Kriebel et al. show that Dictyostelium cells release extracellular vesicles (EVs) that not only contain the chemoattractant cAMP but also actively synthesize and release cAMP to promote chemotaxis. They show that the EV release of cAMP is mediated by the ABCC8 transporter.
Abstract
Chemotactic signals are relayed to neighboring cells through the secretion of additional chemoattractants. We previously showed in Dictyostelium discoideum that the adenylyl cyclase A, which synthesizes the chemoattractant cyclic adenosine monophosphate (cAMP), is present in the intraluminal vesicles of multivesicular bodies (MVBs) that coalesce at the back of cells. Using ultrastructural reconstructions, we now show that ACA-containing MVBs release their contents to attract neighboring cells. We show that the released vesicles are capable of directing migration and streaming and are central to chemotactic signal relay. We demonstrate that the released vesicles not only contain cAMP but also can actively synthesize and release cAMP to promote chemotaxis. Through proteomic, pharmacological, and genetic approaches, we determined that the vesicular cAMP is released via the ABCC8 transporter. Together, our findings show that extracellular vesicles released by D. discoideum cells are functional entities that mediate signal relay during chemotaxis and streaming.
Introduction
Chemotaxis, the process in which cells migrate directionally in response to external chemical cues, is essential in many biological processes, such as immune responses, wound healing, and embryogenesis, as well as during pathological conditions, such as chronic inflammation and metastasis. Although the mechanisms underlying gradient sensing and directed migration have been studied extensively, less is known about how cells amplify chemotactic signals and coordinate their collective movement toward a source of chemoattractant. In this context, the relay of chemotactic signals between neighboring cells is superbly manifested in the social amoebae Dictyostelium discoideum. When placed under starvation conditions, D. discoideum cells enter a developmental program that allows them to chemotax toward secreted cAMP signals, stream in a head-to-tail fashion, and form aggregates that will differentiate into fruiting bodies composed of spores atop a stalk of vacuolated cells (Bagorda et al., 2006; Nichols et al., 2015). cAMP acts as a chemoattractant by specifically binding to a G protein–coupled receptor named cAMP receptor 1 (cAR1). cAMP binding leads to dissociation of the heterotrimeric G protein into Gα and Gβγ subunits and the activation of downstream effectors including the adenylyl cyclase A (ACA), which converts ATP into cAMP. Although part of the cAMP remains inside cells to activate PKA and regulate gene expression, most of the cAMP is secreted to relay chemotactic signals to neighboring cells (Kriebel and Parent, 2004).
We have shown that the enrichment of ACA at the back of polarized cells is essential for cells to align in a head-to-tail fashion and stream during chemotaxis (Kriebel et al., 2003). Indeed, cells lacking ACA or expressing an ACA mutant that is not enriched at the back of cells are unable to stream during chemotaxis. Our studies revealed that ACA is distributed in two distinct cellular pools during chemotaxis: one is restricted to the plasma membrane (PM), and the other is localized on highly dynamic intracellular vesicles that coalesce at the back of cells (Kriebel et al., 2008). Upon closer examination, we also found that actively migrating cells leave behind vesicles enriched in ACA. Ultrastructural immunogold studies revealed that the intracellular pool of ACA partly colocalizes with multivesicular bodies (MVBs), which are often enriched at the back of cells where their content is released in the form of vesicles. Based on the intraluminal localization of the gold particles and the position of the label on ACA, we proposed that the secreted vesicles contain cAMP and represent a mechanism for the sustained release of the chemoattractant during streaming (Kriebel et al., 2008). Remarkably, vesicular packaging of morphogens and chemotactic signals is an evolutionarily conserved process, as it has been reported in Drosophila to propagate Wnt gradients (Entchev and González-Gaitán, 2002) during neutrophil chemotaxis to amplify primary attractant gradients (Majumdar et al., 2016) and to facilitate cancer cell migration (Sung et al., 2015).
In the present study, we set out to establish the nature of the secreted vesicles and to identify their role during D. discoideum chemotaxis and streaming. We purified the secreted vesicles from the supernatants of chemotactic competent cells, identified their proteomic content by mass spectrometry (MS), and assessed their ability to mediate chemotaxis. We show that the vesicles contain and release cAMP through the ABC transporter ABCC8 and that, most remarkably, they have the ability to synthesize cAMP. Together, our findings provide novel insight into the mechanisms that regulate cell–cell communication during chemotaxis and identify extracellular vesicles (EVs) as an active component of the relay machinery.
Results
Migrating cells release vesicles in trails that attract neighboring cells
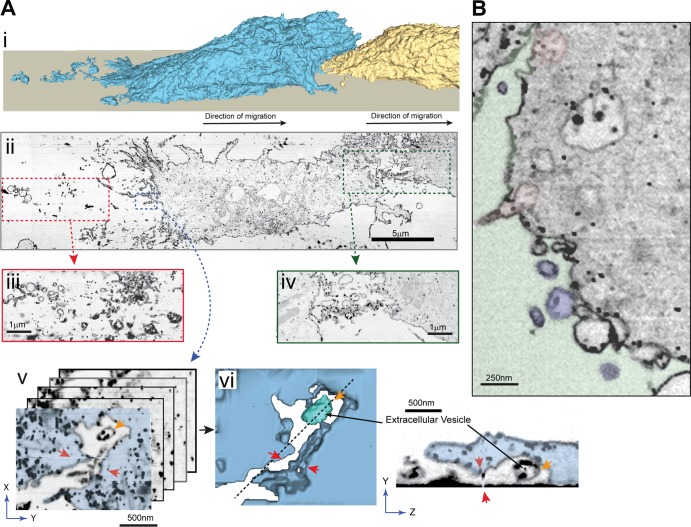
We previously showed that chemotaxing ACA-YFP/aca− cells deposit vesicular structures arising from MVBs containing ACA-YFP (Kriebel et al., 2008). To identify the nature of these ACA-YFP–containing trails and visualize their 3D ultrastructure at nanoscale resolutions, we performed focused ion beam (FIB) scanning EM (SEM) on chemotaxing ACA-YFP/aca− cells labeled with colloidal gold–conjugated antibody marking ACA-YFP. Fixed, stained, and resin-embedded cells were subjected to automated iterations of FIB milling followed by SEM imaging. The resulting image stacks were aligned and processed to generate 3D image volumes that contain entire cells at 15-nm xyz pixel size (1,000 longitudinal sections). The PMs from these volumes were then segmented out to generate a 3D reconstruction of the surface of two cells following each other toward an aggregate (Fig. 1 A [i] and Video 1). Fig. 1 A (ii) shows a single slice from the FIB-SEM image volume, extracted from a plane parallel to and 20–60 nm above the bottom of the coverslip. A trail of colloidal gold–positive EVs (Fig. 1 A [iii]) is readily observed emanating from the back of the distal cell as well as between the two cells (Fig. 1 A [iv]). Several slices from the FIB-SEM image volume of a region inside the distal cell, shown in Fig. 1 A (v) and marked by the blue box in Fig. 1 A (ii), were extracted, and cellular membranes in these slices were segmented and reconstructed in 3D (Fig. 1 A [v and vi]). A vesicle that has been released in the extracellular space is readily evident and depicted in cyan (Fig. 1 A [vi] and Video 1). The vesicle, deposited in the extracellular space, is labeled with colloidal gold on its intraluminal side, where the catalytic site of ACA is located (Kriebel et al., 2008) and cAMP is poised to accumulate. We were also able to identify ACA-YFP–containing MVBs undergoing fusion with the PM and releasing their intraluminal vesicles (Fig. 1 B and Video 2). These data reveal that the ACA-containing vesicles originate from MVBs and are released by a fusion event with the PM.
Figure 1.
D. discoideum cells release vesicular trails during cell migration. (A [i]) 3D reconstruction of the surface of two cells migrating toward an aggregation center. The reconstruction was generated by segmentation of PMs captured in an FIB-SEM image stack that spanned the cells. The leading cell is colored yellow, the following cell is colored blue, and the direction of migration is indicated by black arrows. (A [ii]) A single slice from the reconstructed FIB-SEM image volume of ACA-YFP/aca− cells, orthogonal to the scanning electron microscope imaging plane and parallel to the surface of the coverslip in Ai. Cells were stained with anti-GFP conjugated to colloidal gold. The slice shown is ∼20–60 nm from the surface of the coverslip. (A [iii and iv]) Insets depicting areas of vesicle release between cells (green dotted rectangle) and behind the following cell (red dotted rectangle). For clarity, slices at slightly different heights from the coverslip surface are shown in the inset. (A [v and vi]) Magnified view of the inset (blue) showing an MVB just after fusion with and release from the PM. (A [v]) A stack of slices extracted from the FIB-SEM image volume shown in blue dotted inset. These slices were used to segment out and reconstruct this event in 3D, depicted in A (vi). Red arrows are used as reference points marking the groove containing the released vesicle (shown as cyan, indicated by gold arrow). The cytosol is marked in blue, and the extracellular space is colorless. (Inset) An oblique image plane from the FIB-SEM image volume (indicated by the dotted line in A [vi]) showing the release of an intraluminal body from the PM. (B) A single FIB-SEM slice from Video 2 showing MVB fusion with PM. The extracellular space is overlaid in green, the fusing MVBs in pink, and released vesicles in blue.
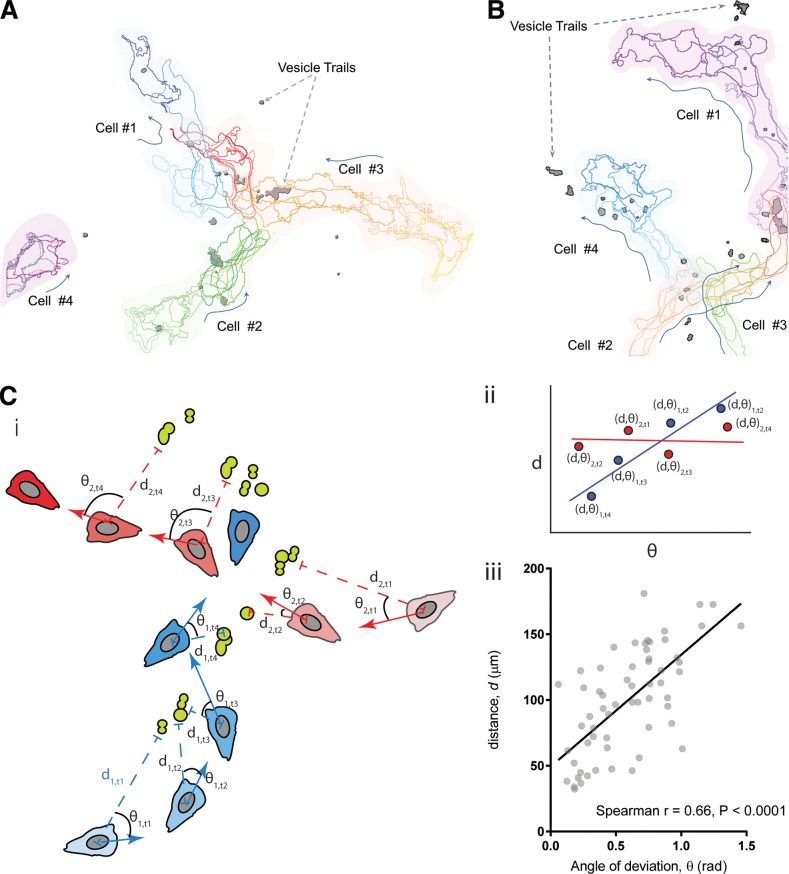
To assess whether the vesicular trails provide cues during cell migration, we tracked the movement of cells relative to the position of the trails. Fig. 2 (A and B) depicts cell tracks extracted from Video 3 (A and B) and shows instances of cells migrating toward or over ACA-YFP trails. The tracks are color coded from light to dark to show time evolution of the cell tracks, and the direction of cell movement for migrating cells for each frame is marked by arrows. In both cases, we found that cells make active directional changes toward the trails. More importantly, the cell tracks are aligned with trails and the cells tend to follow trails released by a preceding cell. For example, in Fig. 2 A and Video 3 A, we observed that trails released by cell 1 serve as a guide for cells 2 and 3. Similarly, in Fig. 2 B and Video 3 B, ACA-YFP trails left by cell 1 appear to guide cells 2 and 3. To quantify the ability of the vesicular trails to provide chemotactic cues, we performed an unbiased correlation between the distance of a cell to the trails and the angle of deviation between the directional vector and the vector to the vesicles as illustrated in Fig. 2 C (i). We measured the distance of a given cell for any given time point and the angle between the direction of the trails and its motion under the assumption that the closer a cell is to the vesicular trails, the higher its chemotactic capability, and hence, the smaller the angle of deviation. This distance would therefore be positively correlated with the angle of deviation only for cells following trails (Fig. 2 C [ii]). We find that the angle of deviation of the cells shown in Video 3 (A and B) is significantly correlated to the distance, with a positive Spearman’s coefficient of 0.66 (Fig. 2 C [iii]). Together, these findings establish that the fusion of MVBs with the PM releases ACA-containing intraluminal vesicles outside actively migrating cells and attracts neighboring cells during streaming.
