Among ERα-positive breast cancers, approximately half fail to respond to endocrine therapy, and the causes of this resistance are unknown. Sampayo et al. show that fibronectin (FN) influences the trafficking of ERα-positive vesicles. FN promotes ERα localization in Rab11+ vesicles, rescues ERα from lysosomal degradation, and reinforces ERα trafficking to the nucleus and transcriptional activity in tumor cells.
Abstract
Estrogen receptor α (ERα) is expressed in tissues as diverse as brains and mammary glands. In breast cancer, ERα is a key regulator of tumor progression. Therefore, understanding what activates ERα is critical for cancer treatment in particular and cell biology in general. Using biochemical approaches and superresolution microscopy, we show that estrogen drives membrane ERα into endosomes in breast cancer cells and that its fate is determined by the presence of fibronectin (FN) in the extracellular matrix; it is trafficked to lysosomes in the absence of FN and avoids the lysosomal compartment in its presence. In this context, FN prolongs ERα half-life and strengthens its transcriptional activity. We show that ERα is associated with β1-integrin at the membrane, and this integrin follows the same endocytosis and subcellular trafficking pathway triggered by estrogen. Moreover, ERα+ vesicles are present within human breast tissues, and colocalization with β1-integrin is detected primarily in tumors. Our work unravels a key, clinically relevant mechanism of microenvironmental regulation of ERα signaling.
Introduction
Estrogen receptor α (ERα) is a transcription factor present in different adult tissues such as mammary gland, ovaries, uterus, and brain (Couse et al., 1997; Han et al., 2013). It regulates cell proliferation, migration, and survival. In the breast in particular, ERα controls mammary development and plays a key role in tumor growth. Therefore, understanding what regulates ERα activation and shutdown is fundamental for cell biology. ERα action can be blocked with tamoxifen (the most widely used selective ER modulator), although one third of breast cancer patients develop resistance, with ERα regaining activity (Nardone et al., 2015; Jeselsohn et al., 2017). The causes of this resistance are still unclear.
So far, the main proposed mechanism for ERα signaling shutdown is estrogen-induced ERα degradation. Estrogen binding to ERα induces its nuclear translocation. Once in the nucleus, ERα binds to its target promoters and is then ubiquitylated and subsequently degraded in cytosolic proteasomes. Therefore, ERα’s half-life decreases from 4 to 2 h in the presence of estrogens. The pool of ERα attached to the plasma membrane by reversible S-palmitoylation on cysteine 447 (Acconcia et al., 2005; Marino et al., 2006; Adlanmerini et al., 2014) has been suggested to follow different degradation dynamics (La Rosa et al., 2012). Whether membrane-bound ERα has transcriptional activity is still a matter of debate (Levin, 2009). Understanding how membrane and cytoplasmic ERα are regulated in breast cancer is crucial to develop strategies to overcome resistance to endocrine therapy.
The ECM plays a key role in cell fate, and evidence is accumulating that it modulates response to therapy in breast cancer as well (Ghajar and Bissell, 2008; Correia and Bissell, 2012). We previously described that ECM components affect the response of breast cancer cells to tamoxifen (Pontiggia et al., 2012). In particular, we found that fibronectin (FN), which correlates with lower survival when levels are increased (Yao et al., 2007; Helleman et al., 2008), induces tamoxifen resistance in breast cancer cells when bound to β1-integrin, its surface receptor. Therefore, we hypothesized that FN–β1-integrin pathway might have a direct effect on ERα signaling, modifying its response to hormone treatment.
We used two well-known cellular models of ERα-positive human breast adenocarcinoma: MCF7 and T47D. These cell lines have been widely used and validated for the study of ERα activity because primary culture of normal or tumor human breast tissues leads to the loss of ERα expression (Graham et al., 2009; Hines et al., 2016). We demonstrate that FN prolongs ERα half-life and strengthens its transcriptional activity. Mechanistically, we show that upon treatment with 17β-estradiol (E2), membrane ERα is endocytosed and travels in these vesicles through the cytoplasm and into the nucleus. In the absence of FN, it is degraded in lysosomes after 60 min of treatment. When FN is present, these endosomes escape lysosomal degradation, and ERα is localized in RAB11+ vesicles, typically involved in recycling. Using superresolution microscopy and coimmunoprecipitation assays, we found that ERα and β1-integrin colocalize at the plasma membrane and are endocytosed together after stimulation with E2. In these vesicles, β1-integrin is also degraded upon 60 min of treatment with E2, unless FN is present. We propose that FN-bound β1-integrin, following its recycling pathway, drags these ERα–β1-integrin+ vesicles back to the plasma membrane, thus bypassing the lysosomal compartment. We show that these endosomes are present in normal and tumor human breast tissues, although only tumor samples showed positive colocalization between ERα and β1-integrin. This indicates that the mechanism of ERα overactivation dependent on its association with FN–β1-integrin pathway would be particularly active within tumors. In light of these findings, we strongly suggest that a novel therapeutic strategy designed to interfere with the cross talk between FΝ and ERα signaling pathways would resensitize patients to endocrine therapy.
Results
FN modulates ERα degradation and transcriptional activity
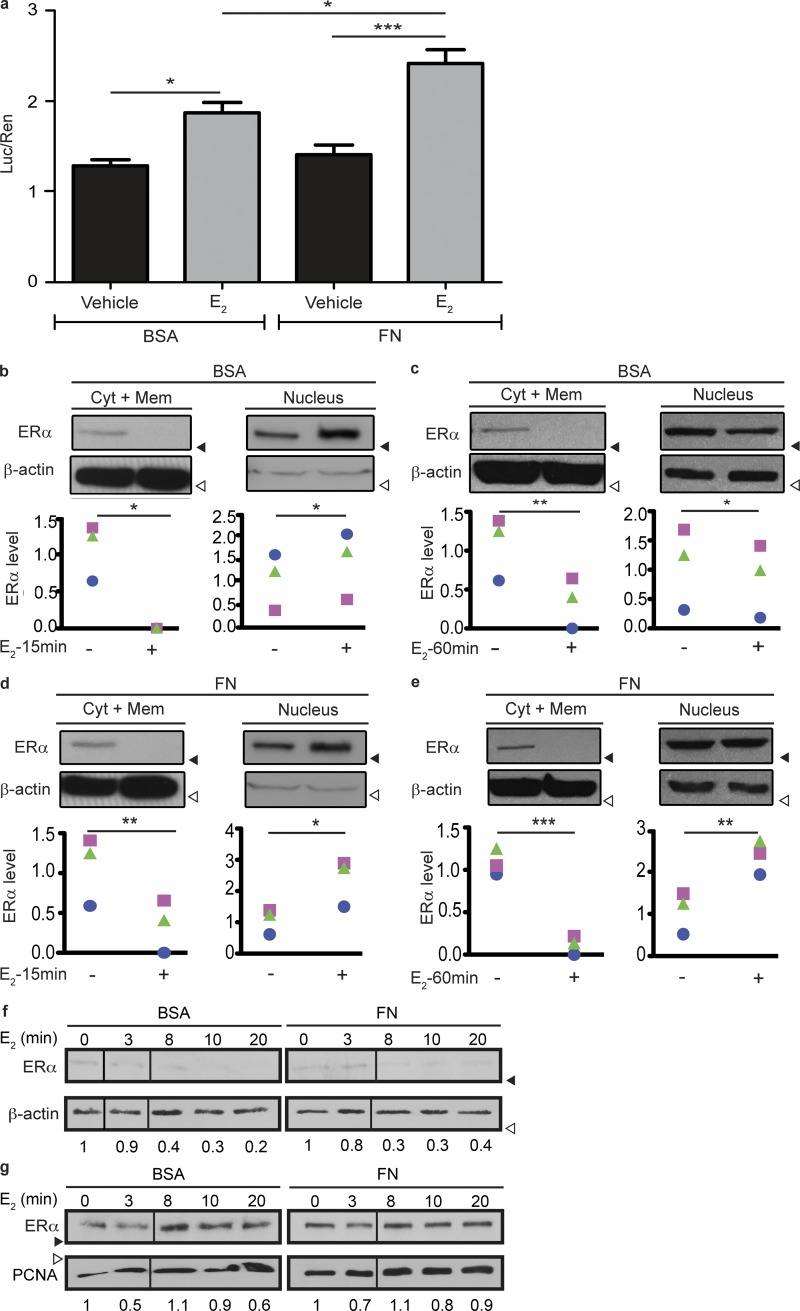
Given that we have previously shown that FN induces resistance to anti-estrogenic therapy (Pontiggia et al., 2012), we wondered whether FN has a direct effect on ERα activity. Research on ERα activity and dynamics in culture is challenging because primary culture of ERα-positive normal tissues and tumors leads to the loss of ERα expression (Hines et al., 2016). Therefore, we used two well-characterized human ERα-positive breast adenocarcinoma cell lines, MCF7 and T47D, that allowed us to modulate and study ERα regulation in culture. We first performed luciferase reporter assays with a construction that allowed us to measure ERα activity mediated by the estrogen response element (ERE). We found that, when cells are seeded on FN, this receptor has a stronger transcriptional activity in the presence of E2, compared with its activity on the control substrate (BSA; Fig. 1 a). To study the mechanism through which FN regulates ERα activity, we analyzed the effect of FN on ERα degradation. We found that when cells are seeded on BSA, ERα completely localizes in the nucleus after 15 min of treatment with E2 (Fig. 1 b). Knowing that E2 triggers ERα degradation, reducing ERα mean expression after 60 min (Reid et al., 2003), we increased the treatment time and found that as expected, total ERα levels drop after stimulation with E2 in cells seeded on BSA (Fig. 1 c). When cells are seeded on FN, ERα is also completely localized in the nucleus after 15 min of treatment with E2 (Fig. 1 d). However, we found that after a longer treatment with the hormone (>60 min), FN inhibits E2-induced ERα degradation (Fig. 1 e). We confirmed these observations by immunofluorescence, showing a more intense signal of nuclear ERα after treatment with E2 in cells seeded on FN compared with BSA (Fig. S1, a and b). Similar results were obtained using T47D cells (Fig. S1, c–f). These data indicate that FN inhibits E2-stimulated ERα degradation. Of note, total ERα levels are increased when cells are seeded on FN even in the absence of E2, indicating that FN might also alter basal ERα degradation dynamics (Reid et al., 2003). Interestingly, when we performed ultracentrifugation to separate cytoplasmic and membrane fractions, we observed that membrane ERα follows a dynamics similar to cytoplasmic ERα (Fig. S1, g–j).
Figure 1.
FN stimulates ERα’s transcriptional activity. (a) Luciferase assay in MCF7 cells transiently transfected with pTK-ERE-Luc and pTK-Renilla, seeded on BSA or FN and treated for 14 h as indicated. Data are represented as mean ± SD. Differences between groups were analyzed by two-way ANOVA followed by Bonferroni contrasts adjusted for multiple comparisons (n = 3 replicates). (b and c) Top: Western blot of a subcellular fractionation of MCF7 cells seeded on BSA and treated for 15 min (b) or 60 min (c) as indicated. Blotting antibodies are shown on the left. Bottom: densitometry. For each subcellular fraction, shown is the ERα/β-actin density ratio normalized to the mean control group. Each symbol represents a different experiment. Differences between groups were analyzed by a one-tailed paired Student’s t test (n = 3 replicates). (d and e) Top: Western blot of a subcellular fractionation of MCF7 cells seeded on FN and treated for 15 (d) or 60 min (e) as indicated. Blotting antibodies are shown on the left. Bottom: Densitometry. For each subcellular fraction, shown is the ERα/β-actin density ratio normalized to the mean control group. Each symbol represents a different experiment. (f) Western blot of the cytosolic + membrane fraction of MCF7 cells seeded on BSA or FN and treated with E2 for the indicated times. Below the blots, the ERα/β-actin density ratio is shown, normalized to the control group. (g) Western blot of the nuclear fraction of MCF7 cells seeded on BSA or FN and treated with E2 for the indicated times. Below the blots, the ERα/PCNA density ratio is shown, normalized to the control group. Differences between groups were analyzed by one-tailed paired Student’s t test (n = 3 replicates). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Shown data are representative of at least three independent experiments. Black arrowheads indicate positions of 50-kD markers. White arrowheads indicate positions of 37-kD markers. Treatments: ethanol (vehicle) or 10−8 M E2.
Moreover, we further tested the effect of FN on ERα shuttling kinetics. As shown in Fig. 1 f, E2 stimulates ERα nuclear localization, reaching its maximum after 8 min of treatment. These kinetics are not affected by the presence of FN (Fig. 1 g). However, upon 20 min of treatment, it can be already observed that degradation of both nuclear and cytoplasmic ERα is reduced when cells are seeded on FN. Altogether, these data indicate that FN modulates ERα degradation but does not alter ERα shuttling dynamics to the nucleus.
ERα is degraded in lysosomes and can be rescued by FN
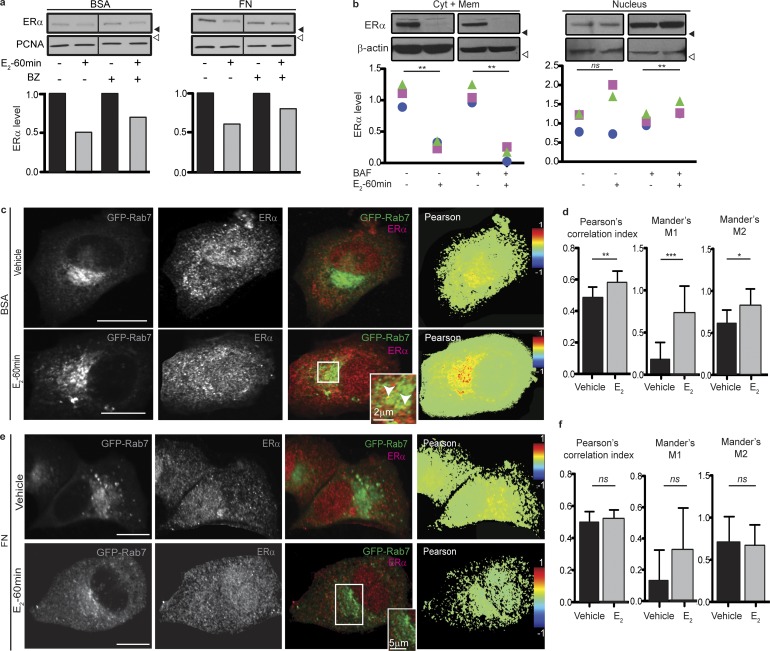
We next tested whether FN was inhibiting E2-triggered proteasomal degradation of ERα, the best-characterized degradation pathway of this receptor. We found that upon E2 stimulation for 60 min, inhibition of the proteasomal pathway with bortezomib (BZ) increases ERα levels even in the presence of FN (Fig. 2 a), suggesting that FN would inhibit a different mechanism of ERα degradation. Because FN has been found to modulate lysosomal degradation of membrane proteins (Caswell et al., 2009; Dozynkiewicz et al., 2012), we next asked whether ERα could be degraded in lysosomes upon E2 stimulation. Blocking the passage of late endosomes to lysosomes by inhibiting V-ATPase with bafilomycin-A1 (BAF; Li et al., 2013) impaired ERα degradation after 60 min of treatment with E2, in cells seeded on BSA (Fig. 2 b). To further test that ERα is degraded in lysosomes after E2 treatment, we expressed GFP-tagged Rab7, a well-known small GTPase that determines the passage of late endosomes to lysosomes (Vanlandingham and Ceresa, 2009). As shown in Fig. 2 c, E2 treatment triggers ERα colocalization with Rab7. Pearson’s and Manders’ correlation coefficients (PCCs and MCCs, respectively) were used to quantify the degree of colocalization observed between these proteins in each analyzed field as previously described (Dunn et al., 2011). The overall significance level of colocalization was calculated from these coefficients for each condition. A shorter treatment with E2 (15 min) revealed an increase in ERα localization closer to the Rab7 compartment, although practically no colocalization with Rab7+ endosomes was observed (Fig. S1 k, top), indicating that a longer treatment is necessary for ERα+ vesicle localization to lysosomes.
