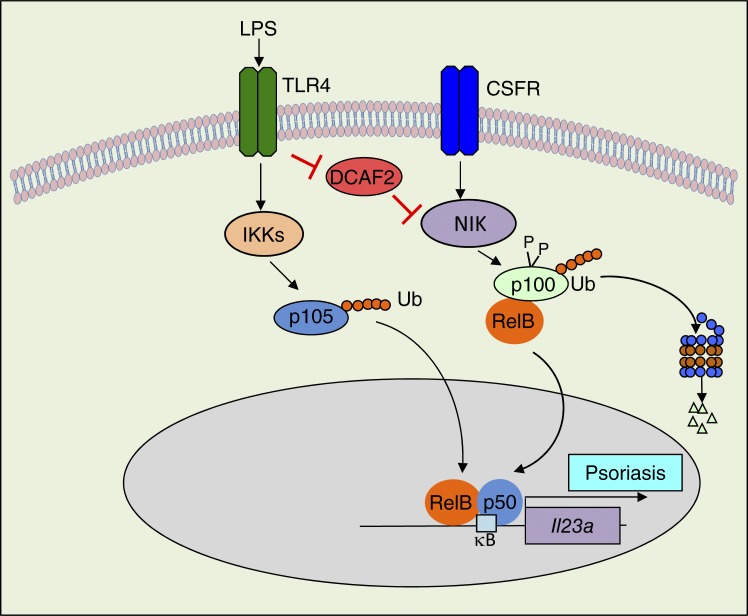
Deregulated NF-κB activation is linked to immunological disorders. Huang et al. revealed CRL4DCAF2 as a negative regulator in controlling NIK stability in DCs. Lower level of DCAF2 is associated with noncanonical NF-κB activation and hyperproduction of IL-23 in psoriasis patients.
Abstract
The E3 ligase CRL4DCAF2 is believed to be a pivotal regulator of the cell cycle and is required for mitotic and S phase progression. The NEDD8-targeting drug MLN4924, which inactivates cullin ring-finger ubiquitin ligases (CRLs), has been examined in clinical trials for various types of lymphoma and acute myeloid leukemia. However, the essential role of CRL4DCAF2 in primary myeloid cells remains poorly understood. MLN4924 treatment, which mimics DCAF2 depletion, also promotes the severity of mouse psoriasis models, consistent with the effects of reduced DCAF2 expression in various autoimmune diseases. Using transcriptomic and immunological approaches, we showed that CRL4DCAF2 in dendritic cells (DCs) regulates the proteolytic fate of NIK and negatively regulates IL-23 production. CRL4DCAF2 promoted the polyubiquitination and subsequent degradation of NIK independent of TRAF3 degradation. DCAF2 deficiency facilitated NIK accumulation and RelB nuclear translocation. DCAF2 DC-conditional knockout mice displayed increased sensitivity to autoimmune diseases. This study shows that CRL4DCAF2 is crucial for controlling NIK stability and highlights a unique mechanism that controls inflammatory diseases.
Graphical Abstract
Introduction
The NF-κB family of transcription factors plays a pivotal role in regulating immune responses, inflammation, and cell growth/survival. Deregulated NF-κB activation is linked to immunological disorders, including autoimmunity and chronic inflammation, as well as various cancers (Vallabhapurapu and Karin, 2009; Ben-Neriah and Karin, 2011; Hayden and Ghosh, 2011). Activation of NF-κB is mediated by two major signaling pathways: the canonical and noncanonical pathways (Sun, 2011). The well-characterized functions of the noncanonical NF-κB pathway include secondary lymphoid organogenesis, thymic epithelial cell differentiation, B cell survival and function, bone metabolism, and dendritic cell (DC) maturation (Razani et al., 2011; Rickert et al., 2011; Onder et al., 2017). DCs are an integral part of the innate immune system and play a crucial role in mediating host defenses against infection and inflammatory responses (Reis e Sousa, 2004). Current evidence has revealed that elevation of either GM-CSF or M-CSF and nuclear translocation of noncanonical NF-κB are observed during innate immune cell development and differentiation (Li et al., 2010; Hofmann et al., 2011; Mouri et al., 2014). DC development is independent of noncanonical NF-κB signaling, as indicated by the normal frequencies of DCs observed in NIKAly/Aly or NIK DC-conditional KO mice compared with their WT littermates (Lind et al., 2008; Katakam et al., 2015). Interestingly, noncanonical NF-κB is critical for DC presentation of foreign antigens or self-antigens in adaptive immunity (Lind et al., 2008). Additionally, activated noncanonical NF-κB signaling negatively regulates TLRs or virus-stimulated type I IFN production (Jin et al., 2014). However, the specific role of noncanonical NF-κB signaling in DCs during autoimmunity initiation remains controversial and incompletely understood.
Although activation of NF-κB is fundamental for effective immune responses, restriction of NF-κB activity is also critical to prevent excessive or prolonged immune activation, which may cause autoimmune diseases and cancers. In contrast to the canonical NF-κB pathway, noncanonical pathway regulation requires extensive elucidation. In cells that are not exposed to noncanonical NF-κB inducers, newly synthesized NIK is rapidly bound by TRAF3 and targeted for degradation, thereby maintaining an extremely low level of NIK to prevent activation of downstream signaling events (Vallabhapurapu et al., 2008; Zarnegar et al., 2008; Sun, 2017). TRAF3 mediates NIK degradation by recruiting NIK to an E3 ubiquitin ligase complex composed of TRAF2 and cIAP1/2 (Sun, 2011). In response to a small subset of TNF receptor family members, noncanonical NF-κB signaling activation results in proteolysis of the inhibitory protein TRAF3 and accumulation of NIK (Sun, 2011, 2017). Recent studies have suggested that OTUD7B (also known as Cezanne) negatively regulates signal-induced noncanonical NF-κB pathway activation by deubiquitylating TRAF3 (Hu et al., 2013). NIK itself is also a target of negative regulators, including IKKα and TANK-binding kinase 1 (TBK1), in response to BAFF receptor (BAFFR) and CD40 signaling (Razani et al., 2010; Jin et al., 2012). However, to date, it is largely unclear how signal-induced NIK ubiquitination is negatively regulated when the upstream signal is turned off. Here, we identified the E3 ubiquitin ligase CRL4DCAF2 as a novel factor that controls NIK stability and noncanonical NF-κB activation via a TRAF3-independent mechanism.
Cullin ring-finger ubiquitin ligase-4 (CRL4) exerts multiple physiological functions by using >90 adaptor proteins, which are also known as DDB1-cullin 4–associated factors (DCAFs). These DCAFs recruit a wide spectrum of substrates to the ubiquitin ligase complex. DCAF2 (also called CDT2 or DTL) is one of the substrate adaptors of CRL4. CRL4DCAF2 promotes the ubiquitin-dependent destruction of the replication initiation protein CDT1, SETD8, and the cyclin-dependent kinase inhibitor p21 (Sansam et al., 2006; Abbas et al., 2008; Terai et al., 2010). Although CRL4DCAF2 is known as a crucial regulator of the cell cycle and various DNA metabolic processes, its in vivo functions are largely unknown due to the lack of viable animal models. DCAF2 expression was significantly suppressed in biopsies from the patients with psoriasis or colitis. By generating DCAF2 DC-conditional KO (DCAF2DKO) mice, we identified CRL4DCAF2 as a pivotal regulator of noncanonical NF-κB signaling and autoimmune responses. Although ablation of DCAF2 had only a moderate effect on the development or homeostasis of DCs, these mutant animals showed hyperproduction of IL-23 in various autoimmune disease models. In vitro studies further revealed that loss of DCAF2 induces noncanonical NF-κB–targeted genes, especially IL-23, which initiated autoimmunity in response to TLR stimulation. DCAF2 deficiency strongly promoted noncanonical NF-κB activation by GM-CSF; however, NF-κB activation by canonical inducers was not affected. Multiple mutant mouse models also supported the conclusion that activation of the noncanonical NF-κB protein RelB is selectively required for IL-23 expression. WT and DCAF2DKO mice with NIK overexpression or NIK KO showed comparable IL-23 production and pathological symptoms of psoriasis-like skin inflammation. DCAF2 deficiency did not promote TRAF3 degradation but resulted in enhanced NIK accumulation. We further showed that DCAF2 binds to NIK and induces NIK degradation independent of the interaction between TRAF3 and NIK. Overall, our studies led to the novel finding that CRL4DCAF2 exerts a nonredundant function in controlling signal-induced noncanonical NF-κB activation, which involves the induction of ubiquitin-dependent NIK degradation. Our data demonstrated that CRL4DCAF2 in DCs helps maintain normal immune homeostasis and responses. Our findings will also contribute to the design of therapeutic strategies for treatment of noncanonical NF-κB–associated diseases.
Results
Activated DCs exhibit low expression of DCAF2
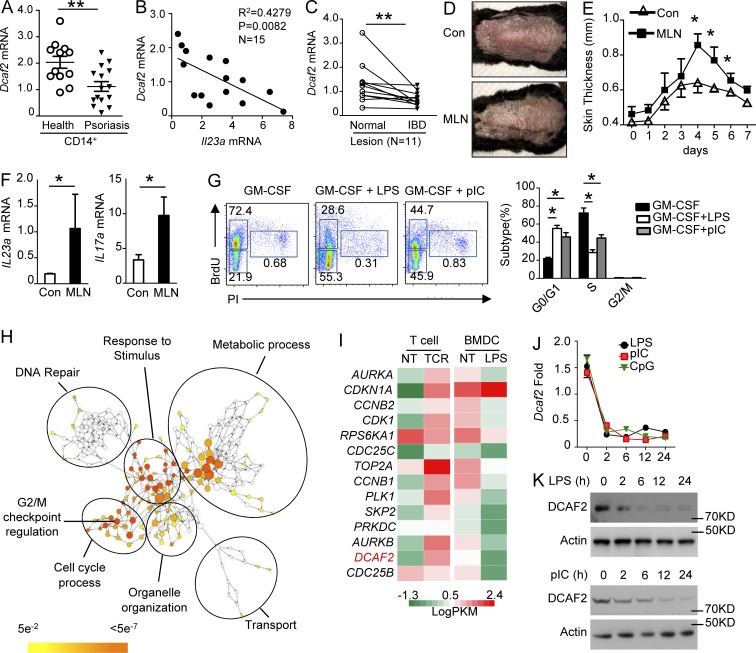
To evaluate the potential importance of DCAF2 in the pathogenesis of autoimmune diseases, we first analyzed DCAF2 mRNA expression in CD14+ peripheral blood mononuclear cells from healthy donors (healthy) and psoriasis patients. As expected, DCAF2 mRNA levels in psoriasis patients were significantly lower than those in healthy controls (Fig. 1 A). Patients with lower DCAF2 mRNA levels had higher expression of Il23a, which is a pathogenic factor for psoriasis (Fig. 1 B). Interestingly, colonic biopsy specimens from inflammatory bowel disease (IBD) patients also showed substantially lower DCAF2 expression than that of normal subjects (Fig. 1 C). These data indicated that DCAF2 expression is negatively associated with inflammatory diseases.
Figure 1.
TLRs restrict cell cycle processes and DCAF2 expression. (A) qPCR analysis of DCAF2 mRNA levels in CD14+ peripheral blood mononuclear cells (PBMCs) from healthy donors and psoriasis patients. (B) Correlation analysis of the DCAF2 mRNA expression with the critical pathogenic cytokine IL-23 in CD14+ PBMCs from psoriasis patients. (C) qRT-PCR analysis of DCAF2 transcripts in IBD inflammatory lesions and normal samples from the same patients (n = 11). (D–F) WT mice (n = 5/group) were i.p. injected with vehicle (Con) or MLN4924 (MLN) for three days before inducing the psoriasis model. (D) Representative photos taken on day 3 post-Aldara treatment. (E) Thickness of back skin lesions on day 4. (F) qRT-PCR analysis of cytokine expression in skin samples on day 5 post-Aldara treatment. (G) WT BMDCs were incubated with BrdU and triggered with GM-CSF plus LPS or pIC for 36 h. DNA synthesis was analyzed by FACS and used to assess the cell cycle. (H) Network visualization of gene ontology enrichment analysis of DEGs in BMDCs in response to LPS for 6 h. (I) Heat map showing G2/M transition–related gene expression in BMDCs activated by LPS for 6 h or T cells stimulated with αCD3/αCD28 for 18 h. (J and K) BMDCs were stimulated with LPS, pIC, or CpG2216 for the indicated time points. DCAF2 expression was monitored by qRT-PCR (J) and immunoblotting (K). All data are representative of three independent experiments. Error bars show the mean ± SEM. Significance was determined by two-tailed Student’s t test. *, P < 0.05; **, P < 0.01.