Figure 2.
Vesicular trails released by migrating cells are capable of attracting other cells. (A and B) Cell outline traces resolved from maximum-intensity fluorescent images of ACA-YFP/aca− cells moving toward a spontaneously formed aggregate of cells, showing the relative position of migrating cells with respect to secreted vesicles containing ACA-YFP (dark gray). Time evolution of the cell outlines are shown from lighter to darker colors for each cell. (C [i]) Cartoon showing the principle used for the analysis of the chemoattractive potential of the released trails. The cartoon shows the different positions of two simulated cells colored in blue and red. The blue cell follows a simulated vesicular trail shown in green, and the red cell moves randomly irrespective of the trail position. Their time evolution (t1–t4) is shown in deepening shades, with darker shades showing the position of cells at later time points. di,t denotes the distance of the ith cell to its nearest vesicle at time point t, whereas θ denotes the angle between its direction of motion and the direction vector to the nearest EV. (C [ii]) A simulated d–θ correlative plot showing the behavior of cells depicted in Ci. (C [iii]) d–θ correlative plot of migrating cells derived from Video 3 (A and B) showing positive correlation between d and θ.
Chemotaxis-competent cells release EVs
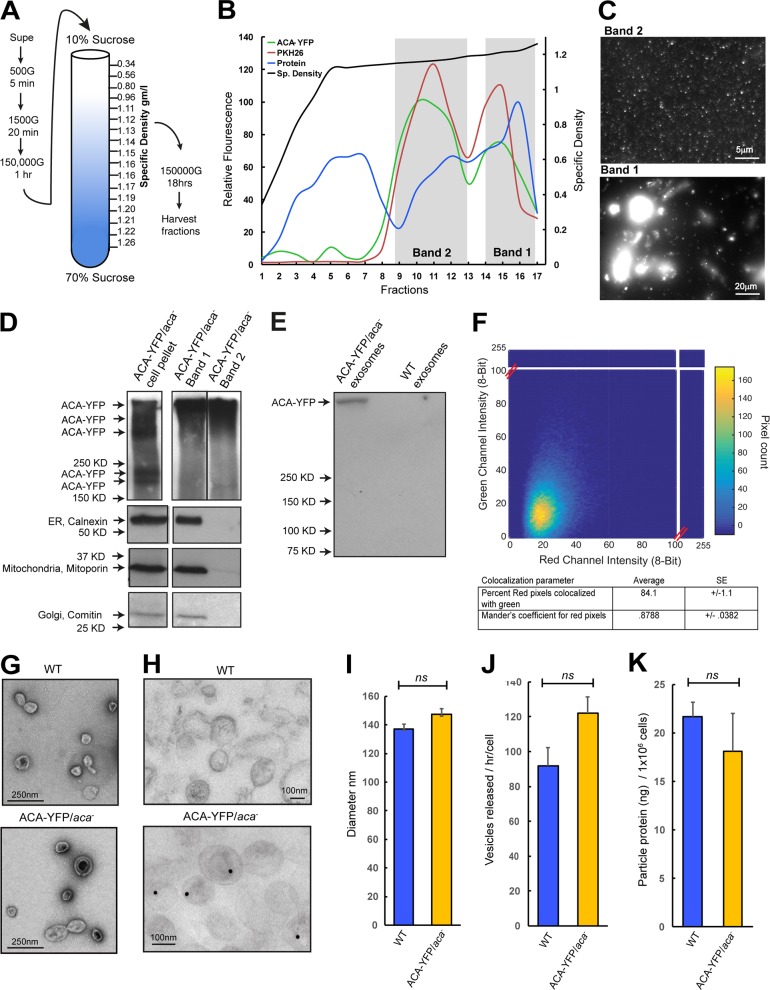
We next sought to purify the secreted vesicles from chemotaxis-competent ACA-YFP/aca− cells. Because our ultrastructural studies showed that these arise from fused MVBs, we followed procedures aimed at purifying exosomes involving differential centrifugation and fractionation on sucrose density gradients (Fig. 3 A). Protein estimation was performed on each fraction, followed by staining with the lipophilic membrane dye PKH26. As seen in Fig. 3 B, although protein was present in all fractions, lipophilic membranes were concentrated in only two major bands: a high-density band (band 1; density 1.20–1.26 g/ml), composed of fractions 14–17, and a low-density band (band 2; density 1.14–1.19 g/ml), composed of fractions 9–13. Although both bands showed strong signal for PKH26 and YFP fluorescence, only band 2 had a density range similar to that of mammalian exosomes (Raposo and Stoorvogel, 2013). Furthermore, YFP fluorescence imaging revealed that whereas band 1 contained large vesicles and membrane aggregates, band 2 contained uniform diffraction limited puncta that were smaller than 220 nm in size (Fig. 3 C). Western analyses confirmed the presence of ACA-YFP in both band 1 and 2, although mitoporin, comitin, and calnexin, which typically reside in mitochondria, Golgi, and ER, respectively, were absent from band 2, further confirming the purity of the preparation (Fig. 3 D). Interestingly, ACA-YFP from both bands 1 and 2 was detected at a higher molecular weight than those from the cell lysates. The content of the high molecular weight ACA complex was confirmed by immunoprecipitation of ACA-YFP from EV fractions and MS analysis (Fig. 3 E), suggesting an altered environment of the enzyme in the vesicles compared with whole cells. To assess the abundance of ACA-YFP vesicles relative to the total secreted population, we labeled the purified vesicle population isolated from ACA-YFP/aca− cells with FM4-64 and generated 2D fluorescence pixel intensity histograms of the labeled vesicles. We found a strong correlation between the FM4-64 and YFP fluorescence, with a high Mander’s coefficient of 0.87 ± 0.0382 (Fig. 3 F), and the percentage of YFP fluorescent pixels positive for red pixels (FM4-64) was 84.1 ± 1.1. These findings indicate that most of the isolated vesicles contain ACA-YFP. To substantiate our findings that the vesicular pool of ACA is exosomal and not composed of EVs budding directly from the PM, we fractionated ACA-YFP/aca− and cAR1-YFP/cAR1/3−/− extracts on sucrose gradients and performed immunoblot analyses for ACA-YFP and cAR1-YFP. We found that whereas ACA-YFP distributes across densities corresponding to bands 1 and 2, cAR1-YFP is specifically present in band 1. We further performed surface biotinylation experiments to mark the PM and found that biotin-labeled membrane components cofractionate with cAR1-YFP (Fig. S1 A).
Figure 3.
Isolation and characterization of D. discoideum EVs. (A) Purification scheme for the isolation of EVs from D. discoideum cell supernatants. See Supplemental methods for details. (B) Graph of the density gradient fractions of isolates from the supernatants of ACA-YFP/aca− cells showing protein concentration (blue), relative lipid levels stained with the lipid dye PKH26 (red), ACA-YFP levels measured by YFP fluorescence (green), and specific density of the gradient (black). Areas marked in gray demarcate bands of high lipid and protein concentration that were characterized in C and D. (C) Diffraction-limited fluorescence images of the vesicular contents of the two bands isolated from a 10–70% sucrose gradient. (D) Western blots of ACA-YFP, calnexin, mitoporin, and comatin highlighting purity of the preparations. The ACA-YFP samples were run on the same high-percentage gel, but not side by side. Results representative of two independent experiments. (E) Immunoprecipitation of ACA-YFP from EV isolates using a GFP specific antibody. Results representative of two independent experiments. (F) 2D correlative histogram of total and ACA-YFP fluorescence of EVs isolated from ACA-YFP/aca− cells. Total EVs were detected by FM4-64 staining. Inset table shows the percentage of red pixels also positive for YFP, calculated from a correlative matrix of individual deconvolved pixels. (G and H) Electron micrographs of whole EVs isolated from ACA-YFP/aca− or WT cells stained with uranyl acetate (G) or colloidal gold for YFP (H). (I–K) Graphs depicting the mean diameter (I), number (J), or protein concentration (K) of EVs isolated from 4.5 × 109 ACA-YFP/aca− or WT cells. All statistics are shown as mean ± SEM of three independent experiments, unless mentioned otherwise. ns, P > 0.05.
Ultrastructural analysis of band 2 isolated from either WT or ACA-YFP/aca− cells revealed that it contains a population of small vesicles with a characteristic cup shape and mean diameter of 102 ± 3.9 or 121.4 ± 4.6 nm, respectively (Fig. 3 G), which is consistent with the size of mammalian exosomes (Raposo and Stoorvogel, 2013). Anti-GFP immunogold labeling of the vesicles also revealed specific labeling in the vesicles derived from the ACA-YFP/aca− cells (Fig. 3 H). To get a more accurate measurement of the diameter of the vesicles and to exclude artifacts created by EM sample preparation, we measured the size of the vesicles using nanoparticle tracking analysis (NanoSight). As depicted in Fig. 3 I, the vesicles isolated from either WT or ACA-YFP/aca− cells were highly homogeneous, with similar mean diameters. Furthermore, we found that both preparations had similar particle counts and total protein content (Fig. 3, J and K), showing that the overexpression of ACA-YFP did not alter the general characteristics of the released vesicles. Although ultrastructural studies as well as the aforementioned characterization of the purified vesicles from band 2 strongly point to the exosomal nature of ACA-YFP vesicles, we cannot discount the presence of nonexosomal EVs in our preparation. Hence, we will refer to our purified preparation as EVs rather than exosomes.
We previously established that nocodazole (Noco) treatment inhibits the enrichment of ACA at the back of cells, as well as streaming, and dramatically reduces the numbers of ACA-positive trails (Kriebel et al., 2008). We have now expanded these findings and measured the recruitment range of Noco-treated cells exposed to a cAMP-containing micropipette. We found that disassembly of the microtubule network inhibits streaming and dramatically reduces the recruitment range (Fig. S1 B; see Video 4 for a control with no cAMP in the micropipette). We also established that Noco-treated cells release 50% fewer particles than DMSO-treated control cells (Fig. S1 C). These effects were not secondary to inhibition of cAMP production, as Noco treatment did not alter the ability of cells to activate ACA through cAR1 (Fig. S1 D). Collectively, these findings show that chemotaxis-competent D. discoideum cells secrete homogeneous exosome-like vesicles that contain ACA and are required for the relay of chemotactic signals during chemotaxis.
