Nutrient starvation or inactivation of TORC1 induces separation of rDNA and nucleolar proteins in yeast. Mostofa et al. report that the rDNA tethering CLIP–cohibin system repositions nucleolar proteins to sites proximal to the nuclear–vacuolar junction (NVJ), where micronucleophagy occurs, whereas rDNA moves to regions distal to the NVJ.
Abstract
Nutrient starvation or inactivation of target of rapamycin complex 1 (TORC1) in budding yeast induces nucleophagy, a selective autophagy process that preferentially degrades nucleolar components. DNA, including ribosomal DNA (rDNA), is not degraded by nucleophagy, even though rDNA is embedded in the nucleolus. Here, we show that TORC1 inactivation promotes relocalization of nucleolar proteins and rDNA to different sites. Nucleolar proteins move to sites proximal to the nuclear–vacuolar junction (NVJ), where micronucleophagy (or piecemeal microautophagy of the nucleus) occurs, whereas rDNA dissociates from nucleolar proteins and moves to sites distal to NVJs. CLIP and cohibin, which tether rDNA to the inner nuclear membrane, were required for repositioning of nucleolar proteins and rDNA, as well as effective nucleophagic degradation of the nucleolar proteins. Furthermore, micronucleophagy itself was necessary for the repositioning of rDNA and nucleolar proteins. However, rDNA escaped from nucleophagic degradation in CLIP- or cohibin-deficient cells. This study reveals that rDNA–nucleolar protein separation is important for the nucleophagic degradation of nucleolar proteins.
Introduction
Macroautophagy degrades cytoplasmic components and organelles in lysosomes/vacuoles, which is a conserved system from yeast to mammalian cells (Nakatogawa et al., 2009; Reggiori and Klionsky, 2013). Newly generated cup-shaped structures, called isolation membranes, expand to encapsulate cellular constituents, and then the edges of the isolation membranes fuse to form double membrane–surrounded autophagosomes. Subsequently, autophagosomes fuse with lysosomes/vacuoles, and the encapsulated cargoes are digested by lysosomal/vacuolar hydrolytic enzymes. Isolation membrane expansion requires various types of autophagy-related (ATG) proteins, including isolation membrane-associated protein Atg8. In contrast, microautophagy degrades cargos by direct lysosomal/vacuolar engulfment of the cytoplasmic cargo without isolation membranes. Cytoplasmic material is trapped in the lysosome/vacuole by the process of membrane invagination. Little is known about microautophagy (Müller et al., 2000; Sattler and Mayer, 2000; Kunz et al., 2004).
Nucleophagy, the process of autophagic degradation of a nonessential portion of the nucleus, including portions of the nuclear membrane and the nucleolus, is found in budding yeast Saccharomyces cerevisiae (Roberts et al., 2003; Kvam and Goldfarb, 2007; Mochida et al., 2015). In macronucleophagy, autophagosomes sequester this nonessential portion of the nucleus and subsequently fuse with lysosomes/vacuoles, resulting in the degradation of their contents. In budding yeast, macronucleophagy is dependent on the outer nuclear membrane receptor Atg39, which promotes preferential engulfment of cargos by isolation membranes (Mochida et al., 2015). In addition, the ER membrane receptor Atg40, which is also located in perinuclear ER membranes (nuclear outer membranes), is partially involved in macronucleophagy (Mochida et al., 2015). Yeast cells lacking Atg39 cannot effectively survive in starvation conditions (Mochida et al., 2015). This suggests that nucleophagy (at least macronucleophagy) is critical for survival in such conditions, although the biological/physiological importance of nucleophagy for survival is unclear.
In contrast, micronucleophagy (also known as piecemeal microautophagy of the nucleus) targets parts of the nucleus for degradation without isolation membranes. The nucleus and the vacuole closely associate via nuclear–vacuolar junctions (NVJs), which involves interactions between the outer nuclear membrane protein Nvj1 and the vacuolar membrane protein Vac8 (Pan et al., 2000; Roberts et al., 2003). The NVJ invaginates toward the vacuolar lumen and evolves into a teardrop-like bleb, which pinched off from the nucleus into the vacuolar lumen. This vesicle contains nuclear material and is degraded inside vacuoles (Roberts et al., 2003). Krick et al. (2008) investigated their involvement using many ATG mutant cells, but the assessment was inaccurate, because macronucleophagy had not been found at that time and defects in nucleophagy resulted from micronucleophagy and/or macronucleophagy. The involvement of ATG proteins in micronucleophagy remains elusive at present.