Figure 2.
ERα is degraded in lysosomes and rescued by FN. (a) Top: Western blot of T47D cells seeded on BSA or FN pretreated with BZ 8nM or its vehicle (saline) for 4 h and treated as indicated. Bottom: Densitometry. For each subcellular fraction, the mean ERα/β-actin density ratio is shown normalized to the mean control group. (b) Top: Western blot of a subcellular fractionation of T47D cells pretreated for 90 min with 25 nM BAF or its vehicle (DMSO) and then treated for 60 min with 10−8 M E2 or its vehicle (ethanol). Blotting antibodies are shown on the left. Bottom: Densitometry. For each experimental condition, the ERα/β-actin density ratio is shown, normalized to the mean control group. Each symbol represents a different experiment. Differences between groups were analyzed by one-tailed paired Student’s t test (n = 3 replicates). (c) Confocal images of T47D cells expressing GFP-Rab7 seeded on BSA treated for 60 min with vehicle or E2, and stained for ERα. In the inset, arrows indicate points of colocalization. (d) Quantification of c. For each experimental condition, Pearson’s correlation index and Manders’ coefficients (M1 and M2) were calculated within the areas of colocalization using Fiji. Data are represented as mean ± SD. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: Pearson’s: nvehicle = 11 fields, nE2 = 12 fields; Manders’: nvehicle = 8 fields, nE2 = 9 fields). (e) Confocal images of T47D cells expressing GFP-Rab7 seeded on FN treated for 60 min with vehicle or E2, and stained for ERα. (f) Quantification of e. For each experimental condition, Pearson’s correlation index and Manders’ coefficients (M1 and M2) were calculated within the areas of colocalization using Fiji. Data are represented as mean ± SD. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: nvehicle = 9 fields, nE2 = 7 fields). Treatments: ethanol (vehicle) or 10−8 M E2, 8 nM BZ. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Shown data are representative of at least three independent experiments. Black arrowheads indicate positions of 50-kD markers. White arrowheads indicate positions of 37-kD markers. Bars, 10 µm (unless otherwise indicated).
We next investigated the effect of FN on E2-induced ERα lysosomal degradation and found that ERα does not colocalize with Rab7 after 60 min (Fig. 2, e and f) or 15 min (Fig. S1 k, bottom) treatment in the presence of FN. These data indicate that FN is rescuing ERα from being degraded in the lysosomal compartment. We confirmed these results with LAMP-1, a lysosomal marker, which shows that after 60 min of treatment with E2, ERα colocalizes with LAMP-1 when cells are seeded on BSA, and this is reverted when cells are seeded on FN (Fig. S1 l).
To ensure that ERα signals observed in these assays correspond in fact with ERα, we checked the specificity of this antibody. We used the epitope this antibody was raised against as a blocking peptide and obtained no ERα signal in Western blot or immunofluorescence assays (Fig. S1, m and n). Moreover, knockdown of ERα significantly reduces the signal obtained with this antibody proving that it specifically recognizes this protein (Fig. S1 o).
ERα is rapidly endocytosed after estrogen treatment
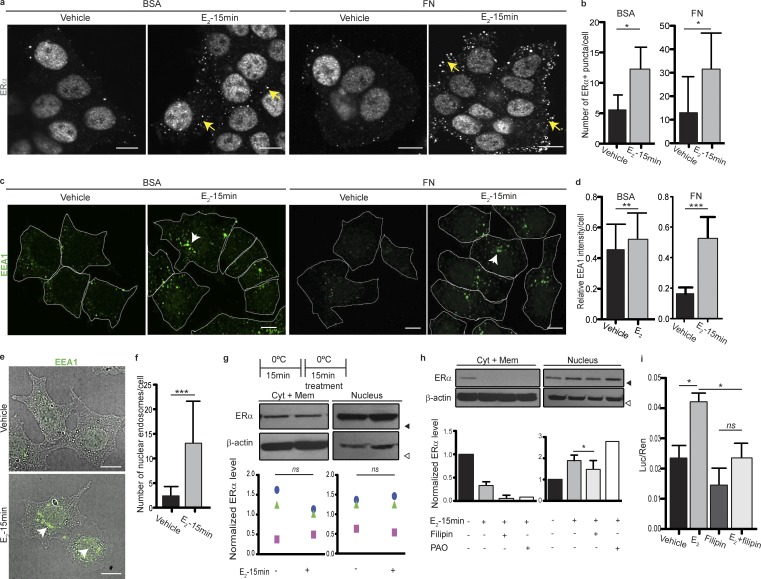
We next asked whether ERα was present in endosomes that could end up in lysosomes upon E2 stimulation and whether this was an FN-induced event. We found that ERα is present in vesicle-like punctae after treatment with E2 for 15 min, regardless of the presence of FN (Fig. 3, a and b). To confirm that E2 was inducing rapid endocytosis in these cells, we stained them for EEA1, an early endosomal marker, and found that the size of EEA1+ vesicles is dramatically increased after a 15-min treatment with E2 (Fig. 3, c and d). Moreover, ERα colocalizes with EEA1 upon E2 treatment (Fig. S2 a). Interestingly, we found that EEA1+ endosomes are strongly localized in the nuclear membrane with this treatment (Fig. 3, e and f). We stained these cells with Lamin B1, a nuclear envelope marker, or with propidium iodide and performed 3D reconstructions to confirm that EEA1 colocalizes with Lamin B1, although EEA1+ endosomes do not seem to enter the nucleus (Fig. S2, b and c). This finding led us to hypothesize that these endosomes could be carrying ERα straight into the nucleus, where it would exert its action, analogous to the way signaling endosomes carry neurotransmitters along neuronal axons (Delcroix et al., 2003; Cosker et al., 2008; Cosker and Segal, 2014). To investigate whether nuclear localization of ERα was in fact endocytosis dependent, we studied the effects of low temperatures on ERα subcellular shuttling. This treatment should immediately block both membrane events: endocytosis and ATP-dependent vesicle trafficking (Letoha et al., 2003). We found that chilling cells to 0°C completely blocks subcellular shuttling of ERα (Fig. 3 g), which is consistent with a static plasma membrane. We confirmed that the effect of low temperatures was reversible because prechilling the cells does not affect ERα shuttling (Fig. S2 d).
Figure 3.
E2 stimulates endocytosis of vesicles containing ERα. (a) Confocal images of MCF7 cells seeded on BSA (left) or FN (right) treated for 15 min as indicated and stained for ERα. Arrows indicate ERα+ endosomes. (b) Quantification of a. For each experimental condition, structures of ∼200-nm diameter (10–15 pixels) were quantified using Fiji. Shown is the number of ERα+ puncta per cell. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: BSA: nvehicle = 81 cells, nE2 = 87 cells; FN: nvehicle = 50 cells, nE2 = 64 cells). (c) Confocal images of MCF7 cells seeded on BSA (left) or FN (right) treated for 15 min as indicated and stained for EEA1. Cells are delineated in white. Arrows indicate early endosomes. (d) Quantification of c. For each experimental condition, shown is EEA1 intensity (mean gray value) per cell calculated using Fiji relative to the highest intensity recorded. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: BSA: nEtOH = 68 cells; nE2 = 39 cells; FN: nEtOH = 94 cells; nE2 = 108 cells). (e) Confocal images of MCF7 cells seeded on FN treated for 15 min as indicated and stained for EEA1. Merges between differential interference contrast (DIC) microscopy and the green channel are shown. Arrows indicate early endosomes present either in the nuclear membrane or inside the nucleus. (f) Quantification of e. For each experimental condition, structures of 10–15 pixels in diameter were quantified using Fiji. Shown is the number of nuclear early endosomes per cell. It was calculated as the total number of EEA1+ vesicles in the nuclear membrane or inside the nucleus, per cell. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: nEtOH = 59 cells; nE2 = 54 cells). (g) Top: Outline of the protocol followed and Western blot of a subcellular fractionation of MCF7 cells treated as indicated. Blotting antibodies are shown on the left. Bottom: Densitometry. For each subcellular fraction, shown is the ERα/β-actin density ratio normalized to the mean control group. Each symbol represents a different experiment. Differences between groups were analyzed by one-tailed paired Student’s t test (n = 3 replicates). (h) Top: Western blot of a subcellular fractionation of MCF7 cells treated for 15 min as indicated. Blotting antibodies are shown on the left. Bottom: Densitometry. For each subcellular fraction, shown is the ERα/β-actin density ratio normalized to the control group. Differences between groups were analyzed by one-tailed paired Student’s t test (n = 3 replicates). (i) Luciferase assay in MCF7 cells transiently transfected with pTK-ERE-Luc and pTK-Renilla and treated for 14 h as indicated. Differences between groups were analyzed by two-way ANOVA followed by Bonferroni contrasts adjusted for multiple comparisons (n = 3 replicates). Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Treatments: ethanol (vehicle) or 10−8 M E2, 2.5 µg/ml filipin, 5 µM PAO. Shown data are representative of at least three independent experiments. Black arrowheads indicate positions of 50-kD markers. White arrowheads indicate positions of 37-kD markers. Bars, 10 µm.
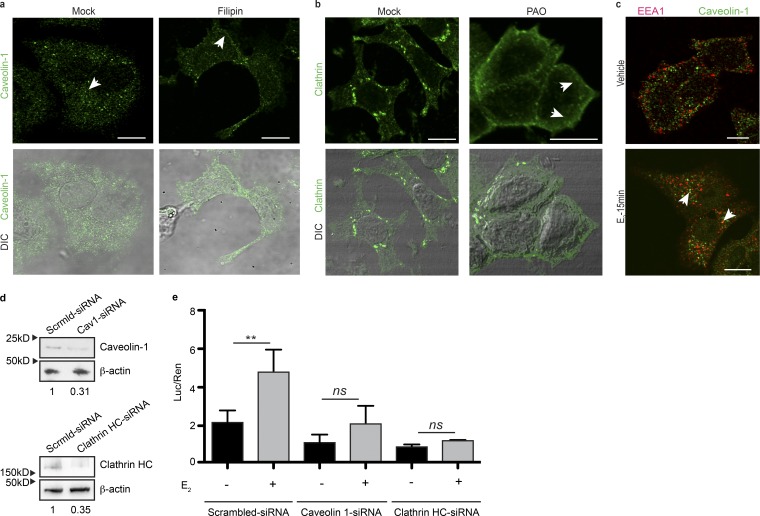
There are two main mechanisms of integrin endocytosis: clathrin dependent and clathrin independent (Mayor and Pagano, 2007). Among clathrin-independent mechanisms, the best described is the caveolin-dependent pathway. Given that ERα is known to interact with caveolin 1 and 3 (Schlegel et al., 1999; Chung et al., 2009), we hypothesized that ERα would be endocytosed through a caveolin-dependent mechanism in the presence of E2. To test this, we used filipin, a specific inhibitor of caveolin-mediated endocytosis/membrane recycling, and found that it inhibits E2-stimulated ERα nuclear translocation (Fig. 3 h). The inhibition of clathrin-mediated endocytosis with phenylarsine oxide (PAO) does not affect ERα shuttling dynamics (Fig. 3 h). We next assessed the effect of filipin treatment on ERα transcriptional activity, performing luciferase reporter assays with a construction that allowed us to measure ERα activity mediated by the ERE. As expected, we found that E2 is not able to induce ERα transcriptional activity in the presence of filipin, supporting the finding that ERα endocytosis has a major effect in gene transcription (Fig. 3 i). We verified the specific action of filipin and PAO through the inhibition of their canonical endocytosis substrates (Fig. 4, a and b). Together with these results, we found that caveolin 1 colocalizes with EEA1 in the cytoplasm upon 15 min of treatment with E2 (Fig. 4 c). We further tested the effect of caveolin 1 knockdown on ERα action and found that it inhibits ERα transcriptional activity, similarly to what we found with its pharmacological inhibitor (Fig. 4, d and e). Interestingly, we found that clathrin knockdown also impairs ERα transcriptional activity (Fig. 4, d and e). This suggests that clathrin might also play at least a partial role in ERα dynamics. Altogether, these data indicate that E2 induces caveolin-mediated ERα endocytosis in cells seeded on BSA or FN.
Figure 4.
ERα is endocytosed through a caveolin 1–dependent pathway. (a) Top: Confocal images of MCF7 cells treated for 15 min as indicated and stained for caveolin 1. Bottom: Merge between caveolin 1 signal and differential interference contrast (DIC) images. Arrows indicate internal or peripheral localization of caveolin 1. (b) Top: Confocal images of MCF7 cells, treated for 15 min as indicated, and stained for clathrin. Bottom: Merge between caveolin 1 signal and DIC images. Arrows indicate internal or peripheral localization of clathrin. (c) Confocal images of MCF7 cells treated for 15 min as indicated and stained for caveolin 1 or EEA1. Arrows indicate regions of colocalization between the two markers. (d) Western blots of MCF7 cells transfected with siRNAs against caveolin 1, clathrin HC, or scrambled for 48 h. Blotting antibodies are shown on the right. Fold change relative to scrambled siRNA is shown on the bottom. (e) Luciferase assay in MCF7 cells transiently transfected with pTK-ERE-Luc and pTK-Renilla and the respective siRNAs and treated for 14 h as indicated. Differences between groups were analyzed by two-way ANOVA followed by Bonferroni contrasts adjusted for multiple comparisons (n = 3 replicates). Data are represented as mean ± SD. **, P < 0.01. Shown data are representative of at least three independent experiments performed. Treatments: ethanol (vehicle) or 10−8 M E2, 2.5 µg/ml filipin, 5 µM PAO. Bars, 10 µm.