Proteomic analysis of purified EVs reveals the presence of cargo involved in vesicle biogenesis and signaling
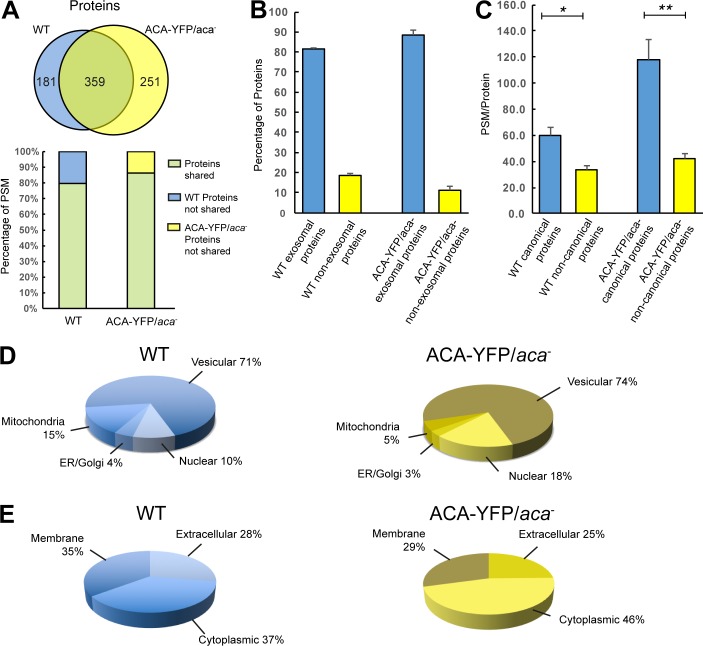
To determine the composition of the secreted vesicles, we analyzed the proteome content of EVs (band 2) isolated from chemotaxis-competent WT or ACA-YFP/aca− cells. Each preparation was analyzed in biological triplicate, and only proteins represented by two or more unique peptides and identified in two or more runs were counted as positive. Using these criteria, we identified a total of 57,271 peptide spectral matches (PSMs) across the three replicates arising from 540 proteins in samples isolated from WT cells, giving an overall resolution of ∼106 peptides/protein. For EVs derived from ACA-YFP/aca− cells, we identified 87,551 PSM across the three replicates arising from 610 proteins, giving an overall resolution of ∼144 peptides/protein. Comparing these findings, we found that ∼60% of the identified proteins were present in both samples, representing 80–85% of peptide identifications, indicating that the overexpression of ACA-YFP did not dramatically change the protein makeup of the secreted vesicles (Fig. 4 A). A comprehensive listing of all proteins identified in the vesicles derived from WT or ACA-YFP/aca− cells is presented in Table S1.
Figure 4.
MS analysis of D. discoideum EVs. (A) Venn diagram (upper image) depicting the number of unique and shared proteins between EVs derived from either ACA-YFP/aca− or WT cells. Split bar graph (lower image) showing the percentage of peptide hits on shared proteins and unshared proteins in EVs derived from either ACA-YFP/aca− or WT cells. (B) Percentage of exosomal and nonexosomal proteins in EVs derived from either ACA-YFP/aca− or WT cells. Results presented as mean ± SEM of three independent experiments. (C) PSM/protein of canonical and noncanonical proteins in EVs derived from either ACA-YFP/aca− or WT cells. Results presented as mean ± SEM of three independent experiments. (D and E) Pie charts depicting the mean percentages of proteins from EVs derived from either WT or ACA-YFP/aca− cells annotated in dictyBase as vesicular, nuclear, mitochondrial, or from the ER/Golgi network or as cytoplasmic, membranous, or extracellular (n = 3).
We next determined the prevalence of exosomal proteins in the samples. Using the EV database ExoCarta (Keerthikumar et al., 2016), all the proteins identified by MS were categorized as either exosomal or nonexosomal. This analysis revealed that >81% of the proteins in either sample are exosomal (Fig. 4 B). Furthermore, canonical exosomal proteins were robustly represented in the MS, exhibiting the largest peptide-to-protein ratios (Fig. 4 C). Additional evidence of a vesicular origin for the purified vesicular preparations was provided by an analysis using dictyBase annotations for organelle subtype (Fey et al., 2013). This analysis showed that >70% of the proteins from either sample are of vesicular origin (Fig. 4 D). We also classified the detected proteins PSMs as membrane bound, cytoplasmic, or extracellular by cross-referencing the protein annotations on dictyBase (Bakthavatsalam and Gomer, 2010; Fig. 4 E). Interestingly, 63–70% of the proteins that were identified as extracellular had no annotated secretory signal peptide. As exosomes and other EVs have been implicated as a mechanism for the unconventional secretion of proteins (Nickel and Rabouille, 2009), the absence of secretory signal peptide sequence in these proteins would suggest that vesicular release represents their primary mode of secretion.
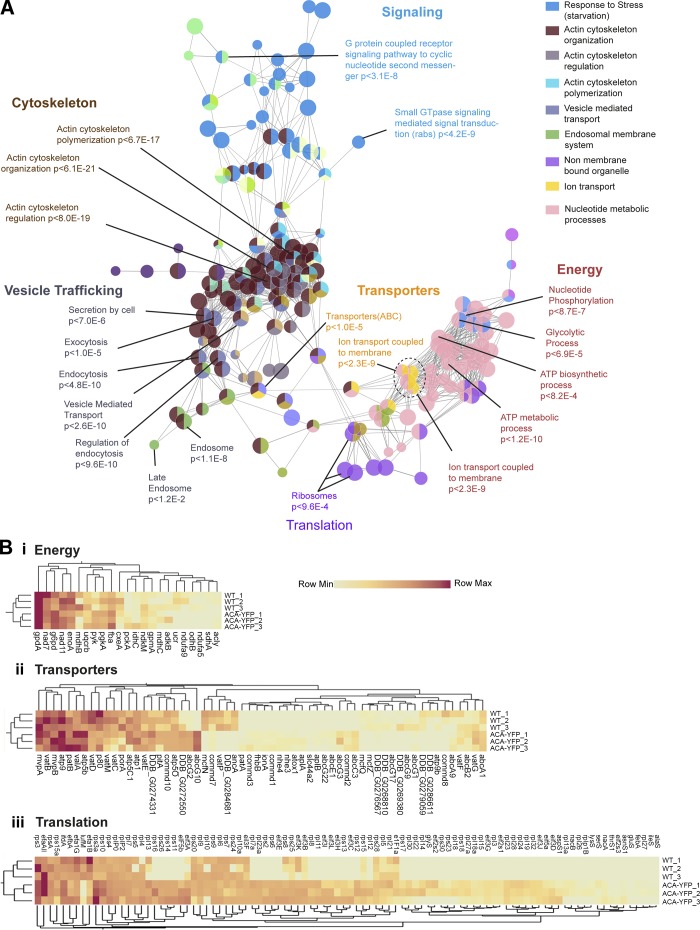
A network-based analysis of gene ontology (GO) terms was performed on all the proteins identified in the MS analysis for WT and ACA-YFP/aca− EVs to understand the physiological consequence of their cargo. As shown in Fig. 5 A, GO network analysis revealed significant enrichment in biological processes related to the cytoskeleton, vesicular trafficking, signaling, energy metabolism, and translation. The overrepresentation of actin-based cytoskeletal proteins was expected, as they have been implicated for the mobilization of secretory vesicles for membrane docking and fusion (Akers et al., 2013). The enrichment of vesicle trafficking machinery was also not surprising, given their involvement in exosome biogenesis and cargo trafficking. For example, vacuolar sorting proteins such as VPS28, VPS2B, and VPS60, involved in exosome biogenesis, and Rab proteins, involved in vesicle trafficking (Piper and Katzmann, 2007), occurred in multiple nodes of the network. GO term functional analysis also identified many proteins involved in the relay of chemotactic signals. The major categories of signaling molecules included multiple Ras proteins, guanine nucleotide exchange factors, and GTPase-activating proteins as well as most heterotrimeric G proteins and elements from the MAP kinase cascade.
Figure 5.
Network analysis and statistical appraisal of EV proteins. (A) Network analysis of GO terms identified from MS of WT EVs. Nodes corresponding to functional groups with high statistical significance and identified in MS analysis are labeled, and their enrichment probabilities relative to the D. discoideum genome are recorded in the label. The legend on the right gives the GO term category for each node color. (B [i–iii]) Heat map and cluster analysis of relative PSMs of proteins involved in ATP generation (B [i]), transport (B [ii]), and translation (B [iii]) derived from three separate MS analyses for EVs from ACA-YFP/aca− and WT cells.
Estimation of relative protein abundance calculated by semiquantitative label-free spectral counting provided additional insight into the similarities or differences in the proteome of ACA-YFP/aca− and WT EVs. No significant enrichment was found for proteins involved in cytoskeletal organization, vesicle trafficking, and signaling between ACA-YFP/aca− cells and WT cells (Fig. S2 A–C). Similarly, transporter and glycolytic proteins such as glycerol-3-phosphate dehydrogenase, glycerol-6-phosphate dehydrogenase, enolase1, and other proteins involved in energy metabolism were equally well represented in the proteome analysis of both ACA-YFP/aca− and WT EVs (Fig. 5 B [i and ii]). Interestingly, proteins involved in translation, RNA processing, and ribosome biogenesis that were overrepresented in the GO analysis and formed significant nodes in the proteomic network (Fig. 5 A) were enriched in EVs derived from ACA-YFP/aca− cells compared with WT cells (Fig. 5 B [iii]).
D. discoideum EVs contain, release, and synthesize cAMP
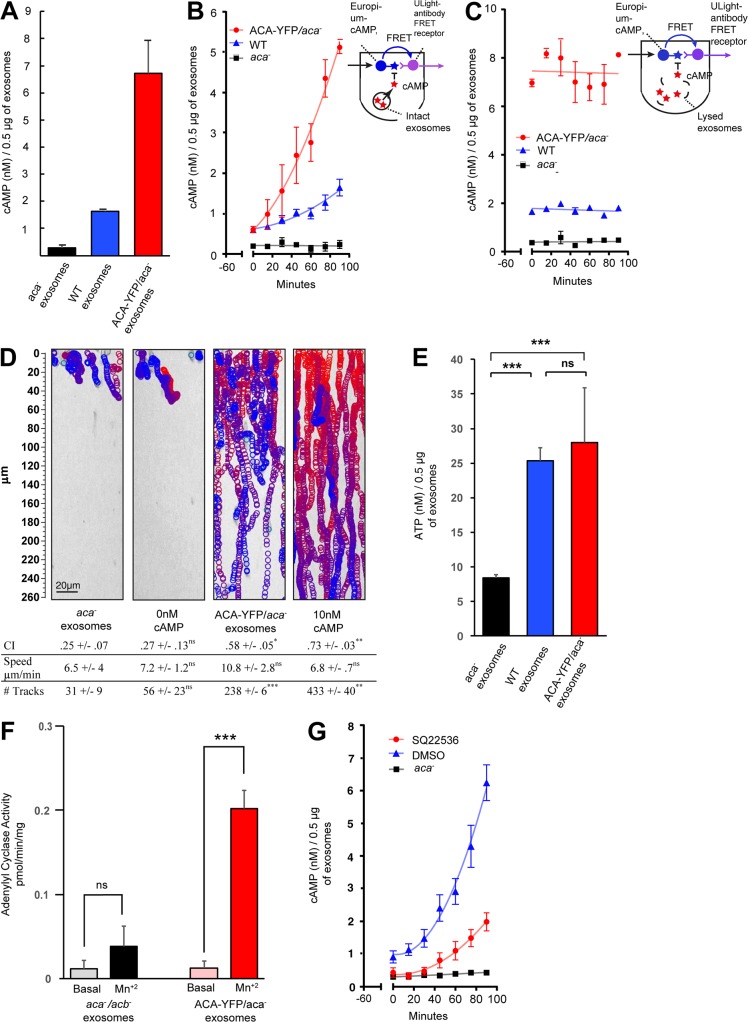
We next assessed if EVs contain cAMP. To do so, we used a competitive cAMP assay using time-resolved (TR) fluorescence resonance energy transfer (FRET), which has higher sensitivity and signal-to-noise ratio than conventional fluorescence-based assays. As shown in Fig. 6 A, lysed EVs isolated from either WT or ACA-YFP/aca− cells contained higher cAMP levels than those isolated from aca− cells. To determine whether cAMP could be secreted from EVs, we performed a cAMP TR-FRET time course in intact vesicles (see cartoon in Fig. 6 B). In this case, intact WT, ACA-YFP/aca−, or aca− EVs were incubated with assay reagents, and the loss of FRET was measured over time. We found that both WT and ACA-YFP/aca− EVs showed a continued increase in extravesicular accumulation of cAMP (Fig. 6 B). To ascertain whether this increase in cAMP was caused by active release from EVs and not other extraneous factors, we monitored cAMP levels from lysed EVs over similar incubation periods. In this case, no increase of cAMP over time was observed for EVs from all three cell types, although the total cAMP levels remained higher for ACA-YFP/aca− and WT EVs (Fig. 6 C). Finally, we determined if the released exosomal cAMP can attract cells. Using the EZ-TAXIScan microfluidic device, we found that aca− cells were able to specifically migrate toward ACA-YFP/aca− EVs, with chemotactic indices (CIs) comparable to those measured in the presence of cAMP alone (Fig. 6 D and Video 5). As expected, EVs from aca− cells recruited much fewer cells, with very low CI values typical for randomly migrating cells exposed to no chemoattractant (0 cAMP).
