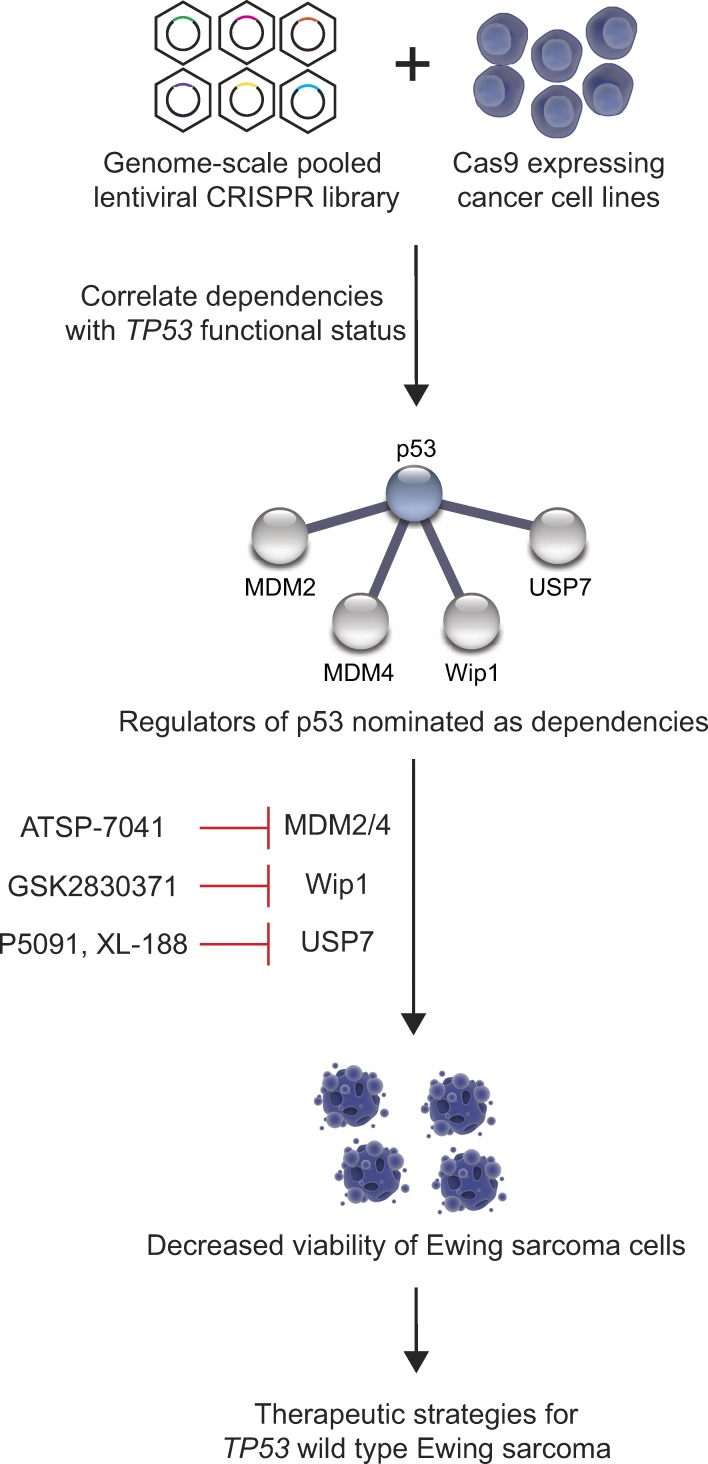
Stolte et al. use genome-scale CRISPR-Cas9 screening technology to identify druggable targets for TP53 wild-type Ewing sarcoma and discover reactivation of p53 through inhibition of MDM2, MDM4, Wip1, or USP7 as therapeutic strategies for the disease.
Abstract
Ewing sarcoma is a pediatric cancer driven by EWS-ETS transcription factor fusion oncoproteins in an otherwise stable genomic background. The majority of tumors express wild-type TP53, and thus, therapies targeting the p53 pathway would benefit most patients. To discover targets specific for TP53 wild-type Ewing sarcoma, we used a genome-scale CRISPR-Cas9 screening approach and identified and validated MDM2, MDM4, USP7, and PPM1D as druggable dependencies. The stapled peptide inhibitor of MDM2 and MDM4, ATSP-7041, showed anti-tumor efficacy in vitro and in multiple mouse models. The USP7 inhibitor, P5091, and the Wip1/PPM1D inhibitor, GSK2830371, decreased the viability of Ewing sarcoma cells. The combination of ATSP-7041 with P5091, GSK2830371, and chemotherapeutic agents showed synergistic action on the p53 pathway. The effects of the inhibitors, including the specific USP7 inhibitor XL-188, were rescued by concurrent TP53 knockout, highlighting the essentiality of intact p53 for the observed cytotoxic activities.
Graphical Abstract
Introduction
Ewing sarcoma is a pediatric small round blue cell tumor that is treated with a combination of interval compressed chemotherapy, radiation, and surgery. While outcomes have improved over the last several decades for patients with localized disease, little progress has been made in the treatment of patients with newly diagnosed metastatic or relapsed disease. Moreover, treatment-related toxicity is significant, and currently, there are no targeted therapies for Ewing sarcoma that are approved by the United States Food and Drug Administration (Balamuth and Womer, 2010; Gaspar et al., 2015).
The defining event in Ewing sarcoma is a somatic chromosomal translocation, most commonly between chromosomes 11 and 22, causing a fusion between the EWSR1 (Ewing sarcoma breakpoint region 1) gene and an ETS family gene FLI1 (Friend leukemia virus integration 1). The resulting fusion protein, EWS/FLI, is an aberrant oncogenic transcription factor (Riggi et al., 2008). Efforts to directly inhibit EWS/FLI have largely been unsuccessful (Gaspar et al., 2015). Several recent massively parallel sequencing efforts revealed that Ewing sarcoma tumors possess remarkably quiet genomes, with few recurrent genetic events and no immediately druggable mutated kinases (Brohl et al., 2014; Crompton et al., 2014; Tirode et al., 2014). While the paucity of genetic events is a challenge for the development of precision medicine approaches using kinase inhibitors, the genomic simplicity may enable other treatment strategies. Indeed, up to 90% of Ewing sarcoma tumors present with wild-type TP53 (Tumor protein 53), allowing for new therapeutic strategies involving p53 activation.
Although the majority of patient tumors retain wild-type TP53, there has been a historic bias against studying p53 dependent genes in this disease. The vast majority of Ewing sarcoma cell lines harbor TP53 mutations (Brohl et al., 2014; Crompton et al., 2014; Tirode et al., 2014), and patient-derived Ewing sarcoma xenografts have only recently been established (Ordóñez et al., 2015). Consequently, models with TP53 mutations have been overrepresented in Ewing sarcoma studies in the past. Therefore, we sought to identify druggable dependencies in TP53 wild-type Ewing sarcoma models, which better recapitulate the more common disease biology.
The use of clustered regularly interspaced short palindromic repeats (CRISPR) paired with the CRISPR-associated nuclease 9 (Cas9) has emerged as a tool to study the biology of mammalian cells (Cong et al., 2013; Mali et al., 2013). Genome-scale CRISPR-Cas9 screening provides a powerful new strategy to identify cancer dependencies (Shalem et al., 2014). Using this approach, we report genetic dependencies specific for TP53 wild-type tumors, including Ewing sarcoma, from analysis of a previously published dataset (Aguirre et al., 2016). We hypothesized that deletion of TP53 by single guide RNA (sgRNA)–guided CRISPR-Cas9 constructs would give a proliferative advantage exclusively in TP53 wild-type cell lines and, therefore, leveraged the data to identify genetic dependencies anti-correlated with TP53 dependency scores. The p53 regulators murine double minute 2 (MDM2), murine double minute 4 (MDM4), ubiquitin specific peptidase 7 (USP7), and protein phosphatase, Mg2+/Mn2+-dependent 1D (PPM1D) were among the top druggable dependencies with strong anti-correlation to TP53 dependency scores. All four were validated in secondary assays to be essential for proliferation of TP53 wild-type Ewing sarcoma cells. Moreover, chemical inhibitors of these targets, including a stapled peptide dual inhibitor of MDM2 and MDM4 (ATSP-7041), an USP7 inhibitor (P5091), and a wild-type p53-induced phosphatase 1 (Wip1; encoded by the PPM1D gene) inhibitor (GSK2830371) reduced the viability of Ewing sarcoma cell lines as single agents. ATSP-7041 showed anti-tumor efficacy in vivo in several Ewing sarcoma models. Consistent with all four targets being highly correlated dependencies in the screening data, combinatorial targeting with these pharmacologic inhibitors showed synergistic activity. Furthermore, ATSP-7041 synergized with some standard-of-care Ewing sarcoma chemotherapeutic agents. To prove that these treatment strategies depend on functional p53, TP53 knockout cell lines were generated. TP53 knockout rescued the cytotoxic effects of CRISPR-Cas9–mediated suppression or pharmacologic inhibition of all four targets. Collectively, these data highlight the therapeutic relevance of the intact p53 regulatory network in Ewing sarcoma tumors and provide preclinical evidence to support the testing of p53 modulators in patients with TP53 wild-type Ewing sarcoma.
Results
Genome-scale CRISPR-Cas9 screening distinguishes between TP53 wild-type and TP53 mutant cell lines
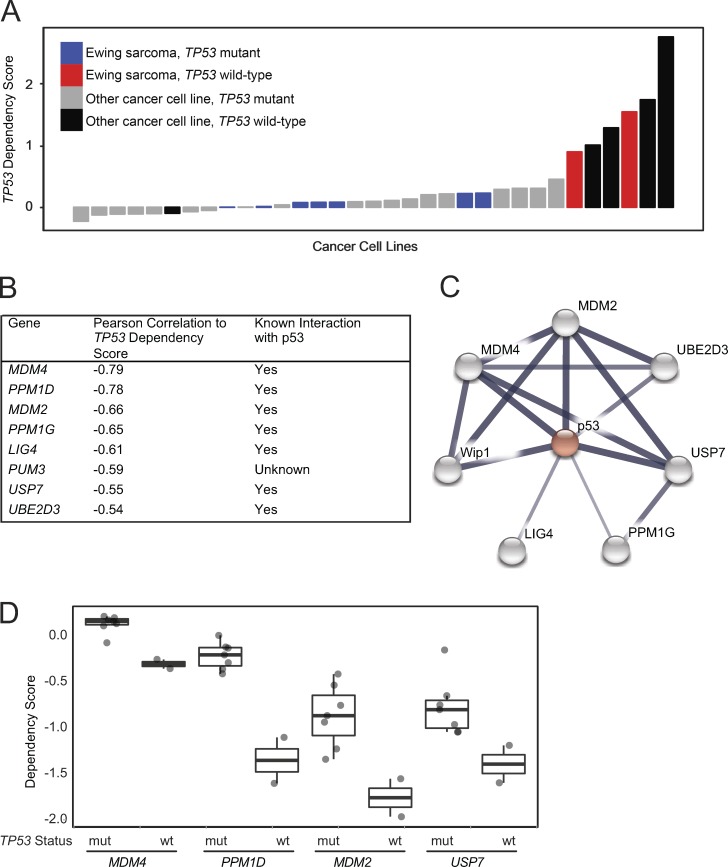
To identify new therapeutic targets for TP53 wild-type Ewing sarcoma, we analyzed the data from our genome-scale CRISPR-Cas9 screen of 33 cancer cell lines, including nine Ewing sarcoma cell lines (Aguirre et al., 2016). We determined that targeting TP53 in this genome-scale screen provided a proliferative advantage in wild-type TP53 cell lines (indicated by positive scores) and very little to no effect in mutant TP53 cell lines (Fig. 1 A). p53 mutation status was assigned by mining published data from several large studies (Barretina et al., 2012; Cancer Cell Line Encyclopedia Consortium; Genomics of Drug Sensitivity in Cancer Consortium, 2015; Klijn et al., 2015), a curated list of mutations (Edlund et al., 2012), and a literature search for cell lines for which no information was available from other sources (Table S1). The response to TP53 disruption was consistent with the annotated mutation status in 97% of cell lines, including all of the Ewing sarcoma cell lines in this screen.
Figure 1.
Genome-scale CRISPR-Cas9 screen of 33 cancer cell lines identifies genetic vulnerabilities negatively correlated with TP53 dependency in TP53 wild-type lines. (A) Waterfall plot of TP53 dependency in 33 cancer cell lines shows positive dependency score in known TP53 wild-type cell lines consistent with the hypothesis that disruption of TP53 in these lines would lead to a proliferation advantage. Based on these data, 6 of 33 lines likely have a functional p53 pathway. A single cell line for which there is no documented TP53 mutation, PANC08.13, behaves like a TP53 mutant line, suggesting it has a nonfunctional p53 pathway. (B) Top eight anti-correlated genetic dependencies to TP53 dependency. (C) Seven of the top eight anti-correlated genes are connected to TP53 in the STRING database indicating putative protein–protein interactions. The widths of the edges correspond to the level of confidence in interactions (medium confidence STRING score of 0.4; high confidence STRING score of 0.7; highest confidence STRING score of 0.9). (D) MDM4, PPM1D, MDM2, and USP7 dependency scores in Ewing sarcoma cell lines in the CRISPR-Cas9 screen stratified by TP53 mutational status (mut, mutant; wt, wild-type).
