2 knockdown MLL-AF9 AML. Table S3 contains drug synergy data, genomic alterations of primary human AML patient samples, and SNP validation of human AML cell lines.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr. Nancy Zeleznik-Le for murine MLL-AF9 leukemia cells. The authors acknowledge the CCHMC Research Flow Cytometry Core, Comprehensive Mouse and Cancer Core, Gene Expression Core, and Transgenic Animal and Genome Editing Core.
The research was supported by Ladies Auxiliary to the Veterans of Foreign Wars (LAVFW) Postdoctoral Fellowship (to S.E.M.), CancerFree KIDS (Loveland, Ohio), University of Cincinnati training grant NIEHS T32ES007250 (S.E. Meyer and D.E. Muench), CCTST Just-In-Time Core Grant Program (University of Cincinnati and CCHMC), Leukemia and Lymphoma Society of America Translational Research Award, Sidney Kimmel Cancer Center NCI Support Grant P30CA056036 (S.E. Meyer), and National Institutes of Health grants R01CA159845 and R01CA196658 (to H.L. Grimes). The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government (E. McGlinn).
The authors declare no competing financial interests.
Author contributions: All authors reviewed and edited the manuscript. S.E. Meyer conceived and performed experiments, analyzed and interpreted data, and wrote the manuscript. D.E. Muench, A.M. Rogers, T.J. Newkold, E. Orr, and E. O’Brien performed experiments and analyzed data. J.G. Doench and J.D. Perentesis planned experiments and interpreted data. B.J. Aronow, N. Salomonis, and S.E. Meyer performed bioinformatic data analyses. A. Lal and J. Lieberman advised pulldown experiments, data analyses, and interpretation. P.J. Morris and C.J. Thomas synthesized 22d compounds and advised drug synergy experiments, data analyses, and interpretation. E. McGlinn performed, analyzed, and interpreted miRNA knockout mouse embryo data. H.L. Grimes conceived experiments, interpreted data, and wrote the manuscript.
References
- Agarwal V., Bell G.W., Nam J.W., and Bartel D.P.. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 4:e05005 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong S.A., Staunton J.E., Silverman L.B., Pieters R., den Boer M.L., Minden M.D., Sallan S.E., Lander E.S., Golub T.R., and Korsmeyer S.J.. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30:41–47. 10.1038/ng765 [DOI] [PubMed] [Google Scholar]
- Ayton P.M., and Cleary M.L.. 2003. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 17:2298–2307. 10.1101/gad.1111603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A., Gurian-West M., Chen X., Kelly-Spratt K.S., Kemp C.J., and Roberts J.M.. 2006. A pathway in quiescent cells that controls p27Kip1 stability, subcellular localization, and tumor suppression. Genes Dev. 20:47–64. 10.1101/gad.1384406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Wilson M., Gabow A., Marks D.S., and Sander C.. 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36(Database):D149–D153. 10.1093/nar/gkm995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., and Peterlin B.M.. 2011. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25:2158–2172. 10.1101/gad.16962311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J.C., Friedmann T., Driever W., Burrascano M., and Yee J.K.. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA. 90:8033–8037. 10.1073/pnas.90.17.8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano A.C., Eytan E., Hershko A., and Pagano M.. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193–199. 10.1038/12013 [DOI] [PubMed] [Google Scholar]
- Ceccarelli D.F., Tang X., Pelletier B., Orlicky S., Xie W., Plantevin V., Neculai D., Chou Y.C., Ogunjimi A., Al-Hakim A., et al. 2011. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 145:1075–1087. 10.1016/j.cell.2011.05.039 [DOI] [PubMed] [Google Scholar]