MLN4924, a small molecule inhibitor of NEDD8-activating enzyme (NAE), inhibits the activity of cullin E3 ligases, thereby stabilizing a vast number of cullin substrates including DCAF2-mediated chromatin licensing and DNA replication factor 1 (CDT1) and SETD8. Generally, inhibition of DCAF2 expression mimicked the pharmacological effects of MLN4924. In vivo, MLN4924 administration potently promoted clinical symptoms and increased skin thickness in an Aldara-induced psoriasis mouse model (Fig. 1, D and E). In inflammatory skin, pathogenic cytokines, such as IL-23 and IL-17A, were significantly elevated after MLN4924 treatment (Fig. 1 F). Furthermore, MLN4924 treatment promoted infiltration of γδT cells into the skin and enhanced the T helper 17 cells (Th17) differentiation (Fig. S1, A and B).
DCs are widely distributed innate immune cells that participate in host defenses against infections and inflammatory responses, such as psoriasis (Reis e Sousa, 2004). Current evidence has shown that proliferation of myeloid cells is severely suppressed by TLR stimulation and p27 (Chow et al., 2005; Hasan et al., 2007). Although GM-CSF facilitates cell cycle entry and proliferation of DCs, BrdU incorporation assays revealed that LPS or poly I:C (pIC) impaired GM-CSF–induced replication of the genome in S phase and subsequent cell cycle processes (Fig. 1 G). To elucidate the molecular mechanism associated with the cell cycle withdrawal mediated by TLR signals, we analyzed the majority of the changes in mRNA abundance. Gene ontology enrichment analysis (Fig. 1 H) indicated that most of the down-regulated genes were associated with regulation of the cell cycle or DNA replication compared with those of the nontreated (NT) group (more than twofold). Consistently, the expression of most of the G2/M transition genes, including DCAF2, was suppressed as revealed by heat map (Fig. 1 I). Quantitative PCR (qPCR) and immunoblot (IB) analyses further confirmed that DCAF2 expression in bone marrow (BM)–derived dendritic cells (BMDCs) was rapidly reduced upon LPS stimulation in vitro (Fig. 1, J and K). Collectively, these data suggest that TLR agonists block cell cycle entry and DCAF2 expression in DCs.
DCAF2 ablation in DCs perturbs T cell homeostasis and causes spontaneous autoimmunity
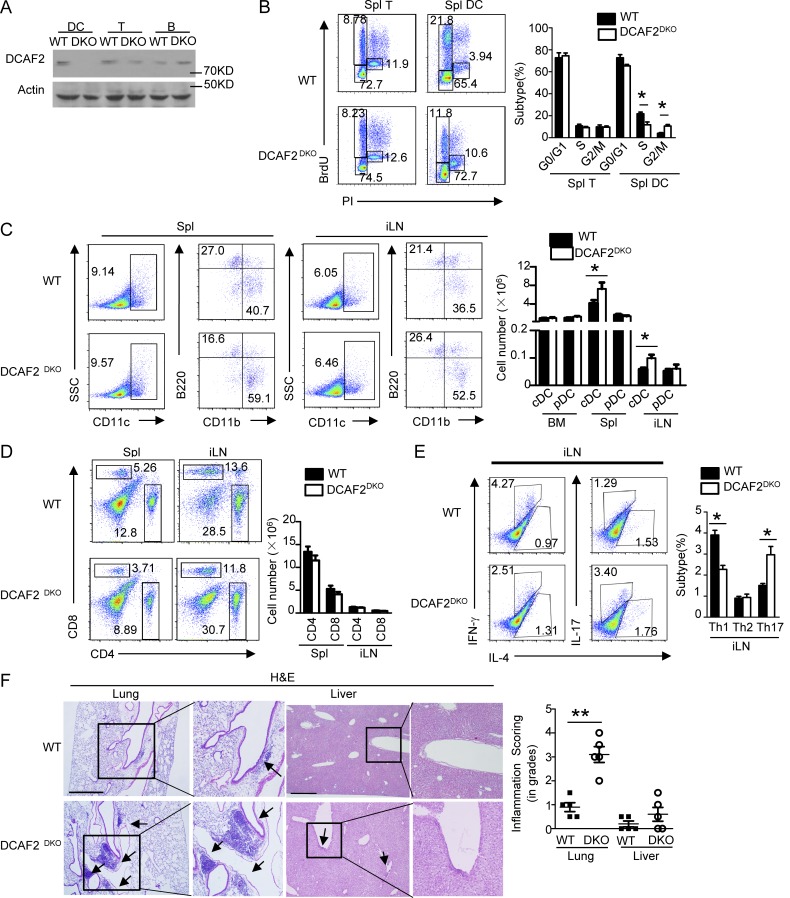
To further assess the physiological function of CRL4DCAF2 in vivo, we generated DCAF2-flox mice, which were further crossed with CD11c-Cre mice to produce DCAF2DKO mice (Fig. 2 A and Fig. S2 A). BrdU incorporation assays demonstrated that DCAF2 deficiency moderately induced DC arrest at G2/M phase (Fig. 2 B). This cell cycle arrest yielded substantial increases in the frequency and absolute cell number of conventional DCs in peripheral lymph nodes, BM, and spleen (Spl; Fig. 2 C). DCs are critical for regulating T cell homeostasis, tolerance, and activation. Although DCAF2DKO mice had lymphocyte subpopulation frequency and absolute cell numbers similar to those of the WT controls (Fig. 2 D and Fig. S2 B), DCAF2 deficiency strikingly elevated the frequency of IL-17–producing CD4+ effector T cells in inguinal lymph nodes (iLNs) of aging mice, but not of young mice (Fig. 2 E and Fig. S2 C). Aging DCAF2DKO mice also had increased levels of anti-nuclear antibody (ANA; Fig. S2 D) in serum, coupled with severe infiltration of lymphocytes to the lung and a weak inflammation in the liver (Fig. 2 F). However, no infiltration of lymphocytes was found in young DCAF2DKO mice (Fig. S2 E). Collectively, these results demonstrated an important role for DC-specific DCAF2 in maintaining immune homeostasis and a dispensable role in regulating DC development.
Figure 2.
DCAF2 in dendritic cells is required for T cell homeostasis. (A) IB analysis of DCAF2 using lyses of BMDCs, T, and B cells from DCAF2DKO mice (n = 3/group). (B) Cell cycle process of T cells or CD11c+ DCs from WT and DCAF2DKO mice (n = 3/group) was evaluated by the DNA synthesis in vivo. (C) Flow cytometric analyses of the subpopulation frequencies of dendritic cells in Spl or iLN from 6-wk-old WT and DCAF2DKO mice (n = 3/group). (D) The absolute cell numbers of CD4+, CD8+ T cells in Spl or iLN. (E) Flow cytometric analysis of the percentage of IFN-γ– and IL-17–producing CD4+ T cells in the Spl of 6-mo-old WT and DCAF2DKO mice (n = 3/group). (F) H&E staining of the indicated tissue sections from 6-mo-old WT and DCAF2DKO mice (n = 3/group), showing immune cell infiltrations (arrows). Inflammation score was quantified by a pathologist blinded to the groups. Bars, 200 µm. All FACS data are presented as a representative plot and summary graph of subpopulation percentage. All data are representative of three independent experiments. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test. *, P < 0.05; ***, P < 0.005.
DCAF2 deficiency in DCs facilitates experimental autoimmunity
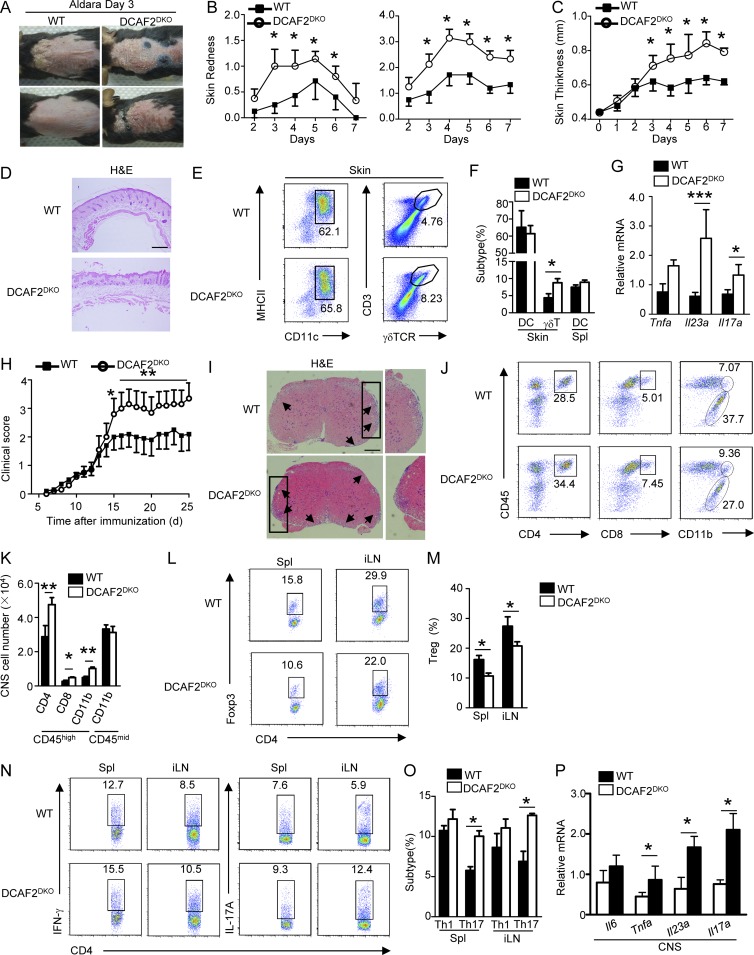
To further investigate the role of CRL4DCAF2 in regulating inflammatory responses, we used tissue-specific autoimmunity by using an Aldara-induced psoriasis-like skin inflammation model. DCAF2DKO mice treated daily with Aldara developed visually apparent erythema and indurations (Fig. 3, A and B). Aldara also significantly increased the epidermal thickness of DCAF2DKO mice (Fig. 3 C and Fig. S2 F). Histological analysis of skin lesions from DCAF2DKO mice showed significant epidermal hyperplasia, increased frequencies of micro-abscesses, and increased acanthosis (Fig. 3 D). Although the levels of resident and migratory DCs and other myeloid cells in the skin were comparable between the WT and DCAF2DKO mice (Fig. 3, E and F; and Fig. S2 G), a higher frequency of total γδ T cells was observed in the psoriatic skin lesions of DCAF2DKO mice (Fig. 3, E and F). However, the frequency of αβ T cells in DCAF2DKO mice was similar to that in their WT littermates (Fig. S2 H). We further evaluated the Aldara-induced keratinocyte proliferation by immunohistochemistry (IHC) analysis. Compared to the WT control, DCAF2 deficiency significantly increased the numbers of proliferating keratinocytes, as assessed based on Ki67 expression (Fig. S2 I). Local cytokine analysis of inflammatory skin also revealed that IL-23 and IL-17A were highly elevated along with DCAF2 deficiency (Fig. 3 G).
Figure 3.