Figure 6.
EVs contain, release, and synthesize cAMP. (A) Bar graph of cAMP TR-FRET data showing cAMP levels from lysed EVs derived from either ACA-YFP/aca−, WT, or aca− cells. 0.5 µg of purified preparation obtained from 16.8 × 108 vesicles was used in the assay. Results presented as mean ± SEM of three independent experiments. (B and C) Time-dependent release of cAMP from intact (B) and lysed (C) EVs derived from ACA-YFP/aca−, WT, or aca− cells. cAMP measurements were made every 15 min for 90 min after a 60-min equilibration incubation. Insets: Illustration depicting the principles of the cAMP TR-FRET assay. Results presented as mean ± SEM of three independent experiments. (D) Tracks of aca− cells migrating toward buffer alone, 10 nM cAMP, ACA-YFP/aca−, or aca− EVs in the EZ-TAXIScan microfluidic chamber. The images show tracks of individual cells migrating as circles (from red to blue with increasing time) overlaid on the final image. The table indicates the chemotaxis index (CI), speed, and number of tracks of cells that responded to the chemoattractant. Results presented as mean ± SD of three independent experiments. (E) Bar graph showing ATP levels from lysed EV preparations derived from ACA-YFP/aca−, WT, or aca− cells. Results presented as mean ± SEM of three independent experiments. (F) Bar graph showing adenylyl cyclase activity levels in lysed EV preparations derived from aca−/acb− or ACA-YFP/aca− cells 10 min after MnSO4 addition. Results presented as mean ± SEM of three independent experiments. (G) Time-dependent release of cAMP from intact EVs isolated from ACA-YFP/aca− cells in the presence or absence of the adenylyl cyclase inhibitor SQ25536 (60 µM). Results presented as mean ± SEM of three independent experiments. All statistics are shown as mean ± SEM, unless mentioned otherwise. ***, **, *, and ns indicate P < 0.0002, P < 0.002, P < 0.03, and P > 0.12, respectively. Significance testing was performed using unpaired t test corrected for multiple comparison (wherever applicable) using Bonferroni–Dunn method.
The fact that extravesicular cAMP levels continue to increase above initial levels in both WT and ACA-YFP/aca− EV preparations (Fig. 6 B) suggested that the ACA packaged in the EVs is catalytically active. To assess if there is enough substrate to drive adenylyl cyclase activity, we first measured ATP content in our EV preparations. We found that EVs derived from all three cell lines contained higher ATP levels than cAMP levels, with EVs derived from aca− cells having the lowest amount (Fig. 6 E). We next directly measured adenylyl cyclase activity in lysed preparations in the presence of MnSO4, which provides a measure of the intrinsic catalytic activity of the enzyme (Hurley, 1999). We found that MnSO4 induced an increase in adenylyl cyclase activity in ACA-YFP/aca− EVs compared with those isolated from cells lacking ACA and ACB (another adenylyl cyclase expressed during D. discoideum aggregation; Kim et al., 1998; Fig. 6 F). Furthermore, pretreatment of vesicles with SQ22536, a noncompetitive inhibitor of adenylyl cyclase, dramatically reduced the release of cAMP from ACA-YFP/aca− EVs over time (Fig. 6 G). Together, these findings establish that EVs have the ability to both synthesize and secrete cAMP and that the released cAMP can promote chemotactic activity.
ABCC transporters mediate the release of cAMP from EVs during streaming
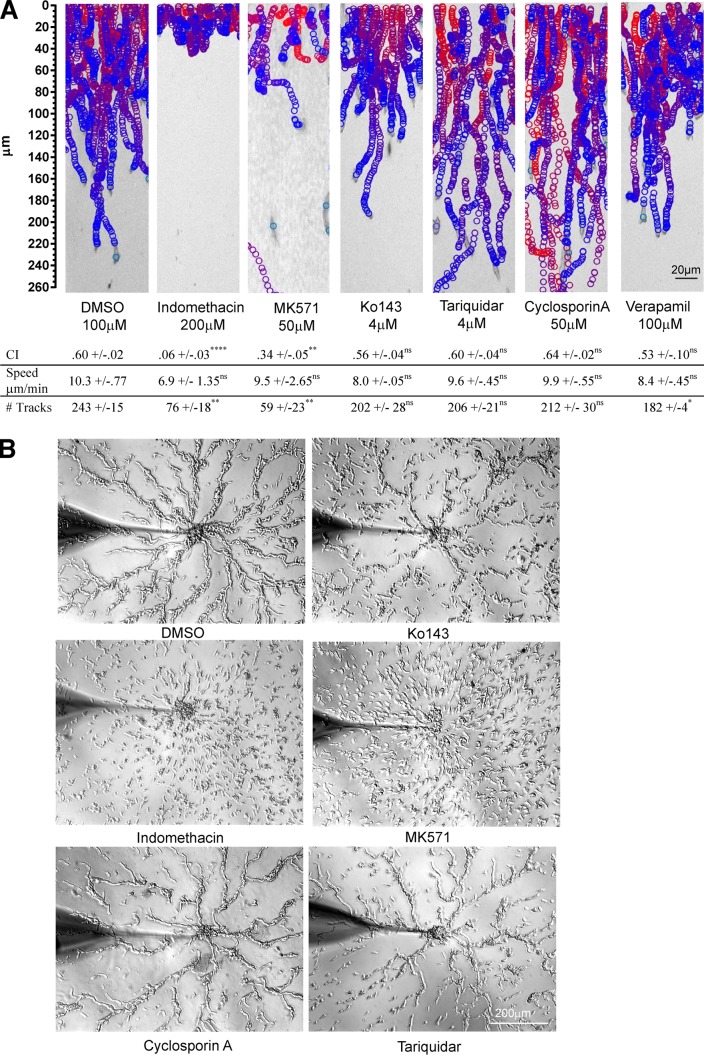
We next set out to determine the mechanism by which cAMP is secreted from EVs. Of the 68 ABC transporters annotated in the D. discoideum genome, MS data identified 13 ABC transporters in D. discoideum EVs (Fig. 5 and Table S2). To assess their role in cAMP release, we treated EVs derived from ACA-YFP/aca− cells with ABC transporter inhibitors that act on different classes of mammalian transporters (Table S3) and assessed their ability to promote the migration of aca− cells using the EZ-TAXIScan microfluidic device. We found that the ABCC transporter inhibitors indomethacin and MK571 specifically inhibited EV-mediated chemotaxis, with dramatically reduced CI values as well as a reduction in cell recruitment (Fig. 7 A and Video 6). Conversely, the ABCG and ABCB inhibitors Ko143, cyclosporine A, and tariquidar had no visible effect on vesicle-mediated chemotaxis and CI values; nor did verapamil, a broad-spectrum inhibitor that inhibits both ABCB and ABCG classes.
Figure 7.
ABCC transporters mediate cAMP release during streaming. (A) Tracks of aca− cells migrating in an EZ-Taxiscan microfluidic device to EVs isolated from ACA-YFP/aca− cells (bottom chamber) treated with DMSO (control), indomethacin, MK571, Ko143, cyclosporine A, or tariquidar. See Table S3 and Fig. 6 D for details. Results presented as mean ± SD of three independent experiments. ****,**, *, and ns indicate P < 0.0001, P < 0.002, P < 0.03, and P > 0.12, respectively. Statistical analysis was performed as described in the legend of Fig. 6. (B) Bright-field images of micropipette streaming assay showing the response of WT cells treated with DMSO, indomethacin, MK571, Ko143, cyclosporine A, and tariquidar to a micropipette filled with 1 µM cAMP. Drugs were added 1 h before the start of the assay and were present during entire assay. Images were taken 20 min after adding the micropipette to the cells. Images representative of three independent experiments.
We next determined whether the inhibitors alter the ability of the cells to stream during chemotaxis. We reasoned that inhibition of cAMP release from EVs will inhibit streaming but not cell migration in the presence of an external cAMP gradient. We pretreated chemotaxis-competent WT cells with ABC transporter inhibitors and assessed streaming using a micropipette assay as previously described (Kriebel et al., 2003). We found that none of the ABC inhibitors tested altered the ability of the cells to chemotax toward the micropipette filled with cAMP. Indeed, even in the presence of the ABCC inhibitors indomethacin and MK-571, cells were able to polarize and move toward the micropipette. However, only ABCC inhibitors gave rise to dramatic streaming defects (Fig. 7 B and Video 7). These data, together with the fact that ABCC class transporters have been shown to transport cAMP in mammalian cells (Jedlitschky et al., 2000; Guo et al., 2003), strongly suggest that ABCC transporters are key meditators of exosomal cAMP secretion and streaming.
EVs isolated from abcC8− cells are defective in cAMP release and have streaming defects
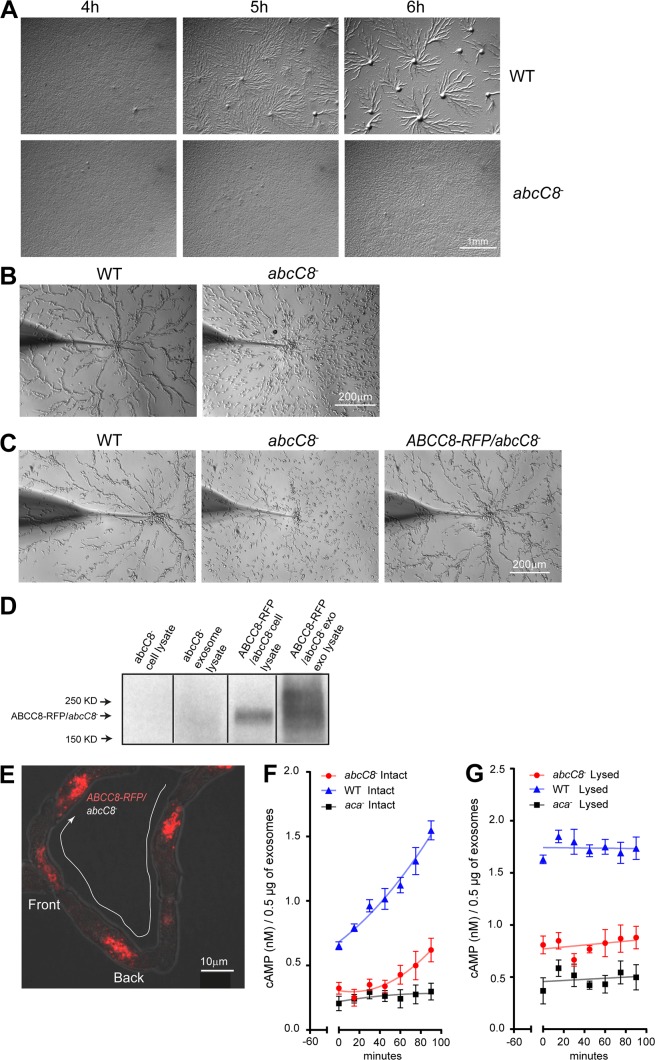
We generated knockouts (KOs) of four ABC transporters identified in our MS analysis (ABCC3, ABCC8, ABCG2, and ABCG10) using gene targeting vectors generated by Miranda et al. (2013) (Fig. S3). We screened two clones of each KO using a micropipette assay and an agar development assay, in which cells are allowed to spontaneously aggregate and differentiate. Although the abcC3−, abcG2−, and abcG10− cells streamed normally using both assays (Fig. S4 and Video 8), the abcC8− cells exhibited dramatic streaming defects in agar development (Fig. 8 A) and micropipette (Fig. 8 B and Video 8) assays. This defect was not a consequence of a general defect in development, as abcC8− cells expressed cAR1 and ACA in a timely manner (Fig. S5, A and B). The chemotactic defect observed in abcC8− cells was rescued by ectopically expressing RFP-tagged ABCC8 (Fig. 8 C). Furthermore, we found that ABCC8-RFP was preferentially enriched in EVs compared with intact cells (Fig. 8 D), strengthening its role in mediating extracellular cAMP release. In streaming cells, ABCC8-RFP was excluded from the PM and showed a punctate distribution that localized at the trailing edge of the migrating cell (Fig. 8 E), similar to ACA.