ERα+ colocalizes with Rab11 in the presence of FN
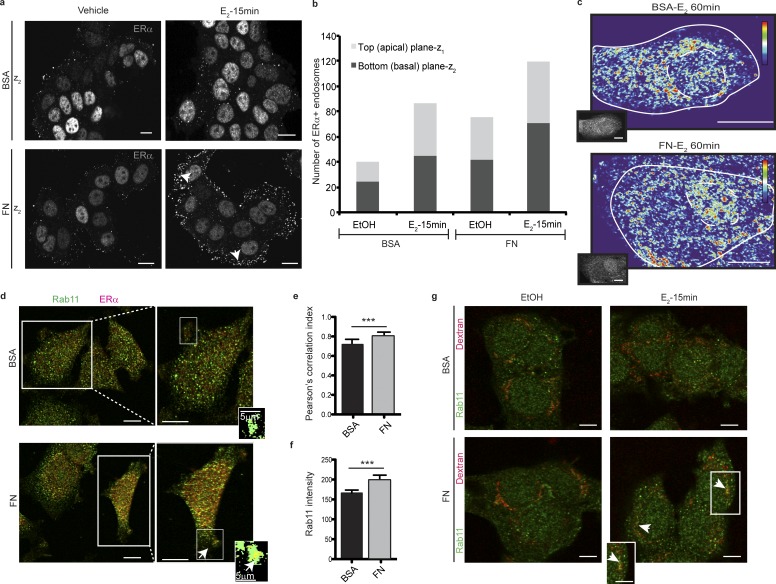
Internalized endosomes typically avoid lysosomal degradation if recycled to the plasma membrane (Gould and Lippincott-Schwartz, 2009). Therefore, we explored whether FN promoted the recycling of ERα+ endosomes, therefore inhibiting its lysosomal degradation induced by E2. We found that, when cells are seeded on FN and treated with E2 for 15 min, there is a larger proportion of ERα+ vesicles closer to the basal plane (ventral membrane) than when cells are seeded on BSA as shown in Figs. 5 (a and b) and S2 e. Consistent with these findings, after a longer treatment with E2, ERα distribution in the cytoplasm is peripheral when cells are seeded on FN compared with a more centered distribution on BSA (Fig. 5 c). These data suggest that in the presence of FN, ERα+ vesicles are more likely to be found closer to the plasma membrane than to the lysosomal–perinuclear compartment.
Figure 5.
ERα+ is localized in Rab11+ vesicles in the presence of FN. (a) Images of confocal microscopy of MCF7 cells seeded on BSA or FN treated for 15 min as indicated and stained for ERα. Panels show the cytoplasmic/plasma membrane (basal) plane (z2). Nuclear/cytoplasmic (apical) plane (z1) is shown in Fig. S3 b. White arrows indicate ERα+ vesicles determined as punctae of 10–15 pixels in diameter (∼200 nm). (b) Quantification of a. Apical (nuclear/cytoplasmic) versus basal (cytoplasmic/plasma membrane) distribution of ERα+ vesicles. Structures of 10–15 pixels in diameter were quantified using Fiji. Mean number of endosomes in each fraction and for each condition is shown. (c) Heatmaps of T47D cells seeded on BSA or FN treated for 60 min as indicated and stained for ERα. Cells are outlined in white. Dashed line outlines the nucleus. Intensity bars are shown on the right (red, maximum pixel intensity; blue, minimum pixel intensity). Original images are shown in the insets. (d) Images of confocal microscopy of MCF7 cells seeded on BSA or FN treated for 60 min with E2 and stained for ERα and Rab11. Arrows indicate areas of colocalization within filopodia. Pearson’s colocalization maps are shown in the insets. (e) Quantification of d. For each experimental condition, Pearson’s correlation index was calculated within filopodia protrusions using Fiji. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: nBSA = 16 fields, nFN = 15 fields). (f) Quantification of d. For each experimental condition, overall Rab11 intensity was calculated using Fiji. Data are represented as mean ± SD. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: nBSA = 5 fields, nFN = 4 fields). (g) Images of confocal microscopy of MCF7 cells seeded on BSA or FN treated for 15 min with E2 in the presence of dextran-CF543 and stained for Rab11. Arrows indicate areas of colocalization between the two fluorophores. Higher magnification is shown in the inset. ***, P < 0.001. Shown data are representative of at least three independent experiments. Treatments: ethanol (vehicle) or 10−8 M E2. Bars, 10 µm (unless otherwise indicated).
To further explore whether ERα+ vesicles are more likely to be redirected to the plasma membrane in the presence of FN, we costained the cells with the recycling marker Rab11 (Grant and Donaldson, 2009). Rab11 is mostly localized in the perinuclear region and is further transported to the cell periphery to participate in membrane fusion when recycling is active (Cox et al., 2000; Takahashi et al., 2012). We found that the degree of colocalization of ERα with Rab11 is highest when cells are seeded on FN, particularly at the membrane tips, indicating that ERα is more likely to be localized in Rab11+ vesicles under these conditions (Fig. 5, d and e). Together with this, overall intensity of Rab11 is higher in the presence of FN, suggesting that this pathway is enhanced either by the presence of more Rab11+ vesicles or by an increased size of these vesicles (Fig. 5, d and f).
We further tested the effect of E2 on endocytosis using dextran (10 kD) conjugated with a red fluorophore. We found that a 15-min treatment with E2 induces strong dextran endocytosis that also colocalizes with EEA1 (Fig. S2, f–h). In addition, we found that dextran colocalizes with Rab11 when cells are seeded on FN, suggesting that it is more frequently localized in Rab11+ vesicles in this condition (Fig. 5 g). Moreover, we measured the amount of dextran present in the supernatant after E2 treatment and found that it is significantly higher when cells are seeded on FN, further suggesting that dextran would be more likely to be recycled in the presence of FN (Fig. S2 i).
ERα is associated to β1-integrin in estrogen-triggered endosomes
To gain insight into the possible mechanism responsible for triggering ERα localization in Rab11+ vesicles on cells seeded on FN after E2 treatment, we explored the possibility that upon endocytosis, ERα+ endosomes might contain integrins that, if engaged with FN, would trigger membrane recycling, therefore making the whole complex avoid lysosomal degradation (Caswell et al., 2009; Sung and Weaver, 2011; Dozynkiewicz et al., 2012; De Franceschi et al., 2015). Because we have previously found that FN-induced endocrine resistance is mediated by β1-integrin (Pontiggia et al., 2012), we asked whether this could be the bona fide integrin associated with ERα at the plasma membrane and, therefore, present in E2-induced endosomes. We performed total internal reflection fluorescence microscopy (TIRFM), which allows the detection only of those fluorophores localized on the ventral plasma membrane, at the cell–substrate interphase. This assay showed that β1-integrin and ERα colocalize at the ventral membrane in MCF7 cells (Fig. 6, a and b; and Video 1). For β1-integrin detection, live-staining technique was used to intensely detect integrin present in the periphery of the cell, although it does not stain cytoplasmic integrin. Therefore, most of the colocalization structures were found at the periphery of the cell, where further colocalization analysis was run as described previously (Dunn et al., 2011). We also found colocalization structures in T47D cells (Fig. S3 a). As a positive control, colocalization between β1-integrin and its well-known partner FAK was assayed with TIRFM, and a similar colocalization pattern was found (Fig. S3 b).
Figure 6.
ERα is spatially associated with β1-integrin and they are endocytosed together. (a) Widefield (top) and TIRFM (bottom) images of a coimmunofluorescence in MCF7 cells, using antibodies against β1-integrin (live-stained) and ERα. In the inset, white arrowheads indicate points of colocalization. Pearson’s correlation maps corresponding with the white box shown on the right. White arrowheads indicate points of positive Pearson’s correlation. (b) Quantification of a. Top left: Polar transformation of TIRFM images was performed using Fiji to align areas of the cell periphery where colocalization is found. For each experimental condition, Pearson’s correlation index and Manders’ coefficients (M1 and M2) were calculated within the areas of colocalization (ROI) and compared with random areas without colocalization (Null), using Fiji. For Pearson’s correlation, datasets are plotted and mean ± SD are shown on the graph. For Manders’ coefficients, the table shows mean and SD for each dataset. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: Pearson’s: nnull = 9 fields, nROI = 9 fields; Manders’: nnull = 14 fields, nROI = 9 fields). (c) Western blot of a coimmunoprecipitation in MCF7 cells, using antibodies against β1-integrin or ERα. Blotting antibodies are shown on the right. Input, whole lysate. IP, immunoprecipitated fraction. (d) ClustalW alignment of the eight β-integrins present in humans. The sequence of SRC1 is shown on top. NR-box motif is indicated in red. On the sequences of β1-integrin and β3-integrin, underlined in black is the region corresponding with their transmembrane domain, and in green is their cytoplasmic domain. The topology was predicted using the algorithm TMpred from the website ExPASy and the algorithm from the website TOPCONS. (e) Cartoon showing β1-integrin structure and putative interaction site with ERα. Black box indicates the localization of NR-box motif (LXXLL) within β1-integrin transmembrane domain. Red dot shows ERα palmitoylation site, and the arrow indicates where its helix 12 would be localized within the AF-2 domain. (f and g) Top: Western blot of total lysates of MCF7 cells, seeded on BSA (f) or FN (g) and treated for 60 min as indicated. Blotting antibodies are shown on the left. Bottom: Densitometry. For each experimental condition, shown is the β1-integrin/β-actin density ratio normalized to the mean control group. Each symbol represents a different experiment. Differences between groups were analyzed by one-tailed paired Student’s t test (n = 3 replicates). (h) Confocal images of MCF7 cells treated for 15 min as indicated and stained for β1-integrin (live stained) and ERα. Arrows indicate points of colocalization. Corresponding Pearson’s correlation maps are shown on the right, respectively. White arrows indicate points of positive Pearson’s correlation. (i) Quantification of h. For each experimental condition, Pearson’s correlation index and Manders’ coefficients (M1 and M2) were calculated within the areas of colocalization (ROI) and compared with random areas without colocalization (Null) using Fiji. For Pearson’s correlation, datasets are plotted and mean ± SD are shown on the graph. For Manders’ coefficients, the table shows mean and SD for each dataset. Differences between groups were analyzed by one-tailed Student’s t test (per replicate: Pearson’s: nnull = 14 fields, nROI = 15 fields; Manders’: nnull = 10 fields, nROI = 11 fields). (j) Confocal images of MCF7 cells seeded on BSA or FN treated with E2 for 15 min and stained for β1-integrin and Rab11. Full images are shown in the insets. (k) Quantification of j. For each experimental condition, Pearson’s correlation index was calculated within the areas of colocalization using Fiji. Data are represented as mean ± SD. Differences between groups were analyzed by a one-tailed Student’s t test (per replicate: nBSA = 4 fields, nFN = 4 fields). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Shown data are representative of at least three independent experiments. Black arrowheads indicate positions of 100-kD markers. White arrowheads indicate positions of 50-kD markers. Treatments: ethanol (vehicle) or 10−8 M E2. Bars, 10 µm (unless otherwise indicated).
Consistent with these results, coimmunoprecipitation experiments showed that ERα and β1-integrin immunoprecipitate together (Figs. 6 c and S3, c and d). Moreover, we found that β1-integrin has a sequence of five amino acids (LXXLL) within the cytoplasmic-proximal region of its transmembrane domain that is present among all steroid hormone receptor coactivators such as steroid receptor coactivator 1 (SRC1; Fig. 6 d; Mak et al., 1999). Indeed, this conserved motif called NR-box is known to be sufficient to mediate the interaction of coactivators with nuclear receptors such as ERα. For ERα in particular, this interaction is established within its helix 12 in the AF-2 domain (Heery et al., 1997; Savkur and Burris, 2004). In addition, we found that only β1- and β3-integrins contain this sequence (Fig. 6 d), and remarkably, these two integrins share several extracellular ligands and moreover are known to have transmembrane and cytoplasmic domains that are functionally interchangeable (Solowska et al., 1991). Fig. 6 e shows our proposed model for ERα–β1-integrin interaction. Further analyses needed to confirm ERα–β1-integrin physical association are being conducted at our laboratory.
We further investigated whether β1-integrin followed the same endocytosis/degradation pathway as ERα. We found that as with ERα, 60-min treatment with E2 generates a strong reduction in β1-integrin levels (Figs. 6 f and S3 e). As shown in Fig. 5 f, E-cadherin levels remain unchanged after this treatment, indicating that E2-induced endocytosis and posterior degradation is specific for certain plasma membrane proteins spatially associated with ERα. As expected, when cells were seeded on FN, this ECM protein rescued β1 integrin from E2-induced degradation (Figs. 6 g and S3 f).
Using the antibody feeding technique, we followed β1-integrin internalization dynamics and found that 15-min treatment with E2 stimulates the internalization of β1-integrin (Fig. S3, g and h). This technique allows the detection of β1-integrin+ endosomes in a cleaner manner, making it possible to see a small fraction of them without the background signal from cytoplasmic β1-integrin. Along with this, β1-integrin and ERα colocalize in a proportion of E2-induced endosomes (Fig. 6, h and i). Moreover, β1-integrin shows a strong colocalization with Rab11 after stimulation with E2 in cells plated on FN, indicating that as with ERα, β1-integrin is largely localized in Rab11+ vesicles under these conditions (Fig. 6, j and k). As another control, cells negative for ERα (such as MDA-MB-231) do not exhibit alterations in β1-integrin levels after prolonged treatment with E2 (Fig. S3, i and j), suggesting that E2-induced β1-integrin degradation is in fact mediated by ERα.
Estrogen treatment stimulates ERα–β1-integrin clustering
To investigate the interaction between ERα and β1-integrin in higher detail, we performed two-color superresolution microscopy using stochastic optical reconstruction microscopy (STORM; Rust et al., 2006; Bates et al., 2007). Fig. 7 a shows representative STORM images taken in regions of the filopodia of MCF7 cells.
Figure 7.