DCAF2 deficiency in DCs aggravates various autoimmune diseases. WT and DCAF2DKO mice (n = 8/group) were treated with Aldara for 6 d. (A–C) Representative photos taken on day 3 after Aldara treatment. Back skin lesions at day 4 represented as percent change in skin redness, scaling (B), and thickness (C). (D) Back skin sections stained with H&E on day 4 after treatment; bar, 200 µm. (E and F) Plots (E) and bar (F) graphs represented flow cytometry analysis of inflamed skin. Cells were gated on CD45+CD11c+ for the presence of skin-resident DCs and skin-infiltrating γδT cells. (G) Real-time quantitative PCR analysis of cytokines expression in skin samples on day 5 after Aldara treatment. (H) Mean clinical scores of WT and DCAF2DKO mice subjected to MOG35-55–induced EAE (n = 10/group). (I) H&E staining of spinal cord sections from WT and DCAF2DKO EAE mice (n = 3/group) for visualizing immune cell infiltration (arrows). Bar, 200 µm. (J and K) Flow cytometry analyses of immune cell infiltration into the CNS (brain and spinal cord) of EAE mice (n = 3/group). (L and M) Flow cytometry analysis of Treg cells frequency in the Spl and draining lymph nodes of EAE mice (n = 3, day 14 post-immunization). (N and O) Flow cytometry analysis of Th1 and Th17 cells gating with CD4+CD45hi in the splenic and draining lymph nodes of EAE mice (n = 3/group). (P) qRT-PCR analysis to determine the relative mRNA expression level of pro-inflammatory genes in spinal cords of EAE mice (n = 3/group). Data were normalized to a reference gene, Actb. All Data are representative of three independent experiments. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
The same immunological mechanisms that mediate local inflammation in skin can also lead to systemic autoimmune diseases. Thus, we used another autoimmunity model, experimental autoimmune encephalomyelitis (EAE), which involves IL-23–driven peripheral Th17 subsets of inflammatory T cells. As expected, DCAF2DKO mice displayed substantially increased sensitivity to EAE induction compared with the WT control mice, as shown by the increased clinical scores and histological features of inflammation (Fig. 3, H and I). These clinical symptoms were associated with increased central nervous system (CNS) infiltration of both total CD4+ T cells and myeloid cells (Fig. 3, J and K). Moreover, the frequencies of regulatory T cells (T reg cells) in the Spl and iLNs of DCAF2DKO EAE mice were significantly reduced (Fig. 3, L and M). Upon exposure to myelin oligodendrocyte glycoprotein (MOG) peptide, splenic T cells from WT and DCAF2DKO mice exhibited a comparable capacity for expansion due to the a recall response (Fig. S2 J). However, DCAF2DKO mice had an increased frequency of Th17 inflammatory effector cells in peripheral lymphoid organs (Fig. 3, N and O). The CNS tissue of the MOG35-55–immunized DCAF2DKO mice had increased inflammatory cell infiltration, which resulted in elevated expression of multiple proinflammatory cytokines including IL-23 and IL-17A (Fig. 3 P). Overall, these results demonstrate an unexpected role for CRL4DCAF2 in DCs in negatively regulating IL-23 expression in autoimmune inflammation.
CRL4DCAF2 specifically regulates IL-23 induction in DCs
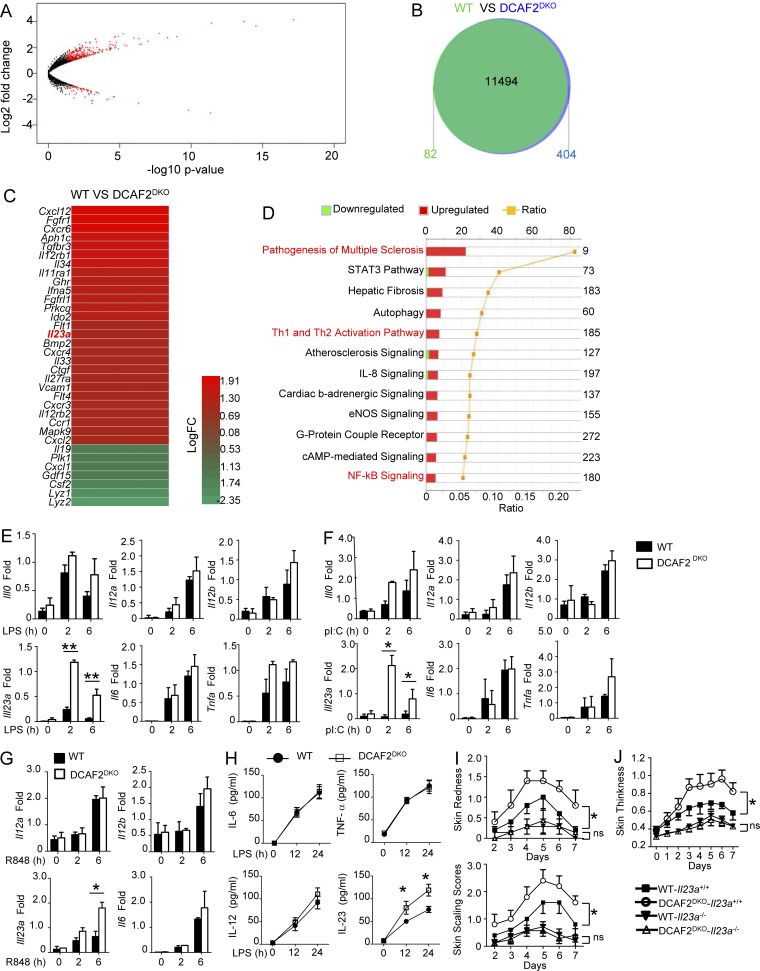
To further examine the underlying mechanism by which CRL4DCAF2 regulates autoimmune inflammation, we first evaluated the ability of DC to activate antigen-specific T cells in vitro. We used an in vitro model involving activation of OTII T cells with DCs pulsed with a specific peptide, chicken OVA 323–339. For quantification of cell proliferation, the OTII T cells were labeled with CFSE. Compared with WT DCs, the DCAF2-deficient DCs did induce stronger proliferation of OTII T cells, as measured by CFSE dilution (Fig. S3 A). Thus, the effects of DCAF2 on gene expression were further monitored following LPS stimulation. Surprisingly, we identified only 487 differentially expressed genes (DEGs) in DCAF2-deficient DCs compared with those of WT littermates (>1.5-fold; Fig. 4, A and B). Consistent with the in vivo phenotype, the proinflammatory cytokine Il23a was selectively elevated in DCAF2-deficient DCs stimulated with LPS for 6 h, as shown by heat map (Fig. 4 C). Ingenuity pathway analysis (IPA) revealed that the major biological processes altered in DCAF2-deficient DCs were associated with helper T cell polarization and pathogenesis of multiple sclerosis (Fig. 4 D). qPCR assays also showed that transcriptional levels of Il23a were significantly elevated in DCAF2-deficient BMDCs compared with WT controls in response to LPS (Fig. 4 E), pIC (Fig. 4 F), or R848 (Fig. 4 G). In addition to the mRNA levels, the protein levels of IL-23 in supernatant from DCAF2-deficient BMDCs were also elevated (Fig. 4 H).
Figure 4.
CRL4DCAF2 negatively regulates IL-23 induction in DCs. Transcriptome analyses of CRL4DCAF2-regulated genes in primary BMDCs when responding to LPS. (A and B) Scatter plots (A) and Venn diagrams (B) illustrating the overlap of differential genes expression between WT and DCAF2 KO BMDCs stimulated with LPS for 6 h. (C) Heat map showed DEG between WT and DCAF2 KO BMDCs. (D) Actual numbers of down- and up-regulated DEG were shown inside bars. Green bars denoted the percentage of down-regulated and red bars performed the percentage of up-regulated genes. The yellow line denotes the likelihood [−log (P value)] that the specific pathway was affected by DCAF2. (E–G) qRT-PCR analysis of the indicated genes using WT or DCAF2 KO BMDCs stimulated with LPS (E), pIC (F), and R848 (G). (H) ELISA of the indicated cytokines in the supernatants of LPS-stimulated WT or DCAF2 KO BMDCs for 12 and 24 h. (I and J) WT or DCAF2DKO mice were crossbred with IL23-KO mice. Skin redness, scaling (I), and thickness (J) of DCAF2WT-Il23a−/− and DCAF2DKO-Il23a−/− mice (n = 5/group) treated with Aldara for 6 d. All data are presented as fold relative to the Actb mRNA level. Data are presented as mean ± SEM values and representative of at least three independent experiments. Statistical analyses represent variations in experimental replicates. *, P < 0.05; **, P < 0.01. ns, not significant.
Current evidence has indicated that the IL-23–IL-17 axis is crucial in the pathogenesis of psoriasis (Miossec and Kolls, 2012; Teng et al., 2015). A clinical trial of anti–IL-23p19 mAb (guselkumab) confirmed the pathogenic role of deregulated IL-23 in psoriasis (Riol-Blanco et al., 2014; Gordon et al., 2015). To evaluate the role of IL-23 in DCAF2 deficiency-mediated inflammation, we crossed DCAF2DKO mice with Il23a−/− (only lacking IL-23) mice. In the Il23a−/− background, a pivotal effect of CRL4DCAF2 on skin pathology was significantly impaired, as indicated by the comparable skin redness; scaling and thickness were compared between WT and DCAF2DKO mice (Fig. 4, I and J). The data collectively suggest a negative role for CRL4DCAF2 in regulating IL-23 expression and associated autoimmune responses.
CRL4DCAF2 negatively regulates noncanonical NF-κB activation
Among the major signaling pathways activated by the TLRs are those leading to the activation of IκB kinase (IKK) and three families of MAP kinases (MAPKs): ERK, JNK, and p38. The MAPKs and IKKs mediate the induction of proinflammatory genes via activation of the transcription factors AP-1 and canonical NF-κB, respectively. The IB results indicated that DCAF2 deficiency in BMDCs had no effect on LPS-mediated activation of MAPKs (Fig. 5 A). STAT3 signaling within the tumor microenvironment induces IL-23 expression (Kortylewski et al., 2009). CRL4DCAF2 is known to diminish H4 methylation at Lys20 by inducing degradation of the methyltransferase SETD8 (Centore et al., 2010; Oda et al., 2010). We thus evaluated whether the H4K20me1 level was up-regulated and enriched in the promoter region of Il23a or Map3k14. The Chromatin immunoprecipitation (ChIP)–seq data revealed that H4K20me1 did not localized at the Il23a promoters. Although H4K20me1 was enriched at the Map3k14 promoter, we did not observe any differences between WT and DCAF2 KO BMDCs (Fig. S3 B).
Figure 5.
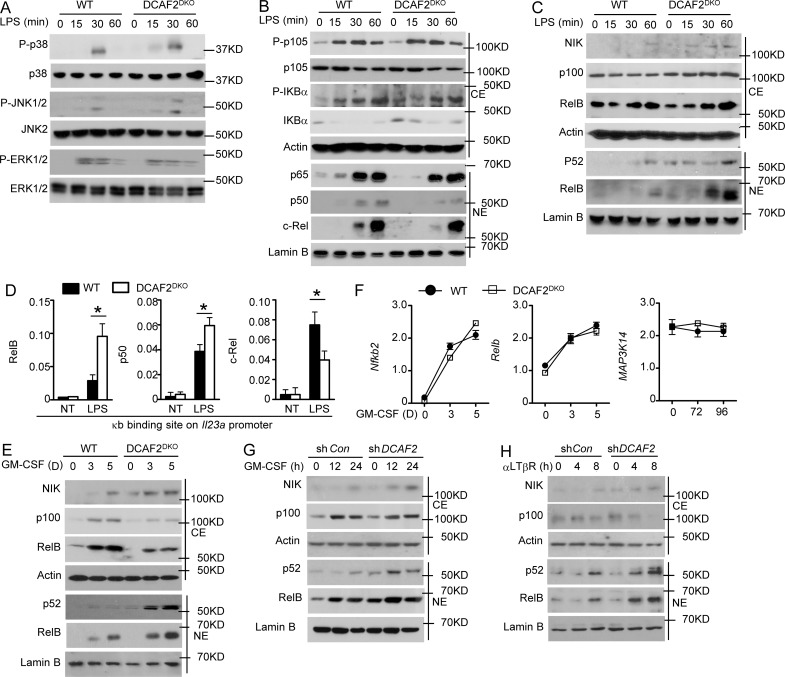
CRL4DCAF2 is a negative regulator of the noncanonical NF-κB pathway. BMDCs generated from WT or DCAF2DKO mice (6 wk old) and stimulated for the indicated times with LPS (1 µg/ml). (A) IB analysis of phosphorylated (P-) or total MAPKs in whole-cell lysates. (B and C) IB analysis of NF-κB members in cytoplasmic (CE) and nuclear (NE) extracts of WT and DCAF2 KO BMDCs stimulated with LPS. (D) ChIP assays to detect the binding of RelB, c-Rel, and p50 to the Il23a promoters in LPS-stimulated WT and DCAF2 KO BMDCs for 3 h. Data are presented as percentage of the total input DNA quantified by qPCR. (E and F) WT and DCAF2 KO BM cells were stimulated with GM-CSF as indicated. IB analyses of the indicated proteins in the CE or NE extracts (E) and qRT-PCR analysis of the indicated genes expression (F). All qPCR data are presented as fold relative to the Actb mRNA level. (G and H) D2SC cell and MEFs were infected with Dcaf2 shRNA, and purficated with GFP expression. GFP+ D2SC were stimulated with GMCSF as indicated (G). GFP+ MEFs were triggered with anti-LTβR (H). IB analyses of the indicated proteins in the CE or NE extracts. Data are presented as mean ± SEM values and representative of at least three independent experiments. Statistical analyses represent variations in experimental replicates. *, P < 0.05.