Figure 8.
abcC8− cells have streaming defects and have impaired cAMP release from EVs. (A) Bright-field mages of WT and abcC8− cells plated on DB agar taken 4, 5, and 6 h after the initiation of starvation. Images representative of three independent experiments. (B) Bright-field images of micropipette streaming assay showing the response of WT and abcC8− cells to a micropipette filled with 1 µM cAMP. Images were taken 20 min after the micropipette was added. Images representative of three independent experiments. (C) Bright-field images of micropipette streaming assay showing the response of WT, abcC8−, and ABCC8-RFP/abcC8− cells to a micropipette filled with 1 µM cAMP. Images were taken 20 min after the micropipette was added. Images representative of three independent experiments. (D) Western blots of ABCC8-RFP expression in various preparations, as depicted. Results representative of three independent experiments. (E) Representative confocal fluorescent maximum-intensity projection image overlaid on bright-field image of differentiated ABCC8-RFP/abcc8− cells streaming toward an aggregate. (F and G) Time-dependent release of cAMP from intact (F) or lysed (G) EVs isolated from WT and abcC8− cells. Results presented as mean ± SEM of three independent experiments.
We next measured cAMP release using the TR-FRET assay on intact WT and abcC8− EVs. As shown in Fig. 8 F, EVs derived from WT cells showed an increased accumulation of extravesicular cAMP compared with EVs derived from abcC8− cells, although lysed abcC8− and WT EVs both showed total cAMP content higher than that of aca− EVs (Fig. 8 G). We nevertheless observed a small amount of cAMP release from abcC8− EVs (Fig. 8 F), which could be attributed to the presence of another redundant cAMP transporter in the EVs. Indeed, we observed that abcC8− cells can partly recover the ability to stream later in development (Fig. S5 C). As many ABCC transporters are expressed in chemotaxis-competent cells and throughout development (Miranda et al., 2013; Rosengarten et al., 2015), the observed streaming recovery could be caused by the later expression of a different ABCC transporter. To assess this, we treated WT and abcC8− cells with ABC transporter inhibitors and followed their development. We found that the ABCC inhibitors indomethacin and MK571 specifically and completely inhibited streaming of the abcC8− cells (Fig. S5 E). Both ABCC inhibitors also dramatically inhibited streaming in WT cells, although some streaming was observed at 8 h (Fig. S5 D). Together, these findings establish that ABCC transporters are the main transporters involved in exosomal cAMP transport during streaming.
Discussion
We previously reported that ACA-YFP is enriched at the trailing edge of chemotaxing D. discoideum cells and is deposited as trails of membranous vesicles during aggregation and streaming (Kriebel et al., 2008). The present study aimed at assessing the nature of the released vesicles. Based on the criteria put forward by the International Society of Extracellular Vesicles (Lötvall et al., 2014), including size; density; endosomal origin; and absence of ER, Golgi, and PM markers, as well as our current data regarding Noco inhibition of vesicle release and EM images, we show that the vesicles in the deposited ACA-YFP trails are derived from MVBs and are capable of synthesizing and releasing cAMP, thereby providing chemotactic cues to cells following in a stream. Because of the lack of D.discoideum–specific antibodies against canonical exosomal markers and the possible presence of vesicles budding from the PM in our preparations, we refer to the purified vesicles as EVs as opposed to exosomes, although we show that the majority of the purified vesicles are exosomal in nature. Exosomes and EVs have been shown to be important agents of cell–cell communication and to induce cell migration, but primarily through the transfer of miRNA (Simons and Raposo, 2009). However, recent evidence from neutrophils (Majumdar et al., 2016) and cancer cells (Chen et al., 2011; Sung et al., 2015; Sung and Weaver, 2017) suggests that cargo present in exosomes can directly control cell migration without the requisite internalization, as we observe in D.discoideum.
Functional analysis of the EV proteome showed nodes with functional pathways also identified in mammalian exosomes. For example, proteomic analysis of exosomes of seven different melanoma cell lines showed proteins that belonged to functional groups such as vesicle formation, intracellular signaling cascade, translation, and cytoskeletal organization, very similar to our own observations with D. discoideum exosomes (Lazar et al., 2015). Moreover, the relative ratios of each functional category were maintained when we compared EVs derived from either WT or ACA-YFP/aca− cells, suggesting that these functions are critical for D. discoideum EVs. Interestingly, heat map analysis revealed that translational proteins were the only functional group that was enriched in vesicles derived from cells overexpressing ACA, compared with WT cells. Although ribosomal RNAs have been reported as exosomal components (Huang et al., 2013; Li et al., 2014), why specific ribosomal components are enriched in EVs derived from ACA-YFP/aca− cells remains to be determined. It is possible that increases in cAMP signaling in ACA-YFP/aca− cells activates target of rapamycin complex 2 (TORC2) activity in a Ras- and RAP1-dependent manner (Bolourani et al., 2008; Khanna et al., 2016), which in turn increases the biogenesis of ribosomal components (Zinzalla et al., 2011). This possibility seems plausible, as other proteins that are directly regulated by small Ras GTPases, such as ERK2 and other associated MAPK members (Knetsch et al., 1996), were also enriched in EVs derived from ACA-YFP/aca− cells.
The presence of glycolytic enzymes in the released vesicles is not surprising and has been reported in studies of mammalian exosomes (Fonseca et al., 2016). However, whether they are passively sequestered from the cytosol during MVB biogenesis or actively recruited in their functional forms for ATP metabolism remains to be determined. ATP-dependent transporters formed central nodes in the proteomic pathway with a high degree of statistical significance. Furthermore, both V-type and F-type transporters involved in ATP synthesis were equally highly represented in EVs derived from ACA-YFP/aca− and WT cells. These transporters are known to maintain pH in the vacuolar system, which is critical for exosome biogenesis (Liégeois et al., 2006). More importantly, ion-coupled ATP synthases use proton gradients for the generation of ATP, the substrate for cAMP production. Although it is probable that ATP is being sequestered from the cytosol during MVB and exosome biosynthesis, it is also possible that ATP can be produced in the vesicles. The presence of F-type ATPases and ion-coupled ATP synthases detected in our MS analysis would present a renewable source of ATP within the EVs. ATP production through glycolytic enzymes has previously been shown for prostate-derived human exosomes (Ronquist et al., 2013). Furthermore, aerobic metabolism and regenerative ATP synthesis has been previously reported in mesenchymal stem cell–derived exosomes (Arslan et al., 2013; Panfoli et al., 2016), and F0/F1 ATPases have been shown to mediate redox functions in urinary exosomes (Bruschi et al., 2015). The possibility of ATP regeneration in D. discoideum EVs is also indicated by the fact that we did not observe saturation of cAMP release in our kinetic assay, which suggests that the system does not suffer from substrate limitation. Moreover, our estimation of ATP within the released vesicles revealed a concentration of ∼50 µM/mg of vesicles, enough to sustain cAMP release for >20 h, further corroborating independence of the system from substrate limitation.
We show that cAMP is stored and synthesized in EVs. Based on the estimated diameter of 120 nm, we estimate the steady-state cAMP concentration levels would be ∼4 µM. Using TR-FRET to assess cAMP released over time, we measured progressive increases in the released exosomal cAMP levels, indicating that the ACA packaged in EVs is catalytically active. Indeed, using an in vitro adenylyl cyclase assay, we showed that exosomal ACA can be activated by MnSO4, which has been shown to form hydration chelates at the active site of adenylyl cyclases and to stabilize active enzyme conformation (Hurley, 1999). Furthermore, the fact that active cAMP synthesis could be blocked by a noncompetitive inhibitor of adenylyl cyclase provides further evidence that exosomal ACA is catalytically active. The mechanism by which the catalytic activity of ACA is regulated in EVs remains to be determined. The chemoattractant-mediated activation of ACA is a G protein–mediated event that requires input from phosphatidyl inositol 3 kinase (PI3K)/cytosolic regulator of ACA (CRAC) and TORC2 (Garcia and Parent, 2008). Although our proteomic analysis showed the presence of Gβγ, RasG/C, Rap1, and PKBR1, we did not find hits for CRAC or any of the TORC2 components. Although the absence of peptides hits does not mean that CRAC or TORC2 components are not present in EVs, we envision that exosomal ACA is activated by an alternate mechanism. We found that ACA is present in a high–molecular weight complex in EVs, perhaps suggesting that the exosomal environment is distinct and gives rise to a conformation of ACA that requires different regulatory inputs for full activation. We envision that the PM and MVB/exosomal pools of ACA are regulated by distinct mechanisms and have separate roles. Indeed, using a cAMP FRET sensor, we previously proposed that although the PM-bound ACA gives rise to intracellular cAMP that is poised to activate PKA and is subject to degradation by phosphodiesterases, the MVB pool produces cAMP that is destined for secretion and signal relay (Bagorda et al., 2009). In the present study, we add more support to the presence of two functionally distinct ACA pools with the observation that Noco treatment does not inhibit adenylyl cyclase activity of the PM-bound ACA but does inhibit streaming by the reduction of vesicle release, indicating the requirement of cAMP released from the EVs for signal relay. Additionally, ABCC8 is localized to organelles inside the cell and not on the PM. It is therefore unlikely that cAMP derived from ACA on the PM is directly released into the extracellular space.
We show that exosomal cAMP is actively transported by ABCC transporters. It has been proposed that ACA, which is topologically related to ABC transporters, could act as a cAMP transporter. However, the fact that aca− cells expressing the structurally unrelated and constitutively active ACG are capable of releasing cAMP showed that an ACA-independent mechanism was involved (Pitt et al., 1993; Wang and Kuspa, 1997). Miranda et al. (2015) recently reported that the ABC transporter, ABCB3, acts as a cAMP transporter in D. discoideum. However, ABCB3 was not identified in our proteomic analysis of EVs, indicating that it may affect cAMP release at a different developmental stage or indirectly. Moreover, ABCB inhibitors (cyclosporine A, verapamil, and tariquidar) did not inhibit the release of cAMP from EVs, nor did they inhibit streaming during chemotaxis. Last, ABCB transporters have not been shown to transport cyclic nucleotides (Cheepala et al., 2013). In fact, several studies have reported that ABCC transporters (ABCC4, ABCC5, and ABCC8) are involved in cAMP transport (Jedlitschky et al., 2000; Guo et al., 2003; Cheepala et al., 2013), and our MS analysis identified ABCC3 and ABCC8 in D. discoideum EVs. Using pharmacological and genetic approaches, we now show that ABCC8 regulates cAMP transport in D. discoideum EVs, as inhibition of ABCC transporters or deletion of abcC8 leads to streaming defects, and most importantly, cAMP release is dramatically reduced in EVs derived from abcc8− cells. However, we did observe that abcC8− cells can partly regain the ability to stream and form fruiting bodies later in development. This phenomenon is most likely a result of the expression of another ABCC transporter. There are 14 known ABCC transporters expressed in D. discoideum at different times during development (Anjard et al., 2002). Of these, three ABCC transporters other than ABCC3 and ABCC8 are up-regulated near the 6-h time point (Miranda et al., 2013). As we showed that treatment of abcC8− cells with ABCC inhibitors abrogates streaming and further development, a combination of ABCC transporters could account for cAMP release later in development where cAMP release is important (Strmecki et al., 2005). We also envision that the regulation of the ABCC transporters will provide another way to control the release of exosomal cAMP during D. discoideum migration.
Together, our findings present a mechanism to establish and maintain the relay of signals where chemoattractants are packaged in MVBs and released as exosomes or allied vesicles, which provide long-lasting beacons to guide cells during chemotaxis and streaming. Similar mechanisms of signaling involving the release of chemoattractants from exosomes exist in mammals. In fact, Lim et al. (2015), using intravital imaging in mice, showed that neutrophils leave trails in tissues that can guide the migration of other neutrophils across actin filaments. Additionally, Majumdar et al. (2016) demonstrated that signal relay between neutrophils involves exosomes that package leukotriene B4 (LTB4) as they migrate toward N-formyl-methionyl-leucyl-phenylalanine. Interestingly, although exosomal ABC transporters are primarily thought to promote multidrug resistance in tumors (Ifergan et al., 2005; Koch et al., 2016), we now report that they are present in EVs and essential in the transport of chemoattractants during signal relay. We envision that such transporters are involved in secreting LTB4 from neutrophil exosomes as well as other signals involved in cell–cell communications.