- Ceppi M., Pereira P.M., Dunand-Sauthier I., Barras E., Reith W., Santos M.A., and Pierre P.. 2009. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA. 106:2735–2740. 10.1073/pnas.0811073106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.H., Morrow J.K., Li C.F., Gao Y., Jin G., Moten A., Stagg L.J., Ladbury J.E., Cai Z., Xu D., et al. 2013. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 154:556–568. 10.1016/j.cell.2013.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., and Mendell J.T.. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40:43–50. 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.W., Koche R.P., Sinha A.U., Deshpande A.J., Zhu N., Eng R., Doench J.G., Xu H., Chu S.H., Qi J., et al. 2015. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 21:335–343. 10.1038/nm.3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu H., Aronow B.J., and Jegga A.G.. 2007. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 8:392 10.1186/1471-2105-8-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Aronow B.J., and Jegga A.G.. 2009a Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 10:73 10.1186/1471-2105-10-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., and Jegga A.G.. 2009b ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37(Web Server):W305-W311. 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Xie W., Kuhn D.J., Voorhees P.M., Lopez-Girona A., Mendy D., Corral L.G., Krenitsky V.P., Xu W., Moutouh-de Parseval L., et al. 2008a Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 111:4690–4699. 10.1182/blood-2007-09-112904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Kumar A.R., Hudson W.A., Li Q., Wu B., Staggs R.A., Lund E.A., Sam T.N., and Kersey J.H.. 2008b Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 13:432–440. 10.1016/j.ccr.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Rodrigues N., Dombkowski D., Stier S., and Scadden D.T.. 2000. Stem cell repopulation efficiency but not pool size is governed by p27(kip1). Nat. Med. 6:1235–1240. 10.1038/81335 [DOI] [PubMed] [Google Scholar]
- Chou T.C. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58:621–681. 10.1124/pr.58.3.10 [DOI] [PubMed] [Google Scholar]
- Corral J., Lavenir I., Impey H., Warren A.J., Forster A., Larson T.A., Bell S., McKenzie A.N., King G., and Rabbitts T.H.. 1996. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 85:853–861. 10.1016/S0092-8674(00)81269-6 [DOI] [PubMed] [Google Scholar]
- Cox M.C., Panetta P., Lo-Coco F., Del Poeta G., Venditti A., Maurillo L., Del Principe M.I., Mauriello A., Anemona L., Bruno A., et al. 2004. Chromosomal aberration of the 11q23 locus in acute leukemia and frequency of MLL gene translocation: results in 378 adult patients. Am. J. Clin. Pathol. 122:298–306. 10.1309/RX27R8GJQM330C22 [DOI] [PubMed] [Google Scholar]
- Daigle S.R., Olhava E.J., Therkelsen C.A., Majer C.R., Sneeringer C.J., Song J., Johnston L.D., Scott M.P., Smith J.J., Xiao Y., et al. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 20:53–65. 10.1016/j.ccr.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-Bailador F., Bonev B., Garcez P., Stanley P., Guillemot F., and Papalopulu N.. 2012. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 15:697–699. 10.1038/nn.3082 [DOI] [PubMed] [Google Scholar]
- Dawson M.A., Prinjha R.K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W.I., Robson S.C., Chung C.W., Hopf C., Savitski M.M., et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 478:529–533. 10.1038/nature10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H., Estey E.H., Amadori S., Appelbaum F.R., Büchner T., Burnett A.K., Dombret H., Fenaux P., Grimwade D., Larson R.A., et al. European LeukemiaNet . 2010. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 115:453–474. 10.1182/blood-2009-07-235358 [DOI] [PubMed] [Google Scholar]
- Ellwanger D.C., Büttner F.A., Mewes H.W., and Stümpflen V.. 2011. The sufficient minimal set of miRNA seed types. Bioinformatics. 27:1346–1350. 10.1093/bioinformatics/btr149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emig D., Salomonis N., Baumbach J., Lengauer T., Conklin B.R., and Albrecht M.. 2010. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 38(suppl_2):W755-W762. 10.1093/nar/gkq405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmrich S., Rasche M., Schöning J., Reimer C., Keihani S., Maroz A., Xie Y., Li Z., Schambach A., Reinhardt D., and Klusmann J.H.. 2014. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 28:858–874. 10.1101/gad.233791.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., and Marks D.S.. 2003. MicroRNA targets in Drosophila. Genome Biol. 5:R1 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfurth F.E., Popovic R., Grembecka J., Cierpicki T., Theisler C., Xia Z.B., Stuart T., Diaz M.O., Bushweller J.H., and Zeleznik-Le N.J.. 2008. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc. Natl. Acad. Sci. USA. 105:7517–7522. 10.1073/pnas.0800090105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., and Bozzoni I.. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 123:819–831. 10.1016/j.cell.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Fong C.Y., Gilan O., Lam E.Y., Rubin A.F., Ftouni S., Tyler D., Stanley K., Sinha D., Yeh P., Morison J., et al. 2015. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 525:538–542. 10.1038/nature14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoth D., Bornstein G., Ko T.K., Larsen B., Tyers M., Pagano M., and Hershko A.. 2001. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat. Cell Biol. 3:321–324. 10.1038/35060126 [DOI] [PubMed] [Google Scholar]