The NF-κB family of transcription factors is crucial for inflammatory cytokine induction. Among the five members of the NF-κB family, c-Rel is specifically required for induction of the proinflammatory cytokines IL-12 and IL-23 (Sanjabi et al., 2000; Hilliard et al., 2002). Surprisingly, the DCAF2 deficiency had no effect on canonical NF-κB activation, as indicated by the comparable IκB degradation and nuclear translocation of p50, p65, and c-Rel (Fig. 5 B). In contrast to canonical NF-κB, noncanonical NF-κB was significantly activated by the loss of DCAF2 in DCs. DCAF2 deficiency resulted in enhanced NIK accumulation at the basal level (Fig. 5 C). ChIP assays revealed that DCAF2 deficiency potently enhanced the recruitment of RelB and p50 to the Il23a promoter following LPS stimulation (Fig. 5 D). Loss of DCAF2 potentially suppressed the recruitment of c-Rel to the Il23a promoter, implying that RelB did not form the transcriptional complex with c-Rel (Fig. 5 D).
Typically, GM-CSF receptor signaling initiates p100 and RelB synthesis in BM progenitor cells that differentiated into DCs (Li et al., 2010; Jin et al., 2014). Therefore, we next analyzed the role of CRL4DCAF2 in GM-CSF–induced noncanonical NF-κB activation. Cultivation of DCAF2-deficient BM in GM-CSF–containing medium led to excess nuclear translocation of p52 and RelB (Fig. 5 E). DCAF2 deficiency-mediated hyperactivation of p52 and RelB was dependent on increased NIK accumulation (Fig. 5 E). qPCR assays revealed that the elevated NIK, p52, and RelB protein abundance was not due to transcriptional enhancement of these genes (Fig. 5 F). To further confirm these findings, we examined the role of CRL4DCAF2 in noncanonical NF-κB activation by GM-CSF in a DC cell line (D2SC). As observed in the primary DCs, GM-CSF strongly elevated the nuclear translocation of p52 and RelB when DCAF2 expression was silenced with shRNA (Fig. 5 G). In addition to GM-CSF, lymphotoxin-b receptor (LTβR) signaling also activated the noncanonical NF-κB pathway in mouse embryonic fibroblasts (MEFs; Keats et al., 2007; Bista et al., 2010). Interestingly, CRL4DCAF2 negatively regulated LTβR-induced noncanonical NF-κB activation in MEFs (Fig. 5 H). Collectively, these data suggest a specific role for the loss of DCAF2 in controlling the signal-induced noncanonical NF-κB activation.
Noncanonical NF-κB is essential for the inflammatory effects of CRL4DCAF2 deficiency
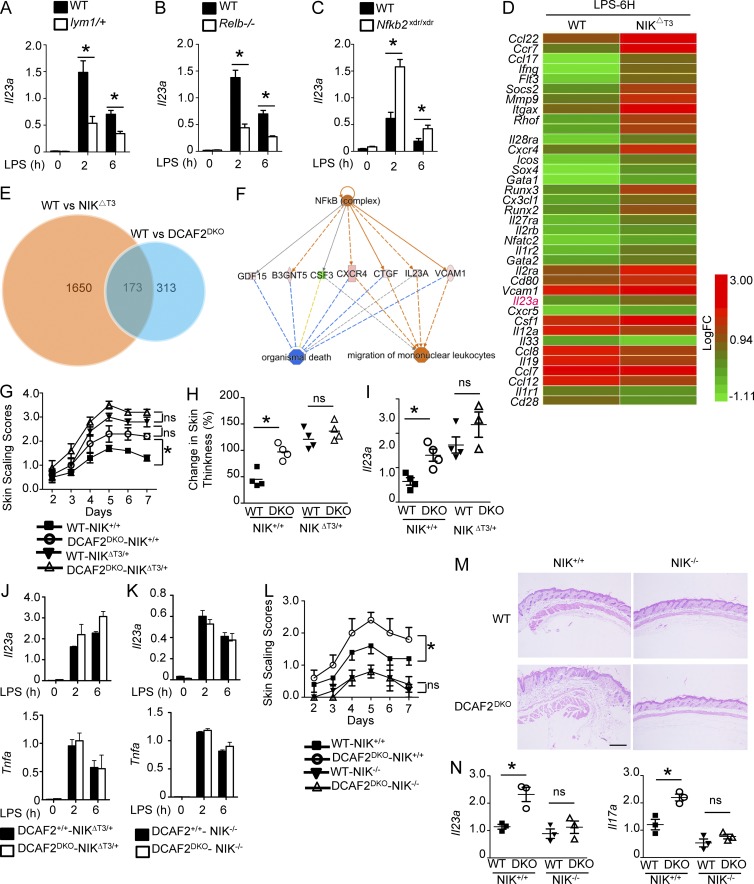
To clarify which noncanonical NF-κB members contribute to DCAF2 deficiency-mediated IL-23 hyperproduction, we assessed various gene-manipulated BMDCs. Nfκb2Lym1 mice express a processing-defective p100 mutant. As indicated in Fig. 6 A, lym1 heterozygous mice showed a severe defect in nuclear p52. As expected, inactivated noncanonical NF-κB severely impaired the induction of Il23a by LPS. To elucidate the specific roles of p52 and RelB in inflammation, we further examined IL-23 expression using BMDCs derived from the Relb-KO or nfkb2-KO (lacking both p100 and p52) mice. As expected, IL-23 induction was impaired in RelB-deficient BMDCs (Fig. 6 B). In addition to its role as a transcription factor, p100 is also an inhibitor of RelB that restricts its nuclear translocation. We also observed that nuclear translocation of RelB was significantly enhanced in nfkb2-KO BMDCs, consistent with previous findings (Fig. 6 C). Interestingly, loss of p100 resulted in abnormal induction of IL-23 (Fig. 6 C). Thus, RelB, but not p52, is a major inducer of TLR-mediated IL-23 production.
Figure 6.
RelB is critical for DCAF2 deficiency-mediated IL-23 hyperproduction. (A–C) Dendritic cells derived from WT, lym1/+ (A), Relb-KO (B) or Nfκb2-KO (C) mice (n = 3/each group) were stimulated with LPS as indicated. qRT-PCR analysis of Il23a induction. (D) Heat map showing DEGs in WT and NIKΔT3 BMDCs in response to LPS stimulation for 6 h. (E) Venn diagrams represent the overlap of DEGs between WT versus DCAF2DKO and WT versus NIKΔT3 BMDCs stimulated with LPS. (F) The pathways affected by overlapped DEGs were revealed by IPA. (G and H) Skin scaling score and change in skin thickness of DCAF2+/+NIKΔT3/+ and DCAF2−/−NIKΔT3/+ mice treated with Aldara for 6 d (n = 5/group). (I) Il23a in skin samples on day 5 post-Aldara treatment. (J) qRT-PCR analysis of the indicated genes in BMDCs generated from DCAF2+/+NIKΔT3/+ and DCAF2−/−NIKΔT3/+ mice. (K–N) DCAF2DKO mice were crossbred with NIKfl/fl mice to generate DCAF2+/+-NIK−/− and DCAF2DKO-NIK−/− mice. (K) qRT-PCR analysis of the indicated genes in BMDCs generated from these mice. DCAF2+/+-NIK−/− and DCAF2DKO-NIK−/− mice were treated with Aldara for 6 d (n = 5/group). (L) Skin scaling scores was measured each day. (M) Back skin sections were assessed with H&E staining on day 4 post-treatment; bar, 200 µm. (N) qRT-PCR analysis of Il23a and Il17a in skin samples on day 5 post-Aldara treatment. All data are presented as the fold change relative to the Actb mRNA level. Data are presented as the mean ± SEM and are representative of at least three independent experiments. Statistical analyses represent variations in experimental replicates. *, P < 0.05. ns, not significant.
To further examine the role of DCAF2-regulated noncanonical NF-κB in inflammation, we crossed NIKΔT3-floxed mice with CD11c-Cre mice to generate NIKΔT3fl/flCD11cCre (hereafter called NIKΔT3) mice. The NIKΔT3 mutant lacks the TRAF3-binding domain and induces near-complete processing of p100 (Sasaki et al., 2008). We performed transcriptomic analysis to identify DEGs in LPS-stimulated WT and NIKΔT3 BMDCs (Fig. 6 D). Both RNA-seq and qPCR assays revealed that NIK overexpression specifically promoted IL-23 expression (Fig. 6 D). We next evaluated the overlap of DEGs between WT versus NIK∆T3 and WT versus DCAF2 KO BMDCs (Fig. 6 E). For these 173 genes, IPA indicated that a broad range of factors were closely associated with NF-κB activation (Fig. 6 F). We then analyzed the correlation coefficient of RNA-seq data, which showed that mRNA levels of overlapped genes between WT versus NIKΔT3 and WT versus DCAF2DKO were quite significantly correlated (Fig. S3 C). The results indicated that DCAF2 deficiency potentially regulated gene expression via noncanonical NF-κB pathway.
We next investigated the involvement of NIK in mediating IL-23 hyperproduction in DCAF2-deficient BMDCs by crossing DCAF2DKO mice with NIKΔT3 mutant mice. Interestingly, NIK overexpression partially eliminated the differences in severity of Aldara-induced skin inflammation between WT and DCAF2DKO mice (Fig. 6 G). Loss of DCAF2 on the NIKΔT3 background did not appreciably promote the change of skin thickness and IL-23 production (Fig. 6, H–J).
To rule out the artificial effect of NIK overexpression on IL-23 induction, we further crossed NIKfl/fl mice with DCAF2DKO mice. NIK deficiency not only attenuated the induction of IL-23 in the WT control, but also abrogated the hyperinduction of IL-23 in DCAF2 KO BMDCs (Fig. 6 K). Furthermore, both WT and DC-specific ablation of DCAF2 on the NIK KO background failed to induce clinical symptoms and infiltrating immune cells in psoriasis-like skin inflammation (Fig. 6, L and M). Cytokine assays also revealed that loss of NIK eliminated the differences in IL-23 and IL-17A increment mediated by DCAF2 deficiency (Fig. 6 N). Our results identified a critical role for NIK and its downstream factor RelB in mediating the IL-23 hyperproduction of DCAF2-deficient DCs.