Materials and methods
Cell culture and constructs
Unless otherwise mentioned, D. discoideum cells were grown in shaking cultures to 4 × 106 cells/ml in HL5 media at 22°C (Devreotes et al., 1987). They were harvested by centrifugation, washed once in developmental buffer (DB; 5 mM Na2HPO4, 5 mM NaH2PO4, pH 6.2, 2 mM MgSO4, and 200 µM CaCl2), and resuspended in DB at 2 × 107 cells/ml. To differentiate cells, 2 × 108 cells were harvested by centrifugation at 500 g for 5 min at RT and washed once in DB. The cells were centrifuged again at 500 g for 5 min at RT, resuspended in 10 ml DB, and placed in a 250-ml glass Erlenmeyer flask. To ensure uniform differentiation, the cells were repeatedly pulsed every 6 min for 4–7 h, depending on the experiment, with 100 µl of cAMP, giving a 75-nM cAMP final concentration in the flask while shaking on an orbital shaker at 100 rpm. The pulses of cAMP were created with a Gilson miniplus 3 peristaltic pump fitted with pump tubing (inner diameter of 0.76 mm) controlled by a Chrontrol Model XT timer (Parent and Devreotes, 1996). The cells were then processed according to the assay used. To generate cell lines lacking abcC3, abcC8, or abcG8, the gene targeting vectors pV20072 (abcC3), pV20066 (abcC8), pV20023 (abcG2), and pV20021 (abcG10); (Miranda et al., 2013) were linearized by SacII and introduced into growing WT AX3 cells by electroporation. The ABC transporter gene targeting vectors were a gift from G. Schaulsky (Baylor College of Medicine, Houston, TX). Transformants were selected in HL5 medium containing 5 µg/ml blasticidin (ICN Biomedicals). After 5–7 d, cells were harvested and plated on SM agar with Klebsiella aerogenes and allowed to form individual colonies. Randomly selected clones were screened for gene disruption by PCR using primers listed in Table S4 and confirmed by Southern blot analysis. The abcC8 gene without its stop codon was synthesized (Gene Scientific) along with the Gateway recombination arm (attL1 and attL2) flanking the coding sequence. The fusion construct ABCC8-RFP was next created through recombination with the mRFPmars containing D. discoideum expression vector PDM354. The PDM354-abcC8-RFP construct was electroporated into abcC8− cells and selected on 20 µg/ml G418.
FIB-SEM
Differentiated ACA-YFP/aca− cells were spotted on a chamber slide, allowed to spontaneously aggregate for 20 min, and processed for EM as previously described (Kriebel et al., 2008). The resin-embedded cells were imaged in a Crossbeam540 FIB-SEM (Zeiss) in conjunction with ATLAS3D software (Fibics). Images of cells were acquired at 15 kV landing energy to locate pairs of trailing cells in close proximity to each other. The FIB operated at 700 pA was used to first deposit a patterned platinum and carbon pad on top of the resin-embedded cells. The resin block in front of the cells and up to the pad was then milled by the FIB at 30-nA current, until the cells appeared on the cliff face. After a fine polish at 1.5 nA, a data run was initiated, with the FIB operated at 700 pA and scanning electron microscope operated at 1.5 kV. Regions of interest were imaged at 1–2-µs dwell time and 2–4× averaging, and backscattered electron signals were recorded using an ESB detector operated with a grid voltage of 900 V. The milling and imaging dimensions were set at 15 and 5 × 5 nm, respectively, with autofocus/autostigmation and drift correction protocols executed on the fly (Narayan et al., 2014). Automated data acquisition runs lasted 2–3 d, resulting in image stacks typically comprising approximately a thousand slices. These image stacks were then processed using in-house scripts and IMOD (Kremer et al., 1996) to yield registered, binned, inverted, and cropped 3D reconstructed volumes at an isotropic pixel sampling of 15 nm3. Features of interest from these image volumes were extracted, segmented, and rendered using 3DSlicer (Fedorov et al., 2012). The FIB-SEM imaging data that support the findings of this study are available in the National Cancer Institute Center for Strategic Scientific Initiatives Data Coordinating Center (https://cssi-dcc.nci.nih.gov/cssiportal/view/5b11980f34b81e6828aea266/).
EM of EVs with negative stain
Purified vesicles (150 µg/ml) were analyzed by a negative stain method that was previously described (Palmer and Martin, 1988). In brief, EV samples (1 µl each) were placed on a carbon-coated formvar-filmed 400-mesh grid. The grid was glow-discharged to remove static charges from the surface. Excess solution was removed by blotting with filter paper, and 5 µl uranyl acetate solution (0.5% vol/wt aqueous solution) was added to each sample. Excess solution was again blotted away, and the sample was allowed to air dry. The grid was examined and imaged with an electron microscope (Hitachi H7650) operated at 80 kv and equipped with a digital camera (model XR41; Advanced Microscopy Techniques).
EM of EVs with colloidal gold stain
EVs (at least 150 µg/ml) were centrifuged at 100,000 g for 45 min and fixed in 0.2% formaldehyde/0.01% glutaraldehyde solution for 2 h. The vesicle pellet was dehydrated using a series of cold ethanol concentrations (35%, 50%, 70%, 95%, and 100%) and infiltrated with a mixture of cold 100% ethanol and LR white embedding resin (2:1 and 1:2) and pure LR white embedding resin (14380; EM Sciences) overnight at 4°C. The pellet was embedded in fresh LR white resin and cured in an oven (55°C) for 24 h. Thin sections (70–80 nm) were mounted on 300-mesh nickel grids. The thin sections were blocked in a blocking solution (26696; EM Sciences) for 20 min and incubated with a primary antibody (rabbit anti-GFP, AB290; Abcam) at 4°C overnight. The grids were washed with 50 mM Tris buffer containing 0.25 M NaCl, 0.1% BSA, and 0.05% Tween 20 for 2 h followed by incubation in a gold-conjugated secondary antibody (25108; EM Sciences) for 1 h at room temperature. After the grids were washed again with Tris wash solution, they were stained in uranyl acetate (0.5% aqueous solution) and lead citrate (Reynold’s solution). The thin sections were stabilized by carbon evaporation, examined, and imaged with the same microscope setup as above.
Purification of ACA-YFP EVs
Vesicles were isolated using a combination of ultracentrifugation and density gradient separation. In brief, supernatants from the differentiated cells were collected by centrifugation at 500 g at 4°C for 5 min, and cell debris was removed by centrifuging the resultant supernatant at 1,500 g at 4°C for 20 min. Total EVs and other membrane components were collected by further centrifuging at 150,000 g at 4°C for 1 h. The resulting pellet was resuspended in phosphate buffer (PB) and overlaid on a 10–70% sucrose gradient created in a SW40 polyallomer tubes using the BioCompIP Gradient Master. The sucrose gradient was spun at 115,000 g using a SW40 rotor at 8°C for 18 h. Seventeen 600-µl fractions were collected and analyzed. To determine the lipid content of the fractions, the EVs were prestained with the lipophilic dye PKH26 (Sigma-Aldrich) before loading on the sucrose gradient, and the fluorescence intensity of the resulting fractions was measured at 567 nm using an LS55 Perkin Elmer fluorimeter. ACA-YFP content was similarly measured by monitoring YFP fluorescence. Protein concentration was measured using the Bradford assay (500-0006; Bio-Rad). Fractions 14–17 and 9–13, henceforth called bands 1 and 2, were pooled based on their protein content and light scattering properties. The pooled samples were further diluted in an equal volume of PB with 100 µM EDTA, centrifuged at 10,000 g at 4°C for 20 min, and washed and resuspended in 1 ml PB. The vesicles were characterized for size and number by nanoparticle tracking using the NanoSight LM 10 (Malvern). For the Western analysis, the antibodies against calnexin and mitoporin were a gift from G. Gerisch (Max Planck Institute of Biochemistry, Martinsried, Germany), and A. Nogel (University of Cologne, Cologne, Germany) provided the antibody against comitin. The GFP antibody was purchased from Abcam (AB290).
EV release assay
WT (AX3) cells were starved and pulsed with cAMP as described. Before the assay, all centrifuge tubes and filters to be used in the assay were coated with BSA (Sigma-Aldrich) 0.25% at room temperature for 1 h. Starved WT cells were washed and resuspended at 107 cells/ml in cold PB with either 60 µM Noco or DMSO and incubated on ice for 30 min. The cells were then washed with cold PB and resuspended in room temperature DB containing either 60 µM Noco or DMSO to 107 cells/ml. 1.2 ml of the cell suspension was added to BSA-precoated 1.5-ml Eppendorf tubes and incubated on a rotator for 1 h at RT. The cells were removed from the rotator and placed on an orbital shaker at 250 rpm for 2 min. The cells were immediately spun at 1,500 g for 20 min at 4°C, and the resulting supernatant was filtered through an FBS-precoated 0.22-µM syringe. 5 µl of 1% BSA, 5 µl of 500 mM EDTA, and 5 µl of 200 mM EGTA was added to each EV preparation. The EV samples were counted with the LM14 Nanosight (Malvern) using NTA3.1 software. As a control for the basal amount of vesicles present at the start of the assay, an aliquot of cells was taken just before the 1-h RT incubation and placed on an orbital shaker at 250 rpm for 2 min, processed, and counted as described. Noco and DMSO EV counts were divided by the basal EV counts to obtain a fold increase in EV release.
MS
Tandem MS was performed on exosomal fractions after TCA precipitation and in-gel tryptic digestion. Each lane was divided into 12 slices that were subjected to in-gel trypsin (Sigma-Aldrich) digestion for 16 h at 37°C, as described (Shevchenko et al., 2006). The peptides were extracted after cleavage and lyophilized. The dried peptides were resuspended in water containing 2% acetonitrile and 0.5% acetic acid. They were then injected onto a 0.2 × 50-mm Magic C18AQ reverse phase column (Michrom Bioresources) using a Paradigm MS4 HPLC (Michrom Bioresources). Peptides were separated at a flow rate of 2 µl/min over a 90-min linear gradient from 98% mobile phase A (2% acetonitrile, 0.5% acetic acid, and 97.5% water) to 42% mobile phase B (10% water, 0.5% acetic acid, and 89.5% acetonitrile) followed by online analysis by tandem MS using an LTQ ion trap mass spectrometer (Thermo Scientific) equipped with an ADVANCE CaptiveSpray ion source (Michrom Bioresources). Peptides were detected in positive ion mode using a data-dependent method in which the nine most abundant ions detected in an initial survey scan were selected for MS/MS analysis. Raw spectral counts were searched against the D. discoideum database using Sequest and analyzed using the Trans-Proteome Pipeline (Deutsch et al., 2010). For inclusion, proteins were required to have a confidence score of at least 0.9 and contain two unique peptides. Network analysis of the detected proteins was performed using Cytoscape software (version 3.4) environment (Shannon et al., 2003) with the ClueGO v2.3 and CluePedia v1.3.3 plug-ins (Bindea et al., 2013) to functionally group GO and identify core nodes of the network. The D. discoideum genome (NCBI taxonomy ID 44689, 352472) was used as a reference set, and both GO Biological and Cellular Process classifications were compared using statistical tests for gene enrichment (right-sided hypergeometric test) with Benjamini–Hochberg multiple test correction. Terms with a P value ≤0.05 are shown, and only the groups with a minimum of three terms were considered. For comparative heat map analysis, one-dimensional unsupervised hierarchical clustering was applied on triplicates of WT and ACA-YFP/aca− EVs using Ward’s minimum variance method coupled with the Hamming distance metric.
cAMP TR-FRET assay
EVs were resuspended in PB containing 480 µM IBMX (I5879; Sigma-Aldrich), 1 mM DTT (D9779; Sigma-Aldrich), and 0.1% BSA (A9576; Sigma-Aldrich). 10 µl EVs at 150 µg/ml were added to 10 µl kit reagents prepared in our assay buffer. The sample and assay solutions were added to a 384-well plate (Opti Plate 384, 6007290; PerkinElmer) that was read on an EnSpire Multimode Plate Reader. FRET excitation was set at 320 nm, and donor and acceptor emissions were simultaneously acquired at 615 and 665 nm, respectively. Acquisition settings were as follows: delay time, 70 µs; number of flashes, 100 Lamp; window, 200 µs; height, 8 mm. Readings were started 60 min after combining reagents with the samples and every 15 min thereafter. cAMP levels in samples were determined using the FRET ratio of 665/615-nm emission by the europium chelate donor and the ULight receptor and comparing them to cAMP standards. EVs were lysed using lysis buffer, which included the hypotonic detergent solution provided in the kit and IBMX, DTT, and BSA in concentrations stated above. The maximum concentration of adenylyl cyclase inhibitor SQ22536 to be used in the assay without interference was first calibrated and set to 60 µM, corresponding to half the reported IC50. To make sure EV ACA activity is inhibited, EVs were pretreated with a saturating dose of SQ22536 during the 1-h spin after harvesting from a sucrose gradient and removed before the assay. For the assay, the EVs were resuspended in our assay buffer containing 60 µM SQ22536 and maintained at that concentration for the duration of the assay. For each experiment, cAMP levels of three concentrations of EVs (7.5, 15, and 30 µg/ml) were measured in the presence and absence of the cAMP antibody to estimate nonspecific FRET contribution from non-cAMP sources. Nonspecific FRET was modeled and factored in, and only signals within the dynamic range were used for cAMP estimation.