Regulators of p53 are anti-correlated with TP53 dependency scores
To identify targets in TP53 wild-type cell lines, we hypothesized that cell lines with the greatest proliferative advantage upon TP53 suppression (presumably due to the presence of a functional p53 pathway) would also be dependent on negative regulators of TP53. The top eight variable genetic dependencies that were anti-correlated to TP53 dependency scores in all 33 cancer cell lines in the screen included MDM2, MDM4, USP7, and PPM1D, as well as other genes with known roles in p53 regulation (Fig. 1 B) and with p53 interaction in the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (Fig. 1 C; Szklarczyk et al., 2015). Prioritizing translatable targets, we focused on the druggable dependencies MDM2, MDM4, USP7, and PPM1D, which have inhibitors in preclinical or clinical evaluation. In Ewing sarcoma, MDM2, MDM4, USP7, and PPM1D were preferential dependencies in the TP53 wild-type cell lines (Fig. 1 D and Fig. S1, A and B).
MDM2 is an E3 ubiquitin ligase that marks p53 for degradation by the proteasome (Wade et al., 2013). MDM4, a structural homologue of MDM2, inhibits p53 by binding to and sequestering its transactivation domain (Wade et al., 2013). USP7 has been implicated in several cellular processes, including deubiquitination of MDM2, which leads to a decrease in p53 (Nicholson and Suresh Kumar, 2011). PPM1D encodes for the serine-phosphatase Wip1 that dephosphorylates and inactivates p53 and other proteins involved in cell stress and DNA damage (Zhu and Bulavin, 2012).
With an eye toward clinical translation, we first focused on MDM2 and MDM4, both of which can be targeted simultaneously with a dual inhibitor currently in clinical trials for adults with TP53 wild-type cancers (Chang et al., 2013; Meric-Bernstam et al., 2017).
Genetic disruption of MDM2 and MDM4 has a selective cytotoxic effect in TP53 wild-type Ewing sarcoma cell lines
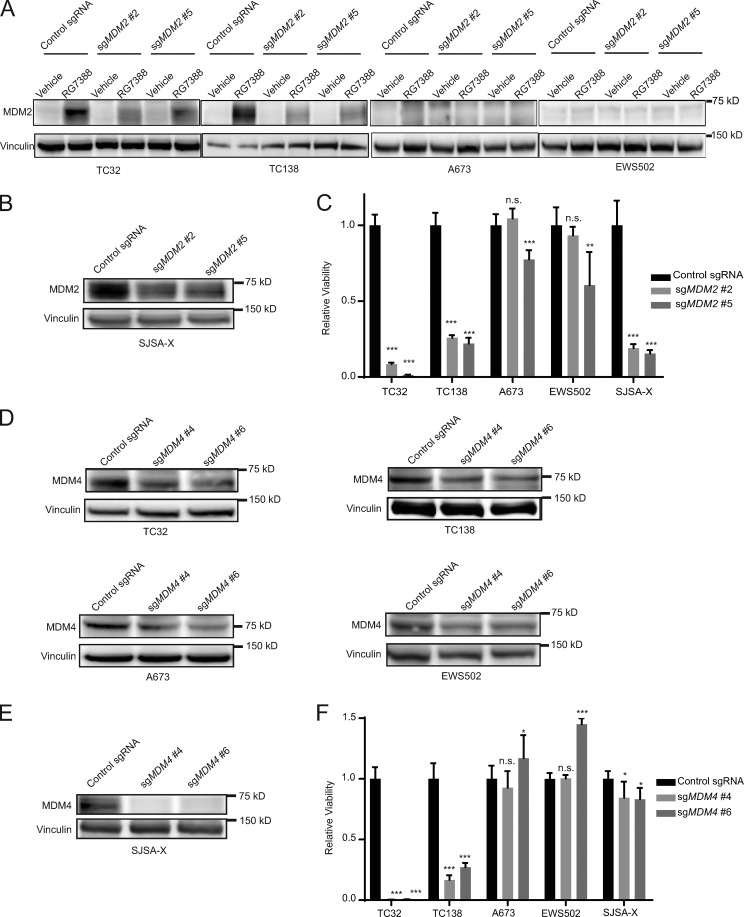
We sought to validate the dependencies on MDM2 and MDM4 by CRISPR-Cas9 in two TP53 wild-type cell lines (TC32 and TC138) and two TP53 mutant cell lines (A673 and EWS502).
To genetically disrupt MDM2, we infected the Ewing sarcoma cell lines with MDM2 sgRNAs and then treated with the MDM2 inhibitor RG7388, which causes an up-regulation of MDM2 through a negative feedback mechanism in response to elevated p53 levels (Ding et al., 2013). TP53 wild-type cells infected with sgRNAs targeting MDM2 exhibited a weaker increase in MDM2 protein levels compared with cells infected with the control sgRNA, consistent with MDM2 knockout in a population of cells in the pool (Fig. 2 A). Given the disrupted p53–MDM2 axis in the TP53 mutant lines, there was no increase in MDM2 protein following RG7388 treatment of A673 and EWS502 cells, irrespective of infection with MDM2 or control sgRNAs (Fig. 2 A). To further validate the on-target activity of the sgRNAs, we infected the osteosarcoma cell line, SJSA-X, engineered to overexpress MDM4 in the context of endogenously elevated MDM2 levels (Wade et al., 2008), with sgRNAs targeting MDM2. Knockout was confirmed by Western blot (Fig. 2 B) and selectively impaired the viability of TP53 wild-type Ewing sarcoma and SJSA-X cells, with little to no effect on the TP53 mutant cell lines (Fig. 2 C), consistent with the CRISPR-Cas9 screening results.
Figure 2.
Validation of MDM2 and MDM4 as dependencies in TP53 wild-type Ewing sarcoma. (A) Western blots demonstrate abrogation of the observed increase in MDM2 protein levels upon RG7388 treatment (1 µM; 4 h) in TP53 wild-type cell lines TC32 and TC138 cells infected with sgRNAs targeting MDM2 compared with a nontargeting control sgRNA and no response to RG7388 treatment in TP53 mutated cell lines A673 and EWS502. (B) Western blot demonstrating decreased protein levels of MDM2 with sgRNAs targeting MDM2 compared with a control guide in the SJSA-X cell line. (C) Relative viability of Ewing sarcoma and SJSA-X cells infected with sgRNAs targeting MDM2 compared with control sgRNAs 14 d after infection. Each data point shows the mean of eight replicates, and data are plotted as mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. (D) Western blots showing decreased protein levels of MDM4 after infection with sgRNAs targeting MDM4 compared with control sgRNAs. (E) Western blot demonstrating decreased protein levels of MDM4 with sgRNAs targeting MDM4 compared with control guides in the SJSA-X cell line. (F) Relative viability of Ewing sarcoma and SJSA-X cells infected with sgRNAs targeting MDM4 or control sgRNAs 14 d after infection. Each data point shows the mean of eight replicates, and data are plotted as mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. Significance was calculated by paired, two-tailed t test: n.s., not significant for P > 0.05; *, P ≤ 0.05; **, for P ≤ 0.01; ***, P ≤ 0.001).
Similarly, we next disrupted MDM4 by CRISPR-Cas9 in Ewing sarcoma cell lines (Fig. 2 D) and demonstrated MDM4 knockout in two TP53 mutant cell lines and SJSA-X cells (Fig. 2 E). As predicted by the screen, and consistent with our MDM2 findings (Fig. 2 C), loss of MDM4 impaired the viability of TP53 wild-type Ewing sarcoma cell lines in a strikingly selective fashion, while SJSA-X, a cell line engineered to overexpress MDM4, does not depend on the gene as previously reported (Wade et al., 2008; Fig. 2 F).
Chemical inhibition of MDM2/MDM4 reduces viability of TP53 wild-type Ewing sarcoma
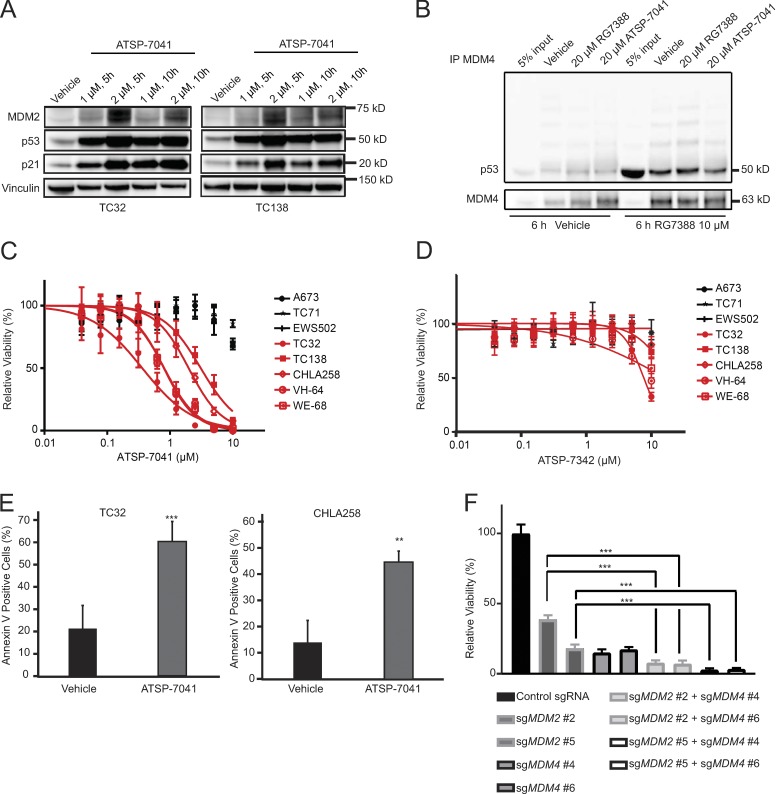
ATSP-7041 is a stapled peptide, dual inhibitor of MDM2 and MDM4 (Chang et al., 2013). Modeled after the p53 transactivation α-helix, stapled p53 peptides engage the p53 binding domain of MDM2 and MDM4 with high affinity and respectively block the degradation and sequestration of p53 (Bernal et al., 2010).
We observed concentration-dependent increases in MDM2, p53, and p21 protein levels after ATSP-7041 treatment of TP53 wild-type Ewing sarcoma cell lines (Fig. 3 A). To verify the mechanism of action of ATSP-7041 in Ewing sarcoma, we performed immunoprecipitation experiments. TC32 cells were pretreated with the MDM2 inhibitor RG7388 to increase p53 protein levels and then cell lysates were treated with vehicle, ATSP-7041 or RG7388, followed by MDM4 immunoprecipitation and p53 and MDM4 Western blot analysis. Whereas RG7388 was unable to dissociate the inhibitory p53–MDM4 complexes formed in response to selective inhibition of MDM2 in Ewing sarcoma cells, ATSP-7041 exposure decreased the level of p53–MDM4 interaction (Fig. 3 B), consistent with the mechanism reported for other cancer cell lines (Bernal et al., 2010; Chang et al., 2013).
Figure 3.
ATSP-7041 reactivates the p53 pathway to induce cell death in TP53 wild-type Ewing sarcoma cell lines. (A) Western blots showing increased protein levels of MDM2, p53, and p21 after ATSP-7041 treatment at the indicated time and concentrations in TP53 wild-type Ewing sarcoma cell lines. (B). Immunoprecipitation experiments show partial disruption of p53–MDM4 complex after treating cellular lysates with ATSP-7041, while RG7388 does not interrupt binding. TC32 cells were treated with RG7388 (last four lanes) to increase p53 protein levels. (C) Ewing sarcoma cells were treated with ATSP-7041 for 3 d. TP53 wild-type Ewing sarcoma cell lines are shown in red. TP53 mutated Ewing sarcoma cell lines are shown in black. Values are normalized to vehicle control. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. (D) Ewing sarcoma cells were treated with negative control peptide ATSP-7342 for 3 d. TP53 wild-type Ewing sarcoma cell lines are shown in red. TP53 mutant Ewing sarcoma cell lines are shown in black. Values are normalized to vehicle control. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. (E) 2-d treatment with ATSP-7041 triggers cell death in TC32 (treated with 2 µM) and CHLA258 (treated with 4 µM) cell lines, as measured by Annexin V staining. Data points represent the mean of five replicates of two experiments and error bars are mean ± standard deviation. (F) Viability effect of dual CRISPR-Cas9 knockout of MDM2 and MDM4 in TC32 cells. Cells were infected with sgRNAs targeting MDM2 and selected with puromycin and sgRNAs targeting MDM4 and selected with blasticidin. Relative viability of eight replicates is shown 11 d post-infection. The experiment was performed twice and data points of one representative experiment are shown. Significance was calculated by paired, two-tailed t test: **, P ≤ 0.01; ***, P ≤ 0.001.
