Rice previews work from McIntosh et al. that reveals the structure of growing microtubule ends.
Abstract
What does the end of a growing microtubule look like? In this issue, McIntosh et al. (2018. J. Cell Biol. https://doi.org/10.1083/jcb.201802138) use electron tomography to provide state-of-the-art three-dimensional images of microtubule ends in cells and in vitro, yielding an unexpected answer to this fundamental question.
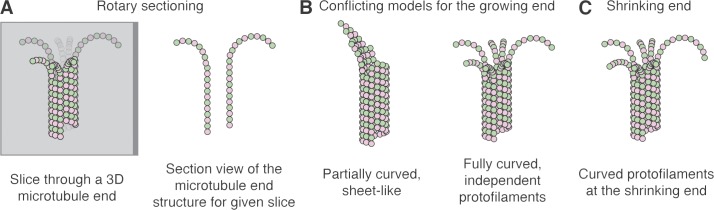
Microtubules are hollow, cylindrical polymers of αβ-tubulin that display dynamic instability, the apparently random switching between phases of growing and shrinking. The dynamic instability of microtubules is essential for formation of the mitotic spindle that organizes chromosome segregation, and for numerous other processes in cell physiology. In this issue, McIntosh et al. use electron tomography to provide 3D snapshots of growing microtubule ends in cells and in vitro. They report that the structures of growing and shrinking microtubule ends are very similar to each other, challenging the current understanding of microtubule dynamics and regulation by contradicting a long-held view in the field (see Fig. 1).
Figure 1.
Illustration of rotary sectioning and the different views of microtubule end structure. (A) Rotary sectioning. Left: Slices through 3D tomographs of microtubule ends are taken at various angles. The αβ-tubulin subunits of the microtubule are represented as pink and green circles, and the gray plane represents one such slice through the volume. Bright subunits are in front of the plane, faint subunits are behind it, and intermediate shaded subunits are in the slice. Right: View of the resulting slice, showing the end structure. The vertical head-to-tail assemblies of αβ-tubulin are called protofilaments. (B) Conflicting models for how microtubules grow. Left: Sheet-like, partially curved extensions on a subset of protofilaments. Right: All protofilaments elongate independently and are fully curved. (C) Cartoon of a shrinking microtubule end, with the ends of protofilaments fully curved.
Microtubule growing and shrinking occurs by the net addition or loss of αβ-tubulins to or from the end of the polymer. Catastrophe, the switch from growing to shrinking, results from the GTPase activity of αβ-tubulins in the polymer. Dynamic instability can be reconstituted in vitro using pure αβ-tubulin and GTP. However, this compositional simplicity belies a sneaky structural complexity: αβ-tubulin subunits adopt a different conformation outside the polymer than they do inside the polymer, and they adopt multiple conformations in the polymer, only three of which are known in atomic detail (Zhang et al., 2015). These different conformations of αβ-tubulin contribute to dynamic instability by modulating tubulin–tubulin interactions (reviewed in Brouhard and Rice, 2018).
The question McIntosh et al. (2018) addressed sounds like a simple one: What does a growing microtubule end look like? It’s an important question because distinctive microtubule end structures can selectively recruit different regulatory factors (reviewed in Akhmanova and Steinmetz, 2015), transitions between different end structures may contribute to force production by microtubules (Grishchuk et al., 2005; Driver et al., 2017), and knowledge of the end structure informs and constrains mechanistic models for microtubule dynamics. If we understood the biochemistry of the αβ-tubulin conformational cycle, we could predict end structure. Conversely, if we knew the end structure, we could infer the biochemistry. But we lack confidence about both the biochemistry and the end structure.
It has been known for some time that growing microtubule ends are structurally heterogeneous and characterized by extensions that curve away from the long axis of the microtubule. However, we lack an atomic resolution view of microtubule ends: their structural features are too small to image by light microscopy and too idiosyncratic for the averaging approaches that have allowed cryoEM to deliver high-resolution structures of αβ-tubulin (Zhang et al., 2015) in the more regular, lattice-like body of the microtubule.
Electron tomography, the technique used by McIntosh et al. (2018), is ideally suited for one-of-a-kind objects like microtubule ends that cannot be averaged together. Electron tomography produces a 3D image of a sample by combining many (2D) transmission electron micrographs in which the same sample is “viewed” from different angles; the different views are obtained by tilting the specimen to different degrees relative to the electron beam. Exposure to the electron beam damages the sample, so the maximum tolerated exposure must be spread among the multiple images required for tomography. As a result of the attendant low signal-to-noise ratio in individual images, and/or because of other cellular components, electron tomograms are often rather noisy, especially when considering “wispy” structures like individual microtubule protofilaments.
McIntosh et al. (2018) obtained tomograms of growing microtubules in cells from six different species, frozen and/or fixed for tomography in a variety of ways. They also obtained tomograms from samples of microtubules in vitro that were plunge-frozen to trap them in the act of growing. The authors then applied “rotary sectioning” (Fig. 1 A) to characterize the structures of individual microtubule ends: they examined “sagittal” sections (parallel to the long axis; Fig. 1 A) at various angles, through hundreds of microtubules. Their approach is admirably rigorous, and the paper has a wonderfully “old school” feel—considerable attention is devoted to the minute workings of the sample preparation and to considering, testing, and excluding possible sources of experimental artifact.