Adenylyl cyclase assay
EVs (100 µg/ml) were resuspended in membrane lysis buffer (250006; Cell Bio Laboratories) and incubated on ice for 2.5 h to lyse the EVs. The samples were then centrifuged at 10,000 g, 4°C for 20 min, and the pellet was resuspended to 250 µg/ml in cyclase assay buffer (10 mM Tris-HCl, 2 mM MgSO4, 0.2 mM EGTA, and 10% glycerol). A 200-µl aliquot of each EV sample was added to 20 µl reaction mix (1.5 µl [32P]ATP; 0132007H; MP Biomedicals), 100 mM DTT, and 100 mM Tris, pH 8.0) with or without 2 mM MnSO4 and incubated at 22°C for 10 min. The [32P]cAMP was purified from the [32P]ATP as previously described (Parent and Devreotes, 1996; Kriebel et al., 2008). In brief, reaction mixtures were added to Dowex AG 50W-X4 (200–400 mesh) columns to trap [32P]cAMP and allowed to drain. Each column was rinsed with 2 ml of 1 mM imidazole. The [32P]cAMP bound to the Dowex column was eluted into alumina columns using 5 ml of 1 mM imidazole. [32P]cAMP was eluted from alumina columns directly into scintillation vials with 3 ml of 100 mM imidazole and counted on a scintillation counter.
The cAMP-mediated activation of ACA in control and Noco-treated cells was performed as previously described (Parent and Devreotes, 1996). In brief, Noco (60 µM) or DMSO (control) was added to differentiated cells, harvested after an hour of incubation, and washed in PB. The cells were kept on ice for 30 min to reduce basal adenylyl cyclase activity. Cells were then added to a cup on an orbital shaker and stimulated with 1 µM cAMP. Samples (200 µl) were taken at 0, 30, 60, 105, 180, and 480 s after stimulation at RT and immediately lysed through a syringe filter (Whatman 110612; 5 µm) and placed into glass tubes containing 20 µl reaction buffer (0.5 µl [32P]ATP [0132007H; MP Biomedicals], 1 mM ATP, 10 mM cAMP, 100 mM DTT, and 100 mM Tris, pH 8.0). The reaction was allowed to proceed for 2 min at RT and stopped with the addition of 100 µl of 2% sodium dodecylsulfate. The samples were brought up to a volume of 1 ml with dH2O and run on the Dowex and Alumina columns as described. The resulting eluate was counted on a scintillation counter. cAMP activity from both cell lysates and EVs were recorded in CPMs and converted to picomoles per minute per milligram using the specific activity of [32P]ATP.
EZ-TAXIScan assay
EVs were resuspended in PB with 1% BSA to a concentration of 150 µg/ml. Differentiated aca− cells were used, as they do not produce cAMP but are able to respond to cAMP (Pitt et al., 1992). The EZ-TAXIScan apparatus was used according to the manufacturer recommendations as previously described, using a chip with a height of 6 µm and a width of 230 µm. aca− cells were introduced into the upper well, and EVs were added to the lower well. Images were taken every 15 s. Automated analysis using Matlab was performed to track and analyze cell migration. Centroids of individual cells were detected after background elimination followed by binarization of the acquired images. Cell tracks were created using an algorithm for finding connected segments from scrambled centroid positions (Crocker and Grier, 1996). The mean image was subtracted to avoid tracking dead cells or other stationary features. Velocities were determined by finding the displacement between smoothed center positions in each frame,
where is the smoothed centroid of cell i at time t, and Δt is the time between frames. Velocity was counted only during a timeframe where cells had a net displacement of 20 µm over a 5-min period. This was done to reduce noise and loss of acquisition tracking. The mean velocity was calculated by
where the brackets indicate averages over all times t and all cells i. The mean CI for cell i at time t is defined as
where is the unit direction vector from cell i at time t to cell i at time t − Δt, and θi(t) is the angle between the motion vector of cell i at time t and t − Δt. A cell moving in a straight line would, hence, have a CI of 1, a CI of 0 if changing direction perpendicular to its previous position, and a CI of −1 if moving back from its previous position. Mean CI is calculated over all times t and all cells i.
Measurement of ATP in EVs
EVs were harvested from aca−, WT, and ACA-YFP/aca− cells as previously outlined. EVs were centrifuged at 150,000 g for 1 h at 4°C, and pellets were then resuspended in cold hypotonic lysis buffer containing detergent to a concentration of 100 µg/100 µl. The EV preparations were incubated for 5 min on ice and then centrifuged at 20,000 g for 20 min at 4°C. Supernatants from each sample were analyzed using the ATP Bioluminescence Assay kit (StayBrite, 791-100; Biovision) and compared with ATP dilution standards. 10 μl of each standard and sample was added to 90 µl of enzyme mix, and bioluminescence was immediately read using a SpectraMax iD3 plate reader set (Molecular Devices) with an exposure of 400 ms and a read height of 4 mm. Three separate experiments were performed, each in triplicate, and the data were analyzed using linear regression from the standards mentioned above.
Micropipette assay
The micropipette assay was performed on differentiated cells as previously described (Kriebel et al., 2003). For assays involving ABC inhibitors, differentiated WT cells were resuspended with ABC inhibitors diluted in PB to the appropriate concentration and incubated 22°C for 1 h while spinning on a rotator. The cells were then spotted on a chambered coverslip and exposed to the micropipette as described. Bright-field images were taken using a Zeiss Axiovert 200 microscope with a Zeiss A Plan 10×, 0.25 aperture, PH1- and Var1-compatible objective, Zeiss Axiovision 4.8.2 software, and using a Photometrics Cool Snap HQ camera every 10 s for 20–30 min. All images were converted to 8-bit black-and-white tiffs and adjusted for brightness and contrast. Time-lapse AVI videos were created from still images using FIGI v.1 software, 1 frame every 20 s. Fluorescent confocal images of differentiated abcC8-RFP/abcC8− cells were obtained using a Zeiss 780 LSM with a Zeiss 63× Plan Apochromat 1.4 aperture objective. Two laser channels were used, one for bright field and one for RFP, and maximum-intensity projections were created from multiple confocal planes, combined in the final image, and adjusted for brightness and contrast as needed using Fiji.
Surface biotinylation and density gradient fractionation
Cells were pulsed for 6 h and harvested as described and resuspended in cold cell lysis buffer (10 mM Tris, 1 mM EDTA, 0.25 M sucrose, and protease inhibitor cocktail) at 109 cells/ml. The cells were lysed using a Balch homogenizer (Isobiotec) by passing them 10 times past a ball bearing allowing 10–18 µm clearance. Total vesicular extracts were prepared by differential centrifugation consisting of 2,000 g for 5 min, 10,000 g for 20 min, and 100,000 g for 1 h. The final 100,000-g pellet was overlaid on a 10–70% sucrose gradient and centrifuged at 115,000 g at 8°C for 18 h. 400-µl fractions were collected and immunoblotted using a mouse anti-GFP antibody (AB290; Abcam). For surface biotinylation, Biotin-NHS (B1513; Invitrogen) was added to cells at 4°C for 15 min according to the manufacturer’s protocol. Unreacted biotin was quenched by adding Tris-Cl and processed as described. Biotin in isolated gradient fractions was visualized using Streptavidin-HRP (1.25 µg/ml).
Agar development assay
Differentiation of vegetative cells on nonnutrient agar was performed as previously described (Kriebel et al., 2003). Images were taken various times after the initiation of starvation using a Lieca MZ125 stereoscope operating with IVision 4.5 software.
Online supplemental material
Fig. S1 shows the assessment of purity and effects of Noco on vesicle release and cell migration. Fig. S2 shows the heat maps and cluster analyses of relative PSMs of proteins involved in signaling, vesicle trafficking, and cytoskeleton. Fig. S3 shows the generation of abcC3, abcC8, abcG10, and abcG2 KO cell lines. Fig. S4 shows agar development and micropipette streaming assays of WT and ABC KO mutants. Fig. S5 shows abcC8− cell phenotypic characterization. Table S1 shows the comprehensive listing of all proteins identified in EVs derived from either WT or ACA-YFP/aca− cells. Table S2 lists the ABC transporters identified in EVs. Table S3 lists the ABC transporter inhibitors used in experiments, with representative reported inhibitory activity against each transporter class. Table S4 lists the primers used for genotyping of ABC KO mutants.
Supplementary Material
Acknowledgments
We thank Drs. Peter Yuen (National Institute of Diabetes and Digestive and Kidney Diseases) and Vanesa Sanchez (Laboratory of Cancer Biology and Genetics) for training and use of the NanoSight, Gad Schaulsky (Baylor College of Medicine) for providing the ABC transporter knockout vectors, Ramiro Iglesias-Bartolome (Laboratory of Cellular and Molecular Biology) for discussions involving the TR-FRET assay, Reema Railkar (Urologic Oncology Branch, Center for Cancer Research) for training and use of the PerkinElmer Enspire plate reader, Matt Hall (Laboratory of Cell Biology) for his input for experiments involving ABC inhibitors, Kunio Nagashima (Center for Cancer Research) for EM work, and Joe Brzostowski and Xi Wen (National Institute of Allergy and Infectious Diseases) for use of their TAXIScan device. We also thank Drs. Gunther Gerisch (Max Planck) and Angelika Nogel (University of Cologne) for providing antibodies used in this study and Lance Shu for his help in pilot studies with D. discoideum EVs. We thank dictyBase for providing annotations for our MS data, and we thank all Parent laboratory members for their excellent discussions and suggestions.
This work was supported in whole or in part with federal funds from the National Cancer Institute (NIH) to C.A. Parent and under contract no. HHSN261200800001E to K. Narayan as well as by NIH grant R01GM084015 to M. Iijima. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
The authors declare no competing financial interests.
Author contributions: P.W. Kriebel and R. Majumdar designed and performed experiments, analyzed and interpreted data, and drafted the manuscript; L.M. Jenkins, H. Senoo, Weiye Wang, S. Chen, and K. Narayan performed experiments and analyzed and interpreted data; S. Ammu performed experiments; M. Iijima designed the experiments and analyzed and interpreted data; and C.A. Parent designed the experiments, analyzed and interpreted data, and drafted the manuscript.