- Ghiaur G., Gerber J., and Jones R.J.. 2012. Concise review: Cancer stem cells and minimal residual disease. Stem Cells. 30:89–93. 10.1002/stem.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grembecka J., He S., Shi A., Purohit T., Muntean A.G., Sorenson R.J., Showalter H.D., Murai M.J., Belcher A.M., Hartley T., et al. 2012. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8:277–284. 10.1038/nchembio.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade D., Hills R.K., Moorman A.V., Walker H., Chatters S., Goldstone A.H., Wheatley K., Harrison C.J., and Burnett A.K.. National Cancer Research Institute Adult Leukaemia Working Group . 2010. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 116:354–365. 10.1182/blood-2009-11-254441 [DOI] [PubMed] [Google Scholar]
- Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., and Minghim R.. 2015. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 16:169 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Huret J.L., Dessen P., and Bernheim A.. 2001. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 15:987–989. 10.1038/sj.leu.2402135 [DOI] [PubMed] [Google Scholar]
- Isono K., Fujimura Y., Shinga J., Yamaki M., O-Wang J., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., and Koseki H.. 2005. Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol. Cell. Biol. 25:6694–6706. 10.1128/MCB.25.15.6694-6706.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N.B., Dimos J.T., Schaniel C., Hackney J.A., Moore K.A., and Lemischka I.R.. 2002. A stem cell molecular signature. Science. 298:601–604. 10.1126/science.1073823 [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., and Marks D.S.. 2004. Human MicroRNA targets. PLoS Biol. 2:e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongen-Lavrencic M., Sun S.M., Dijkstra M.K., Valk P.J., and Löwenberg B.. 2008. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 111:5078–5085. 10.1182/blood-2008-01-133355 [DOI] [PubMed] [Google Scholar]
- Katayose Y., Kim M., Rakkar A.N., Li Z., Cowan K.H., and Seth P.. 1997. Promoting apoptosis: a novel activity associated with the cyclin-dependent kinase inhibitor p27. Cancer Res. 57:5441–5445. [PubMed] [Google Scholar]
- Kent W.J. 2002. BLAT--the BLAST-like alignment tool. Genome Res. 12:656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., and Haussler D.. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., and Segal E.. 2007. The role of site accessibility in microRNA target recognition. Nat. Genet. 39:1278–1284. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., and Orkin S.H.. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 132:1049–1061. 10.1016/j.cell.2008.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., and Orkin S.H.. 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 143:313–324. 10.1016/j.cell.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossatz U., Dietrich N., Zender L., Buer J., Manns M.P., and Malek N.P.. 2004. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 18:2602–2607. 10.1101/gad.321004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818–822. 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- Kumar A.R., Hudson W.A., Chen W., Nishiuchi R., Yao Q., and Kersey J.H.. 2004. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 103:1823–1828. 10.1182/blood-2003-07-2582 [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Lu J., Mercer K.L., Golub T.R., and Jacks T.. 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39:673–677. 10.1038/ng2003 [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Pester R.E., Chen C.Y., Lane K., Chin C., Lu J., Kirsch D.G., Golub T.R., and Jacks T.. 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23:2700–2704. 10.1101/gad.1848209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Thomas M.P., Altschuler G., Navarro F., O’Day E., Li X.L., Concepcion C., Han Y.C., Thiery J., Rajani D.K., et al. 2011. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 7:e1002363 10.1371/journal.pgen.1002363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechman E.R., Gentner B., Ng S.W.K., Schoof E.M., van Galen P., Kennedy J.A., Nucera S., Ciceri F., Kaufmann K.B., Takayama N., et al. 2016. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell. 29:602–606. 10.1016/j.ccell.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., and Burge C.B.. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798. 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., and Bartel D.P.. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li Z., Lu J., Sun M., Mi S., Zhang H., Luo R.T., Chen P., Wang Y., Yan M., Qian Z., et al. 2008. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc. Natl. Acad. Sci. USA. 105:15535–15540. 10.1073/pnas.0808266105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang H., Chen P., He M., Li Y., Arnovitz S., Jiang X., He C., Hyjek E., Zhang J., et al. 2012. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 3:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. 2005. MicroRNA expression profiles classify human cancers. Nature. 435:834–838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Machado I.D., Spatti M., Hastreiter A., Santin J.R., Fock R.A., Gil C.D., Oliani S.M., Perretti M., and Farsky S.H.. 2016. Annexin A1 Is a Physiological Modulator of Neutrophil Maturation and Recirculation Acting on the CXCR4/CXCL12 Pathway. J. Cell. Physiol. 231:2418–2427. 10.1002/jcp.25346 [DOI] [PubMed] [Google Scholar]
- Maragkakis M., Alexiou P., Papadopoulos G.L., Reczko M., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K., Simossis V.A., et al. 2009a Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 10:295 10.1186/1471-2105-10-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M., Reczko M., Simossis V.A., Alexiou P., Papadopoulos G.L., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K., et al. 2009b DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 37(Web Server):W273-276. 10.1093/nar/gkp292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Radmacher M.D., Maharry K., Mrózek K., Ruppert A.S., Paschka P., Vukosavljevic T., Whitman S.P., Baldus C.D., Langer C., et al. 2008. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 358:1919–1928. 10.1056/NEJMoa074256 [DOI] [PubMed] [Google Scholar]
- Mavrakis K.J., Wolfe A.L., Oricchio E., Palomero T., de Keersmaecker K., McJunkin K., Zuber J., James T., Khan A.A., Leslie C.S., et al. 2010. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 12:372–379. 10.1038/ncb2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S.E., Zinser G.M., Stuart W.D., Pathrose P., and Waltz S.E.. 2009. The Ron receptor tyrosine kinase negatively regulates mammary gland branching morphogenesis. Dev. Biol. 333:173–185. 10.1016/j.ydbio.2009.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P.G., Al-Shahrour F., Hartwell K.A., Chu L.P., Järås M., Puram R.V., Puissant A., Callahan K.P., Ashton J., McConkey M.E., et al. 2013. In Vivo RNAi screening identifies a leukemia-specific dependence on integrin beta 3 signaling. Cancer Cell. 24:45–58. 10.1016/j.ccr.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Naldini L., Blömer U., Gage F.H., Trono D., and Verma I.M.. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA. 93:11382–11388. 10.1073/pnas.93.21.11382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.M., Arthur A., Zannettino A.C., and Gronthos S.. 2017. EphA5 and EphA7 forward signaling enhances human hematopoietic stem and progenitor cell maintenance, migration, and adhesion via Rac1 activation. Exp. Hematol. 48:72–78. 10.1016/j.exphem.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Nonne N., Ameyar-Zazoua M., Souidi M., and Harel-Bellan A.. 2010. Tandem affinity purification of miRNA target mRNAs (TAP-Tar). Nucleic Acids Res. 38:e20 10.1093/nar/gkp1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary B., Finn R.S., and Turner N.C.. 2016. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 13:417–430. 10.1038/nrclinonc.2016.26 [DOI] [PubMed] [Google Scholar]
- Odero M.D., Zeleznik-Le N.J., Chinwalla V., and Rowley J.D.. 2000. Cytogenetic and molecular analysis of the acute monocytic leukemia cell line THP-1 with an MLL-AF9 translocation. Genes Chromosomes Cancer. 29:333–338. [DOI] [PubMed] [Google Scholar]
- Oliveros J.C.2015. Venny. An interactive too for comparing lists of Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed April, 2018).