Effect of E2 treatment on the conditional distribution of ERα versus β1-integrin. (a) Images from STORM of filopodia of MCF7 cells treated as indicated for 15 min and stained for ERα or β1-integrin. Insets in the top left corners show the same images taken with widefield microscopy. Inside the zoomed areas, arrows show regions of superposition of the two markers (yellow pixels). Blue squares outline representative areas of 500 × 500 pixels used for subsequent analyses. In the treated cell, arrow inside the blue square shows a region of dense clustering between ERα and β1-integrin. (b) Tables showing the IF calculated as described previously (Bermudez-Hernandez et al., 2017) for 10 representative frames of filopodia of MCF7 cells under control (top) or treated (bottom) conditions. R-G, red–green correlation; G-R, green–red correlation. Red, β1-integrin; green, ERα. (c) IF calculated for one treated cell (frame 7) and for two sub-ROIs of this frame, showing how this index changes between areas of different β1-integrin/ERα densities. Bars 2 µm. (d) Histogram for normalized frequencies of MD between β1-integrin and ERα in filopodia of MCF7 cells among all the analyzed frames for each condition. For each domain detected, centroids were identified, and MDs were calculated from each β1-integrin to its nearest ERα domain throughout each 500 × 500–pixel frame. Frequencies were normalized to the highest value. The graph shows a slight shift toward smaller MDs for treated cells. (e) Mean density covariance between ERα and β1-integrin domains. Each frame was divided into square ROI of different sizes (window lengths ranging from 130 nm [10 pixels] to 2,000 nm [150 pixels] in side length). For each ROI size, the densities of ERα and β1-integrin were obtained, and the correlation coefficient (C) between these densities was calculated for all datasets. The mean of C among all the control (black full circles) or treated (pink empty squares) cells was plotted as a function of the window side length. Light-blue crosses show the z score (defined by the difference between the mean of the control group (for each window) and the mean of the treated group, and further divided by the square root of the sum of the SD of each group normalized by n). Thus, because the z score expresses, in units of SD, the distance between the two distributions, one may safely conclude that here there is no significant difference in density covariance between control and treated cells. (f) Two examples of a Voronoi partition for control (left) or treated (right) cells using the centroids of the ERα domains to compute the transformation. Colors indicated in the color bar on the right represent the size of each Voronoi shell (in square nanometers). Black small dots indicate the location of the centroids for β1-integrin domains. Empty big circles indicate the centroids of ERα domains; red circles denote those ERα that have β1-integrins closer than 160 nm, and blue circles indicate those ERα that have β1-integrins further than 160 nm away on the mean. The examples in these panels reveal a clear difference in ERα–β1-integrin bunching between control and treated conditions. (g) Histogram of frequencies for the ADs from each ERα centroid to the β1-integrins inside its Voronoi shell among all the analyzed data for each condition (note the semilog axis for presentation purposes). The graph shows a shift toward smaller distances for treated cells. Green arrows indicate as an example a region of the plot where the difference between treated and control fields is almost double. The inset shows the histogram for frequencies of the areas of the Voronoi regions among all the analyzed fields for each condition, demonstrating that treated cells present also relatively smaller Voronoi shells. (h) Graph of the bunching index for each cell, which is the ratio between the number of shells (normalized) that contains mean ERα–β1-integrin distances smaller than a threshold value of 160 nm. We named it bunching index as it quantifies the proportion of ERα–β1-integrin complexes among all the domains localized. Control image 1 and E2-treated image 4 are the ones represented in f. Inset shows the z score, which was calculated as the difference between each bunching index for the treated cell and the mean of the bunching indexes for the control group divided by the SD of the control group. Z score results demonstrate for six cells a significant difference (abs[z score] >1) in the bunching index between control and treated cells. In all plots, control cells are represented with black full circles and treated cells with pink empty squares. Treatments: ethanol (vehicle) or 10−8 M E2.
Two-color STORM is a fairly new technique, and therefore there is no consensus yet on the optimal method to quantify correlations between biomolecules. PCC or MCC indices have been used to measure the degree of cooccurrence of the two colors in the same pixel within very small areas of the image where physical colocalization happens (He et al., 2015; Johnson et al., 2016). In principle, the changes in the association between any pair of biomolecules can be estimated by three pairwise quantities: mutual distances between their domains, relative densities, and spatial heterogeneity. We started by analyzing the images using a recently published method (Bermudez-Hernandez et al., 2017) that accounts for two of these quantities: mutual distances and densities. Fig. 7 b shows the results for this interaction factor (IF) between β1-integrin and ERα calculated for control and treated cells. As shown by this index, there are regions with low colocalization (<0.1) and others with higher correlation (>0.4). In fact, this method shows a high variability depending on where the region of interest (ROI) is chosen as shown in Fig. 7 c. Therefore, based on this IF, one conclusion would be that there is no difference between control and treated cells, ignoring any change in β1-integrin/ERα clustering that could have occurred in specific domains of the filopodia of these cells. Indeed, this index, similarly to PCCs and MCCs, is biased toward studying the cooccurrence of the two colors in the same pixel, which makes sense for diffraction-limited microscopy but is not enough for the resolution that STORM offers (20–30 nm).
This calls for alternative ways to incorporate the heterogeneities of the distribution of the proteins in this analysis. Therefore, we implemented a numerical estimation of the changes in the three quantities to describe the interaction between ERα and β1-integrin. To facilitate the analysis, these calculations were done using only the centroids of each domain, a simplification further justified by the fact that the sizes of the domains of both proteins were shown to be invariant between the different conditions analyzed (see Fig. S4 e).
Of the three pairwise quantities, the simplest one is the estimation of the minimum distance (MD) calculated over all pairs of ERα and β1-integrin molecules. The first analyses from these datasets reveal that the overall mean MD between ERα and β1-integrin is ∼100 nm (Fig. S4 a), clearly below the diffraction limit. This indicates that these molecules are probably part of the same subcellular nanodomain and that their localization is not well resolved by conventional diffraction-limited microscopy. Using this approach, we also investigated whether distribution of ERα and β1-integrin was affected by 15-min treatment with E2. Overall, of all frames, we found a small shift toward smaller MD for treated cells (Fig. 7 d). However, when comparing individual frames, we observed that this shift was in the other direction in three of the 10 treated frames analyzed, suggesting that this overall difference is not significant and is highly dependent on the analyzed frame.
The second pairwise quantity estimates the covariation of densities, which is computed as the number of domains per unit area (for each ERα and β1-integrin). The functional association of these two molecules was investigated by computing their covariation as a function of increasing areas. The densities of the two proteins in fact covary, denoted by a positive linear regression coefficient between their densities as shown on the graph of Fig. S4 b. This covariation is shown to be statistically significant when tested against a null model constructed by randomizing the spatial positions of the centroids. This indicates that ERα and β1-integrin exhibit some degree of spatial cooccurrence and that such localization cannot be simply attributed to a random process (as indicated by the values of the z scores in the inset in Fig. S4 b). With this tool, we further sought to explore whether the density covariance was influenced by E2 treatment. In this case, we found no significant difference between control and treated cells (Fig. 7 e).
Finally, the third analysis considers the spatial spread of both molecules, which seems highly heterogeneous. Specifically, we computed the ratio between the two densities (number of ERα centroids over number of β1-integrin centroids) inside a square ROI of a given size. An example of that heterogeneity is presented (for ROI side length = 50 nm) in Fig. S4 c. The revealed heterogeneity calls for caution when reporting overall means because they might not be representative of changes that are very important in one cell but negligible in others. For that reason, we decided to explore a novel measure with the potential to avoid the limitations. The idea is very simple and uses a Voronoi transformation of the ERα receptor centroid positions (Nicovich et al., 2017). This mathematical transformation identifies a “shell” containing all the points in space that are closer to a given ERα protein than to any other ERα protein. After the transformation, the analysis estimates the size of the shells as well as their β1-integrin contents, i.e., the number, distance, and distribution of β1-integrin centroids inside each shell.
Two examples of the Voronoi transformation (control and treated cells) are presented in Fig. 7 f, where the size of the shells are labeled with colors: centroids of each ERα domain with empty circles and locations of the β1-integrin centroids with full black circles. For each frame, we computed in each shell the average distance (AD) of all β1-integrins to the ERα centroid (notice that this is different from the previous computation that only accounted for the MD). From the analysis of such transformations, we found that the AD of all β1-integrins to their corresponding ERαs inside each shell is different between control and treated cells. This is shown in Fig. 7 g, where the AD distributions for vehicle and treatment are plotted. Note that for relatively long AD values, the two estimations are similar; however, shorter ADs are more frequent for the treated cells than for the control ones. In other words, despite the heterogeneity, there are more ERαs having at least one β1-integrin close by. In particular, the region between 50 and 300 nm of ERα-to–β1-integrin distances shows the most significant shift between control and treated cells, revealing that these distances are more frequent after a 15-min treatment with E2. These distances are compatible with the typical sizes of early endosomes (Luzio et al., 2007; Su et al., 2016), which further supports our data showing that ERα and β1-integrin are internalized upon E2 treatment. In addition, the sizes of Voronoi shells are also reduced by E2 treatment, suggesting that ERs are more tightly packed together (Fig. 7 g, inset).
To account for the observed heterogeneity between different trials, particularly in treated cells, we developed an index to measure the statistical relevance of the differences between the mean of the control group and each treated cell individually, in that way revealing the intrinsic differences between treated ones (Fig. 7 h). We computed the ratio of Voronoi shells containing an AD less than a threshold value (we chose 160 nm, suggested by the mode value of AD in Fig. 7 g and also related to the most frequent size of early endosomes between 100 and 200 nm) over the total number of shells. This index, termed “bunching index,” is supposed to reflect for each experiment the tendency seen in Fig. 7 g for short distances.
As Fig. 7 h shows, a number of treated cells present a significantly higher bunching index than the control ones. Interestingly, there are a couple of E2-treated cells that showed the opposite effect. This could represent the different kinetics with which each cell responds to E2. After E2 treatment, ERα–β1-integrin clustering that can be observed in most of the treated cells would be the first step preceding caveolin-dependent internalization as has been extensively shown previously (Mayor et al., 1994; Upla et al., 2004; Bacia et al., 2005). Conversely, faster-responding cells might have already internalized most of the endosomes containing ERα–β1-integrin clusters that would therefore not be present anymore in the membrane region analyzed, explaining why some treated cells showed fewer ERα–β1-integrin clusters than control ones.
Moreover, we studied whether size or number of ERα or β1-integrin domains were affected by the treatment and found there is no significant effect on these variables (Fig. S4, d–f). Interestingly, sizes of these domains are scale free, i.e., they are well represented by a power law distribution, as often happens in many biological systems (Fig. S4 e; Honerkamp-Smith et al., 2009). We also verified that STORM clearly reveals nuclear accumulation of ERα upon E2 treatment as shown in Fig. S5 (a and b).
ERα–β1-integrin complexes are present in tumor and normal human samples
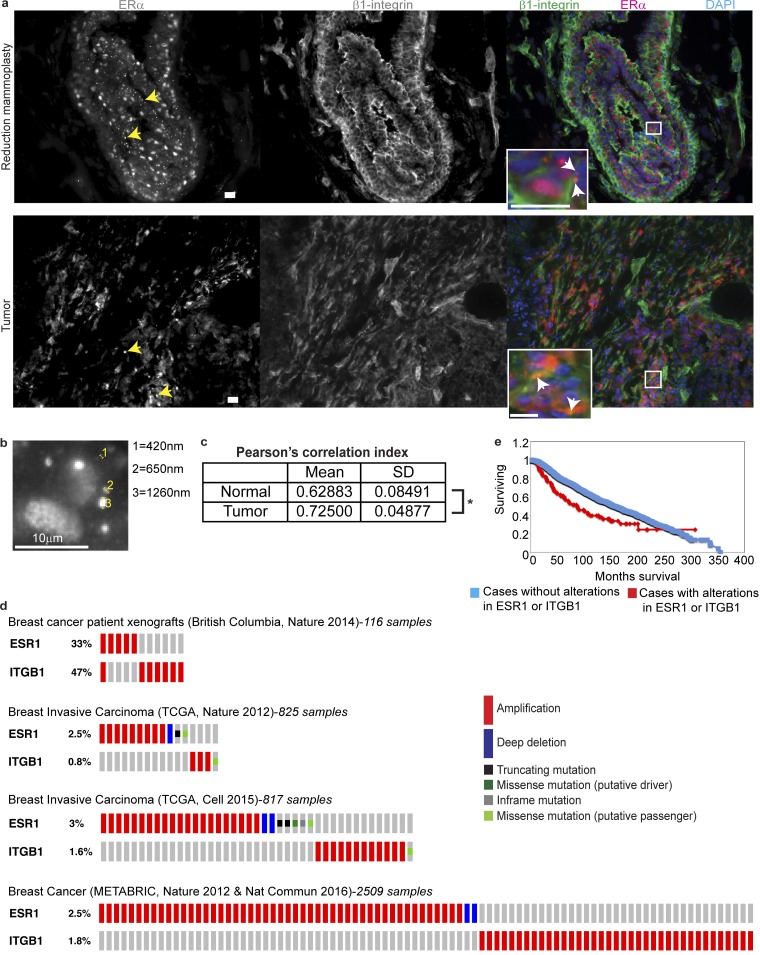
To further explore whether ERα endocytosis takes place within human tissues, we analyzed normal human tissues from reduction mammoplasties and tumor samples from patients with mammary adenocarcinoma. We found that ERα is present in endosome-like bodies in both normal and tumor samples (Fig. 8, a and b). Sizes of the vesicles observed are compatible with early endosomal vesicles (<500 nm), late endosomes (>600 nm), or multivesicular bodies (>1 µm; Luzio et al., 2007; Su et al., 2016). We confirmed these observations using the ERα antibody typically used for clinical analysis (clone SP1) to stain different sections of the same samples used in Fig. 8 a. This antibody reveals ERα+ endosomes in both normal and tumor tissues (Fig. S5, c and d).
Figure 8.