CRL4DCAF2 is a negative regulator of NIK
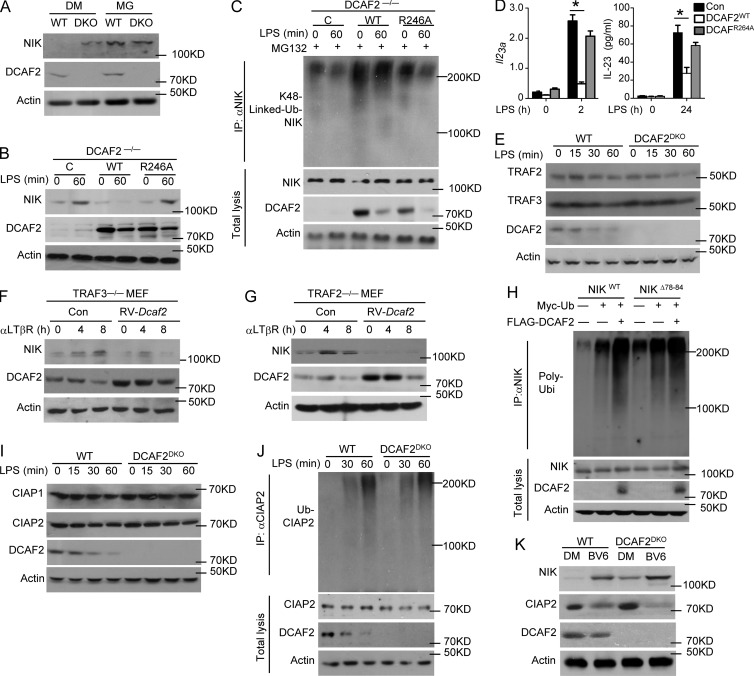
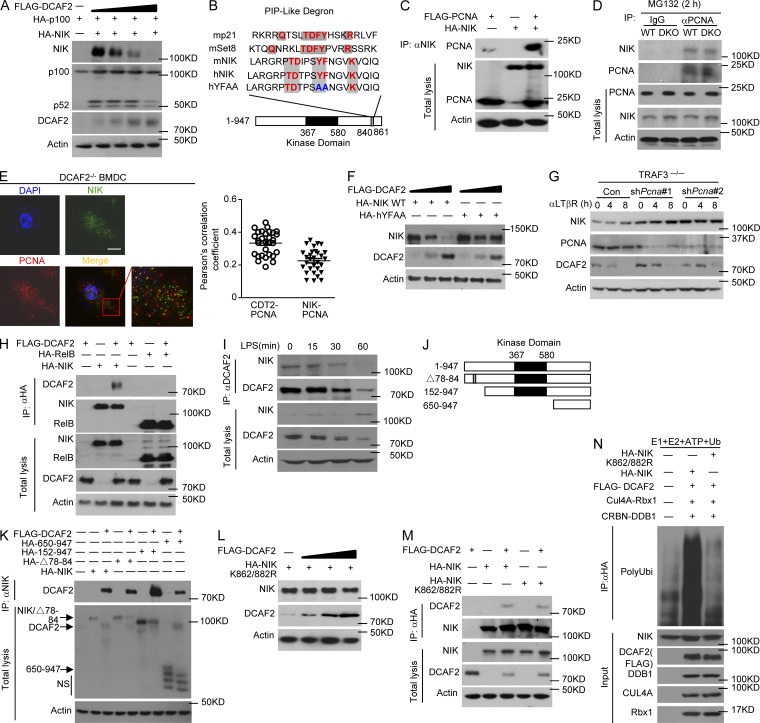
Although our studies clearly demonstrated a role for CRL4DCAF2 in negatively regulating noncanonical NF-κB activation, it remained unclear whether NIK stability was directly affected by DCAF2 deficiency via a proteolytic mechanism. Indeed, upon incubation with MG132 (proteasome inhibitor), the level of NIK in the WT BMDCs was restored to a level similar to that of the DCAF2-deficient cells (Fig. 7 A). This intriguing phenotype was not due to alteration of the GM-CSF receptor expression level (Fig. S4 A). The R246A mutation disrupts the interaction between DCAF2 and DDB1, preventing the assembly of the CRL4-DCAF2 complex (Jin et al., 2006). Compared with WT DCAF2, the R246A mutant lost the ability to mediate NIK degradation (Fig. 7 B). Interestingly, WT DCAF2, but not DCAF2R246A potently promoted polyubiquitination of NIK in DCAF2-deficient BMDCs (Fig. 7 C). The DCAF2R264A mutant did not further suppress Il23a transcription in response to LPS stimuli (Fig. 7 D).
Figure 7.
CRL4DCAF2 induces NIK degradation independent of TRAFs-CIAPs. (A) IB analysis of the NIK levels in BMDCs after pretreatment with DMSO or MG132 for 60 min before harvest. (B–D) IB analysis of NIK level (B) and ubiquitination (C) in LPS-stimulated DCAF2 KO BMDCs reconstituted with empty vector (C), WT DCAF2, or DCAF2 R264A mutant (D; left) and ELISA (right) analysis of Il23a expression in BMDCs as described above in response to LPS stimulation. qRT-PCR data are presented as the fold change relative to the Actb mRNA level. (E) IB analyses of TRAFs expression in whole-cell lysates from WT and DCAF2 KO BMDCs. (F and G) TRAF3- (F) or TRAF2-deficient (G) MEFs reconstituted with empty vector (Con) or WT DCAF2 with retroviral vectors. IB analysis of the indicated proteins in reconstituted MEFs stimulated with αLTβR. (H) HEK293T cells were transfected with Myc-tagged ubiquitin along with the indicated expression plasmids. NIK was isolated by IP using anti-NIK, followed by detection of the ubiquitination level by IB with anti-ubiquitin. Cell lysates were also subjected to direct IBs (bottom three panels). (I and J) IB analyses of CIAP level (I) and LPS-triggered polyubiquitination (J) in WT and DCAF2 KO BMDCs. (K) IB analyses of NIK level in WT and DCAF2 KO BMDCs pretreated with DMSO or the Smac mimetic BV6 for 4 h. Data are presented as the mean ± SEM and are representative of at least three independent experiments. Statistical analyses represent variations in experimental replicates. *, P < 0.05.
TRAF3 is recognized as a negative regulator of the noncanonical NF-κB pathway (Liao et al., 2004; Zarnegar et al., 2008). DCAF2 deficiency did not promote TRAF3 or TRAF2 degradation, which is ever been known to be critical for NIK accumulation (Sasaki et al., 2008; Vallabhapurapu et al., 2008; Bista et al., 2010; Fig. 7 E). To further clarify the roles of TRAFs in CRL4DCAF2-mediated NIK degradation, we evaluated the function of CRL4DCAF2 in TRAF3- or TRAF2-KO MEFs. DCAF2 overexpression in WT, TRAF3−/−, or TRAF2−/− background caused significant NIK degradation (Fig. 7, F and G; and Fig. S4, B and C). The N-terminal △78-84 mutant of NIK destroyed its interaction of TRAF3 and impaired subsequent ubiquitination. However, CRL4DCAF2 still induced ubiquitination of NIK△78-84 (Fig. 7 H), indicating that CRL4DCAF2-mediated NIK degradation is independent of TRAFs.
cIAP1/2 activation is mediated through its K63 ubiquitination by TRAF2 and promotes NIK degradation (Vallabhapurapu et al., 2008). Thus, CRL4DCAF2 may regulate noncanonical NF-κB by promoting cIAP1/2 K63 ubiquitination and activation. DCAF2 deficiency had little or no effect on the protein levels of cIAP1 and cIAP2 in LPS-stimulated BMDCs (Fig. 7 I). Additionally, LPS triggered comparable ubiquitination of cIAP2 between WT and DCAF2 KO BMDCs (Fig. 7 J). Moreover, incubation with BV6, a specific cIAP inhibitor, did not restore NIK accumulation caused by DCAF2 deficiency (Fig. 7 K). These results suggest that CRL4DCAF2 promotes NIK degradation independent of regulating cIAP activity.
CRL4DCAF2 induces NIK ubiquitination and degradation
To further clarify the mechanism by which CRL4DCAF2 regulates NIK, we tested whether CRL4DCAF2 induces NIK degradation directly in HEK293T cells. Indeed, following cotransfection with DCAF2, NIK and processing of p100 were drastically reduced (Fig. 8 A). Similar to p21 and Set8, NIK also contains a “PIP-like degron,” which consists of a noncanonical proliferating cell nuclear antigen (PCNA) interaction motif (Fig. 8 B). Co-immunoprecipitation (coIP) assays both in vitro (Fig. 8 C) and in vivo (Fig. 8 D) readily detected the physical association of NIK with PCNA in HEK 293T cells and DCAF2-deficient DCs, respectively. In confocal images, the analysis further revealed that colocalization of cytoplasmic PCNA and NIK was observed in resting BMDCs, which exhibited quite similar Pearson’s correlation coefficient of interaction between PCNA and DCAF2 (Fig. 8 E). Mutation of the PIP degron (hYFAA) in NIK partially rescued its CRL4DCAF2-mediated degradation (Fig. 8 F). To further clarify the role of PCNA in NIK degradation, TRAF3−/− MEFs were stably transfected with PCNA shRNA to knock down PCNA gene expression, as confirmed by IB analysis. Notably, PCNA shRNA significantly restored NIK reduction, indicating that PCNA play a negative role in regulating NIK protein level (Fig. 8 G). Additionally, PCNA silencing in WT BMDCs led to increment of Il23a expression upon LPS stimulation (Fig. S4 D).
Figure 8.
CRL4DCAF2 promotes PCNA-dependent NIK ubiquitination. (A) HEK293T cells were transfected with (+) or without (−) the indicated expression vectors. After 36 h, whole-cell lysates were subjected to IB to detect the indicated proteins. (B) Alignment of the PIP degron from proteins that are considered targets for destruction by CRL4DCAF2. Canonical degron-like residues are shown in red. Mutated amino acids are shown in blue. (C and D) The interaction between NIK and PCNA was assessed in 293T cells by cotransfection with the indicated gene plasmids (C) or in WT/DCAF2 KO BMDCs (D). Whole-cell lysates were subjected to IP using anti-HA, followed by IB analyses of the associated FLAG-PCNA using anti-FLAG. (E) Confocal microscopy analysis of NIK, PCNA, and DAPI (nuclear staining) and a merged picture in DCAF2 KO BMDCs. Bars, 5 µm. The quantification of PCNA/NIK colocalization was performed as described in Materials and methods using the Pearson's correlation coefficient (n = 30). (F) IB analyses of NIK level in HEK293T cells transfected with the NIK WT or NIK YFAA mutation plasmid along with DCAF2 expression plasmids. (G) TRAF3−/− MEFs infected with pGIPZ lentiviral vectors encoding a nonsilencing control shRNA (C) or two different Pcna-specific shRNAs. GFP+ cells were sorted with FACS and then stimulated with αLTβR. Whole-cell lysates was subjected to IB to detect the NIK level. (H) NIK-DCAF2 interaction assays were performed of HEK293T cells cotransfected with the indicated gene plasmids. After treatment with MG132 for 2 h, whole-cell lysates were subjected to IP using anti-HA, followed by IB analysis of the associated FLAG-DCAF2. (I) WT BMDCs were stimulated with LPS, and whole-cell lysates were subjected to IP using anti-DCAF2, followed by IB analysis of the DCAF2-associated NIK. Cell lysates were also subjected to direct IBs (bottom three panels). (J and K) NIK-DCAF2 interaction analyses in HEK293T cells transfected with HA-NIK or its truncation mutants either in the presence (+) or absence (−) of FLAG-DCAF2 were subjected to coIP analyses. (L and M) Mutated NIK stability (L) and coIP assays (M) were performed by detecting NIK K862/882R expression and its association with DCAF2. (N) In vitro assays were used to detect CRL4DCFA2-mediated NIK polyubiquitination. In samples with a mixture of E1, E2, ATP, Flag-DCAF2, Cul4A-Rbx1 and CRBN-DDB1, HA-NIK, or NIK K862/882R ubiquitination was detected by IB after IP with anti-HA. Data are representative of at least three independent experiments.
Our findings indicated that CRL4DCAF2 directly regulates NIK ubiquitination and stability. The NIK-DCAF2 physical interaction was confirmed in cotransfected HEK293T cells, which also showed that only NIK, but not RelB, bound to DCAF2 (Fig. 8 H). In the absence of a stimulus, DCAF2 physically associated with endogenous NIK in BMDCs; however, upon LPS stimulation, the decrease in the amount of co-immunoprecipitating NIK was caused by the reduction in DCAF2 expression (Fig. 8 I). Domain mapping analyses revealed that the 650-C terminal region of NIK was required for its physical interaction with DCAF2 (Fig. 8, J and K). The 650-C truncation further suggested that this region contains specific ubiquitinated sites of NIK mediated by CRL4DCAF2 (Fig. 8 K). Interestingly, the NIK point mutation K862/882R retained its ability to interact with DCAF2, but was resistant to CRL4DCAF2-induced degradation, indicating these sites were critical for NIK stability (Fig. 8, L and M). Additionally, in vitro ubiquitination assays demonstrated that CRL4DCAF2 promoted the ubiquitination of WT NIK, but not its K862/882R mutant, in the presence of E1, E2, and ATP (Fig. 8 N). Collectively, these data suggested that CRL4DCAF2 serves as a direct E3 ligase and induces the PCNA-dependent degradation of NIK.
Discussion
DCs function as essential APCs that link innate and adaptive immunity and are involved in T cell responses (Guermonprez et al., 2002; Bousso, 2008). When exposed to microbial components, DCs are activated to produce various proinflammatory cytokines and mediate potent anti-infectious functions (Guermonprez et al., 2002). Type 1 DCs are characterized by the production of high levels of proinflammatory mediators, such as IL-12, IL-23, and inducible nitric oxide synthase (iNOS; Kaliński et al., 1999; Segura and Amigorena, 2013). Deregulated IL-23 in activated DCs contribute to the pathogenesis of various autoimmune and inflammatory disorders (Teng et al., 2015). Therefore, elucidation of the mechanisms underlying the regulation of IL-23 in innate immune cells is important for therapeutic approaches for autoimmune diseases.