ATSP-7041 selectively reduced the viability of five TP53 wild-type Ewing sarcoma cell lines at low micromolar concentrations (Fig. 3 C), whereas TP53 mutated Ewing sarcoma cell lines were resistant, mirroring the results of our genetic perturbation studies. ATSP-7342, a negative control stapled peptide that bears an inactivating F19A point mutation and thus exhibits impaired MDM2/MDM4 binding activity (Chang et al., 2013), was essentially ineffective at these concentrations (Fig. 3 D). Annexin V staining likewise demonstrated induction of cell death at the corresponding concentrations of ATSP-7041 in TP53 wild-type Ewing sarcoma cell lines (Fig. 3 E).
To study a genetic model of ATSP-7041 that allows for the comparison of MDM2/4 dual inhibition to MDM2 inhibition alone, we performed dual-knockout experiments with CRISPR-Cas9 constructs targeting MDM2 and MDM4. TC32 cells were infected with CRISPR-Cas9 constructs either targeting MDM2 alone, MDM4 alone, or one construct each for targeting MDM2 and MDM4 simultaneously. Overall, knockout of both MDM2 and MDM4 decreased viability more effectively than loss of either alone at this time point (Fig. 3 F), validating the potential therapeutic benefit of dual inhibition of MDM2 and MDM4 over inhibiting MDM2 alone.
ATSP-7041 reactivates the p53 transcriptional pathway and suppresses Ewing sarcoma growth in vivo
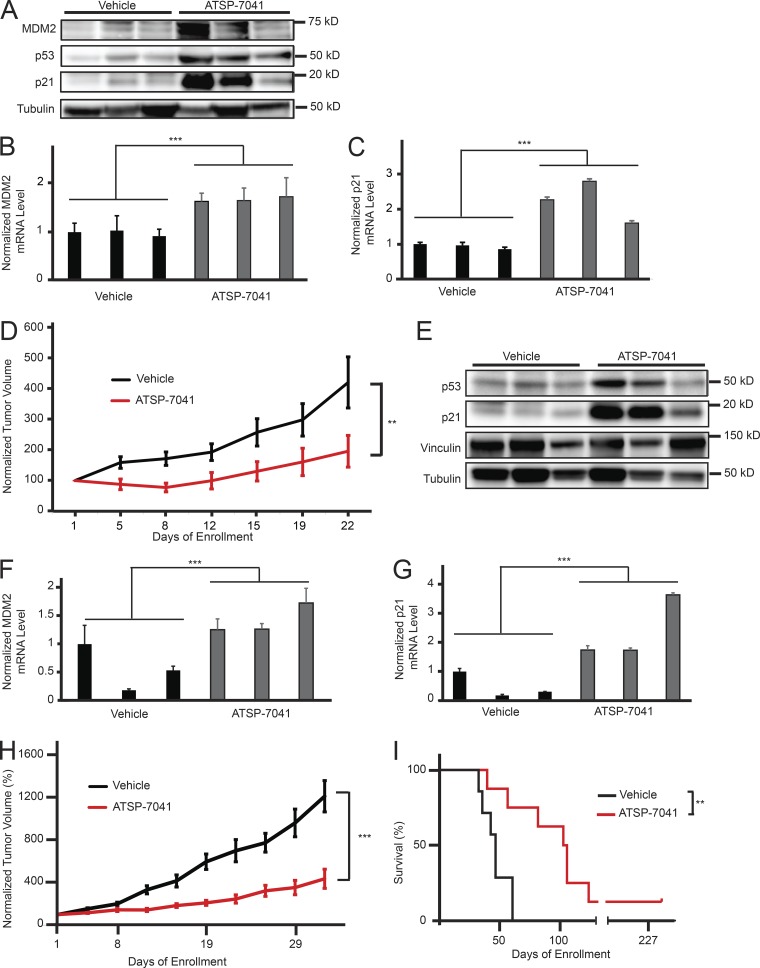
We next sought to evaluate the activity of ATSP-7041 Ewing sarcoma in vivo. TC32 Ewing sarcoma cells were implanted subcutaneously in nude mice. Following tumor engraftment with tumor volumes of >100 mm3, mice were dosed with 30 mg/kg IV ATSP-7041 or vehicle every other day for 6 d. 8 h after the last dose, mice were sacrificed and tumors were collected for comparative analysis of p53 pathway reactivation. We found that ATSP-7041 treatment led to both an increase in MDM2, p53, and p21 protein levels (Fig. 4 A) and MDM2 and p21 mRNA levels (Fig. 4, B and C) in tumor tissue. After validating the on-mechanism activity of ATSP-7041 in vivo, we next sought to assess anti-tumor efficacy. Treatment of TC32 Ewing sarcoma xenografted mice with 30 mg/kg ATSP-7041 or vehicle IV every other day for 20 d significantly suppressed tumor growth, sustaining a reduction in tumor progression throughout the 22-d evaluation period (Fig. 4 D).
Figure 4.
ATSP-7041 shows anti-tumor efficacy in Ewing sarcoma models in vivo. (A) Western blot showing an increase of MDM2, p53, and p21 protein levels in TC32 xenograft tumor tissues after ATSP-7041 treatment in vivo. After tumor engraftment, mice were treated with three doses of 30 mg/kg q.o.d. ATSP-7041 or vehicle and sacrificed 8 h after the last dose. Each lane represents an individual mouse tumor. (B) Quantitative PCR showing comparative MDM2 mRNA levels with vehicle (black) or ATSP-7041 (gray) treatment of TC32 xenograft cells in vivo. Values were normalized to the first vehicle-treated sample. Each bar represents an individual mouse tumor; error bars represent standard deviation of three technical replicates. Significance was calculated by paired, two-tailed t test: ***, P ≤ 0.001. (C) Quantitative PCR showing comparative p21 mRNA levels with vehicle (black) or ATSP-7041 (gray) treatment of TC32 xenograft cells in vivo. Values were normalized to the first vehicle-treated sample. Each bar represents an individual mouse tumor; error bars represent standard deviation of three technical replicates. Significance was calculated by paired, two-tailed t test: ***, P ≤ 0.001. (D) Normalized average tumor volume from mice bearing TC32 xenograft tumors treated with 30 mg/kg ATSP-7041 q.o.d. (red, n = 8), or vehicle q.o.d. (black, n = 7). Mice were treated with 10 doses. Tumor volume from each mouse was normalized to the tumor volume at the day of enrollment. Error bars represent standard deviation. Significance was calculated by two-way ANOVA analysis: **, P ≤ 0.01. (E) Western blot showing an increase of MDM2, p53, and p21 protein levels in PDX tumor tissues after ATSP-7041 treatment in vivo. After tumor engraftment, mice were treated with three doses of 30 mg/kg q.o.d. ATSP-7041 or vehicle and sacrificed 8 h after the last dose. Each lane represents an individual mouse tumor. (F) Quantitative PCR showing comparative MDM2 mRNA levels with vehicle (black) or ATSP-7041 (gray) treatment of PDX cells in vivo. Values were normalized to the first vehicle-treated sample. Each bar represents an individual mouse tumor; error bars represent standard deviation of three technical replicates. Significance was calculated by paired, two-tailed t test: ***, P ≤ 0.001. (G) Quantitative PCR showing comparative p21 mRNA levels with vehicle (black) or ATSP-7041 (gray) treatment of PDX cells in vivo. Values were normalized to the first vehicle-treated sample. Each bar represents an individual mouse tumor; error bars represent standard deviation of three technical replicates. Significance was calculated by paired, two-tailed t test: ***, P ≤ 0.001. (H) Normalized average tumor volume from mice bearing PDX tumors treated with 30 mg/kg ATSP-7041 q.o.d. (red, n = 8), or vehicle q.o.d. (black, n = 7). Mice were treated with 10 doses. Tumor volume for each mouse was normalized to the tumor volume at the day of enrollment. Error bars represent standard deviation. Significance was calculated by two-way ANOVA analysis: ***, P ≤ 0.001. (I) Survival of mice bearing PDX tumors. One mouse treated with ATSP-7041 had complete tumor regression without recurrence over the observed time frame. Significance was calculated by Log-rank (Mantel-Cox) test: **, P ≤ 0.01.
To test ATSP-7041 in a model that more closely recapitulates patient tumors, we studied a patient-derived xenograft (PDX) model of Ewing sarcoma (HSJD-ES-002), which was derived from a tumor resected from the fibula of a 12-yr-old patient at diagnosis (Ordóñez et al., 2015). In vivo treatment of mice bearing HSJD-ES-002 tumors with ATSP-7041 increased intratumoral p53 and p21 protein levels (Fig. 4 E) and increased MDM2 and p21 mRNA levels (Fig. 4, F and G). Tumor growth was slowed significantly after 10 doses of ATSP-7041 (Fig. 4 H). The survival of ATSP-7041–treated mice was significantly extended, and remarkably, one mouse was cured of disease, showing complete tumor regression without recurrence over the observed time frame of 227 d (Fig. 4 I).
Genetic disruption of USP7 and PPM1D is selectively cytotoxic to Ewing sarcoma lines bearing wild-type TP53
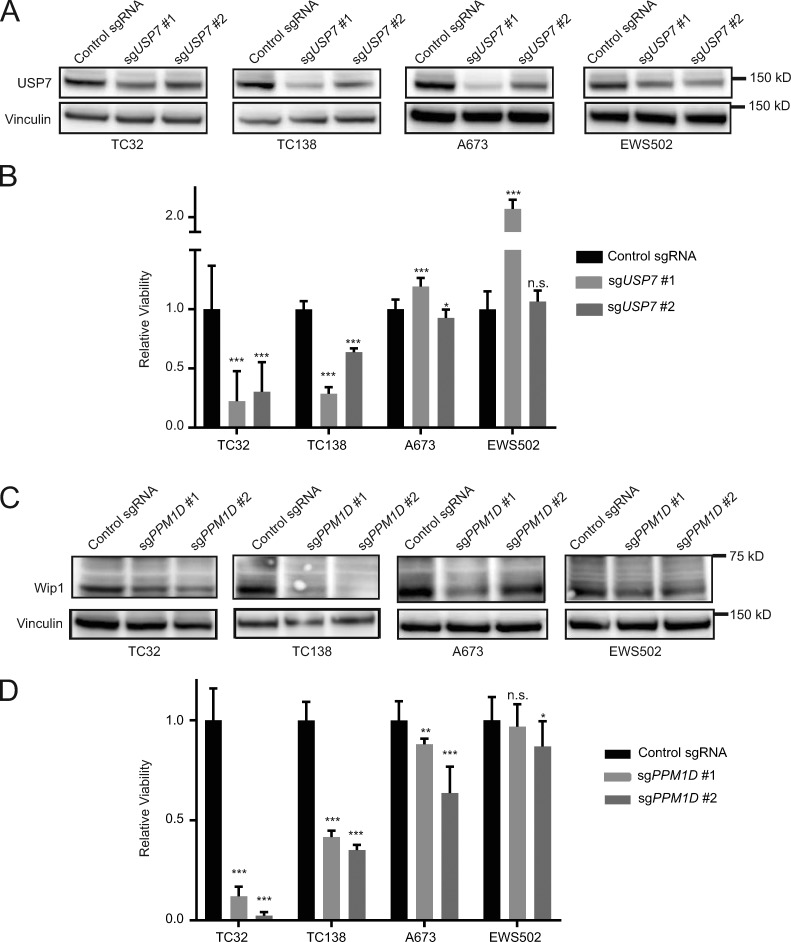
After validating MDM2 and MDM4 as gene dependencies in TP53 wild-type Ewing sarcoma, we next evaluated USP7 and PPM1D. We disrupted USP7 by CRISPR-Cas9 in TC32, TC138, A673, and EWS502 cells (Fig. 5 A) and observed reduced viability of TP53 wild-type compared with mutant Ewing sarcoma cell lines (Fig. 5 B).
Figure 5.
Validation of PPM1D and USP7 as dependencies in TP53 wild-type Ewing sarcoma. (A) Western blots showing decreased protein levels of USP7 after infection with sgRNAs targeting USP7 compared with control sgRNAs. (B) Relative viability of Ewing sarcoma cells infected with sgRNAs targeting USP7 or control sgRNAs 14 d after infection. Each data point shows the mean of eight replicates; data are plotted as mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. (C) Western blots showing decreased protein levels of Wip1 after infection with sgRNAs targeting PPM1D compared with control sgRNAs. (D) Relative viability of Ewing sarcoma cells infected with sgRNAs targeting PPM1D or control sgRNAs 14 d after infection. Each data point shows the mean of eight replicates, and data are plotted as mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. Significance was calculated by paired, two-tailed t test: not significant (n.s.) for P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Similarly, the disruption of PPM1D by CRISPR-Cas9 in TC32, TC138, A673, and EWS502 led to reduced protein levels of Wip1 (Fig. 5 C) and a decrease in viability of the TP53 wild-type compared with the TP53 mutant Ewing sarcoma cell lines (Fig. 5 D).