Endosomes containing ERα are present in normal and tumor human breast tissues. (a) Top: Confocal images of a normal human breast tissue (reduction mammoplasty; sample N211) stained for ERα (HC-20 clone), β1-integrin, and DAPI. In the inset, arrows indicate the presence of ERα+ endosomes. Similar results were obtained in the four different specimens analyzed. Bottom: Confocal images of a human breast tumor (Luminal A subtype adenocarcinoma; sample T171) stained for ERα (HC-20 clone), β1-integrin, and DAPI. Yellow arrows indicate ERα+ endosomes. In the inset, arrows indicate the presence of ERα+ endosomes. Similar results were obtained in the three different specimens analyzed. (b) Magnification from the inset shown in panel a (top). Diameters of ER+ vesicles are shown on the right. (c) Table showing mean and SD of Pearson’s correlation index calculated for the overall colocalization between ERα and β1-integrin. Differences between groups were analyzed by two-tailed Student’s t test (per replicate: nnormal = six fields; ntumor = seven fields). (d) OncoPrint from http://www.cbioportal.org (Cerami et al., 2012; Gao et al., 2013) showing the alterations found in ERα (ESR1) and β1-integrin (ITGB1) genes in different patients obtained from the search in four different datasets: British Columbia, Nature 2014 (Eirew et al., 2015); TCGA, Nature 2012 (Cancer Genome Atlas Network, 2012); TCGA, Cell 2015 (Ciriello et al., 2015); and Nature 2012 and Nature Communications 2016 (Pereira et al., 2016). (e) Kaplan–Meier plot of the overall survival of patients with alterations in ESR1 or ITGB1 genes using thelargest and newest dataset available in http://www.cbioportal.org (Breast Cancer-METABRIC; Cerami et al., 2012; Gao et al., 2013). Significance level after the log-rank test is shown in the plot. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 20 µm (unless otherwise indicated). The results shown in this study are in whole or part based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Interestingly, ERα colocalizes with β1-integrin in several areas of the analyzed tumor samples, showing a higher degree of colocalization compared with normal tissues (Fig. 8, a and c). Remarkably, membrane localization of ERα in the tumor samples is higher than in normal tissues; this might account for the increased degree of colocalization with β1-integrin within these samples.
To further explore the clinical relevance of ERα–β1-integrin association, we analyzed TCGA data through cBioPortal (Cerami et al., 2012; Gao et al., 2013) and found that alterations in ERα (ESR1) and β1-integrin (ITGB1) genes present a tendency to be mutually exclusive in breast cancer (Fig. 8 d). As has been intensely studied, alterations that affect the same pathway tend to not co-occur in the same patient (Cancer Genome Atlas Network, 2012; Ciriello et al., 2012). Therefore, mutual exclusivity would be further evidence indicating that ERα and β1-integrin signaling have a close relationship. Moreover, breast cancer patients with genetic alterations in ESR1 or ITGB1 have decreased survival (Fig. 8 e).
These preliminary clinical findings reveal that even though ERα+ vesicles are present in both normal and tumor tissues, ERα and β1-integrin might only be co-endocytosed within tumors. Therefore, FN-induced strengthening of ERα signaling would be a tumor-specific phenomenon, which further suggests this pathway as a target for new antitumor therapies.
Discussion
In this study, we demonstrate by biochemical approaches and high-resolution microscopy that E2 induces endocytosis of ERα by a mechanism involving caveolin 1. In the presence of FN, ERα avoids lysosomal degradation and is localized in Rab11+ recycling endosomes. We found that ERα is functionally associated with β1-integrin at the plasma membrane of breast tumor cells. We show that β1-integrin follows the same endocytosis/degradation dynamics in the presence of E2 and would be responsible for dragging ERα to Rab11+ vesicles in the presence of FN, avoiding lysosomal degradation. In this context, FN has a direct, positive impact on ERα’s transcriptional activity. ERα+ vesicles are present within human breast tissues, and colocalization with β1-integrin is detected primarily in tumors. The mechanism we describe in this study unravels a new level of regulation of cancer cell signaling by the ECM and provides a putative target for new treatments directed to resensitize patients to endocrine therapy.
We found that upon E2 treatment, activated membrane ERα is endocytosed in a caveolin-dependent manner and travels in endosomes through the cytoplasm and into the nucleus. In addition, inhibition of clathrin also impairs ERα transcriptional activity, suggesting that the clathrin-dependent pathway might be also, at least partially, involved in ERα endocytosis. The shuttling of plasma membrane proteins into the nucleus through endosomes has been described for other endosomal proteins (Chaumet et al., 2015) and transmembrane receptors such as FGF (Małecki et al., 2004). A mechanism for E2-induced ERα endocytosis has been proposed for ERα-positive neurons (Kisler et al., 2013). We propose that binding of E2 to ERα would occur at the plasma membrane so that at least a proportion of hormones could exert their action without actually crossing through the membrane. We show in this study that E2-induced endosomes containing ERα can also be targeted to lysosomes, where ERα is degraded. Therefore, we propose that E2-induced lysosomal degradation of ERα is a relevant mechanism of desensitization to E2. However, this mechanism is lost when cells are in a FN-rich matrix, where ERα escapes lysosomal degradation and its transcriptional activity is enhanced. We present evidence indicating that FN promotes ERα localization in Rab11+ vesicles that would therefore inhibit its lysosomal degradation.
As with other membrane proteins such as caveolin 1 and membrane-associated proteins such as integrin-linked kinase (Schlegel et al., 1999; Acconcia et al., 2006; Chung et al., 2009), ERα colocalizes with β1-integrin in plasma membrane structures. Through high-resolution microscopy and coimmunoprecipitation analyses, we show evidence of a close association between these proteins that would be mediated by the NR-box that we identified within the β1-integrin sequence. Ongoing experiments in our laboratory are aiming to confirm the physical association between these two proteins and the domains involved.
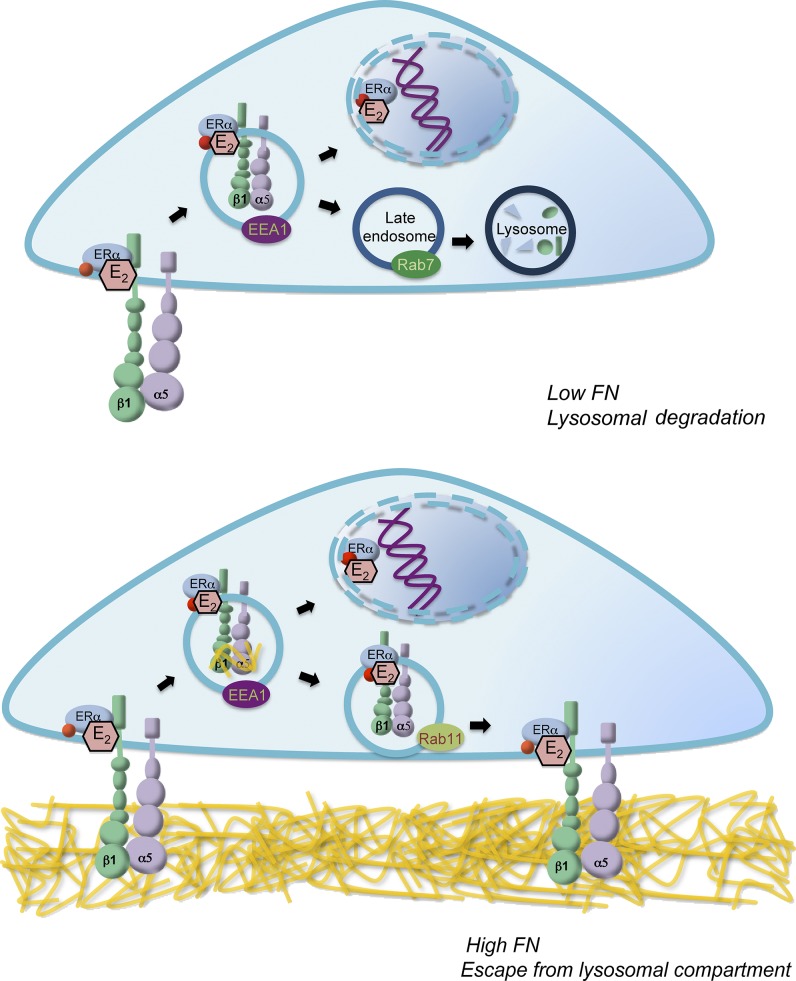
Through STORM superresolution microscopy, we showed that ERα and β1-integrin are present in the same nanodomains within the filopodia and, moreover, are organized in structures of size consistent with early endosomes. In addition, STORM revealed that ERα–β1-integrin clustering is incremented upon E2 treatment, further supporting the presented evidence that indicates that ERα–β1-integrin complexes are endocytosed together upon E2 treatment. Membrane β1-integrin could be in both its active (high-affinity) or inactive (low-affinity) conformations, each of which normally undergo different endocytosis/recycling pathways (De Franceschi et al., 2015). Inactive β1-integrins are rapidly recycled to the cell membrane to form protrusions such as lamellipodia and filopodia, following a fast recycling pathway. Active β1-integrins are less efficiently recycled and are targeted to the Rab7 compartment (Arjonen et al., 2012). However, in the presence of FN, ligand-occupied active β1-integrins are rapidly recycled from the lysosomal compartment to the rear of the cell, keeping their active conformation. Interestingly, this process occurs specifically in cancer cells and is a way by which cells remodel their ECM (Dozynkiewicz et al., 2012). Therefore, we propose that E2 induces internalization of at least active β1-integrins associated with ERα (Fig. 9). In the absence of FN, active β1-integrin and ERα are directed to the lysosomal compartment, where both proteins are degraded. However, when FN is present, ligand-occupied active β1-integrin would be recycled to the plasma membrane in Rab11+ vesicles, carrying ERα with it and inhibiting its lysosomal degradation (Fig. 9). The role of integrins as masters of endosomal trafficking has been also demonstrated for other receptors and cargos such as VEGFR2 and lipid rafts (Caswell et al., 2009). In this context, an FN-rich matrix represents a double advantage for breast tumor cell survival because it triggers proliferative signals transduced through β1-integrin (Han and Roman, 2006; Moreno-Layseca and Streuli, 2014) and also intensifies E2 signaling. Ongoing experiments in our laboratory are aiming to determine whether E2 affects active and inactive β1-integrins differently.
Figure 9.
Model for endocytic transport of ERα and β1-integrin regulated by FN in breast cancer cells. Estrogens induce rapid endocytosis of membrane ERα–β1-integrin complexes, generating EEA1+ vesicles. In the absence of FN, vesicles containing β1-integrin and ERα could either fuse to the nuclear membrane where ERα exerts its action or follow the lysosomal pathway, where ERα colocalizes with Rab7. After 60 min, ERα and β1-integrin are degraded in lysosomes and the signal ends. In the presence of FN, ERα and β1-integrin are localized in Rab11+ vesicles, suggesting that they might be recycled and therefore avoid the lysosomal pathway. ERα and β1-integrin levels are maintained over time and the cycle continues, keeping ERα transcriptionally active.
Several functions have been associated with membrane ERα and are mainly related to nonclassic (extranuclear) ERα signaling pathways (Levin, 2009). However, some authors have begun to suggest that there is a direct link between membrane ERα and its classic nuclear activity (Pedram et al., 2002; La Rosa et al., 2012). Our results provide a link between genomic and nongenomic effects of E2 through the activity of membrane-bound ERα. We show evidence suggesting that membrane ERα travels in endosomes into the nucleus, where it would also have transcriptional (genomic) activity. The evidence of the signaling pathway shown in this study is, to our knowledge, the first demonstration of the previously suggested hypothesis of an active mechanism responsible for E2 shuttling into the nucleus (Pietras and Szego, 1984; Razandi et al., 2002). Further analyses are needed to unravel whether membrane ERα dimerizes within these endosomes and at which stage of its subcellular shuttling.
Endocytosis and subsequent degradation of ERα in lysosomes described in this study is a novel pathway of subcellular signaling and negative feedback induced by E2. Totta et al. (2014, 2015) recently proposed that membrane-bound ERα could be degraded in lysosomes as well as in the proteasomal compartment. However, the authors did not demonstrate what membrane events take place in response to E2, leading to ERα internalization and further lysosomal degradation. In this study, we show robust evidence of this phenomenon, describing how this process is regulated and what is the response of the membrane proteins involved. However, it still remains to be elucidated how ERα enters the lysosomal lumen for its degradation. In this sense, it has been proposed that the molecular pump LAMP-2, which has been shown to interact with ERα, would be responsible for allowing its uptake into the lysosomal lumen (Totta et al., 2014; Wang and Robbins, 2014). The signal that triggers ERα lysosomal degradation after prolonged exposure to E2 and whether this mechanism requires ERα’s previous translocation to the nucleus are still unknown.
The finding that FN strengthens ERα activity suggests that tumor-like stromas would enhance ERα’s activity. FN has been shown to be incremented within cancer cells (Nam et al., 2010; Bae et al., 2013) and is known to contact breast epithelial cells when the normal structure of the basement membrane is disrupted, which occurs during malignant transformation (Ghajar and Bissell, 2008; Lu et al., 2011, 2012). Within tumors, not only ECM composition changes but also its stiffness (Acerbi et al., 2015). Therefore, it is tempting to speculate that not only the presence of FN but also the distorted tissue architecture in stiff substrates (i.e., more cell-substrate contacts, loss of cell apical-basal polarity) might affect ERα degradation in tumors. The loss of normal cellular polarity and subcellular compartmentalization might lead to a different frequency of ERα–β1-integrin interactions in tumor cells. Indeed, we show that although ERα+ endosomes are present in normal as well as tumor human samples, ERα colocalizes with β1-integrin fundamentally in tumors. These data are consistent with studies showing that high levels of FN and β1-integrin in breast tumors are associated with lower survival (Yao et al., 2007; Helleman et al., 2008). Moreover, clinical data of breast cancer patients from TCGA databases show that alterations in ERα and β1-integrin genes are mutually exclusive, suggesting that they might be implicated in the same signaling pathway. In addition, alterations in these genes correlate with decreased survival. The findings presented in this study have direct therapeutic implications for breast cancer as blocking FN-dependent activation of ERα, potentially by inhibiting the interaction between ERα and β1-integrin, arises as a novel target for new therapies. This would be a breakthrough approach to overcome endocrine resistance induced by the ECM in breast cancer.
Materials and methods
Cell culture
MCF7 and T47D cell lines were purchased from ATCC and regularly checked for mycoplasma. These cell lines were routinely maintained in DMEM/F12 cell culture medium (Sigma-Aldrich) supplemented with 10% FBS (Internegocios) and gentamicin, in a humidified 5% CO2/air atmosphere. Serial passages were performed by treatment of 80% confluent monolayers with 0.25% trypsin (Invitrogen) and 0.02% EDTA in Ca2+-free and Mg2+-free PBS.
Reagents
E2 was purchased from Sigma-Aldrich; BAF and BSA from Santa Cruz Biotechnology; and FN from EMD Millipore. Filipin and PAO, both from Sigma-Aldrich, were provided by C. Davio (Universidad de Buenos Aires, Buenos Aires, Argentina). BZ was purchased from Velcade; Lipofectamine 2000 from Thermo Fisher Scientific; LyoVec transfection reagent InvivoGen; phalloidin from Sigma-Aldrich, and DAPI from Research Organics.