Similar to other autophagic processes, macronucleophagy and micronucleophagy are both induced by nutrient starvation and inactivation of target of rapamycin complex 1 (TORC1) kinase (Roberts et al., 2003; Mochida et al., 2015). Nucleophagy was monitored using the processing of several proteins fused to GFP, including Nvj1-GFP and Nop1-GFP; Nop1 (fibrillarin in mammals) is a nucleolar ribosome biogenesis/maturation (Ribi) protein (Krick et al., 2008; Dawaliby and Mayer, 2010; Mochida et al., 2015). Free GFP is produced from these fusion proteins during autophagic processes, because Nvj1 and Nop1 are degraded by vacuolar proteases during the autophagic process, whereas GFP is a stably folded protein and relatively resistant to vacuolar proteases.
In contrast to the nucleolus, in yeast, chromosomal DNA is excluded from nucleophagy through an undefined mechanism (Roberts et al., 2003; Millen et al., 2009). This indicates that ribosomal DNA (rDNA; encoding rRNA) also escapes from nucleophagy, even though it is a core component of the nucleolus. How does nucleophagy preferentially degrade nucleolar components? How does rDNA escape from nucleophagy? In this study, we addressed these questions and found that after TORC1 inactivation, rDNA and nucleolar proteins were dynamically relocated in opposing manners and were thereby separated from each other. In addition, the rDNA-tethering CLIP–cohibin system and nucleophagy are required for these motions. Thus, this study revealed key events for selective degradation of the specific nuclear component.
Results
TORC1 inactivation triggers intranuclear repositioning of nucleolar proteins and rDNA
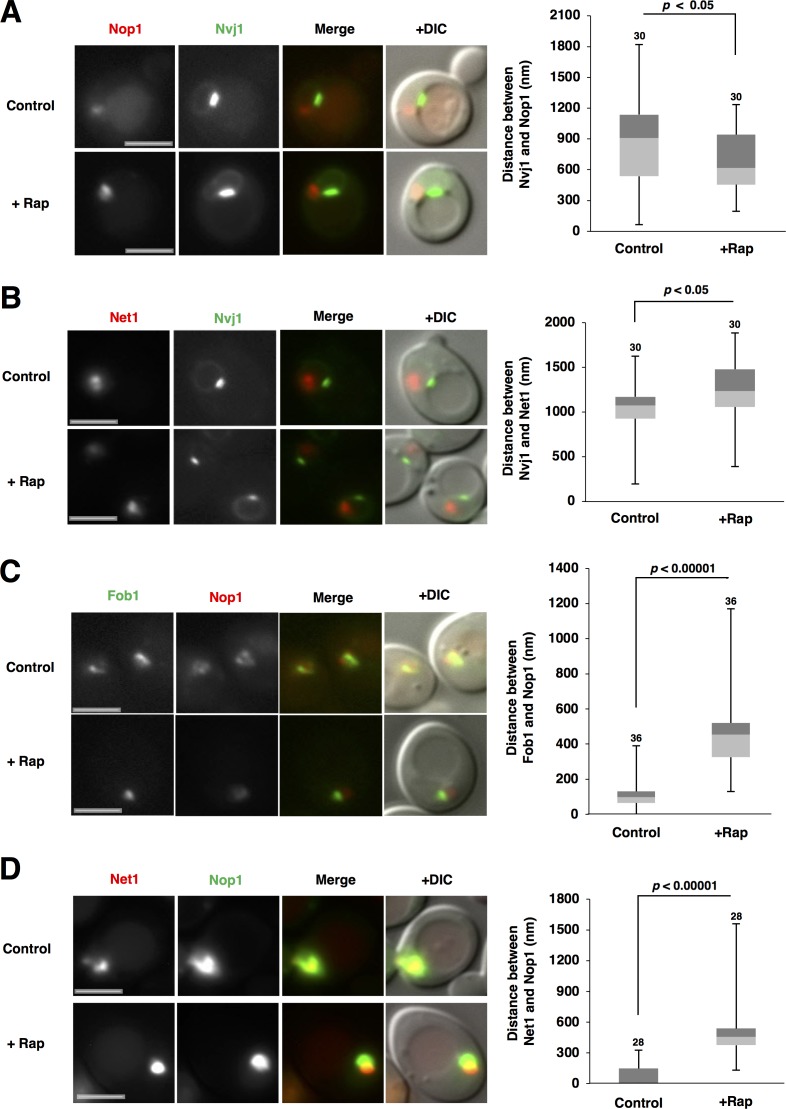
Nucleophagy preferentially degrades nucleolar proteins. Micronucleophagy occurs in a limited region, namely the NVJ, but it is unknown where macronucleophagy occurs on the nuclear membrane. We wondered whether nucleolar proteins are localized at an intranuclear region suitable for nucleophagy, namely at or near the NVJ. To test this idea, we observed the relative positions of nucleolar proteins (monitored using RFP-Nop1) and the NVJ (monitored using Nvj1-GFP). Nvj1 accumulated at the NVJ in addition to slight distribution throughout the nuclear envelope (Pan et al., 2000; Fig. 1 A). Nucleolar proteins were localized near the nuclear envelope, but not near the NVJ in normal conditions (Fig. 1 A, control). However, we found that nucleolar proteins accessed the NVJ after rapamycin treatment (Fig. 1 A, +Rap). We measured the distance between nucleolar proteins and the NVJ before and after rapamycin treatment (Fig. S1 A) and found that the distance decreased after rapamycin treatment (Fig. 1 A, right). Similar observations were also obtained using another nucleolar protein Nop58 (data not shown). These findings indicated that TORC1 inactivation promoted access of nucleolar proteins to the NVJ, which should be necessary for the preferential degradation of nucleolar proteins by nucleophagy.