Chemical inhibition of USP7 and Wip1 impairs the viability of TP53 wild-type Ewing sarcoma cells
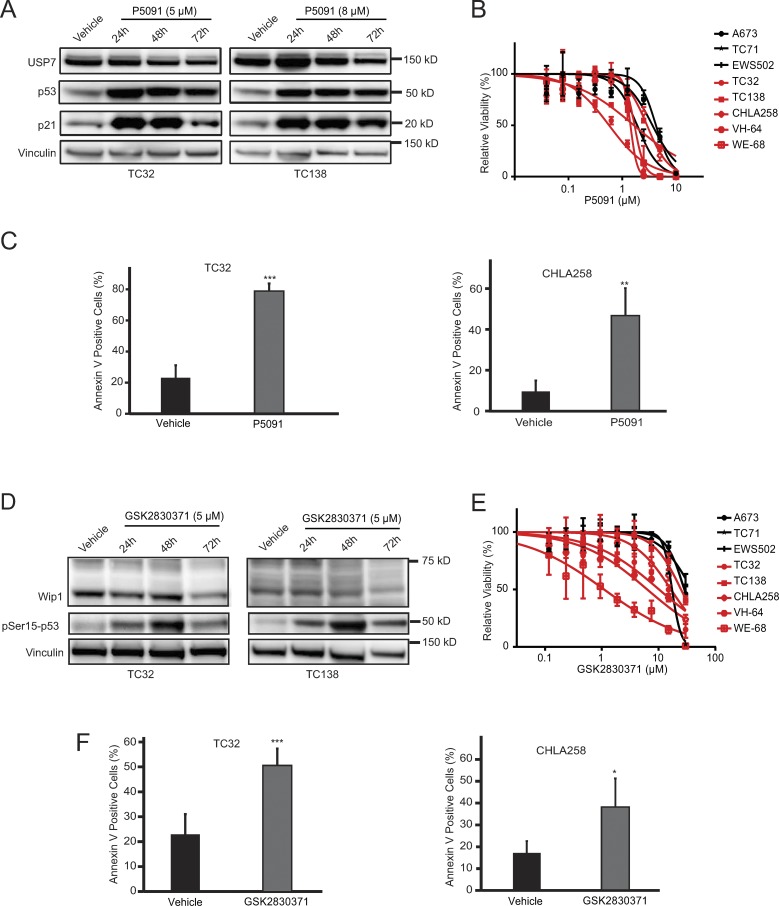
Given the selective effects of genetic disruption of USP7 and PPM1D on the viability of wild-type TP53 Ewing sarcoma, we tested the pharmacologic activities of their respective small inhibitors, P5091 (Chauhan et al., 2012) and GSK2830371 (Gilmartin et al., 2014). P5091 increased p53 and p21 protein levels in a time-dependent fashion in the wild-type TP53 cells, TC32 and TC138 (Fig. 6 A). The inhibitor appeared to be relatively more cytotoxic to a subset of TP53 wild-type Ewing sarcoma lines compared with TP53 mutant cells (Fig. 6 B). Annexin V staining likewise demonstrated P5091-induced cell death in TP53 wild-type Ewing sarcoma cell lines (Fig. 6 C).
Figure 6.
GSK2830371 and P5091 reduce viability and induce cell death in TP53 wild-type Ewing sarcoma cell lines. (A) Western blots showing increase in p53 and p21 protein levels with P5091 treatment in TP53 wild-type Ewing sarcoma cell lines. (B) Ewing sarcoma cells were treated with P5091 for 3 d. TP53 wild-type Ewing sarcoma cell lines are shown in red; TP53 mutant Ewing sarcoma cell lines are shown in black. Values were normalized to vehicle controls. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. (C) 2-d treatment with P5091 triggers cell death in TC32 (treated with 6.5 µM) and CHLA258 cells (treated with 8 µM) as measured by Annexin V staining. Data points represent the mean of five replicates of two experiments, and error bars are mean ± standard deviation. (D) Western blots showing decreased protein levels of Wip1 and increased pSer15-p53 upon GSK2830371 treatment at the indicated time and concentration. (E) Ewing sarcoma cells were treated with GSK2830371 for 3 d. TP53 wild-type Ewing sarcoma cell lines are shown in red. TP53 mutated Ewing sarcoma cell lines are shown in black. Values were normalized to vehicle controls. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice, and data points of one representative experiment are shown. (F) 3-d treatment with GSK2830371 triggers cell death in TC32 and CHLA258 (both treated with 15 µM) cell lines, as measured by Annexin V staining. Data points represent the mean of five replicates of two experiments, and error bars are mean values ± standard deviation. Significance was calculated by paired, two-tailed t test: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
GSK2830371 reduced the protein levels of Wip1 in a time-dependent manner and triggered a surge in phosphorylation of Serine 15 of p53, the primary p53 dephosphorylation target site of Wip1 (Fig. 6 D). There was a notable increase in susceptibility of TP53 wild-type Ewing sarcoma cell lines to micromolar concentrations of GSK2830371, as compared with the TP53 mutated cell lines (Fig. 6 E). GSK2830371-induced cell death in TP53 wild-type Ewing sarcoma cell lines was likewise observed by Annexin V staining (Fig. 6 F).
ATSP-7041 synergizes with chemical inhibition of USP7 and Wip1
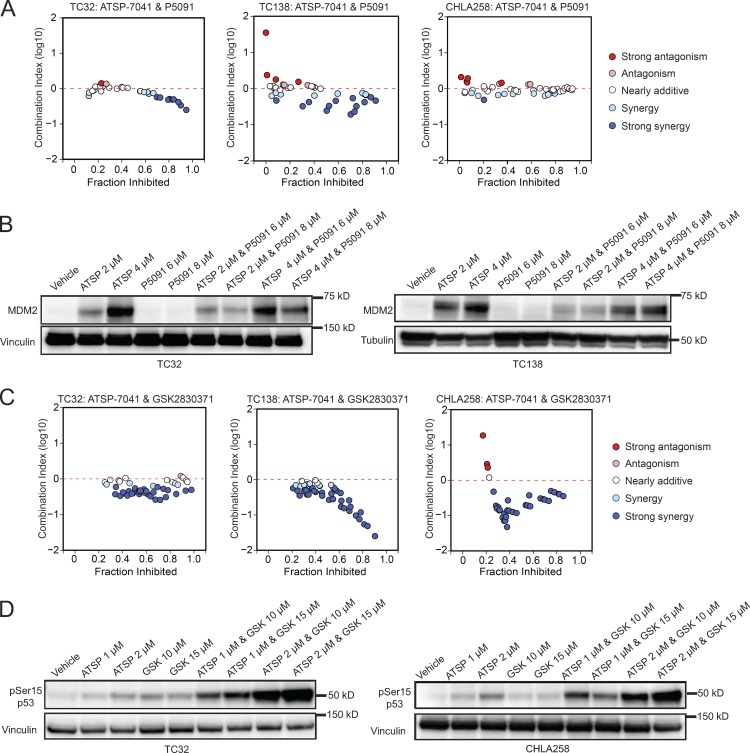
Given the promising single-agent activity of GSK2830371 and P5091 in reactivating the p53 pathway, we next evaluated the therapeutic potential of combining these molecules with ATSP-7041. Because MDM2, MDM4, USP7, and PPM1D all scored as top dependencies in TP53 wild-type Ewing sarcoma, we reasoned that chemically inhibiting these proteins in combination could provide the most effective mechanism to trigger p53-mediated cell death in Ewing sarcoma. Indeed, we found that the combination of ATSP-7041 with P5091 exhibited synergy in TC32 and TC138 and additivity in CHLA258 cells, as assessed by the Chou-Talalay combination index for Loewe additivity model (Fig. 7 A; Chou, 2006, 2010). We hypothesized that the heightened therapeutic response could be due to synergistic action on the p53 axis. Western blot analysis revealed that P5091 decreased the level of MDM2 protein that is otherwise induced by ATSP-7041 as a result of the surge in p53 and counter–up-regulation of MDM2 (Fig. 7 B). These data suggest that pharmacologic blockade of USP7 may counteract the natural up-regulation of MDM2 in response to elevated p53 levels, thereby maximizing the p53 response to ATSP-7041.
Figure 7.
ATSP-7041 synergizes with GSK2830371 and P5091. (A) CI plots for the combination of ATSP-7041 with P5091 in TC32, TC138, and CHLA258 cells after 5 d of treatment. (B) Western blots showing decreased MDM2 protein levels in TC32 and TC138 cells treated with a combination of ATSP-7041 and P5091 compared with ATSP-7041 treatment alone. Cells were treated at the indicated concentrations for 2 d (ATSP, ATSP-7041). (C) CI plots for the combination of ATSP-7041 with GSK2830371 in TC32, TC138, and CHLA258 cells after 3 d of treatment. (D) Western blots showing increased phospho-Serine15-p53 protein levels with combination treatment of ATSP-7041 and GSK2830371 in TC32 and CHLA258 cells. Cells were treated at the indicated concentrations for 2 d (ATSP, ATSP-7041; GSK, GSK2830371).
The combination of ATSP-7041 with GSK2830371 exhibited strong synergy across a broad concentration range in TC32, TC138, and CHLA258 cells (Fig. 7 C). Western blot assays revealed that the combination of ATSP-7041 and GSK2830371 increased the phosphorylation of p53 at Serine 15 in two Ewing sarcoma cell lines (Fig. 7 D). These data suggest that phosphorylation of p53 may underlie the synergism of these two drugs and provides mechanistic evidence that Wip1 acts as a phosphatase of p53 in Ewing sarcoma.
After identifying MDM2, MDM4, USP7, and PPM1D as co-dependencies in the CRISPR-Cas9 screen, we tested the combination of pharmacologic inhibitors targeting these proteins. We observed synergistic cytotoxicity, suggesting that a subset of correlated genetic dependencies may indeed predict the synergy of inhibitor combinations.
ATSP-7041 synergizes with standard of care chemotherapeutics in Ewing sarcoma
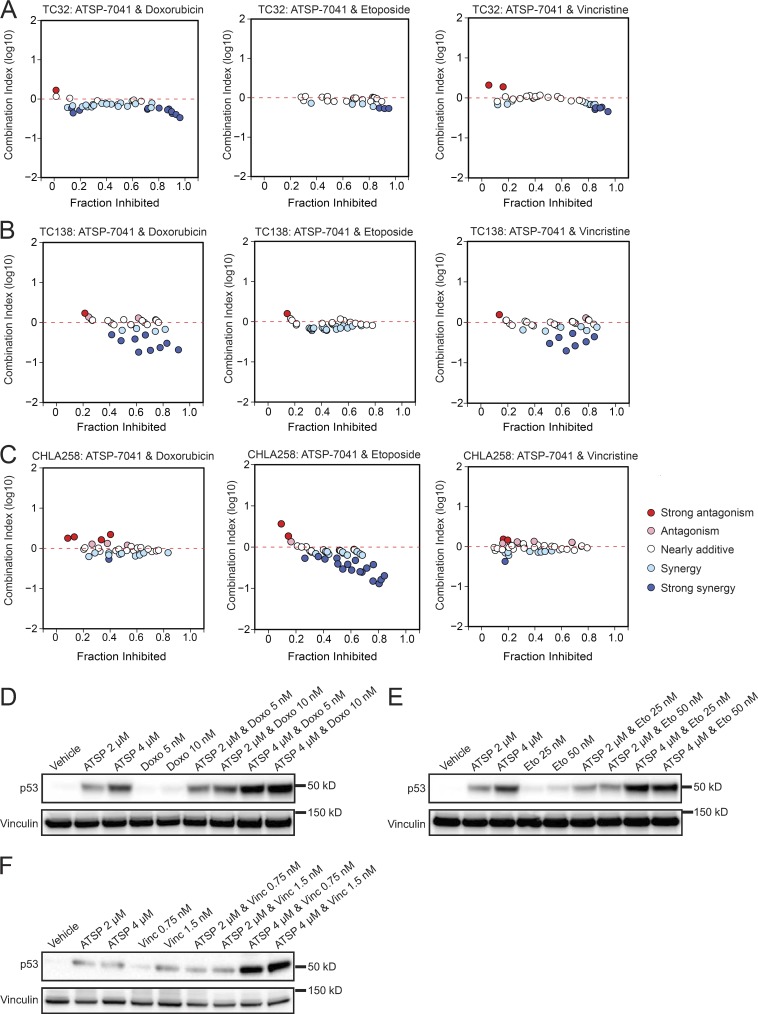
Since an MDM2/4 dual inhibitor stapled peptide is currently in Phase 1 and Phase 2 testing in adult cancers bearing wild-type TP53, we sought to test for synergistic activity of ATSP-7041 with approved treatment regimens for Ewing sarcoma. Thus, we combined ATSP-7041 with doxorubicin, etoposide, or vincristine, three drugs used clinically in the treatment of Ewing sarcoma (Gaspar et al., 2015). All drug combinations demonstrated additivity or synergy at several concentrations, as assessed by the Chou-Talalay combination index for Loewe additivity model (Fig. 8, A–C).