Both in cells and in vitro, McIntosh et al. (2018) observed short, curved extensions on the ends of growing microtubules (Fig. 1 B, right). Manually tracing these extensions using rotary sectioning identified about as many curved extensions as protofilaments, and the extensions were evenly spaced around the microtubule. McIntosh et al. (2018) conclude that the extensions are in fact curled protofilaments growing independently of one another. The curvature observed was highly variable, which was taken to indicate that the curled protofilaments are flexible in the plane of curvature. The average curvature was comparable in magnitude to that seen in head-to-tail assemblies modeled from atomic structures of unpolymerized αβ-tubulin, so it likely reflects the intrinsic curvature of unpolymerized, GTP-bound αβ-tubulin.
That growing and shrinking microtubule ends have markedly different structures is practically axiomatic in the current understanding of microtubule dynamics (Fig. 1, B and C). But in cells and in vitro, McIntosh et al. (2018) describe remarkably similar structures for growing and shrinking ends. Their results are therefore quite provocative, contradicting a long-held belief and challenging models built on that belief. Their work has potential implications for how we think about the molecular mechanisms of microtubule dynamics and regulation and for understanding processes like kinetochore–microtubule attachment, where assumptions about different end structures figure prominently (Schmidt et al., 2012).
The poor signal-to-noise ratio of tomographic reconstructions makes robust annotation of fine features challenging. The long-held belief that growing and shrinking microtubule ends have very different structures is itself based on a landmark cryoEM study (Chrétien et al., 1995) that revealed tapered and partially curved sheet-like structures at growing microtubule ends that were obviously distinct from the more curled, independent protofilaments on shrinking ends. Two recent tomography studies also show curved and partially curved extensions at the growing end (Guesdon et al., 2016; Atherton et al., 2017). However, this other work (in which rotary sectioning was not applied) was interpreted as showing that many of the curved protofilaments make lateral contacts to another protofilament (as in a sheet), contrary to the independent protofilaments described by McIntosh et al. (2018).
The McIntosh team has set a new standard in the quest to define the molecular features of the growing microtubule end. In doing so, they introduce a dissenting view about what the growing end looks like and about how and where curved αβ-tubulins are added to the microtubule end. The conflict revolves around whether the curved extensions are laterally connected, and whether “pioneer” elongation from a subset of protofilaments occurs. These different scenarios, which might in principle coexist on the same microtubule end, have implications for our understanding of the biochemistry of microtubule growth: Do curved tubulins interact tightly enough with individual protofilament ends for the protofilaments to elongate independently, or is simultaneous interaction with more than one protofilament (sheet-like growth) required for efficient elongation? A definitive resolution to this conflict has implications for microtubule regulation and for how growing and shrinking ends are recognized, and will likely come in the not too distant future as ongoing developments in the hardware and software for cryoEM and tomography are expected to improve the signal-to-noise ratio achievable. A clearer view of growing microtubule end structure looks to be around the curve!
Acknowledgments
L.M. Rice is the Thomas O. Hicks Scholar in Medical Research. Work in his laboratory is supported by the National Institutes of Health (R01-GM098543), the National Science Foundation (MCB-1615938), and the Robert A. Welch Foundation (I-1908).
The author declares no competing financial interests.
References
- Akhmanova A., and Steinmetz M.O.. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16:711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Atherton J., Jiang K., Stangier M.M., Luo Y., Hua S., Houben K., van Hooff J.J.E., Joseph A.-P., Scarabelli G., Grant B.J., et al. . 2017. A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat. Struct. Mol. Biol. 24:931–943. 10.1038/nsmb.3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., and Rice L.M.. 2018. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 19:451–463. 10.1038/s41580-018-0009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D., Fuller S.D., and Karsenti E.. 1995. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 129:1311–1328. 10.1083/jcb.129.5.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver J.W., Geyer E.A., Bailey M.E., Rice L.M., and Asbury C.L.. 2017. Direct measurement of conformational strain energy in protofilaments curling outward from disassembling microtubule tips. eLife. 6:576 10.7554/eLife.28433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Molodtsov M.I., Ataullakhanov F.I., and McIntosh J.R.. 2005. Force production by disassembling microtubules. Nature. 438:384–388. 10.1038/nature04132 [DOI] [PubMed] [Google Scholar]
- Guesdon A., Bazile F., Buey R.M., Mohan R., Monier S., García R.R., Angevin M., Heichette C., Wieneke R., Tampé R., et al. . 2016. EB1 interacts with outwardly curved and straight regions of the microtubule lattice. Nat. Cell Biol. 18:1102–1108. 10.1038/ncb3412 [DOI] [PubMed] [Google Scholar]
- McIntosh J.R., O’Toole E., Morgan G., Austin J., Ulyanov E., Ataullakhanov F., and Gudimchuk N.. 2018. Microtubules grow by the addition of bent guanosine triphosphate tubulin to the tips of curved protofilaments. J. Cell Biol. 10.1083/jcb.201802138 10.1083/jcb.201802138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. . 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Alushin G.M., Brown A., and Nogales E.. 2015. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 162:849–859. 10.1016/j.cell.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]