DNA constructs and RNA interference sequences
GFP-Rab7 expression construct was a gift from C. Arregui (Universidad de San Martin, Buenos Aires, Argentina; Hernández et al., 2006). Plasmid pTK-ERE-luc containing five copies of the ERE upstream of the luciferase cassette was a gift from C. Jordan (University of Texas, Houston, TX). pTK-renilla was purchased from Promega.The constructs were verified by sequencing. siRNA/Stealth against ERα was purchased from Invitrogen as the following sequences: sense 5′-CAGAGGCUCUCAAACUAUAAAGAAA-3′, and antisense 5′-UUUCUUUAUAGUUUGAGAGCCUCUG-3′. siRNA against caveolin 1 (sc-29241), siRNA against clathrin–heavy chain (HC; sc-35067), and scrambled siRNA (sc-37007) were purchased from Santa Cruz Biotechnology.
Antibodies
The following antibodies were used in this study and were purchased from Santa Cruz Biotechnology unless otherwise noted (including dilutions/amounts used for immunofluorescence, Western blot [WB], and immunoprecipitation [IP]): ERα (HC-20 rabbit; 1:100 immunofluorescence, 1:200 WB; 3 µg IP), ERα (F-10 mouse; 3 µg IP), β1-integrin (LM534 mouse; 1:100 immunofluorescence; EMD Millipore), β1-integrin (M-106 rabbit; 1:300 WB; 3 µg IP), E-cadherin (H-108 rabbit; 1:1,000 WB), β-actin (C4 mouse; 1:10,000 WB), Rab11 (H-87 rabbit; 1:200 WB), Rab7 (sc-376362 mouse; 1:100 immunofluorescence), and caveolin 1 (sc-53564 mouse; 1:600 immunofluorescence; 1:200 WB). LAMP-1 (ab25630 mouse; 1:20 immunofluorescence), clathrin (ab2731 mouse; 1:500 immunofluorescence), and Lamin B1 (ab133741 rabbit; 1:243 immunofluorescence) were purchased from Abcam; and clathrin-HC (clone 23 mouse; 610500; 1:1,000 WB) was purchased from BD. HC-20 peptide was purchased from Santa Cruz Biotechnology. Secondary antibodies used for WB (1:5,000) were goat anti–mouse HRP-conjugated (AP308P) and goat anti–rabbit HRP conjugated (AP132P) purchased from EMD Millipore. Secondary antibodies used for immunofluorescence (1:500) were goat anti–mouse and goat anti–rabbit Alexa Fluor 488–, 555–, and 647–conjugated antibodies, all purchased from Thermo Fisher Scientific.
Subcellular fractionation
1,000,000 cells were seeded in 60-mm cell culture dishes (Greiner-Bio-One) coated with FN (2 µg/cm2 in PBS) in regular culture medium. Culture dishes coated with BSA (2 µg/cm2 in PBS) were used as control. After 16 h, cells were washed three times with PBS, and culture medium was changed to phenol red–free DMEM/F12 plus 1% charcoal-stripped FBS and left in the incubator for another 24 h. Cells were then treated with 10−8 M E2 at 37°C for the indicated times and washed twice with PBS, and protein extracts were prepared by homogenizing fresh cells on ice in subcellular fractionation buffer (250 mM sucrose, 20 mM Hepes, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, and 1 mM EGTA; Abcam). The Abcam subcellular fractionation protocol was followed. Briefly, cells were centrifuged at 720 g to obtain the nuclear pellet, and the supernatant was recentrifuged at 3,000 g to obtain the cytoplasmic and membrane fraction. After two further ultracentrifugations, cytosolic and membrane fractions were obtained. These fractions were subsequently analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies. Efficiency of nuclear/cytoplasmic separation was verified using nuclear-specific protein H2A.X (Fig. S5 e). E-cadherin was used to verify the efficiency of membrane purification (Fig. S1, g–j).
Western blot
Protein extracts from whole cells were prepared by scraping the culture dishes on ice with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 8.0, containing 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, and 1% NP-40) containing protease inhibitors (40 µM phenylmethylsulfonyl fluoride, 5 µg/ml leupeptin, 50 µg/ml aprotinin, and 200 µM orthovanadate). Protein extracts form subcellular fractions were obtained as described above. Protein concentration was measured using the Bradford method (Bradford, 1976). After adding sample buffer containing β-mercaptoethanol, samples were heated at 95°C for 5 min. 50 µg of each sample was then run in SDS-PAGE mini-gels and transferred to polyvinylidene difluoride membranes (Amersham Biosciences). Membranes were blocked for 1 h at RT in 5% fat-free milk in Tris-buffered saline plus 0.1% Tween-20 (TBST). Primary antibodies were prepared in blocking medium and incubated at 4°C overnight. After washing with TBST, membranes were incubated with secondary antibodies for 1 h at RT. Signal was detected with an enhanced chemiluminescence kit (ECL; Amersham Biosciences). Densitometry was performed using the gel analyzer plugin of Fiji (ImageJ; National Institutes of Health; Schindelin et al., 2012) and the plugin Gels. The standard process with this software was to select the ROI in each lane individually and then plot the intensity measurements. The area under the curve was quantified, which represents the final intensity for each band. The intensities were further analyzed as has been extensively reported previously (Degasperi et al., 2014; McDonough et al., 2015). We then normalized each intensity value to the intensity measured for β-actin in the corresponding lane for each one of the three repetitions of the experiment. The three results for the density ratio of the control group were then averaged, and each ratio was normalized to the control mean, so that the control value will be 1 with its correspondent SD.
Membrane fluidity reduction, endocytosis/membrane recycling blockade, and lysosomal inhibition
For membrane fluidity reduction, a 15-min pretreatment of chilling cells at 0°C followed by a treatment with E2 at 0°C or 37°C was performed. For endocytosis/membrane recycling blockade, filipin (2.5 µg/ml), or PAO (5 µM) were administrated together with E2 for the indicated times. For lysosomal inhibition, a 90-min pretreatment with BAF (25 nM) at 37°C was done. After these treatments, subcellular fractionation and Western blot were performed as described above.
Dextran endocytosis assay
We followed the protocol described previously for substrate endocytosis/recycling (Gillespie et al., 2013). Briefly, cells were seeded at 80% confluence in 24-well plates. After 16 h, cells were washed three times with PBS and serum-starved for 24 h. Cells were then treated with 10−8 M E2 or its vehicle for 15 min at 37°C. Then 10 µg/ml dextran-CF543 (80111; Biotium) was added, and cells were left at 37°C for another 10 min. Subsequently, cells were washed once with cold serum-free medium and twice with 0.2 M sodium acetate, pH 4.5, and washed again once with cold serum-free medium. For the chase, cells were incubated another 15 min at 37°C with serum-free medium. After the chase, the medium was recovered before washing one more time with 0.2 M sodium acetate, pH 4.5. Fluorescence from acid washes and medium was measured on a FilterMax F3 MultiMode Microplate-Reader (Molecular Devices) at 535/595-nm absorption/emission.
IP
The protocol described by Bonifacino et al. (2001) was followed with slight modifications. Briefly, fresh cells were lysed with weak RIPA buffer. 3 µg antibody was preincubated with protein A/G agarose beads (Santa Cruz Biotechnology) with gentle mixing for 1 h at RT. Antibody–bead complexes were then mixed overnight at 4°C with 500 µg protein. After several washes with weak RIPA, samples were analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies.
Luciferase assay
100,000 cells per well were seeded in 48-well culture dishes coated or not coated with FN (2 µg/cm2) in the presence of LyoVec–DNA complexes (50:1). DNA constructs pTK-Renilla and pTK-ERE-Luc were used in a 10:1 ratio. After 18 h, cells were washed three times with PBS, and culture medium was changed to phenol red–free DMEM/F12 plus 1% charcoal-stripped FBS and left in the incubator for another 24 h. Cells were then incubated in the presence of 10−8 M E2 at 37°C for 14 h. Dual-luciferase reporter assay system kit (Promega) was used to reveal luciferase or renilla signals, following the instructions described by the manufacturer.
Immunofluorescence and confocal microscopy
50,000 cells were seeded on glass coverslips (Marienfeld) in 24-well plates coated with BSA or FN (2 µg/cm2) in regular culture medium. After 16 h, cells were washed three times with PBS, and culture medium was changed to phenol red–free DMEM/F12 plus 1% charcoal-stripped FBS and left in the incubator for 24 h. Cells were then treated with 10−8 M E2 for the indicated times. When cells were transfected before this treatment, 3 × 104 cells were seeded on glass coverslips in 24-well plates covered or uncovered with FN (2 µg/cm2) in regular culture medium. After 16 h, cells were transfected using Lipofectamine 2000, following the protocol described by the manufacturer. Briefly, culture medium was replaced by OptiMEM (Thermo Fisher Scientific), and cells were incubated for 24 h with lipofectamine–DNA (5:1) complexes. Treatment was then administered as described above. For immunofluorescence staining, the protocol described by Debnath et al. (2003) was followed with slight modifications. In brief, cells were fixed for 20 min at RT with 4% PFA in PBS and permeabilized with 0.1% Triton X-100 in PBS for 10 min at 4°C, then blocked for 90 min at RT with immunofluorescence buffer (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaN3, 0.1% BSA, 0.2% Triton X-100, and 0.05% Tween-20) plus 10% goat serum. They were subsequently stained with the indicated primary antibodies (prepared in blocking medium) overnight at 4°C, followed by incubation with the secondary antibodies for 1 h at RT. For β1-integrin staining, when indicated, cells were live-stained: antibody was prepared in the culture medium, and cells were incubated with it for 1 h in the incubator. They were subsequently fixed and further stained as described above. For β1-integrin endocytosis assay, after live-staining, cells were treated with 10−8 M E2 for 15 min and then fixed and permeabilized with 0.1% Triton X-100 in PBS for 20 min at 37°C. Secondary antibodies were incubated for 1 h at RT. Coverslips were mounted using Mowiol 4–88 (Sigma-Aldrich). Widefield images were acquired using a Nikon Eclipse TE 2000-S inverted microscope with a Nikon Plan Fluor 20×/0.50-NA objective at RT using a Nikon Digital Sight DS-Fi1 camera and the acquisition software NIS-Elements (Nikon) and processed with Fiji. Confocal images were acquired using an Olympus FV-1000 inverted confocal microscope with an Olympus 60×/1.20-NA UPLAN Apochromat water objective, at RT, using the acquisition software Olympus FluoView v.5.0 and processed with Fiji. For colocalization analyses, PCCs and MCCs were calculated within the desired areas of the images as described previously (Dunn et al., 2011). Briefly, PCC is a useful statistic to quantify colocalization, with values range from 1 for two images whose fluorescence intensities are perfectly and linearly related to −1 for two images whose fluorescence intensities are perfectly but inversely related to one another. Values near zero reflect distributions of probes that are uncorrelated with one another. In the images shown, red color indicates a value of 1 for the PCC, and blue indicates a value of −1. The formula for PCC for a typical image consisting of red and green channels is
where Ri and Gi refer to the intensity values of the red and green channels, respectively, of pixel i, and R̄ and Ḡ refer to the mean intensities of the red and green channels, respectively, across the entire image. We calculated it for each analyzed frame using the Colocalization colormap plugin for Fiji. Although PCC provides an effective statistic for measuring overall association of two probes in an image, MCC is useful to measure the fraction of one protein that colocalizes with a second protein. For two probes, denoted as R and G, two different MCC values are derived, M1, the fraction of R in compartments containing G, and M2, the fraction of G in compartments containing R. These coefficients are simply calculated as
where Ri,colocal = Ri if Gi > 0 and Ri,colocal = 0 if Gi = 0 and
where Gi,colocal = Gi if Ri > 0 and Gi,colocal = 0 if Ri = 0. We calculated these coefficients for each analyzed frame using the Coloc 2 plugin for Fiji.
3D rendering from z stacks was performed using Fiji and the plugin 3D Viewer. This plugin performs the 3D reconstitution from which we recorded the videos and images shown in this paper.
Verification of antibody specificity
Every antibody used in this paper was chosen based on its wide usage in the literature. In particular, the antibody used to detect ERα has been used in >200 papers (Gao et al., 2015; Arnal et al., 2017). β1-integrin antibodies used throughout this paper have also been widely used in the literature (Tiwari et al., 2011; Waxmonsky and Conner, 2013; Long et al., 2016). In the case of the antibody used to detect ERα, the most thoroughly used in the present paper, its specificity was tested for Western blot and immunofluorescence. For immunoneutralization assays, ERα antibody (clone HC-20; Santa Cruz Biotechnology) was preincubated for 90 min at 4°C with HC-20 peptide (Santa Cruz Biotechnology) or its control before using it in the blotting membranes from MCF7 cell lysates or for immunofluorescence of these cells. Considering that each antibody has two HC-20 epitopes and that an excess (at least 10-fold) of the peptide is needed to ensure efficient blocking (Skliris et al., 2009), the amount of blocking peptide used was calculated as
However, the specificity of this antibody was further confirmed by knocking down endogenous ERα in MCF7 cells, with the specific siRNA described above, followed by its detection through Western blot to evaluate the loss of the ERα signal.
TIRFM
This technique is based in the formation of an evanescent electromagnetic field generated when the incident light is totally internally reflected at the glass–water interface. The evanescent field decays exponentially from the interface and thus only illuminates fluorophores that are close to the glass. Therefore, it is used to selectively detect molecules localized in the ventral plasma membrane, within a radius of ∼100 nm from the glass surface (Ambrose, 1956; Axelrod, 1981, 2001). 300,000 cells were seeded on BSA- or FN-coated (2 µg/cm2) 25-mm coverslips in six-well plates. After 16 h, cells were live-stained for β1-integrin as explained for confocal microscopy, or fixed with 4% PFA/4% sucrose and stained with the antibody against ERα. Coverslips were mounted in PBS on the stage of a fully motorized Nikon TE2000-E inverted microscope equipped for widefield and TIRFM. In all cases, cells were visualized through a TIRF 60×/1.45-NA water objective, at RT, and imaged using an ORCA II ER charge-coupled device (CCD) camera controlled by the MetaMorph software (Molecular Devices). Images were then processed with the software Fiji. For TIRFM, cells were illuminated using a 488-nm argon laser. Evanescent wave penetration depth was calculated to be ∼210 nm using the following parameters: 488 nm as λ, 1.52 as n1, 1.33 as n2, and 62° as the incident light beam angle. For image processing, when indicated, polar transformation was performed using Fiji and the plugin Polar Transformer (https://imagej.nih.gov/ij/plugins/polar-transformer.html). This transformation takes an input image in a Cartesian space and transforms it into polar coordinates. This transformation is useful for “unwrapping” images with a generally round object. As a consequence of this transformation, if the image has a visible background, the four angles of the original can be often seen as four triangle-like shapes to the right of the transformed image (giving rise to five black arches, which is the color of the underlying background).