Figure 8.
ATSP-7041 synergizes with chemotherapy agents. (A–C) CI plots for the combination of ATSP-7041 with doxorubicin, etoposide, and vincristine after 3-d treatment of (A) TC32, (B) TC138, and (C) CHLA258 cells. (D) Western blots showing increased p53 protein levels in TC32 cells treat with combinations of ATSP-7041 and doxorubicin. Cells were treated at indicated concentrations for 2 d (ATSP, ATSP-7041; Doxo, doxorubicin). (E) Western blots showing increased p53 protein levels in TC32 cells treated with combinations of ATSP-7041 and etoposide. Cells were treated at the indicated concentrations for 2 d (ATSP, ATSP-7041; Eto, etoposide). (F) Western blots showing increased p53 protein levels in TC32 cells treated with combinations of ATSP-7041 and vincristine. Cells were treated at the indicated concentrations for 2 d (ATSP, ATSP-7041; Vinc, vincristine).
As cytotoxic chemotherapeutic agents are well known to induce pro-apoptotic signals in cancer cells, we investigated the effect of combining ATSP-7041 and chemotherapy agents on p53 protein levels. Combination treatments of ATSP-7041 with etoposide, doxorubicin, or vincristine greatly increased p53 protein levels in Ewing sarcoma cells, suggesting that the observed synergy is due to combinatorial action on the p53 signaling axis (Fig. 8, D–F).
These data support further consideration of adding a stapled peptide dual inhibitor of MDM2/MDM4 to standard chemotherapy regimens in patients with TP53 wild-type Ewing sarcoma.
Loss of TP53 rescues the effects of MDM2, MDM4, PPM1D, and USP7 inhibition
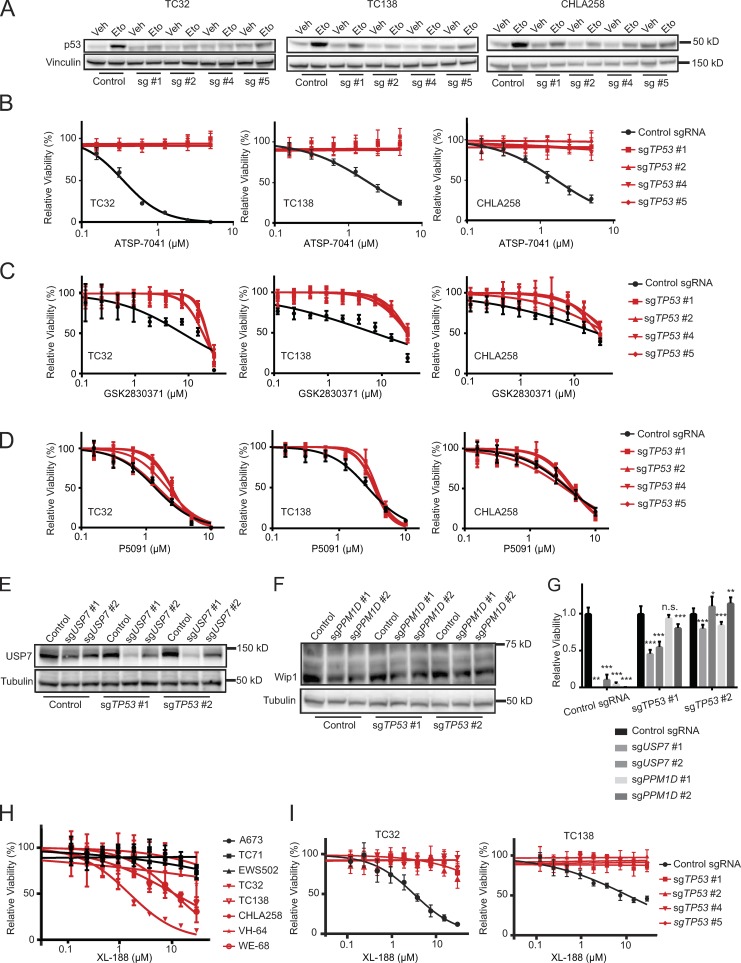
While these data suggest that TP53 wild-type Ewing sarcoma cancer cell lines are more sensitive to loss of MDM2, MDM4, PPM1D, and USP7 than TP53 mutated ones, we next generated isogenic cell lines with TP53 loss to more definitively support this hypothesis. Three TP53 wild-type cell lines were infected with CRISPR-Cas9 constructs targeting TP53, and loss of TP53 was demonstrated by diminished increases of p53 protein levels in response to etoposide treatment (Fig. 9 A). Treatment of TP53 knockout cells revealed that loss of TP53 fully rescues the cytotoxic effect of ATSP-7041, indicating on-target activity of the drug (Fig. 9 B) and dependency on intact p53 for the response to MDM2/MDM4 inhibition. Similarly, the Wip1 inhibitor GSK2830371 was less effective in TP53 knockout cells than control cells, which indicates on-target activity of GSK2830371 (Fig. 9 C).
Figure 9.
Loss of PPM1D and USP7 is rescued by concurrent TP53 loss. (A) Western blots showing attenuated increase of p53 protein levels in TC32, TC138, and CHLA258 cells infected with sgRNAs targeting TP53 after etoposide treatment (Control, control sgRNA; sg #1, sgTP53 #1; sg #2, sgTP53 #2; sg #4, sgTP53 #4; sg #5, sgTP53 #5). Cells were treated with vehicle or 50 μM etoposide for one hour (Veh, vehicle; Eto, etoposide). (B) TP53 knockout cells were treated with ATSP-7041 for 3 d. Values were normalized to vehicle controls. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. (C) TP53 knockout cells were treated with GSK2830371 for 3 d. Values were normalized to vehicle controls. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice, and data points of one representative experiment are shown. (D) TP53 knockout cells were treated with P5091 for 3 d. Values were normalized to vehicle controls. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice, and data points of one representative experiment are shown. (E) Western blots showing decreased protein levels of USP7 after infection with sgRNAs targeting USP7 in TC32 TP53 knockout cells. (F) Western blots showing decreased protein levels of Wip1 after infection with sgRNAs targeting PPM1D in TC32 TP53 knockout cells. (G) Relative viability of TC32 TP53 knockout cells infected with sgRNAs targeting USP7 or PPM1D or control sgRNAs 14 d after infection. Each data point shows the mean of eight replicates, and data are plotted as mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown. Significance was calculated by paired, two-tailed t test: not significant (n.s.) for P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (H) Ewing sarcoma cells were treated with XL-188 for 3 d. TP53 wild-type Ewing sarcoma cell lines are shown in red. TP53 mutated Ewing sarcoma cell lines are shown in black. Values were normalized to vehicle controls. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice, and data points of one representative experiment are shown. (I) TP53 knockout cells were treated with XL-188 for 3 d. Values were normalized to vehicle controls. Each data point shows the mean of eight replicates; error bars are mean values ± standard deviation. The experiment was performed twice and data points of one representative experiment are shown.
TP53 knockout, however, did not rescue cells from the effects of P5091, suggestive of either a p53 independent mechanism or the possibility of an off-target effect(s) of the molecule (Fig. 9 D). To address this question, we undertook both genetic and chemical analyses. First, we infected TP53 knockout cells with CRISPR-Cas9 constructs targeting USP7. The concurrent loss of TP53 effectively rescued the cytotoxic effect of USP7 knockout, as also observed for PPM1D knockout (Fig. 9, E–G).
Several new USP7 inhibitors have been described in recent publications. We chose the highly selective XL-188 molecule (Lamberto et al., 2017) to examine the effect of a more refined USP7 inhibitor on Ewing sarcoma susceptibility. XL-188 reduced viability predominantly in TP53 wild-type Ewing sarcoma cell lines, with an especially robust effect observed in TC32 cells (Fig. 9 H). Strikingly, TP53 knockout completely reversed the cytotoxic effect of XL-188 (Fig. 9 I), supporting the requirement of functional p53 for the observed response to USP7 inhibition in Ewing sarcoma. Collectively, these data validate the hypothesis that in Ewing sarcoma, MDM2, MDM4, PPM1D, and USP7 dependencies are mediated by functional p53 and exert their cytotoxic effects, singly and in combination, by reactivating the p53 tumor suppressor pathway.
Discussion
TP53 is a potent tumor suppressor gene critical to cellular homeostasis (Lane, 1992). Loss of p53, either by genetic deletion, mutation, or protein interaction–based suppression, is a key oncogenic event in tumorigenesis (Hanahan and Weinberg, 2011). Whereas TP53 is mutated in ∼50% of human tumors (Leroy et al., 2014), a large subset of pediatric cancers exhibit a low frequency of TP53 mutations (Malkin et al., 1994; Kato et al., 1996; Hendy et al., 2009; Hof et al., 2011; Ognjanovic et al., 2012), implicating negative regulation of p53 through protein interactions as a pathogenic mechanism. Indeed, several recent studies indicate that the TP53 mutation rate in Ewing sarcoma is very low (Brohl et al., 2014; Crompton et al., 2014; Tirode et al., 2014). Given the urgent need for new therapeutic approaches to treat Ewing sarcoma, we sought to identify drug targets in the context of TP53 wild-type disease.
To discover druggable candidates in TP53 wild-type Ewing sarcoma, we mined the data of a previously published CRISPR-Cas9 screen (Aguirre et al., 2016). The large number of heterogeneous cell lines with diverse molecular features allows for analysis of disease-specific dependencies or dependencies correlated with specific gene mutations. Here, we analyzed the dataset to study the differences between TP53 wild-type and TP53 mutated cell lines. As expected, cell lines with functional p53 proliferate faster upon TP53 knockout (leading to a positive dependency score), while cell lines with mutated TP53 show little to no effect. We leveraged data from 33 cancer cell lines and identified MDM2, MDM4, PPM1D, and USP7 as anti-correlated with TP53 dependency scores in Ewing sarcoma and across all cancer cell lines in the dataset. A similar approach was recently used in the context of acute myeloid leukemia (Wang et al., 2017).
Each of the proteins, MDM2, MDM4, Wip1, and USP7, has been implicated in p53 regulation. MDM2 is an E3 ubiquitin ligase that targets p53 for degradation and is induced by p53 in a negative feedback loop. MDM2-deficient murine embryos are nonviable, a phenotype that can be rescued by concurrent TP53 loss (Wade et al., 2013). In cancer, MDM2 can act as an oncogene whose overexpression promotes malignancy by inhibiting the tumor suppressor function of p53 (Wade et al., 2013). Indeed, MDM2 was found to be amplified in Ewing sarcoma patient samples, highlighting the importance of the gene in this disease (Ladanyi et al., 1995). MDM4 is a structural homologue of MDM2 that inhibits p53 by binding and blocking its transactivation domain. Similar to MDM2, MDM4 deficiency is embryonic lethal in mice and can be rescued by TP53 loss (Wade et al., 2013). The MDM4 gene is located on chromosome 1q, which is found to have copy number gains in a subset of Ewing sarcoma patient samples (Crompton et al., 2014). One study reported that 50% of Ewing sarcoma tumors contained greater than threefold amplification of MDM4 (Ito et al., 2011). PPM1D encodes the phosphatase wild-type p53–induced phosphatase 1 (Wip1) that has several functions as an anti-apoptotic regulator, including dephosphorylation of p53 at serine 15 and deactivation. Wip1 has also been suggested to target ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3 related (ATR), checkpoint kinase 1 (CHK1), and checkpoint kinase 2 (CHK2), as well as MDM2 and MDM4 (Lu et al., 2007; Zhu and Bulavin, 2012). High Wip1 levels or PPM1D amplification have been found to correlate with poor prognosis in a variety of cancer types (Saito-Ohara et al., 2003; Castellino et al., 2008; Tan et al., 2009; Lambros et al., 2010; Ma et al., 2014; Peng et al., 2014; Xu et al., 2016; Zhao et al., 2016). USP7 is a deubiquitinating enzyme involved in a variety of cellular processes and is implicated in the regulation of MDM2, MDM4, and p53, as well as several other targets (Nicholson and Suresh Kumar, 2011).