- Papadopoulos G.L., Alexiou P., Maragkakis M., Reczko M., and Hatzigeorgiou A.G.. 2009. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 25:1991–1993. 10.1093/bioinformatics/btp299 [DOI] [PubMed] [Google Scholar]
- Parant J., Chavez-Reyes A., Little N.A., Yan W., Reinke V., Jochemsen A.G., and Lozano G.. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29:92–95. 10.1038/ng714 [DOI] [PubMed] [Google Scholar]
- Petersen M., and Wengel J.. 2003. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 21:74–81. 10.1016/S0167-7799(02)00038-0 [DOI] [PubMed] [Google Scholar]
- Placke T., Faber K., Nonami A., Putwain S.L., Salih H.R., Heidel F.H., Krämer A., Root D.E., Barbie D.A., Krivtsov A.V., et al. 2014. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood. 124:13–23. 10.1182/blood-2014-02-558114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Kato J.Y., Solomon M.J., Sherr C.J., Massague J., Roberts J.M., and Koff A.. 1994. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 8:9–22. 10.1101/gad.8.1.9 [DOI] [PubMed] [Google Scholar]
- Popovic R., Riesbeck L.E., Velu C.S., Chaubey A., Zhang J., Achille N.J., Erfurth F.E., Eaton K., Lu J., Grimes H.L., et al. 2009. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 113:3314–3322. 10.1182/blood-2008-04-154310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H., Bardelli A., Lengauer C., Kinzler K.W., Vogelstein B., and Velculescu V.E.. 2002. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 418:934 10.1038/418934a [DOI] [PubMed] [Google Scholar]
- Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D., and Kent W.J.. 2014. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 30:1003–1005. 10.1093/bioinformatics/btt637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., Nelson B., Vranizan K., Pico A.R., Hanspers K., Kuchinsky A., Ta L., Mercola M., and Conklin B.R.. 2009. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLOS Comput. Biol. 5:e1000553 10.1371/journal.pcbi.1000553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., Schlieve C.R., Pereira L., Wahlquist C., Colas A., Zambon A.C., Vranizan K., Spindler M.J., Pico A.R., Cline M.S., et al. 2010. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl. Acad. Sci. USA. 107:10514–10519. 10.1073/pnas.0912260107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte D., Lange-Turenhout E.A., Stumpel D.J., Stam R.W., Buijs-Gladdines J.G., Meijerink J.P., Pieters R., and Den Boer M.L.. 2010. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 95:1675–1682. 10.3324/haematol.2010.023481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Sran A., Carroll D.C., Huang J., Tsvetkov L., Zhou X., Sheung J., McLaughlin J., Issakani S.D., Payan D.G., and Shaw S.J.. 2015. Developing structure-activity relationships from an HTS hit for inhibition of the Cks1-Skp2 protein-protein interaction. Bioorg. Med. Chem. Lett. 25:5199–5202. 10.1016/j.bmcl.2015.09.067 [DOI] [PubMed] [Google Scholar]
- So C.W., Karsunky H., Wong P., Weissman I.L., and Cleary M.L.. 2004. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 103:3192–3199. 10.1182/blood-2003-10-3722 [DOI] [PubMed] [Google Scholar]
- Somervaille T.C., and Cleary M.L.. 2006. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 10:257–268. 10.1016/j.ccr.2006.08.020 [DOI] [PubMed] [Google Scholar]
- Somervaille T.C., Matheny C.J., Spencer G.J., Iwasaki M., Rinn J.L., Witten D.M., Chang H.Y., Shurtleff S.A., Downing J.R., and Cleary M.L.. 2009. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 4:129–140. 10.1016/j.stem.2008.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck C., Strohmaier H., Watson M., Smith A.P., Ryan A., Krek T.W., and Reed S.I.. 2001. A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol. Cell. 7:639–650. 10.1016/S1097-2765(01)00210-6 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlüty H., Chatelain E., Marti A., Wirbelauer C., Senften M., Müller U., and Krek W.. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207–214. 10.1038/12027 [DOI] [PubMed] [Google Scholar]