IL-23 production is triggered by innate immune receptors, particularly pattern-recognition receptors (PRRs), which detect various molecular patterns associated with invading pathogens. The downstream signals activated by the PRRs include canonical NF-κB and MAPKs. Activation of NF-κB by cytoplasmic IKKs plays a central role in inflammation via the production of proinflammatory cytokines. Among the five members of the NF-κB family, p65 and c-Rel are crucial for TLR-stimulated expression of proinflammatory cytokines, including IL-12 and IL-23 (Sanjabi et al., 2000). ChIP assays also demonstrated that RelB was recruited to the promoter regions of the Tnf and Il23a genes after DCs were activated by CpG (Shih et al., 2012). RelB is potentially translocated into the nucleus and activated when signal-induced noncanonical NF-κB kinase NIK degradation occurs (Sun, 2011). Recent evidence has revealed that disordered RelB in different cell types is generally associated with various autoimmune and inflammatory conditions, including IgA nephropathy (Jin et al., 2012), rheumatoid arthritis (Pettit et al., 2000), EAE (Ginwala et al., 2017), systemic lupus erythematosus (Wu et al., 2016), and psoriasis (Barton et al., 2000). However, to date, it remains largely unclear how RelB and the upstream kinase NIK are negatively regulated in myeloid cells. In the present study, we identified an E3 ubiquitin ligase, CRL4DCAF2, which was significantly suppressed in DCs isolated from patients with autoimmune diseases and negatively regulated the noncanonical NK-κB pathway. CRL4DCAF2 is believed to be a critical mediator that regulates cell cycle progression (Sansam et al., 2006), but its in vivo functions, especially those in the immune system, are poorly understood due to the embryonic lethality of conventional DCAF2 KO mice. Here, we found that DC-specific DCAF2 ablation resulted in aberrant IL-23 hyperproduction, coupled with enhanced autoimmune disease symptoms. Thus, our data strongly suggest an important role for CRL4DCAF2 in controlling IL-23 production and indicated that it may function in inflammatory diseases.
In several human malignancies, amplified CRL4DCAF2 targets various critical cell cycle regulators during S phase and prevents the reinitiation of DNA replication (Sansam et al., 2006; Kim et al., 2008; Abbas and Dutta, 2011). Thus far, the physiological role of CRL4DCAF2 in the inflammatory response remains elusive. In vivo, administration of MLN4924, an inhibitor of cullin neddylation, mimicked the physiological effects of DCAF2 and accentuated the disease severity of TNBS-induced colitis. Indeed, MLN4924 increased mortality of the inflammatory response (Curtis et al., 2014, 2015). However, the underlying mechanism remains controversial. Our data also revealed that MLN4924 potentially promoted Aldara-induced psoriasis and increased IL-23/IL-17 cytokine production. These results establish CRL4DCAF2 as a novel regulator of the innate immune response and proinflammatory cytokine production. Surprisingly, CRL4DCAF2 has no or only a slight effect on the development or homeostasis of DCs. Recently, several studies indicated that chronic inflammation acts on genetic and epigenetic changes involved in the regulation of critical cell cycle checkpoints (Altznauer et al., 2004; Hoodless et al., 2016). However, there is little evidence concerning whether cell cycle checkpoints also contribute to inflammatory regulation. Recently, ataxia telangiectasia mutated kinase (ATM) was shown to inhibit IL-23 release via XBP1, a key component of the endoplasmic reticulum stress response (Wang et al., 2013). However, their mechanistic model did not clarify their observation of elevated Il23a transcription, which was independent of ER function. Here, our studies revealed that deficiency of the cell cycle regulator CRL4DCAF2 promoted IL-23 induction and Th17 polarization by enhancing NIK accumulation. DCAF2 deficiency had no effect on canonical NF-κB activation but strikingly enhanced GM-CSF–induced activation of noncanonical NF-κB. Our data establish a pivotal role of cell cycle dysregulation in triggering inflammatory responses.
In contrast to the TRAF3 deficiency, loss of DCAF2 did not cause constitutive noncanonical NF-κB activation, suggesting a specific role for CRL4DCAF2 in controlling signal-induced noncanonical NF-κB activation during the cell cycle process. Interestingly, CRL4DCAF2 did not affect signal-induced TRAF3 ubiquitination and degradation in response to GM-CSF and LPS stimulation. Our results indicated that CRL4DCAF2 negatively regulated signal-induced noncanonical NF-κB signaling by physically interacting with NIK and promoted NIK degradation. Additionally, CRL4DCAF2 directly ubiquitinated NIK at K862/882, but did not control the activation of the TRAF3-specific E3 ubiquitin ligase cIAP1/2. Notably, recent studies have suggested the involvement of both TRAF3/cIAP-dependent and independent mechanisms in regulating NIK stability in malignant B cells. Our studies suggest that CRL4DCAF2 may be responsible for TRAF3/cIAP-independent NIK degradation. Therefore, our findings are crucial for illustrating the molecular mechanism that controls noncanonical NF-κB signaling. These results not only greatly advance the field, but also have profound implications for therapeutic approaches for autoimmune diseases.
Materials and methods
Mice
Mice were maintained under specific pathogen-free conditions in a controlled environment of 20–22°C, with a 12/12 h light/dark cycle and 50–70% humidity; food and water were provided ad libitum. Dcaf2fl/fl mice with B6 background were generated by the Model Animal Resource Information Platform, Model Animal Research Center of Nanjing University. Embryonic stem cells used to generate this mouse strain were purchased from the European Conditional Mouse Mutagenesis Program (embryonic stem cell clone EPD0842_C05). The Dcaf2fl/fl mice were further crossed with Lyz2-Cre mice (all from Jackson Laboratory; C57BL/6 background) to generate myeloid cell conditional DCAF2 KO (Dcaf2f/fLyz2Cre/+) and dendritic cell conditional DCAF2 KO (Dcaf2f/fCD11cCre/+) mice. Il23a-KO mice were provided by C. Dong (Tsinghua University, Beijing, China). Nfκb2Lym1 mutant mice (Tucker et al., 2007) were provided by S.-C. Sun (University of Texas MD Anderson Cancer Center, Houston, TX) and generated by R. Starr (St. Vincent’s Institute, Fitzroy, Victoria, Australia) and The Walter and Eliza Hall Institute of Medical Research (Parkville, Victoria, Australia). We used the Lym1/+ and age-matched WT controls in the experiments. The Nfkb2xdr/xdr mice (Miosge et al., 2002) were provided by S.-C. Sun and the Australian National University and are available from the Australian Phenome Bank, ID 93. Relb-KO mice were gifted form S.-C. Sun and Bristol-Myers Squibb Pharmaceutical Research Institute. NIKΔT3-floxed mice (Jackson Lab) were crossed with CD11c-Cre transgenic mice to produce age-matched NIKΔT3fl/fl-CD11c+/+ (termed WT) and NIKΔT3fl/fl-CD11cCre/+ (termed NIKΔT3) mice for experiments. The NIKfl/fl mice were provided by H. Hu (West China Hospital, Sichuan University, and Collaborative Innovation Center of Biotherapy, Chengdu, China). Heterozygous mice were bred to generate littermate controls and conditional KO mice for experiments. In the animal studies, the WT and multiple KO mice are randomly grouped. Outcomes of animal experiments were collected blindly and recorded based on ear tag numbers of 6–8-wk-old experimental mice. Mice were genotyped by PCR using genomic tail DNA. All animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Zhejiang University.
Human specimen analysis
Patient samples for this study were recruited from Zhejiang University Sir Run Run Shaw Hospital. Samples from IBD or psoriasis patients were collected and analyzed after informed consent was obtained. The diagnosis was based on a standard combination of clinical, endoscopic, histological, and radiological criteria. The use of pathological specimens, as well as the review of all pertinent patient records, was approved by the Ethics Review Board of Zhejiang University Sir Run Run Shaw Hospital.
Antibodies, plasmids, and reagents
Antibodies targeting IKBα (C-21; 1:1,000), p65 (C-20; 1:1,000), Lamin B (C-20; 1:1,000), ERK (K-23; 1:2,000), phospho-ERK (E-4; 1:1,000), JNK2 (C-17; 1:1,000), p38 (H-147; 1:1,000), ubiquitin (P4D1; 1:1,000), p105/p50 (C-19; 1:1,000), RelB (C-19; 1:1,000), NIK (H248; 1:1,000), TRAF2 (C-20; 1:1,000), TRAF3 (C-20; 1:1,000), cIAP1 (H83; 1:1,000), cIAP2 (H85; 1:1,000), and c-Rel (sc-71; 1:1,000), as well as a control rabbit IgG (sc-2027), were from Santa Cruz Biotechnology. Phospho-IκBα (Ser32; 14D4; 1:1,000), phospho-JNK (Thr180/Tyr185; 9251; 1:1,000), phospho-p38 (Thr180/Tyr182; 3D7; 1:1,000), phospho-p105 (Ser933; 18E6; 1:1,000), p100/p52 (4882; 1:1,000), and K48-linkage specific polyubiquitin (D9D5; 1:8,000) were purchased from Cell Signaling Technology. Anti-actin (C-4; 1:10,000) was from Sigma-Aldrich. HRP-conjugated anti-HA (HA-7) and anti-FLAG (M2) were from Sigma-Aldrich. DCAF2 (184548; 1:1,000) and PCNA (PC10; 1:1,000) were from Abcam. Anti-NIK (1:1,000) was homemade. Fluorescence-labeled antibodies are listed in the section describing the flow cytometry and cell sorting procedures.
Mouse Dcaf2 and Relb cDNA were amplified from splenic mouse mRNA using PCR and inserted into the pCLXSN(GFP) retroviral vector. HA-Ub, HA-NIK plasmids were provided by S.-C. Sun. pGIPZ lentiviral vectors encoding a nonsilencing shRNA control and two different Pcna shRNAs were designed and produced by Invitrogen.
LPS (derived from Escherichia coli strain 0127:B8) and CpG (2216) were from Sigma-Aldrich. R848 and pIC was from Amersham, and recombinant murine GM-CSF was from Peprotech.
Flow cytometry, cell sorting, and intracellular cytokine staining
Single-cell suspensions from B16 melanoma, Spls, or draining lymph nodes were subjected to flow cytometry using CytoFlex (Beckman Coulter) and the following fluorescence-labeled antibodies from eBioscience: PB-conjugated anti-CD4 and anti-CD11c; PE-conjugated anti-B220, anti-IL-17, and anti-F4/80; PerCP5.5-conjugated anti–Gr-1 (Ly6G); APC-conjugated anti-CD62L; APC-CY7–conjugated anti-CD11b and anti-CD8; and FITC-conjugated anti–IFN-γ, anti-CD44, and anti-Foxp3. DAPI was from Life Technologies, and MitoSpy Orange CMTMRos was from BioLegend.
For intracellular cytokine staining, the tumor-infiltrating T cells were stimulated for 4 h with PMA plus ionomycin in the presence of monensin and then subjected to intracellular IFN-γ and subsequent flow cytometry analysis. All FACS data were analyzed by FlowJo 7.6.1.
IB, immunoprecipitation (IP), and kinase assays
Whole-cell lysates or subcellular extracts were prepared and subjected to IB and IP assays as described. The samples were resolved by 8.25% SDS-PAGE. After electrophoresis, separated proteins were transferred onto polyvinylidene difluoride membrane (Millipore). For immunoblotting, the polyvinylidene difluoride membrane was blocked with 5% nonfat milk. After incubation with specific primary antibody, HRP-conjugated secondary antibody was applied. The positive immune reactive signal was detected by enhanced chemiluminescence (Amersham Biosciences). All relative density of Western blot results were quantified and tested statistical significance in Fig. S5.