Prior studies have targeted MDM2 in Ewing sarcoma and found anti-tumor efficacy in vitro and in vivo (Pishas et al., 2011; Sonnemann et al., 2011; Carol et al., 2013). MDM2 inhibition by RG7112 and RG7388 is being investigated in clinical trials as single agents or combination treatments for several malignancies. While early clinical trials testing RG7112 in patients with leukemia (Andreeff et al., 2016) and advanced solid tumors (Patnaik et al., 2015) have shown promise, coexpression of MDM4 can cause resistance (Hu et al., 2006; Patton et al., 2006; Wade et al., 2008; Chapeau et al., 2017). In addition, our finding that alternative negative regulators can coexist to thwart wild-type TP53 signaling suggests that inhibitory strategies beyond selective MDM2 targeting may be required to achieve maximal reactivation of p53. The notion of dual targeting of MDM2 and MDM4 is further supported by dual-knockout experiments of MDM2 and MDM4 presented here, where we demonstrate that loss of both MDM2 and MDM4 decreases viability of Ewing sarcoma cells more effectively than loss of either target alone. Of note, these latter experiments must be interpreted within the technical limitations of dual knockout of two strong dependencies, where we were unable to obtain adequate protein for confirmation of equivalent knockout across these conditions because of the rapid onset of cell death.
Because MDM4 overexpression has been established as a mechanism of resistance to MDM2 inhibitors, the development of MDM4 inhibitors has been of special interest (Bernal et al., 2010). Whereas a putative small molecule MDM4 inhibitor, XI-006, has been reported previously (Wang et al., 2011), TP53 mutational status was not a biomarker for XI-006 sensitivity in Ewing sarcoma and breast cancer models (Pishas et al., 2015), suggesting potential off-target effects. ATSP-7041 is a mechanistically validated stapled peptide inhibitor of both MDM2 and MDM4 (Chang et al., 2013; Wachter et al., 2017). The ATSP-7041 derivative, ALRN-6924, is the first clinical grade stapled peptide to target intracellular protein interactions in human cancer, prompting its advancement to clinical testing. ALRN-6924 is currently in phase 1/2 evaluation for TP53 wild-type solid tumors and lymphomas in adults (NCT02264613) and in phase 1 evaluation for acute myeloid leukemia and for myelodysplastic syndrome (NCT02909972). Thus, stapled p53 peptides offer the unique opportunity to target two of the highest scoring dependencies in TP53 wild-type Ewing sarcoma cell lines simultaneously. We have shown that both MDM2 and MDM4 are strong dependencies in Ewing sarcoma. The therapeutic potential of ATSP-7041 is shown by its ability to decrease tumor growth in two different Ewing sarcoma models, including a study with an aggressive PDX model, where tumor progression was slowed in all mice, and one of eight mice achieved complete and sustained remission of disease. Therefore, we consider a dual MDM2/MDM4 inhibitor strategy to potentially be the most effective and rapidly translatable approach to reactivate p53 in patients with Ewing sarcoma.
Since single agent therapies rarely cure cancer, we sought to identify agents that could be used in combination with ATSP-7041. To evaluate the most readily translatable combinations, we combined ATSP-7041 with standard-of-care Ewing sarcoma cytotoxic chemotherapeutics and observed additivity or synergy with doxorubicin, etoposide, and vincristine. We have shown that the combination of ATSP-7041 with chemotherapeutic agents induces a stronger p53 response than these agents achieve individually. These mechanistic data support the addition of ATSP-7041 or other p53 reactivating agents to chemotherapy regimens. The synergistic induction of pro-apoptotic signals in cancer cells might allow for reduced doses of chemotherapy, thereby decreasing adverse effects. Our preclinical data support this notion and provide a compelling rationale for additional testing of ATSP-7041 with cytotoxic chemotherapy regimens in TP53 wild-type Ewing sarcoma models. Such studies will help prioritize combinations for testing in clinical trials.
Furthermore, the combination of ATSP-7041 with the Wip1 inhibitor GSK2830371, and the USP7 inhibitor P5091, strongly synergizes in TP53 wild-type Ewing sarcoma cell lines. These results support the hypothesis that inhibiting several members of the p53 regulatory network could be therapeutically beneficial (Pechackova et al., 2016; Sriraman et al., 2016). For example, in the case of ATSP-7041 and P5091 treatment, the addition of P5091 suppresses the counter up-regulation of MDM2. The combination of ATSP-7041 with GSK2830371 increases the level of pSer15 p53 more than with either molecule alone. Future studies will evaluate the efficacy of these combination treatments in vivo.
This study highlights the potential of genetic screening approaches to predict synergistic drug combinations, as MDM2, MDM4, PPM1D, and USP7 were highly correlated dependencies in our analysis of the CRISPR-Cas9 screen. Based on our findings that inhibitors of these targets have synergistic anti-cancer activity, we suggest that systematic analysis of correlated dependencies in genetic screens can inform new, effective, and potentially rapidly translatable drug combinations. Additionally, these results suggest that analysis of genetic screens for biomarker-specific dependencies (in this case TP53 status) can reveal proteins involved in the homeostasis of that biomarker (in this case MDM2, MDM4, Wip1, and USP7). This methodology may be applicable to a wide variety of clinical contexts.
USP7 has received increasing attention as a target in cancer and recent publications report new, selective inhibitors of the enzyme (Kategaya et al., 2017; Lamberto et al., 2017; Turnbull et al., 2017; Gavory et al., 2018). A series of p53-independent molecular targets and functions of USP7 have been proposed in different cancer types, such as the regulation of RAD18 in DNA damage response in hematologic malignancies (Agathanggelou et al., 2017), regulation of wingless-type MMTV integration site (Wnt) family signaling in colorectal cancer (An et al., 2017), Geminin deregulation in breast cancer (Hernández-Pérez et al., 2017), regulation of the Sonic Hedgehog pathway in medulloblastomas (Zhan et al., 2017), and stabilization of MYCN in neuroblastoma (Tavana et al., 2016). Here, we show that in Ewing sarcoma a key target of USP7 is the p53 pathway, as demonstrated by TP53 knockout experiments and the notable synergism of ATSP-7041 and P5091. The identified role of USP7 in Ewing sarcoma is distinct from that reported in other cancer types and warrants further exploration in this disease. Given the superior selectivity of the small-molecule inhibitor XL-188, as compared with previous generation molecules such as P5091, the clinical translation of our USP7 findings may be achievable.
Our results also indicate a p53 dependent mechanism for Wip1 in Ewing sarcoma. While the phosphatase has been shown to target a variety of proteins in different disease contexts, it appears to act through p53 in Ewing sarcoma, as indicated by TP53 knockout experiments and the synergistic elevation of phosphorylated p53 at serine 15 when GSK2830371 was combined with ATSP-7041. This finding advances our understanding of Wip1 activity in Ewing sarcoma and warrants further evaluation of Wip1 as a drug target in this disease.
In summary, we used CRISPR-Cas9 screening data to identify dependencies specific for TP53 wild-type cancers, including Ewing sarcoma, and discovered the four p53 regulators MDM2, MDM4, PPM1D, and USP7 as top hits. Validation of these targets using genetic and pharmacologic approaches confirmed their dependencies in Ewing sarcoma via a p53-dependent mechanism of action. We further demonstrated the in vivo activity of ATSP-7041 in two mouse models of Ewing sarcoma and highlight potential synergistic combinations for clinical translation using standard cytotoxic drugs and small molecule inhibitors of Wip1 and USP7, which warrant further development.
Materials and methods
CRISPR-Cas9 screening
The CRISPR-Cas9 screen was performed using the Broad Institute’s GeCKO library (Sanjana et al., 2014; Aguirre et al., 2016). 33 cancer cell lines (including nine Ewing sarcoma lines) were screened with the GeCKO library, containing ∼95,000 guides and an average of six guides per gene (Sanjana et al., 2014; Aguirre et al., 2016). The library contains ∼1,000 negative control guides that do not target any location in the reference genome. The library also included guides with more than one perfect match in the reference genome allowing us to computationally correct for the previously described cutting toxicity associated with multiple Cas9 cuts in the genome (Aguirre et al., 2016).
Cancer cell lines were transduced with Cas9 using a lentiviral system (Aguirre et al., 2016). Cell lines that met quality control criteria, including Cas9 activity measured using a GFP reporter, and other parameters, were then screened with the CRISPR library. A pool of guides was transduced into a population of cells. The cells were cultured for ∼21 d in vitro, and at the end of the assay, barcodes for each guide were sequenced for each cell line in replicate. Reads per kilobase were calculated for each replicate and then the log2 fold change compared with the initial plasmid pool was calculated for each guide. Samples with poor replicate reproducibility, as well as guides that have low representation in the initial plasmid pool, were removed from the analysis. Next, the guides from multiple replicates for each sample were used to collapse the data into gene scores using the CERES algorithm (Meyers et al., 2017), which models the cutting effect of each guide correcting for multiple cuts in the genome to produce a score that reflects the effect of disruption of the gene. After the dependency scores were calculated using the CERES algorithm, the scores for each cell line were scaled so that mean of negative controls was 0 and the mean of a subset of positive controls was −1.
For all of our analysis, the data were filtered and only the set of genetic dependencies with variable dependency scores that had standard deviations two sigma above the mean standard deviation across all genes were used. This resulted in 705 dependencies. Pearson correlations were then computed between the dependency gene score for TP53 and all other variable dependencies in the screen. The top eight anti-correlated genes were used for subsequent analysis.
Cell lines and chemical compounds
Cell lines were obtained from the American Type Culture Collection (ATCC), except for VH-64 and WE-68, which were provided by J. Sonnemann (Universitätsklinikum Jena, Jena, Germany); TC138 and CHLA258, which were purchased from the COG Cell Line and Xenograft Repository; and SJSA-X, which was provided by G. Wahl (The Salk Institute for Biological Studies, La Jolla, CA). Cell line identity was confirmed by Short Tandem Repeat (STR) profiling. ATSP-7041 and ATSP-7342 were synthesized according to established methods (Bird et al., 2011; Chang et al., 2013). XL-188 was synthesized according to established methods (Lamberto et al., 2017). RG7388 (ApexBio Technology), GSK2830371 (Selleck Chemicals), P5091 (Sigma-Aldrich), doxorubicin (Cell Signaling), etoposide (Selleck Chemicals), and vincristine (Selleck Chemicals) were solubilized in DMSO.
Lentivirus production and transduction
Lentivirus was produced by transfecting HEK-293T cells with the pLentiV2 vector (Addgene plasmid 52961) and the packaging plasmids pCMV8.9 and pCMV-VSVG according to the FuGENE 6 (Roche) protocol. For lentiviral transduction, Ewing sarcoma cells were incubated with 2 ml of virus and 8 µg/ml of polybrene (Sigma-Aldrich). Cells were selected in puromycin (Sigma-Aldrich) 48 h after infection for single knockout experiments. For dual knockout experiments PPM1D, USP7, and MDM4 sgRNA sequences were cloned into a LentiV2 vector with a blasticidin selection marker (Addgene plasmid 83480).
sgRNA sequences
sgRNAs were designed using the Broad Institute’s sgRNA designer tool. The following sequences were used as control or to target the respective genes: control sgRNA, 5′-GTAGCGAACGTGTCCGGCGT-3′; sgMDM2 2: 5′-AGTTACTGTGTATCAGGCAG-3′; sgMDM2 5: 5′-AGACACTTATACTATGAAAG-3′; sgMDM4 4: 5′-AGATGTTGAACACTGAGCAG-3′; sgMDM4 6: 5′-AAGAATTCCACTGAGTTGCA-3′; sgUSP7 1: 5′-AGATGTATGATCCCAAAACG-3′; sgUSP7 2: 5′-ACCATACCCAAATTATTCCG-3′; sgPPM1D 1: 5′-CTGAAGAAAAGCCCTCGCCG-3′; sgPPM1D 2: 5′-CAGGTGATTTGTGGAGCTAT-3′; sgTP53 1: 5′-GCTTGTAGATGGCCATGGCG-3′; sgTP53 2: 5′-TCCTCAGCATCTTATCCGAG-3′; sgTP53 4: 5′-GCAGTCACAGCACATGACGG-3′; and sgTP53 5: 5′-GTAGTGGTAATCTACTGGGA-3′.
Protein extraction and immunoblotting
Whole-cell lysates were extracted in cell lysis buffer (Cell Signaling) supplemented with EDTA-free protease inhibitors and PhosSTOP phosphatase inhibitors (Roche). Western immunoblotting was performed using standard techniques. Primary antibodies used included anti-MDM2 (ab178938; Abcam), anti-MDM2 (86934; Cell Signaling), anti-MDM4 (A300-287A; Bethyl Laboratories), anti-p53 (2527S; Cell Signaling), anti-p21 (2946S; Cell Signaling), anti-Vinculin (18058; Abcam), anti-Wip1 (A300-664A; Bethyl Laboratories), anti-pSer15-p53 (9284; Cell Signaling), anti-USP7 (A300-033A; Bethyl Laboratories), and anti-Tubulin (cp06; CalBiochem).