STORM
Setup
The STORM microscope is custom-built on an Olympus IX-73 inverted microscope. Two continuous-wave lasers of wavelength 642 nm (2RU-VFL-P-1500-642; MPB Communications) and 532 nm (Ventus 532; Laser Quantum) and output power of 1.5 W are used for fluorescence excitation/deactivation (van de Linde et al., 2011), and a 405-nm continuous-wave, 50-mW laser (RGB Photonics Lambda Mini) is used for fluorescence reactivation. The lasers are combined with dichroic mirrors (LM01-552-25 and LM01-427-25; Semrock), magnified, and then focused to the back focal plane of the oil-immersion objective Olympus Plan Apochromat 60× NA 1.42. A multiband dichroic mirror (Di03-R 405/488/532/635-t1 25 × 36; Semrock) is used for decoupling of the fluorescence emission of the sample from the laser excitation. Further blocking of the illumination lasers is performed with a multi-edge notch filter (NF03-405/488/532/635E-25; Semrock). The emission light is further divided into two channels with a longpass dichroic (zt647rdc; Chroma) and expanded with a 2× telescope so that the pixel size of the electron-multiplying CCD camera (Andor iXon3 897) matches an optimal value for single-molecule localization, in this case a pixel size of 133 nm. Both channels are filtered with appropriate emission dichroics for Alexa Fluor 565 and 647 (Semrock 582/75 BrightLine HC and Chroma ET700/75m) and imaged side by side into the same electron-multiplying CCD camera (Andor ixon3 897) by using a D-shaped mirror.
A motorized platform is used to laterally displace the illumination (two mirrors and lens), focusing at the back focal plane of the objective. This allows to switch among conventional widefield, HILO, and TIRF illumination. To prevent defocusing within the relatively long STORM imaging acquisition, the setup is equipped with a custom-built focus stabilization system that senses the total internal reflection of an IR diode laser at the interface between the coverslip and the sample and then actuates by mechanically correcting the objective’s axial position. The camera, lasers, motorized parts of the microscope, and focus stabilization system are controlled and integrated with Tormenta, open-source, free Python software for fluorescence microscopy control and measurements (Barabas et al., 2016).
Data acquisition
Cells cultured on 18-mm coverslips were placed in a holder, and imaging was performed in 50 mM Tris, pH 8, 10 mM NaCl buffer, at RT. The imaging buffer was supplemented with 10% wt/vol glucose, 100 mM 2-mercaptoethylamine, 1 µg/ml glucose oxidase (Sigma-Aldrich), and 0.5 µg/ml catalase (Sigma-Aldrich) as oxygen scavenging system.
Before STORM imaging, conventional fluorescence images of the ROI were acquired by setting the excitation laser intensity to 1–5 W cm−2. STORM data acquisition was then started by changing the excitation lasers’ intensity to 5–15 kW cm−2, thus inducing on-off switching of the fluorescent marker in the tens of milliseconds time range. A camera rate of 20 Hz was found appropriate considering both Alexa Fluor 647 and 565 switching times. Throughout the whole acquisition, the activation 405-nm laser power (1–10 µW cm−2) was increased manually in steps whenever the density of single-molecule events decreased below ∼1–2 molecules per frame. Typically, 15,000 frames were recorded to assure a high density of localizations.
To correct for chromatic aberrations, the two channels were calibrated using fluorescent nanoparticles emitting in both channels (TetraSpeck microspheres; Thermo Fisher Scientific). An affine matrix was computed as the best transformation that matches the location of the beads in both channels (Hartley and Zisserman, 2004). Only calibrations resulting in an error <10 nm were used. A preprocessing background subtraction step is performed to match the background level of both channels for subsequent analysis. The localization analysis and the rendering of the final superresolved image were performed with ThunderSTORM software (Ovesný et al., 2014).
Data analysis
MatLab software (release 2014a; MathWorks) was used to analyze all the images using codes developed by our laboratory to study distribution of distances, densities, and architecture of the proteins imaged by STORM as described for each corresponding figure.
Human breast tissues
Breast tissues from reduction mammoplasties and tumors were acquired from the Cooperative Human Tissue Network, a program funded by the National Cancer Institute. All specimens were collected with patient consent; reduction mammoplasties were reported negative for proliferative breast disease by board-certified pathologists. Use of anonymous samples was granted exemption status by the University of California, Berkeley, Institutional Review Board in accordance with the Code of Federal Regulations 45 CFR 46.101.
Statistical analysis
All statistical analyses were performed using SPSS (IBM SPSS Statistics for Mac OS X, v.23.0; IBM Corp.) or Prism 5 (v.5.0c for Mac OS X; GraphPad Software). To detect differences between media, we used Student’s t test when comparing between two media. For multiple contrasts, we used two-way ANOVA followed by Bonferroni contrast adjusted for multiple comparisons. Statistical tests and the corresponding contrasts used for each assay are indicated in the figure legends.
Online supplemental material
Fig. S1 shows that FN stabilizes ERα and verifies specificity of the antibodies used. Fig. S2 provides further evidence that ERα is endocytosed in the presence of E2. Fig. S3 confirms that ERα and β1-integrin colocalize at the plasma membrane and are internalized upon treatment with E2 in both MCF7 and T47D cells. Fig. S4 shows the conditional distribution of ERα and β1-integrin and its statistical properties from superresolution microscopy analyses. Fig. S5 shows the nuclear localization of ERα through STORM and provides evidence for ERα+ endosomes in human breast tissues using the antibody typically used for clinical analysis (clone SP1). Video 1 provides a 3D render to show that ERα and β1-integrin colocalize in membrane structures in breast tumor cells.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Carlos Arregui for the gift of GFP-Rab7 expression construct. We also acknowledge Dr. Carlos Davio for providing us with the endocytosis inhibitors. We thank the scientific and technical support provided by the members of the Institute of Oncology “Ángel H. Roffo.”
This work was supported by the following grants to M. Simian: Susan G. Komen for the Cure (BCTR0600341), Agencia Nacional de Promoción Científica y Tecnológica (PICT2008-0325/Préstamo BID), Instituto Nacional del Cáncer Ministerio de Salud de la Nación (2015), Fundación Florencio Fiorini, and the Consejo Nacional de Investigaciones Científicas y Técnicas. In addition, the work was supported by a grant from U.S. Department of Defense to M.J. Bissell (W81XWH0810736) and the Breast Cancer Research Foundation to M.J. Bissell as well as a Department of Defense grant to V.M. Weaver (W81XWH-13-1-0216) and a grant from the National Institutes of Health/National Cancer Institute to V.M. Weaver (R01CA192914; Weaver/Seewaldt). We also thank Consejo Nacional de Investigaciones Científicas y Técnicas for the doctoral fellowship awarded to R.G. Sampayo and the Fulbright Association and the Fundación Bunge y Born for the fellowship awarded to R.G. Sampayo.
The authors declare no competing financial interests.
Author contributions: R.G. Sampayo and M. Simian conceived the project; R.G. Sampayo and M. Simian designed experimental approaches; R.G. Sampayo performed most of the experiments and analyzed all the data; R.G. Sampayo and A.M. Toscani developed TIRFM and confocal microscopy analyses; A.M. Toscani also helped in the experimental design; A. Cáceres provided expertise in high-resolution microscopy and the tools to develop TIRFM experiments; M.G. Rubashkin and J.N. Lakins provided technical support for developing confocal and high-resolution microscopy; K. Thi and W.C. Hines performed immunofluorescence staining and imaging of human samples; W.C. Hines also provided key advice for manuscript preparation; F.C. Leskow provided experimental advice and insightful ideas on data interpretation and contributed to the confocal microscopy analyzes; L.A. Masullo, I.L. Violi, and F.D. Stefani provided the necessary equipment to perform STORM, helped to carry out the measurements, and contributed in the elaboration of the superresolution section of this manuscript; D.R. Chialvo provided the numerical tools applied in this study to analyze superresolution microscopy images, performed the analysis with R.G. Sampayo, and contributed in the elaboration of the superresolution section of this manuscript; M.J. Bissell and V.M. Weaver provided expertise in ECM biology, conceptual advice, and experimental support; V.M. Weaver provided expertise in biophysics of cell–matrix interaction; R.G. Sampayo and M. Simian wrote the paper. All authors read and extensively critiqued the manuscript.
References
- Acconcia F., Ascenzi P., Bocedi A., Spisni E., Tomasi V., Trentalance A., Visca P., and Marino M.. 2005. Palmitoylation-dependent estrogen receptor alpha membrane localization: Regulation by 17beta-estradiol. Mol. Biol. Cell. 16:231–237. 10.1091/mbc.e04-07-0547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acconcia F., Manavathi B., Mascarenhas J., Talukder A.H., Mills G., and Kumar R.. 2006. An inherent role of integrin-linked kinase-estrogen receptor alpha interaction in cell migration. Cancer Res. 66:11030–11038. 10.1158/0008-5472.CAN-06-2676 [DOI] [PubMed] [Google Scholar]
- Acerbi I., Cassereau L., Dean I., Shi Q., Au A., Park C., Chen Y.Y., Liphardt J., Hwang E.S., and Weaver V.M.. 2015. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 7:1120–1134. 10.1039/C5IB00040H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlanmerini M., Solinhac R., Abot A., Fabre A., Raymond-Letron I., Guihot A.L., Boudou F., Sautier L., Vessières E., Kim S.H., et al. 2014. Mutation of the palmitoylation site of estrogen receptor α in vivo reveals tissue-specific roles for membrane versus nuclear actions. Proc. Natl. Acad. Sci. USA. 111:E283–E290. 10.1073/pnas.1322057111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose E.J. 1956. A surface contact microscope for the study of cell movements. Nature. 178:1194 10.1038/1781194a0 [DOI] [PubMed] [Google Scholar]
- Arjonen A., Alanko J., Veltel S., and Ivaska J.. 2012. Distinct recycling of active and inactive β1 integrins. Traffic. 13:610–625. 10.1111/j.1600-0854.2012.01327.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal J.F., Lenfant F., Metivier R., Flouriot G., Henrion D., Adlanmerini M., Fontaine C., Gourdy P., Chambon P., Katzenellenbogen B., and Katzenellenbogen J.. 2017. Membrane and nuclear estrogen receptor alpha actions: From tissue specificity to medical implications. Physiol. Rev. 97:1045–1087. 10.1152/physrev.00024.2016 [DOI] [PubMed] [Google Scholar]
- Axelrod D. 1981. Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 89:141–145. 10.1083/jcb.89.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D. 2001. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2:764–774. 10.1034/j.1600-0854.2001.21104.x [DOI] [PubMed] [Google Scholar]
- Bacia K., Schwille P., and Kurzchalia T.. 2005. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc. Natl. Acad. Sci. USA. 102:3272–3277. 10.1073/pnas.0408215102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y.K., Kim A., Kim M.K., Choi J.E., Kang S.H., and Lee S.J.. 2013. Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum. Pathol. 44:2028–2037. 10.1016/j.humpath.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Barabas F.M., Masullo L.A., and Stefani F.D.. 2016. Tormenta: An open source Python-powered control software for camera based optical microscopy. Rev. Sci. Instrum. 87:126103 10.1063/1.4972392 [DOI] [PubMed] [Google Scholar]
- Bates M., Huang B., Dempsey G.T., and Zhuang X.. 2007. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 317:1749–1753. 10.1126/science.1146598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Hernandez K., Keegan S., Whelan D.R., Reid D.A., Zagelbaum J., Yin Y., Ma S., Rothenberg E., and Fenyö D.. 2017. A method for quantifying molecular interactions using stochastic modelling and super-resolution microscopy. Sci. Rep. 7:14882 10.1038/s41598-017-14922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Dell’Angelica E.C., and Springer T.A.. 2001. Immunoprecipitation. Curr. Protoc. Immunol. Chapter 8:Unit 8 3. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P.T., Vadrevu S., and Norman J.C.. 2009. Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10:843–853. 10.1038/nrm2799 [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumet A., Wright G.D., Seet S.H., Tham K.M., Gounko N.V., and Bard F.. 2015. Nuclear envelope-associated endosomes deliver surface proteins to the nucleus. Nat. Commun. 6:8218 10.1038/ncomms9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T.H., Wang S.M., Liang J.Y., Yang S.H., and Wu J.C.. 2009. The interaction of estrogen receptor alpha and caveolin-3 regulates connexin43 phosphorylation in metabolic inhibition-treated rat cardiomyocytes. Int. J. Biochem. Cell Biol. 41:2323–2333. 10.1016/j.biocel.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Ciriello G., Cerami E., Sander C., and Schultz N.. 2012. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 22:398–406. 10.1101/gr.125567.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A., Zhang H., McLellan M., Yau C., Kandoth C.; TCGA Research Network, et al. 2015. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 163:506–519. 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia A.L., and Bissell M.J.. 2012. The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist. Updat. 15:39–49. 10.1016/j.drup.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker K.E., and Segal R.A.. 2014. Neuronal signaling through endocytosis. Cold Spring Harb. Perspect. Biol. 6:a020669 10.1101/cshperspect.a020669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker K.E., Courchesne S.L., and Segal R.A.. 2008. Action in the axon: Generation and transport of signaling endosomes. Curr. Opin. Neurobiol. 18:270–275. 10.1016/j.conb.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse J.F., Lindzey J., Grandien K., Gustafsson J.A., and Korach K.S.. 1997. Tissue distribution and quantitative analysis of estrogen receptor-alpha (ERalpha) and estrogen receptor-beta (ERbeta) messenger ribonucleic acid in the wild-type and ERalpha-knockout mouse. Endocrinology. 138:4613–4621. 10.1210/endo.138.11.5496 [DOI] [PubMed] [Google Scholar]
- Cox D., Lee D.J., Dale B.M., Calafat J., and Greenberg S.. 2000. A Rab11-containing rapidly recycling compartment in macrophages that promotes phagocytosis. Proc. Natl. Acad. Sci. USA. 97:680–685. 10.1073/pnas.97.2.680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., and Brugge J.S.. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268. 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- De Franceschi N., Hamidi H., Alanko J., Sahgal P., and Ivaska J.. 2015. Integrin traffic: The update. J. Cell Sci. 128:839–852. 10.1242/jcs.161653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degasperi A., Birtwistle M.R., Volinsky N., Rauch J., Kolch W., and Kholodenko B.N.. 2014. Evaluating strategies to normalise biological replicates of Western blot data. PLoS One. 9:e87293 10.1371/journal.pone.0087293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcroix J.D., Valletta J.S., Wu C., Hunt S.J., Kowal A.S., and Mobley W.C.. 2003. NGF signaling in sensory neurons: Evidence that early endosomes carry NGF retrograde signals. Neuron. 39:69–84. 10.1016/S0896-6273(03)00397-0 [DOI] [PubMed] [Google Scholar]