- Terwijn M., Zeijlemaker W., Kelder A., Rutten A.P., Snel A.N., Scholten W.J., Pabst T., Verhoef G., Löwenberg B., Zweegman S., et al. 2014. Leukemic stem cell frequency: a strong biomarker for clinical outcome in acute myeloid leukemia. PLoS One. 9:e107587 10.1371/journal.pone.0107587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima H., and Hunter T.. 1994. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 78:67–74. 10.1016/0092-8674(94)90573-8 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S.L.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov L.M., Yeh K.H., Lee S.J., Sun H., and Zhang H.. 1999. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9:661–664. 10.1016/S0960-9822(99)80290-5 [DOI] [PubMed] [Google Scholar]
- van Rhenen A., Feller N., Kelder A., Westra A.H., Rombouts E., Zweegman S., van der Pol M.A., Waisfisz Q., Ossenkoppele G.J., and Schuurhuis G.J.. 2005. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin. Cancer Res. 11:6520–6527. 10.1158/1078-0432.CCR-05-0468 [DOI] [PubMed] [Google Scholar]
- Velu C.S., Chaubey A., Phelan J.D., Horman S.R., Wunderlich M., Guzman M.L., Jegga A.G., Zeleznik-Le N.J., Chen J., Mulloy J.C., et al. 2014. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Invest. 124:222–236. 10.1172/JCI66005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlach J., Hennecke S., and Amati B.. 1997. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 16:5334–5344. 10.1093/emboj/16.17.5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Nichogiannopoulou A., Wu L., Sun L., Sharpe A.H., Bigby M., and Georgopoulos K.. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 5:537–549. 10.1016/S1074-7613(00)80269-1 [DOI] [PubMed] [Google Scholar]
- Wang J., Han F., Wu J., Lee S.W., Chan C.H., Wu C.Y., Yang W.L., Gao Y., Zhang X., Jeong Y.S., et al. 2011. The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal. Blood. 118:5429–5438. 10.1182/blood-2010-10-312785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V.E., Schmidt T., Chen J., Sharp P.A., and Tantin D.. 2004. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell. Biol. 24:1022–1032. 10.1128/MCB.24.3.1022-1032.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gorospe M., Huang Y., and Holbrook N.J.. 1997. p27Kip1 overexpression causes apoptotic death of mammalian cells. Oncogene. 15:2991–2997. 10.1038/sj.onc.1201450 [DOI] [PubMed] [Google Scholar]
- Winandy S., Wu P., and Georgopoulos K.. 1995. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 83:289–299. 10.1016/0092-8674(95)90170-1 [DOI] [PubMed] [Google Scholar]
- Wong D.J., Liu H., Ridky T.W., Cassarino D., Segal E., and Chang H.Y.. 2008. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2:333–344. 10.1016/j.stem.2008.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.F., Agarwal V., Mansfield J.H., Denans N., Schwartz M.G., Prosser H.M., Pourquié O., Bartel D.P., Tabin C.J., and McGlinn E.. 2015. Independent regulation of vertebral number and vertebral identity by microRNA-196 paralogs. Proc. Natl. Acad. Sci. USA. 112:E4884–E4893. 10.1073/pnas.1512655112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynendaele J., Böhnke A., Leucci E., Nielsen S.J., Lambertz I., Hammer S., Sbrzesny N., Kubitza D., Wolf A., Gradhand E., et al. 2010. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 70:9641–9649. 10.1158/0008-5472.CAN-10-0527 [DOI] [PubMed] [Google Scholar]
- Yagi T., Morimoto A., Eguchi M., Hibi S., Sako M., Ishii E., Mizutani S., Imashuku S., Ohki M., and Ichikawa H.. 2003. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 102:1849–1856. 10.1182/blood-2003-02-0578 [DOI] [PubMed] [Google Scholar]
- Yang J.H., Li J.H., Shao P., Zhou H., Chen Y.Q., and Qu L.H.. 2011. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 39(suppl_1):D202–D209. 10.1093/nar/gkq1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.C., Di C., Hu B., Zhou M., Liu Y., Song N., Li Y., Umetsu J., and Lu Z.J.. 2015. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics. 16:51 10.1186/s12864-015-1273-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., and Bartel D.P.. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596. 10.1126/science.1097434 [DOI] [PubMed] [Google Scholar]
- Zhang J., Seet C.S., Sun C., Li J., You D., Volk A., Breslin P., Li X., Wei W., Qian Z., et al. 2013. p27kip1 maintains a subset of leukemia stem cells in the quiescent state in murine MLL-leukemia. Mol. Oncol. 7:1069–1082. 10.1016/j.molonc.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 478:524–528. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray and RNA-seq data can be accessed in the Gene Expression Omnibus accession no. GSE75843.