Cell viability assays
Cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Viability assays were performed after Ewing sarcoma cell lines were infected with sgRNAs targeting TP53, MDM2, MDM4, PPM1D, or USP7 or treated with ATSP-7041, ATSP-7342, GSK2830371, P5091, XL-188, or vehicle control.
Immunoprecipitation experiments
Five million TC32 cells were treated with either 10 µM RG7388 or vehicle control for 4 h. Cells were lysed in buffer A (150 mM NaCl, 50 mM Tris, and 0.5% NP-40, pH 7.4) and combined with anti-MDM4 antibody (A300-287A; Bethyl Laboratories) in the presence of 20 µM RG7388, ATSP-7041, or vehicle control in a total volume of 1 ml, rotating at 4°C for 16 h. Subsequently, 50 µl washed Protein AG beads (sc-2003; Santa Cruz) were added and the mixture was incubated for 1 h rotating at 4°C. Beads were washed three times in buffer A, and protein complexes were eluted by boiling in NuPage LDS Sample Buffer (NP0007; Invitrogen) supplemented with DTT. Samples were analyzed by Western blot with antibodies against p53 DO-1 (sc-126; Santa Cruz) and MDM4 (A300-287A; Bethyl Laboratories).
Annexin V staining
Ewing sarcoma cells lines were assessed for induction of cell death after 2 d of treatment with ATSP-7041 or P5091, or after 3 d of treatment with GSK2830371. Cell death was measured using flow cytometric analysis of Annexin V staining according to the manufacturer’s instructions (eBioscience). Data analysis was completed using Flowjo 7.6 software (Treestar).
Quantitative PCR
RNA was extracted from cells with the RNeasy kit and on-column DNA digestion (Qiagen). cDNA was prepared using M-MLV reverse transcription (ThermoFisher Scientific). TaqMan Gene Expression Master Mix (Applied Biosystems) was used per the manufacturer’s protocol. TaqMan probes included RPL13A (Hs04194366_g1; ThermoFisher Scientific), CDKN1A (Hs00355782_m1; Thermo Fisher Scientific), and MDM2 (Hs01066930_m1; ThermoFisher Scientific). Data were collected in triplicate and analyzed using the ΔΔCT method.
Ewing sarcoma xenograft studies
For anti-tumor efficacy studies, tumor xenografts were established in 15 nude female mice by implanting three million TC32 cells into the right flank. Animals were randomized to either 20 d of treatment with 30 mg/kg q.o.d. IV ATSP-7041 (n = 8) or vehicle (n = 7) for a total of 10 doses. Treatment was started when tumors reached 100–200 mm3. Tumor volumes were measured with calipers twice a week. For PDX studies, tumor fragments were implanted into the right flank of nude female mice by minor surgery. After tumor engraftment, studies were performed as described for TC32 xenograft studies.
For pharmacodynamics studies, tumor xenografts were established in six nude female mice by implanting three million TC32 cells or PDX tumor fragments into the right flank. Animals were randomized to either ATSP-7041 (n = 3) or vehicle treatment (n = 3). Mice were treated with 30 mg/kg ATSP-7041 IV or vehicle every other day for three total doses and were sacrificed 8 h after the third dose. Tumor tissue was flash frozen for protein or RNA extraction using standard methods.
For in vivo studies, ATSP-7041 was prepared using the following protocol: mPEG-DSPE (Nanocs) was dissolved in chloroform and dried by a rotary evaporator. ATSP-7041 was dissolved in 1 M NaOH and diluted 100-fold in 10 mM histidine-buffered saline to a final concentration of 3 mg/ml. This mixture was added to the dried lipid film to a final mPEG-DSPE concentration of 50 mg/ml and final pH of 7. The film was rehydrated by brief sonication and heating in a 50°C water bath. The mixture was then subjected to five freeze-thaw cycles in liquid nitrogen and 40°C water, respectively, and the solution passed 10 times through an Avanti Mini-Extruder Set (Avanti Polar Lipids) equipped with a 800-nm filter (Whatman).
All animal studies were conducted under the auspices of protocols approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
Drug synergy analysis Chou-Talalay combination index for Loewe additivity
Loewe Additivity is a dose-effect approach that estimates the effect of combining two drugs based on the concentration of each individual drug that produces the same quantitative effect (Goldoni and Johansson, 2007). Chou and Talalay (Chou, 2006, 2010) showed that Loewe equations are valid for enzyme inhibitors with similar mechanisms of action, either competitive or noncompetitive toward the substrate. They introduced the combination index (CI) scores to estimate the interaction between the two drugs. If CI < 1, the drugs have a synergistic effect, and if CI > 1, the drugs have an antagonistic effect. CI = 1 means the drugs have an additive effect.
Online supplemental material
Supplemental material includes two items providing additional information on the CRISPR-Cas9 screen: TP53 mutation annotations of cancer cell lines (Table S1) and correlation of TP53 dependency with top scoring genes (Fig. S1).
Supplementary Material
Acknowledgments
The authors thank Jaume Mora for providing the PDX model HSJD-ES-002. The authors thank Jürgen Sonnemann for providing VH-64 and WE-68 Ewing sarcoma cell lines and Geoff Wahl for SJSA-X cells. The authors thank Franziska Wachter for insightful discussions.
B. Stolte was supported by a DAAD (Deutscher Akademischer Austauschdienst) fellowship in the thematic network “Research for Rare Diseases and Personalized Medicine.“ A. Balboni Iniguez is a Damon Runyon-Sohn Pediatric Fellow supported by the Damon Runyon Cancer Research Foundation (grant DRSG-12-15). N.V. Dharia is supported by the National Institutes of Health (NIH) training grant T32 CA136432. A.M. Morgan is supported by the NIH grant F30 CA221087, with prior support from NIH training grants T32 GM007753 and T32 GM008313. G.H. Bird is supported by NIH grant R50 CA211399. S.J. Buhrlage was supported by NIH grant R01 CA211681. This work was supported by a Quantum Award from Hyundai Hope on Wheels and a REACH grant from the Alex’s Lemonade Stand Foundation to L.D. Walensky and K. Stegmaier, Leukemia and Lymphoma Society Scholar Awards to L.D. Walensky and K. Stegmaier, and by the Brian MacIsaac Sarcoma Foundation, St. Baldrick’s Foundation Robert J. Arceci Innovation Award and National Cancer Institute grant R35 CA210030 to K. Stegmaier. This work was also supported by a generous gift from the family of Ivo Coll and the Cubans Curing Children’s Cancers (4C’s Fund).
L.D. Walensky is a scientific advisory board member and consultant for Aileron Therapeutics. The authors declare no further competing financial interests.
B. Stolte, A. Balboni Iniguez, N.V. Dharia, A.M. Morgan, G.H. Bird, A. Tsherniak, F. Vazquez, N.J. Schauer, X. Liu, S.J. Buhrlage, L.D. Walensky, and K. Stegmaier designed the experiments. B. Stolte, A.M. Morgan, A.L. Robichaud, and A. Conway Saur acquired the data. N.V. Dharia, G. Alexe, and B. Stolte performed statistical analysis, biostatistics, and computational analysis. B. Stolte, A. Balboni Iniguez, N.V. Dharia, A.L. Robichaud, A. Saur Conway, A.M. Morgan, G. Alexe, G.H. Bird, A. Tsherniak, F. Vazquez, N.J. Schauer, X. Liu, S.J. Buhrlage, L.D. Walensky, and K. Stegmaier wrote and revised the manuscript. K. Stegmaier and L.D. Walensky supervised the study.
References
- Agathanggelou A., Smith E., Davies N.J., Kwok M., Zlatanou A., Oldreive C.E., Mao J., Da Costa D., Yadollahi S., Perry T., et al. 2017. USP7 inhibition alters homologous recombination repair and targets CLL cells independently of ATM/p53 functional status. Blood. 130:156–166. 10.1182/blood-2016-12-758219 [DOI] [PubMed] [Google Scholar]
- Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.Z., Ben-David U., Cook A., Ha G., Harrington W.F., Doshi M.B., et al. 2016. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 6:914–929. 10.1158/2159-8290.CD-16-0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An T., Gong Y., Li X., Kong L., Ma P., Gong L., Zhu H., Yu C., Liu J., Zhou H., et al. 2017. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem. Pharmacol. 131:29–39. 10.1016/j.bcp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Andreeff M., Kelly K.R., Yee K., Assouline S., Strair R., Popplewell L., Bowen D., Martinelli G., Drummond M.W., Vyas P., et al. 2016. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin. Cancer Res. 22:868–876. 10.1158/1078-0432.CCR-15-0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamuth N.J., and Womer R.B.. 2010. Ewing’s sarcoma. Lancet Oncol. 11:184–192. 10.1016/S1470-2045(09)70286-4 [DOI] [PubMed] [Google Scholar]
- Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D., et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 483:603–607. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F., Wade M., Godes M., Davis T.N., Whitehead D.G., Kung A.L., Wahl G.M., and Walensky L.D.. 2010. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 18:411–422. 10.1016/j.ccr.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G.H., Crannell W.C., and Walensky L.D.. 2011. Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting. Curr. Protoc. Chem. Biol. 3:99–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohl A.S., Solomon D.A., Chang W., Wang J., Song Y., Sindiri S., Patidar R., Hurd L., Chen L., Shern J.F., et al. 2014. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 10:e1004475 10.1371/journal.pgen.1004475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Cell Line Encyclopedia Consortium. Genomics of Drug Sensitivity in Cancer Consortium . 2015. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 528:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol H., Reynolds C.P., Kang M.H., Keir S.T., Maris J.M., Gorlick R., Kolb E.A., Billups C.A., Geier B., Kurmasheva R.T., et al. 2013. Initial testing of the MDM2 inhibitor RG7112 by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer. 60:633–641. 10.1002/pbc.24235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino R.C., De Bortoli M., Lu X., Moon S.H., Nguyen T.A., Shepard M.A., Rao P.H., Donehower L.A., and Kim J.Y.. 2008. Medulloblastomas overexpress the p53-inactivating oncogene WIP1/PPM1D. J. Neurooncol. 86:245–256. 10.1007/s11060-007-9470-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.S., Graves B., Guerlavais V., Tovar C., Packman K., To K.H., Olson K.A., Kesavan K., Gangurde P., Mukherjee A., et al. 2013. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. USA. 110:E3445–E3454. 10.1073/pnas.1303002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapeau E.A., Gembarska A., Durand E.Y., Mandon E., Estadieu C., Romanet V., Wiesmann M., Tiedt R., Lehar J., de Weck A., et al. 2017. Resistance mechanisms to TP53-MDM2 inhibition identified by in vivo piggyBac transposon mutagenesis screen in an Arf-/- mouse model. Proc. Natl. Acad. Sci. USA. 114:3151–3156. 10.1073/pnas.1620262114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D., Tian Z., Nicholson B., Kumar K.G., Zhou B., Carrasco R., McDermott J.L., Leach C.A., Fulcinniti M., Kodrasov M.P., et al. 2012. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 22:345–358. 10.1016/j.ccr.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58:621–681. 10.1124/pr.58.3.10 [DOI] [PubMed] [Google Scholar]
- Chou T.C. 2010. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 70:440–446. 10.1158/0008-5472.CAN-09-1947 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton B.D., Stewart C., Taylor-Weiner A., Alexe G., Kurek K.C., Calicchio M.L., Kiezun A., Carter S.L., Shukla S.A., Mehta S.S., et al. 2014. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 4:1326–1341. 10.1158/2159-8290.CD-13-1037 [DOI] [PubMed] [Google Scholar]
- Ding Q., Zhang Z., Liu J.J., Jiang N., Zhang J., Ross T.M., Chu X.J., Bartkovitz D., Podlaski F., Janson C., et al. 2013. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J. Med. Chem. 56:5979–5983. 10.1021/jm400487c [DOI] [PubMed] [Google Scholar]
- Edlund K., Larsson O., Ameur A., Bunikis I., Gyllensten U., Leroy B., Sundström M., Micke P., Botling J., and Soussi T.. 2012. Data-driven unbiased curation of the TP53 tumor suppressor gene mutation database and validation by ultradeep sequencing of human tumors. Proc. Natl. Acad. Sci. USA. 109:9551–9556. 10.1073/pnas.1200019109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar N., Hawkins D.S., Dirksen U., Lewis I.J., Ferrari S., Le Deley M.C., Kovar H., Grimer R., Whelan J., Claude L., et al. 2015. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 33:3036–3046. 10.1200/JCO.2014.59.5256 [DOI] [PubMed] [Google Scholar]