Fluorescence microscopy
BMDCs (5 × 105) were collected and seeded on 12-well plates containing 70% alcohol-pretreated slides for starvation overnight. Cells treated with or without stimulation as indicated were fixed with 4% paraformaldehyde (PFA) for 20 min. Then, the cells were washed with PBS three times and stained with 10 µg/ml DAPI and FITC-conjugated anti-DCAF2. All the samples were imaged on a LSM710 (Carl Zeiss) confocal microscope outfitted with a Plan-Apochromat 63× oil immersion objective lens (Carl Zeiss). The data were collected using Carl Zeiss software ZEN 2010. Colocalization analyses were performed on 30 cells, was expressed as a Pearson's coefficient (R).
Generation of BMDCs and BMDMs
WT, myeloid cell–, or dendritic cell–specific KO mice were sacrificed by cervical dislocation and removal of both femurs. After removing the muscle and fat with a pair of scissors, the femurs were put in 5 ml DMEM (7% FCS and PS) on ice. The ends of the bone were cut with a sharp pair of scissors in the tissue culture hood and then the BM were flushed out with a 10-ml syringe (25-G needle) and DMEM media (7% FCS, PS). BM cells were collected in a 50-ml tube, and centrifuge at 1,600 rpm for 5 min. These BM cells were cultured in DMEM or RPMI 1640 medium containing 20% FBS supplemented with M-CSF (10 ng/ml) or GM-CSF (10 ng/ml) for 5–8 d, respectively. 2 d later, these cells were changed media and cultured for two more days. The macrophages were harvested and replated to obtain even number of cells on different plates. The cells can be split 1:2 or higher dilution based on time schedule. To harvest macrophages, these cells were washed once with cold PBS, incubate in 4 ml PBS containing 10 mM EDTA and 20% FCS at 37°C for 5 min. The differentiated DCs were stained with Pacific blue–conjugated anti-CD11c and isolated with a FACS sorter.
BMDMs and BMDCs were starved overnight in medium supplemented with 0.5% FCS before being stimulated with LPS (1 µg/ml for IB experiments and 100 ng/ml for cytokine induction experiments), pIC (20 µg/ml), CpG (2216, 5 µM), and murine recombinant IL-1β (20 ng/ml). Total and subcellular extracts were prepared for IB assays, and total RNA was prepared for qRT-PCR assays.
Histopathology
Organs were removed from WT or DCAF2DKO mice, fixed in 10% neutral buffered formalin, embedded in paraffin and sectioned for staining with H&E. Slides were numbered randomly and evaluated blindly by two different investigators to determine inflammation scores. The quantity of perivascular or peribronchial inflammation was assessed as described previously (Jin et al., 2012), with some modifications. In brief, a grade of 0 was assigned when no inflammation was detectable, a grade 1 for occasional cuffing with inflammatory cells, and grades 2–4 when most bronchi or vessels were surrounded by a thin layer (one to three cells), a moderate layer (four to five cells), or a thick layer (more than five cells deep) of inflammatory cells, respectively. An increment of 0.5 was given if the inflammation fell between two grades. Total inflammation score was calculated by addition of both peribronchial and perivascular inflammation scores.
Aldara-induced psoriasis-like skin inflammation
Mice at 7–10 wk of age received a daily topical dose of 62.5 mg of commercially available Imiquimod (IMQ) cream (5%; Aldara; 3M Pharmaceuticals) on the shaved back for five consecutive days, and then translated in a daily dose of 3.125 mg of the active compound. Aldara (IMQ) cream was obtained from Valeant (3M Health Care Limited). Samples of back skin and Spls from these mice were collected immediately upon sacrifice on day 6, flash frozen, and stored at −80°C before isolation of total RNA. Skin adjacent to that collected for RNA was placed into formalin and processed for histology.
Induction and assessment of EAE
For active EAE induction, age- and sex-matched mice were immunized s.c. with MOG35-55 peptide (300 µg) mixed in CFA (Sigma-Aldrich) containing 5 mg/ml heat-killed Mycobacterium tuberculosis H37Ra (Difco). Pertussis toxin (200 ng, List Biological Laboratories) in PBS was administered i.v. on days 0 and 2. Mice were examined daily and scored for disease severity using the standard scale: 0, no clinical signs; 1, limp tail; 2, paraparesis (weakness, incomplete paralysis of one or two hind limbs); 3, paraplegia (complete paralysis of two hind limbs); 4, paraplegia with forelimb weakness or paralysis; 5, moribund or death. After the onset of EAE, food and water were provided on the cage floor. For visualizing CNS immune cell infiltration and demyelination, spinal cords of the EAE-induced mice were collected on day 30 and subjected to H&E staining, respectively. Mononuclear cells were prepared from the CNS (brain and spinal cord) of EAE-induced mice as described (Jin et al., 2009) and analyzed by flow cytometry.
shRNA knockdown
Lentiviral particles were produced by transfecting HEK293T cells (using the calcium-phosphate method) with a pGIPZ lentiviral vector encoding either a nonsilencing shRNA or Pcna-specific shRNA, along with the packaging vectors psPAX2 and pMD2. BMDMs, differentiated using GM-CSF–supplemented medium for 8 d, were infected with the lentiviruses for 8 h. After 72 h, the infected cells were enriched via flow cytometric cell sorting (based on GFP expression) and subsequently used for experiments.
RNA-seq analysis
Primary BMDCs from WT and DCAF2DKO mice (6–8-wk old) were stimulated with LPS. 6 h later, BMDCs were used for total RNA isolation with Trizol (Invitrogen), and subjected to RNA-seq analysis. RNA sequencing was performed by the Life Science Institute Sequencing and Microarray Facility using an Illumina sequencer. The raw reads were aligned to the mm10 reference genome (build mm10), using Tophat2 RNA-seq alignment software. The mapping rate was 70% overall across all the samples in the dataset. HTseq-Count was used to quantify the gene expression counts from Tophat2 alignment files. Differential expression analysis was performed on the count data using R package DESeq2. P values obtained from multiple binomial tests were adjusted using false discovery rate with Benjamini and Hochberg method. Significant genes are defined by a Benjamini and Hochberg corrected P value of cut-off of 0.05 and fold-change of at least 1.5.
ELISA and qRT-PCR
Supernatants of in vitro cell cultures were analyzed via ELISA using a commercial assay system (eBioScience). For qRT-PCR, total RNA was isolated using TRIzol reagent (Molecular Research Center, Inc.) and subjected to cDNA synthesis using RNase H-reverse transcription (Invitrogen) and oligo (dT) primers. qRT-PCR was performed in triplicate using an iCycler Sequence Detection System (Bio-Rad) and iQTM SYBR Green Supermix (Bio-Rad). The expression of individual genes was calculated with a standard curve and normalized to the expression of Actb. The gene-specific PCR primers (all for mouse genes) are shown in Table 1.
Table 1. Gene-specific primers used in qRT-PCR experiments.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Il6 | CACAGAGGATACCACTCCCAACA | TCCACGATTTCCCAGAGAACA |
| Tnf | CATCTTCTCAAAATTCGAGTGACAA | CCAGCTGCTCCTCCACTTG |
| Il12p35 | ACTAGAGAGACTTCTTCCACAACAAGAG | GCACAGGGTCATCATCAAAGAC |
| Il12p40 | GGAGACACCAGCAAAACGAT | TCCAGATTCAGACTCCAGGG |
| Il10 | CCAGAGCCACATGCTCCTAGA | GGTCCTTTGTTTGAAAGAAAGTCTTC |
| Il23p19 | GCCAAGAAGAC CATTCCCGA | TCAGTGCTACAATCTTCTTCAGAGGACA |
| Arg1 | TTTTTCCAGCAGACCAGCTT | AGAGATTATCGGAGCGCCTT |
| Mrc1 | CAGGTGTGGGCTCAGGTAGT | TGGCATGTCCTGGAATGAT |
| Nfkb2 | CATCCATGACAGCAAGTCTC | TCCTCATAGAACCGAACCTC |
| Relb | GAATGTCGTCAGGATCTGC | TGGTGGACTTCTTGTCGTAG |
| Map3k14 | GTACTTCCGAGGCCCGCTTT | GTGGACGGGCTCTTTCCTCA |
Ubiquitination assays
Cells were pretreated with MG132 for 2 h and then lysed with a Nonidet P-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, and 1 mM DTT) containing 6 M urea and protease inhibitors. IRF1 or ChIP was isolated by IP with antibodies targeting NIK (H-248) and CIAP2 (H85). The ubiquitinated proteins were detected by IB using an anti-ubiquitin (P4D1; Santa Cruz) or anti–K48-linked ubiquitin (05-1307; Millipore) antibody.
Statistical analysis
Statistical analysis was performed using Prism software. No data are excluded from the analyses. Two-tailed unpaired t tests were performed. P values <0.05 were considered significant, and the level of significance is indicated as *, P < 0.05 and **, P < 0.01. In the animal studies, a minimum of four mice were required for each group based on the calculated number necessary to achieve a 2.3-fold change (effect size) in a two-tailed t test with 90% power and a significance level of 5%. All statistical tests are justified as appropriate, and the data meet the assumptions of the tests. The variance is similar between the statistically compared groups.
Data availability
Sequence data that support the findings of this study are available from the authors and have been deposited in the National Center for Biotechnology Information. For ChIP-seq data, the primary accession code is PRJNA359723. For RNA-seq data, the primary code is PRJNA417890.
Online supplemental material
Fig. S1 shows that MLN4924 treatment enhances the production of IL-17 in γδ T cells under inflammatory condition. Fig. S2 shows DCAF2 deficiency promotes autoimmune symptom in psoriasis mice or nontreated aging mice. Fig. S3 shows that DCAF2 negatively regulates noncanonical NF-κB, but no effect on capacity of antigen presentation in DCs. Fig. S4 shows that DCAF2 does not affect the protein level of GM-CSF receptor, but regulates NIK stability independent of TRAF2 or TRAF3. Fig. S5 shows the relative density of all Western blot results.
Supplementary Material
Acknowledgments
We thank Dr. Shao-cong Sun and Dr. Chen Dong for sharing the mice.
This study was supported by the National Key Research and Development Program of China (grant 2018YFD0500100), the National Natural Science Foundation of China (grants 81572651 and 81771675), Fundamental Research Funds for the Central Universities (grant 2018LZA6015), the Thousand Young Talents Plan of China, and the Zhejiang University Special Fund for Fundamental Research.
The authors declare no competing financial interests.
Author contributions: T. Huang and Z. Gao performed the research, prepared the figures, and wrote the manuscript; Y. Zhang, K. Fan, F. Wang, Y. Li., and J. Zhong contributed to the experiments; H.Y. Fan, Q. Cao, J. Zhou, Y. Xiao, and H. Hu contributed to the experimental design; J. Jin supervised the work, prepared the figures, and wrote the manuscript.