References
- Akers J.C., Gonda D., Kim R., Carter B.S., and Chen C.C.. 2013. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 113:1–11. 10.1007/s11060-013-1084-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C., Dictyostelium Sequencing Consortium, and Loomis W.F.. 2002. Evolutionary analyses of ABC transporters of Dictyostelium discoideum. Eukaryot. Cell. 1:643–652. 10.1128/EC.1.4.643-652.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan F., Lai R.C., Smeets M.B., Akeroyd L., Choo A., Aguor E.N., Timmers L., van Rijen H.V., Doevendans P.A., Pasterkamp G., et al. 2013. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. (Amst.). 10:301–312. 10.1016/j.scr.2013.01.002 [DOI] [PubMed] [Google Scholar]
- Bagorda A., Mihaylov V.A., and Parent C.A.. 2006. Chemotaxis: Moving forward and holding on to the past. Thromb. Haemost. 95:12–21. [PubMed] [Google Scholar]
- Bagorda A., Das S., Rericha E.C., Chen D., Davidson J., and Parent C.A.. 2009. Real-time measurements of cAMP production in live Dictyostelium cells. J. Cell Sci. 122:3907–3914. 10.1242/jcs.051987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatsalam D., and Gomer R.H.. 2010. The secreted proteome profile of developing Dictyostelium discoideum cells. Proteomics. 10:2556–2559. 10.1002/pmic.200900516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Galon J., and Mlecnik B.. 2013. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 29:661–663. 10.1093/bioinformatics/btt019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P., Spiegelman G.B., and Weeks G.. 2008. Rap1 activation in response to cAMP occurs downstream of ras activation during Dictyostelium aggregation. J. Biol. Chem. 283:10232–10240. 10.1074/jbc.M707459200 [DOI] [PubMed] [Google Scholar]
- Bruschi M., Ravera S., Santucci L., Candiano G., Bartolucci M., Calzia D., Lavarello C., Inglese E., Petretto A., Ghiggeri G., and Panfoli I.. 2015. The human urinary exosome as a potential metabolic effector cargo. Expert Rev. Proteomics. 12:425–432. 10.1586/14789450.2015.1055324 [DOI] [PubMed] [Google Scholar]
- Cheepala S., Hulot J.-S., Morgan J.A., Sassi Y., Zhang W., Naren A.P., and Schuetz J.D.. 2013. Cyclic nucleotide compartmentalization: Contributions of phosphodiesterases and ATP-binding cassette transporters. Annu. Rev. Pharmacol. Toxicol. 53:231–253. 10.1146/annurev-pharmtox-010611-134609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Guo J., Yang M., Zhu X., and Cao X.. 2011. Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft-dependent pathway and act as efficient tumor vaccine. J. Immunol. 186:2219–2228. 10.4049/jimmunol.1002991 [DOI] [PubMed] [Google Scholar]
- Crocker J.C., and Grier D.G.. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179:298–310. 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Deutsch E.W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., et al. 2010. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 10:1150–1159. 10.1002/pmic.200900375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P., Fontana D., Klein P., Sherring J., and Theibert A.. 1987. Transmembrane signaling in Dictyostelium. Methods Cell Biol. 28:299–331. 10.1016/S0091-679X(08)61653-2 [DOI] [PubMed] [Google Scholar]
- Entchev E.V., and González-Gaitán M.A.. 2002. Morphogen gradient formation and vesicular trafficking. Traffic. 3:98–109. 10.1034/j.1600-0854.2002.030203.x [DOI] [PubMed] [Google Scholar]
- Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J.C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., et al. 2012. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging. 30:1323–1341. 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P., Dodson R.J., Basu S., and Chisholm R.L.. 2013. One stop shop for everything Dictyostelium: DictyBase and the Dicty Stock Center in 2012. Methods Mol. Biol. 983:59–92. 10.1007/978-1-62703-302-2_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca P., Vardaki I., Occhionero A., and Panaretakis T.. 2016. Metabolic and signaling functions of cancer cell-derived extracellular vesicles. Int. Rev. Cell Mol. Biol. 326:175–199. 10.1016/bs.ircmb.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Garcia G.L., and Parent C.A.. 2008. Signal relay during chemotaxis. J. Microsc. 231:529–534. 10.1111/j.1365-2818.2008.02066.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Kotova E., Chen Z.S., Lee K., Hopper-Borge E., Belinsky M.G., and Kruh G.D.. 2003. MRP8, ATP-binding cassette C11 (ABCC11), is a cyclic nucleotide efflux pump and a resistance factor for fluoropyrimidines 2′,3′-dideoxycytidine and 9′-(2′-phosphonylmethoxyethyl)adenine. J. Biol. Chem. 278:29509–29514. 10.1074/jbc.M304059200 [DOI] [PubMed] [Google Scholar]
- Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R.L., Liu Y., Liang M., et al. 2013. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 14:319 10.1186/1471-2164-14-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. 1999. Structure, mechanism, and regulation of mammalian adenylyl cyclase. J. Biol. Chem. 274:7599–7602. 10.1074/jbc.274.12.7599 [DOI] [PubMed] [Google Scholar]
- Ifergan I., Scheffer G.L., and Assaraf Y.G.. 2005. Novel extracellular vesicles mediate an ABCG2-dependent anticancer drug sequestration and resistance. Cancer Res. 65:10952–10958. 10.1158/0008-5472.CAN-05-2021 [DOI] [PubMed] [Google Scholar]
- Jedlitschky G., Burchell B., and Keppler D.. 2000. The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J. Biol. Chem. 275:30069–30074. 10.1074/jbc.M005463200 [DOI] [PubMed] [Google Scholar]
- Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N., et al. 2016. ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 428:688–692. 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A., Lotfi P., Chavan A.J., Montaño N.M., Bolourani P., Weeks G., Shen Z., Briggs S.P., Pots H., Van Haastert P.J., et al. 2016. The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci. Rep. 6:25823 10.1038/srep25823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Chang W.T., Meima M., Gross J.D., and Schaap P.. 1998. A novel adenylyl cyclase detected in rapidly developing mutants of Dictyostelium. J. Biol. Chem. 273:30859–30862. 10.1074/jbc.273.47.30859 [DOI] [PubMed] [Google Scholar]
- Knetsch M.L.W., Epskamp S.J.P., Schenk P.W., Wang Y., Segall J.E., and Snaar-Jagalska B.E.. 1996. Dual role of cAMP and involvement of both G-proteins and ras in regulation of ERK2 in Dictyostelium discoideum. EMBO J. 15:3361–3368. [PMC free article] [PubMed] [Google Scholar]
- Koch R., Aung T., Vogel D., Chapuy B., Wenzel D., Becker S., Sinzig U., Venkataramani V., von Mach T., Jacob R., et al. 2016. Nuclear trapping through inhibition of exosomal export by indomethacin increases cytostatic efficacy of doxorubicin and pixantrone. Clin. Cancer Res. 22:395–404. 10.1158/1078-0432.CCR-15-0577 [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. [DOI] [PubMed] [Google Scholar]
- Kriebel P.W., and Parent C.A.. 2004. Adenylyl cyclase expression and regulation during the differentiation of Dictyostelium discoideum. IUBMB Life. 56:541–546. 10.1080/15216540400013887 [DOI] [PubMed] [Google Scholar]
- Kriebel P.W., Barr V.A., and Parent C.A.. 2003. Adenylyl cyclase localization regulates streaming during chemotaxis. Cell. 112:549–560. 10.1016/S0092-8674(03)00081-3 [DOI] [PubMed] [Google Scholar]
- Kriebel P.W., Barr V.A., Rericha E.C., Zhang G., and Parent C.A.. 2008. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J. Cell Biol. 183:949–961. 10.1083/jcb.200808105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar I., Clement E., Ducoux-Petit M., Denat L., Soldan V., Dauvillier S., Balor S., Burlet-Schiltz O., Larue L., Muller C., and Nieto L.. 2015. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res. 28:464–475. 10.1111/pcmr.12380 [DOI] [PubMed] [Google Scholar]
- Li M., Zeringer E., Barta T., Schageman J., Cheng A., and Vlassov A.V.. 2014. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130502 10.1098/rstb.2013.0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liégeois S., Benedetto A., Garnier J.M., Schwab Y., and Labouesse M.. 2006. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 173:949–961. 10.1083/jcb.200511072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K., Hyun Y.-M., Lambert-Emo K., Capece T., Bae S., Miller R., Topham D.J., and Kim M.. 2015. Neutrophil trails guide influenza-specific CD8+ T cells in the airways. Science. 349:aaa4352 10.1126/science.aaa4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P., et al. 2014. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 3:26913 10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R., Tavakoli Tameh A., and Parent C.A.. 2016. Exosomes mediate LTB4 release during neutrophil chemotaxis. PLoS Biol. 14:e1002336 10.1371/journal.pbio.1002336 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Miranda E.R., Zhuchenko O., Toplak M., Santhanam B., Zupan B., Kuspa A., and Shaulsky G.. 2013. ABC transporters in Dictyostelium discoideum development. PLoS One. 8:e70040 10.1371/journal.pone.0070040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda E.R., Nam E.A., Kuspa A., and Shaulsky G.. 2015. The ABC transporter, AbcB3, mediates cAMP export in D. discoideum development. Dev. Biol. 397:203–211. 10.1016/j.ydbio.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K., Danielson C.M., Lagarec K., Lowekamp B.C., Coffman P., Laquerre A., Phaneuf M.W., Hope T.J., and Subramaniam S.. 2014. Multi-resolution correlative focused ion beam scanning electron microscopy: Applications to cell biology. J. Struct. Biol. 185:278–284. 10.1016/j.jsb.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J.M., Veltman D., and Kay R.R.. 2015. Chemotaxis of a model organism: Progress with Dictyostelium. Curr. Opin. Cell Biol. 36:7–12. 10.1016/j.ceb.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Nickel W., and Rabouille C.. 2009. Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10:148–155. 10.1038/nrm2617 [DOI] [PubMed] [Google Scholar]
- Palmer E.L., and Martin M.L.. 1988. Electron microscopy in viral diagnosis. CRC Press, Boca Raton, FL. [Google Scholar]
- Panfoli I., Ravera S., Podestà M., Cossu C., Santucci L., Bartolucci M., Bruschi M., Calzia D., Sabatini F., Bruschettini M., et al. 2016. Exosomes from human mesenchymal stem cells conduct aerobic metabolism in term and preterm newborn infants. FASEB J. 30:1416–1424. 10.1096/fj.15-279679 [DOI] [PubMed] [Google Scholar]
- Parent C.A., and Devreotes P.N.. 1996. Constitutively active adenylyl cyclase mutant requires neither G proteins nor cytosolic regulators. J. Biol. Chem. 271:18333–18336. 10.1074/jbc.271.31.18333 [DOI] [PubMed] [Google Scholar]
- Piper R.C., and Katzmann D.J.. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519–547. 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt G.S., Milona N., Borleis J., Lin K.C., Reed R.R., and Devreotes P.N.. 1992. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 69:305–315. 10.1016/0092-8674(92)90411-5 [DOI] [PubMed] [Google Scholar]
- Pitt G.S., Brandt R., Lin K.C., Devreotes P.N., and Schaap P.. 1993. Extracellular cAMP is sufficient to restore developmental gene expression and morphogenesis in Dictyostelium cells lacking the aggregation adenylyl cyclase (ACA). Genes Dev. 7:2172–2180. 10.1101/gad.7.11.2172 [DOI] [PubMed] [Google Scholar]
- Raposo G., and Stoorvogel W.. 2013. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 200:373–383. 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist K.G., Ek B., Stavreus-Evers A., Larsson A., and Ronquist G.. 2013. Human prostasomes express glycolytic enzymes with capacity for ATP production. Am. J. Physiol. Endocrinol. Metab. 304:E576–E582. 10.1152/ajpendo.00511.2012 [DOI] [PubMed] [Google Scholar]
- Rosengarten R.D., Santhanam B., Fuller D., Katoh-Kurasawa M., Loomis W.F., Zupan B., and Shaulsky G.. 2015. Leaps and lulls in the developmental transcriptome of Dictyostelium discoideum. BMC Genomics. 16:294 10.1186/s12864-015-1491-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., and Mann M.. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Simons M., and Raposo G.. 2009. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21:575–581. 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- Strmecki L., Greene D.M., and Pears C.J.. 2005. Developmental decisions in Dictyostelium discoideum. Dev. Biol. 284:25–36. 10.1016/j.ydbio.2005.05.011 [DOI] [PubMed] [Google Scholar]
- Sung B.H., and Weaver A.M.. 2017. Exosome secretion promotes chemotaxis of cancer cells. Cell Adhes. Migr. 11:187–195. 10.1080/19336918.2016.1273307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung B.H., Ketova T., Hoshino D., Zijlstra A., and Weaver A.M.. 2015. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 6:7164 10.1038/ncomms8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., and Kuspa A.. 1997. Dictyostelium development in the absence of cAMP. Science. 277:251–254. 10.1126/science.277.5323.251 [DOI] [PubMed] [Google Scholar]
- Zinzalla V., Stracka D., Oppliger W., and Hall M.N.. 2011. Activation of mTORC2 by association with the ribosome. Cell. 144:757–768. 10.1016/j.cell.2011.02.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.