- Gavory G., O’Dowd C.R., Helm M.D., Flasz J., Arkoudis E., Dossang A., Hughes C., Cassidy E., McClelland K., Odrzywol E., et al. 2018. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 14:118–125. 10.1038/nchembio.2528 [DOI] [PubMed] [Google Scholar]
- Gilmartin A.G., Faitg T.H., Richter M., Groy A., Seefeld M.A., Darcy M.G., Peng X., Federowicz K., Yang J., Zhang S.Y., et al. 2014. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat. Chem. Biol. 10:181–187. 10.1038/nchembio.1427 [DOI] [PubMed] [Google Scholar]
- Goldoni M., and Johansson C.. 2007. A mathematical approach to study combined effects of toxicants in vitro: evaluation of the Bliss independence criterion and the Loewe additivity model. Toxicol. In Vitro. 21:759–769. 10.1016/j.tiv.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hendy O.M., Elghannam D.M., El-Sharnouby J.A., Goda E.F., El-Ashry R., and Al-Tonbary Y.. 2009. Frequency and prognostic significance of murine double minute protein-2 overexpression and p53 gene mutations in childhood acute lymphoblastic leukemia. Hematology. 14:335–340. 10.1179/102453309X12473408860389 [DOI] [PubMed] [Google Scholar]
- Hernández-Pérez S., Cabrera E., Salido E., Lim M., Reid L., Lakhani S.R., Khanna K.K., Saunus J.M., and Freire R.. 2017. DUB3 and USP7 de-ubiquitinating enzymes control replication inhibitor Geminin: molecular characterization and associations with breast cancer. Oncogene. 36:4802–4809. 10.1038/onc.2017.21 [DOI] [PubMed] [Google Scholar]
- Hof J., Krentz S., van Schewick C., Körner G., Shalapour S., Rhein P., Karawajew L., Ludwig W.D., Seeger K., Henze G., et al. 2011. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 29:3185–3193. 10.1200/JCO.2011.34.8144 [DOI] [PubMed] [Google Scholar]
- Hu B., Gilkes D.M., Farooqi B., Sebti S.M., and Chen J.. 2006. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J. Biol. Chem. 281:33030–33035. 10.1074/jbc.C600147200 [DOI] [PubMed] [Google Scholar]
- Ito M., Barys L., O’Reilly T., Young S., Gorbatcheva B., Monahan J., Zumstein-Mecker S., Choong P.F., Dickinson I., Crowe P., et al. 2011. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin. Cancer Res. 17:416–426. 10.1158/1078-0432.CCR-10-2050 [DOI] [PubMed] [Google Scholar]
- Kategaya L., Di Lello P., Rougé L., Pastor R., Clark K.R., Drummond J., Kleinheinz T., Lin E., Upton J.P., Prakash S., et al. 2017. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 550:534–538. 10.1038/nature24006 [DOI] [PubMed] [Google Scholar]
- Kato M.V., Shimizu T., Ishizaki K., Kaneko A., Yandell D.W., Toguchida J., and Sasaki M.S.. 1996. Loss of heterozygosity on chromosome 17 and mutation of the p53 gene in retinoblastoma. Cancer Lett. 106:75–82. 10.1016/0304-3835(96)04305-4 [DOI] [PubMed] [Google Scholar]
- Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J., et al. 2015. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33:306–312. 10.1038/nbt.3080 [DOI] [PubMed] [Google Scholar]
- Ladanyi M., Lewis R., Jhanwar S.C., Gerald W., Huvos A.G., and Healey J.H.. 1995. MDM2 and CDK4 gene amplification in Ewing’s sarcoma. J. Pathol. 175:211–217. 10.1002/path.1711750209 [DOI] [PubMed] [Google Scholar]
- Lamberto I., Liu X., Seo H.S., Schauer N.J., Iacob R.E., Hu W., Das D., Mikhailova T., Weisberg E.L., Engen J.R., et al. 2017. Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7. Cell Chem. Biol. 24:1490–1500.e11. 10.1016/j.chembiol.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambros M.B., Natrajan R., Geyer F.C., Lopez-Garcia M.A., Dedes K.J., Savage K., Lacroix-Triki M., Jones R.L., Lord C.J., Linardopoulos S., et al. 2010. PPM1D gene amplification and overexpression in breast cancer: a qRT-PCR and chromogenic in situ hybridization study. Mod. Pathol. 23:1334–1345. 10.1038/modpathol.2010.121 [DOI] [PubMed] [Google Scholar]
- Lane D.P. 1992. p53, guardian of the genome. Nature. 358:15–16. 10.1038/358015a0 [DOI] [PubMed] [Google Scholar]
- Leroy B., Anderson M., and Soussi T.. 2014. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum. Mutat. 35:672–688. 10.1002/humu.22552 [DOI] [PubMed] [Google Scholar]
- Lu X., Ma O., Nguyen T.A., Jones S.N., Oren M., and Donehower L.A.. 2007. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 12:342–354. 10.1016/j.ccr.2007.08.033 [DOI] [PubMed] [Google Scholar]
- Ma D., Zhang C.J., Chen Z.L., and Yang H.. 2014. Prognostic value of PPM1D in 800 gastric cancer patients. Mol. Med. Rep. 10:191–194. 10.3892/mmr.2014.2165 [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D., Sexsmith E., Yeger H., Williams B.R., and Coppes M.J.. 1994. Mutations of the p53 tumor suppressor gene occur infrequently in Wilms’ tumor. Cancer Res. 54:2077–2079. [PubMed] [Google Scholar]
- Meric-Bernstam F., Saleh M.N., Infante J.R., Goel S., Falchook G.S., Shapiro G., Chung K.Y., Conry R.M., Hong D.S., Wang J.S.-Z., et al. 2017. Phase I trial of a novel stapled peptide ALRN-6924 disrupting MDMX- and MDM2-mediated inhibition of WT p53 in patients with solid tumors and lymphomas. J. Clin. Oncol. 35:2505–2505. [Google Scholar]
- Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S., et al. 2017. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49:1779–1784. 10.1038/ng.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B., and Suresh Kumar K.G.. 2011. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem. Biophys. 60:61–68. 10.1007/s12013-011-9185-5 [DOI] [PubMed] [Google Scholar]
- Ognjanovic S., Martel G., Manivel C., Olivier M., Langer E., and Hainaut P.. 2012. Low Prevalence of TP53 Mutations and MDM2 Amplifications in Pediatric Rhabdomyosarcoma. Sarcoma. 2012:492086 10.1155/2012/492086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordóñez J.L., Amaral A.T., Carcaboso A.M., Herrero-Martín D., del Carmen García-Macías M., Sevillano V., Alonso D., Pascual-Pasto G., San-Segundo L., Vila-Ubach M., et al. 2015. The PARP inhibitor olaparib enhances the sensitivity of Ewing sarcoma to trabectedin. Oncotarget. 6:18875–18890. 10.18632/oncotarget.4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A., Tolcher A., Beeram M., Nemunaitis J., Weiss G.J., Bhalla K., Agrawal M., Nichols G., Middleton S., Beryozkina A., et al. 2015. Clinical pharmacology characterization of RG7112, an MDM2 antagonist, in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 76:587–595. 10.1007/s00280-015-2830-8 [DOI] [PubMed] [Google Scholar]
- Patton J.T., Mayo L.D., Singhi A.D., Gudkov A.V., Stark G.R., and Jackson M.W.. 2006. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 66:3169–3176. 10.1158/0008-5472.CAN-05-3832 [DOI] [PubMed] [Google Scholar]
- Pechackova S., Burdova K., Benada J., Kleiblova P., Jenikova G., and Macurek L.. 2016. Inhibition of WIP1 phosphatase sensitizes breast cancer cells to genotoxic stress and to MDM2 antagonist nutlin-3. Oncotarget. 7:14458–14475. 10.18632/oncotarget.7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T.S., He Y.H., Nie T., Hu X.D., Lu H.Y., Yi J., Shuai Y.F., and Luo M.. 2014. PPM1D is a prognostic marker and therapeutic target in colorectal cancer. Exp. Ther. Med. 8:430–434. 10.3892/etm.2014.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishas K.I., Al-Ejeh F., Zinonos I., Kumar R., Evdokiou A., Brown M.P., Callen D.F., and Neilsen P.M.. 2011. Nutlin-3a is a potential therapeutic for ewing sarcoma. Clin. Cancer Res. 17:494–504. 10.1158/1078-0432.CCR-10-1587 [DOI] [PubMed] [Google Scholar]
- Pishas K.I., Adwal A., Neuhaus S.J., Clayer M.T., Farshid G., Staudacher A.H., and Callen D.F.. 2015. XI-006 induces potent p53-independent apoptosis in Ewing sarcoma. Sci. Rep. 5:11465 10.1038/srep11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N., Suvà M.L., Suvà D., Cironi L., Provero P., Tercier S., Joseph J.M., Stehle J.C., Baumer K., Kindler V., and Stamenkovic I.. 2008. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 68:2176–2185. 10.1158/0008-5472.CAN-07-1761 [DOI] [PubMed] [Google Scholar]
- Saito-Ohara F., Imoto I., Inoue J., Hosoi H., Nakagawara A., Sugimoto T., and Inazawa J.. 2003. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 63:1876–1883. [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnemann J., Palani C.D., Wittig S., Becker S., Eichhorn F., Voigt A., and Beck J.F.. 2011. Anticancer effects of the p53 activator nutlin-3 in Ewing’s sarcoma cells. Eur. J. Cancer. 47:1432–1441. 10.1016/j.ejca.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Sriraman A., Radovanovic M., Wienken M., Najafova Z., Li Y., and Dobbelstein M.. 2016. Cooperation of Nutlin-3a and a Wip1 inhibitor to induce p53 activity. Oncotarget. 7:31623–31638. 10.18632/oncotarget.9302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43(D1):D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D.S., Lambros M.B., Rayter S., Natrajan R., Vatcheva R., Gao Q., Marchiò C., Geyer F.C., Savage K., Parry S., et al. 2009. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin. Cancer Res. 15:2269–2280. 10.1158/1078-0432.CCR-08-2403 [DOI] [PubMed] [Google Scholar]
- Tavana O., Li D., Dai C., Lopez G., Banerjee D., Kon N., Chen C., Califano A., Yamashiro D.J., Sun H., and Gu W.. 2016. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 22:1180–1186. 10.1038/nm.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F., Surdez D., Ma X., Parker M., Le Deley M.C., Bahrami A., Zhang Z., Lapouble E., Grossetête-Lalami S., Rusch M., et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project and the International Cancer Genome Consortium . 2014. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 4:1342–1353. 10.1158/2159-8290.CD-14-0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull A.P., Ioannidis S., Krajewski W.W., Pinto-Fernandez A., Heride C., Martin A.C.L., Tonkin L.M., Townsend E.C., Buker S.M., Lancia D.R., et al. 2017. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 550:481–486. 10.1038/nature24451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter F., Morgan A.M., Godes M., Mourtada R., Bird G.H., and Walensky L.D.. 2017. Mechanistic validation of a clinical lead stapled peptide that reactivates p53 by dual HDM2 and HDMX targeting. Oncogene. 36:2184–2190. 10.1038/onc.2016.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M., Rodewald L.W., Espinosa J.M., and Wahl G.M.. 2008. BH3 activation blocks Hdmx suppression of apoptosis and cooperates with Nutlin to induce cell death. Cell Cycle. 7:1973–1982. 10.4161/cc.7.13.6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M., Li Y.C., and Wahl G.M.. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer. 13:83–96. 10.1038/nrc3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma X., Ren S., Buolamwini J.K., and Yan C.. 2011. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol. Cancer Ther. 10:69–79. 10.1158/1535-7163.MCT-10-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Yu H., Hughes N.W., Liu B., Kendirli A., Klein K., Chen W.W., Lander E.S., and Sabatini D.M.. 2017. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 168:890–903.e15. 10.1016/j.cell.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Cao C., Xia H., Shi S., Hong L., Wei X., Gu D., Bian J., Liu Z., Huang W., et al. 2016. Protein phosphatase magnesium-dependent 1δ is a novel tumor marker and target in hepatocellular carcinoma. Front. Med. 10:52–60. 10.1007/s11684-016-0433-3 [DOI] [PubMed] [Google Scholar]
- Zhan M., Sun X., Liu J., Li Y., Li Y., He X., Zhou Z., and Lu L.. 2017. Usp7 promotes medulloblastoma cell survival and metastasis by activating Shh pathway. Biochem. Biophys. Res. Commun. 484:429–434. 10.1016/j.bbrc.2017.01.144 [DOI] [PubMed] [Google Scholar]
- Zhao M., Zhang H., Zhu G., Liang J., Chen N., Yang Y., Liang X., Cai H., and Liu W.. 2016. Association between overexpression of Wip1 and prognosis of patients with non-small cell lung cancer. Oncol. Lett. 11:2365–2370. 10.3892/ol.2016.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.H., and Bulavin D.V.. 2012. Wip1-dependent signaling pathways in health and diseases. Prog. Mol. Biol. Transl. Sci. 106:307–325. 10.1016/B978-0-12-396456-4.00001-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.