References
- Abbas T., and Dutta A.. 2011. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle. 10:241–249. 10.4161/cc.10.2.14530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., and Dutta A.. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22:2496–2506. 10.1101/gad.1676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altznauer F., Martinelli S., Yousefi S., Thürig C., Schmid I., Conway E.M., Schöni M.H., Vogt P., Mueller C., Fey M.F., et al. 2004. Inflammation-associated cell cycle-independent block of apoptosis by survivin in terminally differentiated neutrophils. J. Exp. Med. 199:1343–1354. 10.1084/jem.20032033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D., HogenEsch H., and Weih F.. 2000. Mice lacking the transcription factor RelB develop T cell-dependent skin lesions similar to human atopic dermatitis. Eur. J. Immunol. 30:2323–2332. [DOI] [PubMed] [Google Scholar]
- Ben-Neriah Y., and Karin M.. 2011. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12:715–723. 10.1038/ni.2060 [DOI] [PubMed] [Google Scholar]
- Bista P., Zeng W., Ryan S., Bailly V., Browning J.L., and Lukashev M.E.. 2010. TRAF3 controls activation of the canonical and alternative NFkappaB by the lymphotoxin beta receptor. J. Biol. Chem. 285:12971–12978. 10.1074/jbc.M109.076091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousso P. 2008. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nat. Rev. Immunol. 8:675–684. 10.1038/nri2379 [DOI] [PubMed] [Google Scholar]
- Centore R.C., Havens C.G., Manning A.L., Li J.-M., Flynn R.L., Tse A., Jin J., Dyson N.J., Walter J.C., and Zou L.. 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell. 40:22–33. 10.1016/j.molcel.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow E.K., O’connell R.M., Schilling S., Wang X.F., Fu X.Y., and Cheng G.. 2005. TLR agonists regulate PDGF-B production and cell proliferation through TGF-β/type I IFN crosstalk. EMBO J. 24:4071–4081. 10.1038/sj.emboj.7600867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis V., Ehrentraut S., Campbell E., Glover L., Bowers B., Bayless A., Kelly C., Kominsky D., and Colgan S.. 2014. Contribution of human deneddylase-1/SENP8 to the mucosal inflammatory response (488.1). FASEB J. 28:1.24385568 [Google Scholar]
- Curtis V., Ehrentraut S., Campbell E., Glover L., Bayless A., Kelly C., Kominsky D., and Colgan S.. 2015. The Influence of Neddylation on the Mucosal Inflammatory Response. FASEB J. 29:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginwala R., McTish E., Moore P., Revuri N., Raman C., Singh N.P., Nagarkatti M., Nagarkatti P., Kranz V.A., Houle J.D., et al. 2017. Nutraceutical Apigenin regulates DC function in a RelB-dependent manner during neuroinflammation. J. Immunol. 198:219.2. [Google Scholar]
- Gordon K.B., Duffin K.C., Bissonnette R., Prinz J.C., Wasfi Y., Li S., Shen Y.-K., Szapary P., Randazzo B., and Reich K.. 2015. A Phase 2 Trial of Guselkumab versus Adalimumab for Plaque Psoriasis. N. Engl. J. Med. 373:136–144. 10.1056/NEJMoa1501646 [DOI] [PubMed] [Google Scholar]
- Guermonprez P., Valladeau J., Zitvogel L., Théry C., and Amigorena S.. 2002. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 20:621–667. 10.1146/annurev.immunol.20.100301.064828 [DOI] [PubMed] [Google Scholar]
- Hasan U.A., Caux C., Perrot I., Doffin A.-C., Menetrier-Caux C., Trinchieri G., Tommasino M., and Vlach J.. 2007. Cell proliferation and survival induced by Toll-like receptors is antagonized by type I IFNs. Proc. Natl. Acad. Sci. USA. 104:8047–8052. 10.1073/pnas.0700664104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., and Ghosh S.. 2011. NF-κB in immunobiology. Cell Res. 21:223–244. 10.1038/cr.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard B.A., Mason N., Xu L., Sun J., Lamhamedi-Cherradi S.-E., Liou H.-C., Hunter C., and Chen Y.H.. 2002. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J. Clin. Invest. 110:843–850. 10.1172/JCI0215254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J., Mair F., Greter M., Schmidt-Supprian M., and Becher B.. 2011. NIK signaling in dendritic cells but not in T cells is required for the development of effector T cells and cell-mediated immune responses. J. Exp. Med. 208:1917–1929. 10.1084/jem.20110128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoodless L.J., Robb C.T., Felton J.M., Tucker C.S., and Rossi A.G.. 2016. Models for the Study of the Cross Talk Between Inflammation and Cell Cycle. In Cyclin-Dependent Kinase (CDK) Inhibitors: Methods and Protocols. M. Orzáez, M. Sancho Medina, and E. Pérez-Payá, editors. Humana, NY: pp. 179–209. 10.1007/978-1-4939-2926-9_15 [DOI] [PubMed] [Google Scholar]
- Hu H., Brittain G.C., Chang J.-H., Puebla-Osorio N., Jin J., Zal A., Xiao Y., Cheng X., Chang M., Fu Y.-X., et al. 2013. OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature. 494:371–374. 10.1038/nature11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Arias E.E., Chen J., Harper J.W., and Walter J.C.. 2006. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 23:709–721. 10.1016/j.molcel.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Jin J., Xiao Y., Chang J.-H., Yu J., Hu H., Starr R., Brittain G.C., Chang M., Cheng X., and Sun S.-C.. 2012. The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-κB signaling. Nat. Immunol. 13:1101–1109. 10.1038/ni.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Hu H., Li H.S., Yu J., Xiao Y., Brittain G.C., Zou Q., Cheng X., Mallette F.A., Watowich S.S., and Sun S.-C.. 2014. Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity. Immunity. 40:342–354. 10.1016/j.immuni.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Zhou X.F., Yu J., Cheng X., and Sun S.C.. 2009. Regulation of Th17 cell differentiation and EAE induction by MAP3K NIK. Blood. 113:6603–6610. 10.1182/blood-2008-12-192914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliński P., Hilkens C.M.U., Wierenga E.A., and Kapsenberg M.L.. 1999. T-cell priming by type-1 and type-2 polarized dendritic cells: the concept of a third signal. Immunol. Today. 20:561–567. 10.1016/S0167-5699(99)01547-9 [DOI] [PubMed] [Google Scholar]
- Katakam A.K., Brightbill H., Franci C., Kung C., Nunez V., Jones C. III, Peng I., Jeet S., Wu L.C., Mellman I., et al. 2015. Dendritic cells require NIK for CD40-dependent cross-priming of CD8+ T cells. Proc. Natl. Acad. Sci. USA. 112:14664–14669. 10.1073/pnas.1520627112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats J.J., Fonseca R., Chesi M., Schop R., Baker A., Chng W.-J., Van Wier S., Tiedemann R., Shi C.-X., Sebag M., et al. 2007. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 12:131–144. 10.1016/j.ccr.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Starostina N.G., and Kipreos E.T.. 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22:2507–2519. 10.1101/gad.1703708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M., Xin H., Kujawski M., Lee H., Liu Y., Harris T., Drake C., Pardoll D., and Yu H.. 2009. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 15:114–123. 10.1016/j.ccr.2008.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Morgan M.J., Choksi S., Zhang Y., Kim Y.-S., and Liu Z.G.. 2010. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol. 11:799–805. 10.1038/ni.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G., Zhang M., Harhaj E.W., and Sun S.-C.. 2004. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279:26243–26250. 10.1074/jbc.M403286200 [DOI] [PubMed] [Google Scholar]
- Lind E.F., Ahonen C.L., Wasiuk A., Kosaka Y., Becher B., Bennett K.A., and Noelle R.J.. 2008. Dendritic cells require the NF-kappaB2 pathway for cross-presentation of soluble antigens. J. Immunol. 181:354–363. 10.4049/jimmunol.181.1.354 [DOI] [PubMed] [Google Scholar]
- Miosge L.A., Blasioli J., Blery M., and Goodnow C.C.. 2002. Analysis of an ethylnitrosourea-generated mouse mutation defines a cell intrinsic role of nuclear factor kappaB2 in regulating circulating B cell numbers. J. Exp. Med. 196:1113–1119. 10.1084/jem.20020959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miossec P., and Kolls J.K.. 2012. Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 11:763–776. 10.1038/nrd3794 [DOI] [PubMed] [Google Scholar]
- Mouri Y., Nishijima H., Kawano H., Hirota F., Sakaguchi N., Morimoto J., and Matsumoto M.. 2014. NF-κB-inducing kinase in thymic stroma establishes central tolerance by orchestrating cross-talk with not only thymocytes but also dendritic cells. J. Immunol. 193:4356–4367. 10.4049/jimmunol.1400389 [DOI] [PubMed] [Google Scholar]
- Oda H., Hübner M.R., Beck D.B., Vermeulen M., Hurwitz J., Spector D.L., and Reinberg D.. 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell. 40:364–376. 10.1016/j.molcel.2010.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder L., Mörbe U., Pikor N., Novkovic M., Cheng H.-W., Hehlgans T., Pfeffer K., Becher B., Waisman A., Rülicke T., et al. 2017. Lymphatic Endothelial Cells Control Initiation of Lymph Node Organogenesis. Immunity. 47:80–92.e4. 10.1016/j.immuni.2017.05.008 [DOI] [PubMed] [Google Scholar]
- Pettit A.R., MacDonald K.P.A., O’Sullivan B., and Thomas R.. 2000. Differentiated dendritic cells expressing nuclear RelB are predominantly located in rheumatoid synovial tissue perivascular mononuclear cell aggregates. Arthritis Rheum. 43:791–800. [DOI] [PubMed] [Google Scholar]
- Razani B., Zarnegar B., Ytterberg A.J., Shiba T., Dempsey P.W., Ware C.F., Loo J.A., and Cheng G.. 2010. Negative feedback in noncanonical NF-kappaB signaling modulates NIK stability through IKKalpha-mediated phosphorylation. Sci. Signal. 3:ra41 10.1126/scisignal.2000778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B., Reichardt A.D., and Cheng G.. 2011. Non-canonical NF-κB signaling activation and regulation: principles and perspectives. Immunol. Rev. 244:44–54. 10.1111/j.1600-065X.2011.01059.x [DOI] [PubMed] [Google Scholar]
- Reis e Sousa C. 2004. Activation of dendritic cells: translating innate into adaptive immunity. Curr. Opin. Immunol. 16:21–25. 10.1016/j.coi.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Rickert R.C., Jellusova J., and Miletic A.V.. 2011. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol. Rev. 244:115–133. 10.1111/j.1600-065X.2011.01067.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riol-Blanco L., Ordovas-Montanes J., Perro M., Naval E., Thiriot A., Alvarez D., Paust S., Wood J.N., and von Andrian U.H.. 2014. Nociceptive sensory neurons drive interleukin-23-mediated psoriasiform skin inflammation. Nature. 510:157–161. 10.1038/nature13199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S., Hoffmann A., Liou H.-C., Baltimore D., and Smale S.T.. 2000. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc. Natl. Acad. Sci. USA. 97:12705–12710. 10.1073/pnas.230436397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam C.L., Shepard J.L., Lai K., Ianari A., Danielian P.S., Amsterdam A., Hopkins N., and Lees J.A.. 2006. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20:3117–3129. 10.1101/gad.1482106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y., Calado D.P., Derudder E., Zhang B., Shimizu Y., Mackay F., Nishikawa S., Rajewsky K., and Schmidt-Supprian M.. 2008. NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc. Natl. Acad. Sci. USA. 105:10883–10888. 10.1073/pnas.0805186105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., and Amigorena S.. 2013. Inflammatory dendritic cells in mice and humans. Trends Immunol. 34:440–445. 10.1016/j.it.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Shih V.F.S., Davis-Turak J., Macal M., Huang J.Q., Ponomarenko J., Kearns J.D., Yu T., Fagerlund R., Asagiri M., Zuniga E.I., and Hoffmann A.. 2012. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways. Nat. Immunol. 13:1162–1170. 10.1038/ni.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-C. 2011. Non-canonical NF-κB signaling pathway. Cell Res. 21:71–85. 10.1038/cr.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-C. 2017. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17:545–558. 10.1038/nri.2017.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng M.W.L., Bowman E.P., McElwee J.J., Smyth M.J., Casanova J.-L., Cooper A.M., and Cua D.J.. 2015. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med. 21:719–729. 10.1038/nm.3895 [DOI] [PubMed] [Google Scholar]
- Terai K., Abbas T., Jazaeri A.A., and Dutta A.. 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell. 37:143–149. 10.1016/j.molcel.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker E., O’Donnell K., Fuchsberger M., Hilton A.A., Metcalf D., Greig K., Sims N.A., Quinn J.M., Alexander W.S., Hilton D.J., et al. 2007. A novel mutation in the Nfkb2 gene generates an NF-kappa B2 “super repressor”. J. Immunol. 179:7514–7522. 10.4049/jimmunol.179.11.7514 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S., and Karin M.. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P.-H., Keats J.J., Wang H., Vignali D.A.A., Bergsagel P.L., and Karin M.. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 9:1364–1370. 10.1038/ni.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Franks H.A., Lax S.J., El Refaee M., Malecka A., Shah S., Spendlove I., Gough M.J., Seedhouse C., Madhusudan S., et al. 2013. The ataxia telangiectasia mutated kinase pathway regulates IL-23 expression by human dendritic cells. J. Immunol. 190:3246–3255. 10.4049/jimmunol.1201484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Lo Y., Chan A., Law K.S., and Mok M.Y.. 2016. Rel B-modified dendritic cells possess tolerogenic phenotype and functions on lupus splenic lymphocytes in vitro. Immunology. 149:48–61. 10.1111/imm.12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar B.J., Wang Y., Mahoney D.J., Dempsey P.W., Cheung H.H., He J., Shiba T., Yang X., Yeh W.C., Mak T.W., et al. 2008. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9:1371–1378. 10.1038/ni.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study are available from the authors and have been deposited in the National Center for Biotechnology Information. For ChIP-seq data, the primary accession code is PRJNA359723. For RNA-seq data, the primary code is PRJNA417890.