- Dozynkiewicz M.A., Jamieson N.B., Macpherson I., Grindlay J., van den Berghe P.V., von Thun A., Morton J.P., Gourley C., Timpson P., Nixon C., et al. 2012. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell. 22:131–145. 10.1016/j.devcel.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K.W., Kamocka M.M., and McDonald J.H.. 2011. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300:C723–C742. 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P., Steif A., Khattra J., Ha G., Yap D., Farahani H., Gelmon K., Chia S., Mar C., Wan A., et al. 2015. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 518:422–426. 10.1038/nature13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W.W., Xiao R.Q., Peng B.L., Xu H.T., Shen H.F., Huang M.F., Shi T.T., Yi J., Zhang W.J., Wu X.N., et al. 2015. Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation. Proc. Natl. Acad. Sci. USA. 112:E3327–E3336. 10.1073/pnas.1509658112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghajar C.M., and Bissell M.J.. 2008. Extracellular matrix control of mammary gland morphogenesis and tumorigenesis: Insights from imaging. Histochem. Cell Biol. 130:1105–1118. 10.1007/s00418-008-0537-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie E.J., Ho C.L., Balaji K., Clemens D.L., Deng G., Wang Y.E., Elsaesser H.J., Tamilselvam B., Gargi A., Dixon S.D., et al. 2013. Selective inhibitor of endosomal trafficking pathways exploited by multiple toxins and viruses. Proc. Natl. Acad. Sci. USA. 110:E4904–E4912. 10.1073/pnas.1302334110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould G.W., and Lippincott-Schwartz J.. 2009. New roles for endosomes: From vesicular carriers to multi-purpose platforms. Nat. Rev. Mol. Cell Biol. 10:287–292. 10.1038/nrm2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.D., Mote P.A., Salagame U., Balleine R.L., Huschtscha L.I., and Clarke C.L.. 2009. Hormone-responsive model of primary human breast epithelium. J. Mammary Gland Biol. Neoplasia. 14:367–379. 10.1007/s10911-009-9160-6 [DOI] [PubMed] [Google Scholar]
- Grant B.D., and Donaldson J.G.. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608. 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.W., and Roman J.. 2006. Fibronectin induces cell proliferation and inhibits apoptosis in human bronchial epithelial cells: Pro-oncogenic effects mediated by PI3-kinase and NF-kappa B. Oncogene. 25:4341–4349. 10.1038/sj.onc.1209460 [DOI] [PubMed] [Google Scholar]
- Han X., Aenlle K.K., Bean L.A., Rani A., Semple-Rowland S.L., Kumar A., and Foster T.C.. 2013. Role of estrogen receptor α and β in preserving hippocampal function during aging. J. Neurosci. 33:2671–2683. 10.1523/JNEUROSCI.4937-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley R.I., and Zisserman A.. 2004. Multiple View Geometry in Computer Vision. Chapter 4. Cambridge University Press, Cambridge, UK. [Google Scholar]
- He Y., Ren Y., Wu B., Decourt B., Lee A.C., Taylor A., and Suter D.M.. 2015. Src and cortactin promote lamellipodia protrusion and filopodia formation and stability in growth cones. Mol. Biol. Cell. 26:3229–3244. 10.1091/mbc.e15-03-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven E., Hoare S., and Parker M.G.. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 387:733–736. 10.1038/42750 [DOI] [PubMed] [Google Scholar]
- Helleman J., Jansen M.P., Ruigrok-Ritstier K., van Staveren I.L., Look M.P., Meijer-van Gelder M.E., Sieuwerts A.M., Klijn J.G., Sleijfer S., Foekens J.A., and Berns E.M.. 2008. Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin. Cancer Res. 14:5555–5564. 10.1158/1078-0432.CCR-08-0555 [DOI] [PubMed] [Google Scholar]
- Hernández M.V., Sala M.G., Balsamo J., Lilien J., and Arregui C.O.. 2006. ER-bound PTP1B is targeted to newly forming cell-matrix adhesions. J. Cell Sci. 119:1233–1243. 10.1242/jcs.02846 [DOI] [PubMed] [Google Scholar]
- Hines W.C., Kuhn I., Thi K., Chu B., Stanford-Moore G., Sampayo R., Garbe J.C., Stampfer M., Borowsky A.D., and Bissell M.J.. 2016. 184AA3: A xenograft model of ER+ breast adenocarcinoma. Breast Cancer Res. Treat. 155:37–52. 10.1007/s10549-015-3649-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honerkamp-Smith A.R., Veatch S.L., and Keller S.L.. 2009. An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Biochim. Biophys. Acta. 1788:53–63. 10.1016/j.bbamem.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeselsohn R., De Angelis C., Brown M., and Schiff R.. 2017. The evolving role of the estrogen receptor mutations in endocrine therapy-resistant breast cancer. Curr. Oncol. Rep. 19:35 10.1007/s11912-017-0591-8 [DOI] [PubMed] [Google Scholar]
- Johnson J.L., He J., Ramadass M., Pestonjamasp K., Kiosses W.B., Zhang J., and Catz S.D.. 2016. Munc13-4 is a Rab11-binding protein that regulates Rab11-positive vesicle trafficking and docking at the plasma membrane. J. Biol. Chem. 291:3423–3438. 10.1074/jbc.M115.705871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisler K., Chow R.H., and Dominguez R.. 2013. Fluorescently-labeled estradiol internalization and membrane trafficking in live N-38 neuronal cells visualized with total internal reflection fluorescence microscopy. J. Steroids Horm. Sci. 10.4172/2157-7536.S12-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa P., Pesiri V., Leclercq G., Marino M., and Acconcia F.. 2012. Palmitoylation regulates 17β-estradiol-induced estrogen receptor-α degradation and transcriptional activity. Mol. Endocrinol. 26:762–774. 10.1210/me.2011-1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoha T., Gaál S., Somlai C., Czajlik A., Perczel A., and Penke B.. 2003. Membrane translocation of penetratin and its derivatives in different cell lines. J. Mol. Recognit. 16:272–279. 10.1002/jmr.637 [DOI] [PubMed] [Google Scholar]
- Levin E.R. 2009. Plasma membrane estrogen receptors. Trends Endocrinol. Metab. 20:477–482. 10.1016/j.tem.2009.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Garrity A.G., and Xu H.. 2013. Regulation of membrane trafficking by signalling on endosomal and lysosomal membranes. J. Physiol. 591:4389–4401. 10.1113/jphysiol.2013.258301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K., Moss L., Laursen L., Boulter L., and ffrench-Constant C.. 2016. Integrin signalling regulates the expansion of neuroepithelial progenitors and neurogenesis via Wnt7a and Decorin. Nat. Commun. 7:10354 10.1038/ncomms10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Takai K., Weaver V.M., and Werb Z.. 2011. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 3:a005058 10.1101/cshperspect.a005058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Weaver V.M., and Werb Z.. 2012. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 196:395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio J.P., Pryor P.R., and Bright N.A.. 2007. Lysosomes: Fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632. 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- Mak H.Y., Hoare S., Henttu P.M., and Parker M.G.. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895–3903. 10.1128/MCB.19.5.3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małecki J., Wesche J., Skjerpen C.S., Wiedłocha A., and Olsnes S.. 2004. Translocation of FGF-1 and FGF-2 across vesicular membranes occurs during G1-phase by a common mechanism. Mol. Biol. Cell. 15:801–814. 10.1091/mbc.e03-08-0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino M., Ascenzi P., and Acconcia F.. 2006. S-palmitoylation modulates estrogen receptor alpha localization and functions. Steroids. 71:298–303. 10.1016/j.steroids.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Mayor S., and Pagano R.E.. 2007. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8:603–612. 10.1038/nrm2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Rothberg K.G., and Maxfield F.R.. 1994. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 264:1948–1951. 10.1126/science.7516582 [DOI] [PubMed] [Google Scholar]
- McDonough A.A., Veiras L.C., Minas J.N., and Ralph D.L.. 2015. Considerations when quantitating protein abundance by immunoblot. Am. J. Physiol. Cell Physiol. 308:C426–C433. 10.1152/ajpcell.00400.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Layseca P., and Streuli C.H.. 2014. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 34:144–153. 10.1016/j.matbio.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Nam J.M., Onodera Y., Bissell M.J., and Park C.C.. 2010. Breast cancer cells in three-dimensional culture display an enhanced radioresponse after coordinate targeting of integrin alpha5beta1 and fibronectin. Cancer Res. 70:5238–5248. 10.1158/0008-5472.CAN-09-2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone A., De Angelis C., Trivedi M.V., Osborne C.K., and Schiff R.. 2015. The changing role of ER in endocrine resistance. Breast. 24(Suppl 2):S60–S66. 10.1016/j.breast.2015.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicovich P.R., Owen D.M., and Gaus K.. 2017. Turning single-molecule localization microscopy into a quantitative bioanalytical tool. Nat. Protoc. 12:453–460. 10.1038/nprot.2016.166 [DOI] [PubMed] [Google Scholar]
- Ovesný M., Křížek P., Borkovec J., Svindrych Z., and Hagen G.M.. 2014. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 30:2389–2390. 10.1093/bioinformatics/btu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A., Razandi M., Aitkenhead M., Hughes C.C., and Levin E.R.. 2002. Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J. Biol. Chem. 277:50768–50775. 10.1074/jbc.M210106200 [DOI] [PubMed] [Google Scholar]
- Pereira B., Chin S.F., Rueda O.M., Vollan H.K., Provenzano E., Bardwell H.A., Pugh M., Jones L., Russell R., Sammut S.J., et al. 2016. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7:11479 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras R.J., and Szego C.M.. 1984. Specific internalization of estrogen and binding to nuclear matrix in isolated uterine cells. Biochem. Biophys. Res. Commun. 123:84–91. 10.1016/0006-291X(84)90383-8 [DOI] [PubMed] [Google Scholar]
- Pontiggia O., Sampayo R., Raffo D., Motter A., Xu R., Bissell M.J., Joffé E.B., and Simian M.. 2012. The tumor microenvironment modulates tamoxifen resistance in breast cancer: A role for soluble stromal factors and fibronectin through β1 integrin. Breast Cancer Res. Treat. 133:459–471. 10.1007/s10549-011-1766-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razandi M., Oh P., Pedram A., Schnitzer J., and Levin E.R.. 2002. ERs associate with and regulate the production of caveolin: Implications for signaling and cellular actions. Mol. Endocrinol. 16:100–115. 10.1210/mend.16.1.0757 [DOI] [PubMed] [Google Scholar]
- Reid G., Hübner M.R., Métivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., and Gannon F.. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell. 11:695–707. 10.1016/S1097-2765(03)00090-X [DOI] [PubMed] [Google Scholar]
- Rust M.J., Bates M., and Zhuang X.. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–795. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur R.S., and Burris T.P.. 2004. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 63:207–212. 10.1111/j.1399-3011.2004.00126.x [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A., Wang C., Katzenellenbogen B.S., Pestell R.G., and Lisanti M.P.. 1999. Caveolin-1 potentiates estrogen receptor alpha (ERalpha) signaling. J. Biol. Chem. 274:33551–33556. 10.1074/jbc.274.47.33551 [DOI] [PubMed] [Google Scholar]
- Skliris G.P., Rowan B.G., Al-Dhaheri M., Williams C., Troup S., Begic S., Parisien M., Watson P.H., and Murphy L.C.. 2009. Immunohistochemical validation of multiple phospho-specific epitopes for estrogen receptor alpha (ERalpha) in tissue microarrays of ERalpha positive human breast carcinomas. Breast Cancer Res. Treat. 118:443–453. 10.1007/s10549-008-0267-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solowska J., Edelman J.M., Albelda S.M., and Buck C.A.. 1991. Cytoplasmic and transmembrane domains of integrin β 1 and β 3 subunits are functionally interchangeable. J. Cell Biol. 114:1079–1088. 10.1083/jcb.114.5.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q.P., Du W., Ji Q., Xue B., Jiang D., Zhu Y., Lou J., Yu L., and Sun Y.. 2016. Vesicle size regulates nanotube formation in the cell. Sci. Rep. 6:24002 10.1038/srep24002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung B.H., and Weaver A.M.. 2011. Regulation of lysosomal secretion by cortactin drives fibronectin deposition and cell motility. Bioarchitecture. 1:257–260. 10.4161/bioa.1.6.19197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Kubo K., Waguri S., Yabashi A., Shin H.W., Katoh Y., and Nakayama K.. 2012. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 125:4049–4057. 10.1242/jcs.102913 [DOI] [PubMed] [Google Scholar]
- Tiwari A., Jung J.J., Inamdar S.M., Brown C.O., Goel A., and Choudhury A.. 2011. Endothelial cell migration on fibronectin is regulated by syntaxin 6-mediated alpha5beta1 integrin recycling. J. Biol. Chem. 286:36749–36761. 10.1074/jbc.M111.260828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totta P., Pesiri V., Marino M., and Acconcia F.. 2014. Lysosomal function is involved in 17β-estradiol-induced estrogen receptor α degradation and cell proliferation. PLoS One. 9:e94880 10.1371/journal.pone.0094880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totta P., Pesiri V., Enari M., Marino M., and Acconcia F.. 2015. Clathrin heavy chain interacts with estrogen receptor α and modulates 17β-estradiol signaling. Mol. Endocrinol. 29:739–755. 10.1210/me.2014-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upla P., Marjomäki V., Kankaanpää P., Ivaska J., Hyypiä T., Van Der Goot F.G., and Heino J.. 2004. Clustering induces a lateral redistribution of alpha 2 beta 1 integrin from membrane rafts to caveolae and subsequent protein kinase C-dependent internalization. Mol. Biol. Cell. 15:625–636. 10.1091/mbc.e03-08-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Linde S., Löschberger A., Klein T., Heidbreder M., Wolter S., Heilemann M., and Sauer M.. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6:991–1009. 10.1038/nprot.2011.336 [DOI] [PubMed] [Google Scholar]
- Vanlandingham P.A., and Ceresa B.P.. 2009. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284:12110–12124. 10.1074/jbc.M809277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., and Robbins J.. 2014. Proteasomal and lysosomal protein degradation and heart disease. J. Mol. Cell. Cardiol. 71:16–24. 10.1016/j.yjmcc.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxmonsky N.C., and Conner S.D.. 2013. Αvβ3-integrin-mediated adhesion is regulated through an AAK1L- and EHD3-dependent rapid-recycling pathway. J. Cell Sci. 126:3593–3601. 10.1242/jcs.122465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao E.S., Zhang H., Chen Y.Y., Lee B., Chew K., Moore D., and Park C.. 2007. Increased beta1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 67:659–664. 10.1158/0008-5472.CAN-06-2768 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.