Figure 1.
TORC1 inactivation triggers intranuclear repositioning of nucleolar proteins and rDNA. (A) Exponentially growing cells of strain SCU3287 (NVJ1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with 200 ng/ml rapamycin for 1 h. Cell images with GFP and RFP signals were captured using a fluorescence microscope. Bars, 2.5 µm. The distance between Nvj1-GFP and RFP-Nop1 peaks, measured as described in Materials and methods (also see Fig. S1 A), is shown in the box plot. Numbers above the bars are sample sizes. P-values were calculated using a two-tailed Mann–Whitney U test. (B) Cells of strain SCU4433 (NVJ1-GFP NET1-mRuby2) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Nvj1-GFP and Net1-RFP peaks is shown in the box plot. (C) Cells of strain SCU359 (FOB1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between Fob1-GFP and RFP-Nop1 peaks, measured as described in Materials and methods (also see Fig. S1 C), is shown in the box plot. (D) Cells of strain SCU4449 (NET1-mRuby2) harboring plasmid pSCU740 (pNOP1-GFP) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Nop1-GFP and Net1-RFP peaks is shown in the box plot.
In contrast, DNA is not degraded by nucleophagy (Roberts et al., 2003; Millen et al., 2009), although rDNA is a core structure of the nucleolus. This allowed us to speculate that after rapamycin treatment, rDNA might show a different movement from that found in nuclear proteins. rDNA (monitored using rDNA-binding proteins Net1 and Rpa43 [a subunit of rRNA polymerase I] tagged with fluorescent protein) was also resident adjacent to the nuclear membrane, but not at an NVJ-proximal site in normal conditions, like nucleolar proteins (Figs. 1 B and S2 A). Surprisingly, rDNA was located farther away from the NVJ after rapamycin treatment, in sharp contrast to nucleolar proteins: the distance between the rDNA and the NVJ increased after rapamycin treatment. Thus, TORC1 inactivation promoted the relocation of rDNA to an NVJ-distal site, which probably contributed to the escape of rDNA from nucleophagic degradation. Collectively, TORC1 inactivation evoked the opposing movements of nucleolar proteins and rDNA to NVJ-proximal and -distal sites, respectively. We confirmed this idea using 3D image analysis (Fig. S5, A and B).
It might be expected that the opposing movements of nucleolar proteins and rDNA after TORC1 inactivation should result in rDNA dissociation from nucleolar proteins. rDNA (monitored using GFP-tagged Fob1, an rDNA-binding protein) was embedded in nucleolar proteins (monitored using RFP-Nop1) in normal conditions (Figs. 1 C and S5 C). However, rDNA partially dissociated from nucleolar proteins after rapamycin treatment. Similar tendencies were observed using another rDNA marker, the Fob1-binding protein Net1 (Fig. 1 D) or another nucleolar protein Nop58 (not depicted). Of note, these rDNA-binding proteins Fob1 and Net1 were still associated with rDNA even after rapamycin treatment (Fig. S3 A), indicating that these rDNA-binding proteins are still suitable markers for the rDNA position after TORC1 inactivation. In addition, a similar rDNA–nucleolar protein separation after rapamycin treatment was observed using the rDNA-binding protein Rpa43 (Fig. S2 B) and rDNA-lacO/lacI-GFP system, which directly marks rDNA by GFP signals (Miyazaki and Kobayashi, 2011; Figs. S3 B), which were still colocalized with the rDNA-binding protein Net1 (namely, Rpa43 and rDNA-lacO/lacI-GFP were still associated with rDNA) even after rapamycin treatment (Figs. S2 C and S3 C). These observations indicated that TORC1 inactivation stimulates rDNA–nucleolar protein separation.
Nitrogen starvation causes TORC1 inactivation and is a natural trigger of autophagy induction (Loewith and Hall, 2011). We again found that nitrogen starvation promoted the access of nucleolar proteins to the NVJ, escape of rDNA from the NVJ, and rDNA–nucleolar protein separation (Fig. S4, A–C). These findings demonstrated that nitrogen starvation-induced TORC1 inactivation stimulated the repositioning of nucleolar proteins and rDNA, probably contributing to the preferential autophagic degradation of nucleolar proteins and escape of rDNA from nucleophagic degradation.
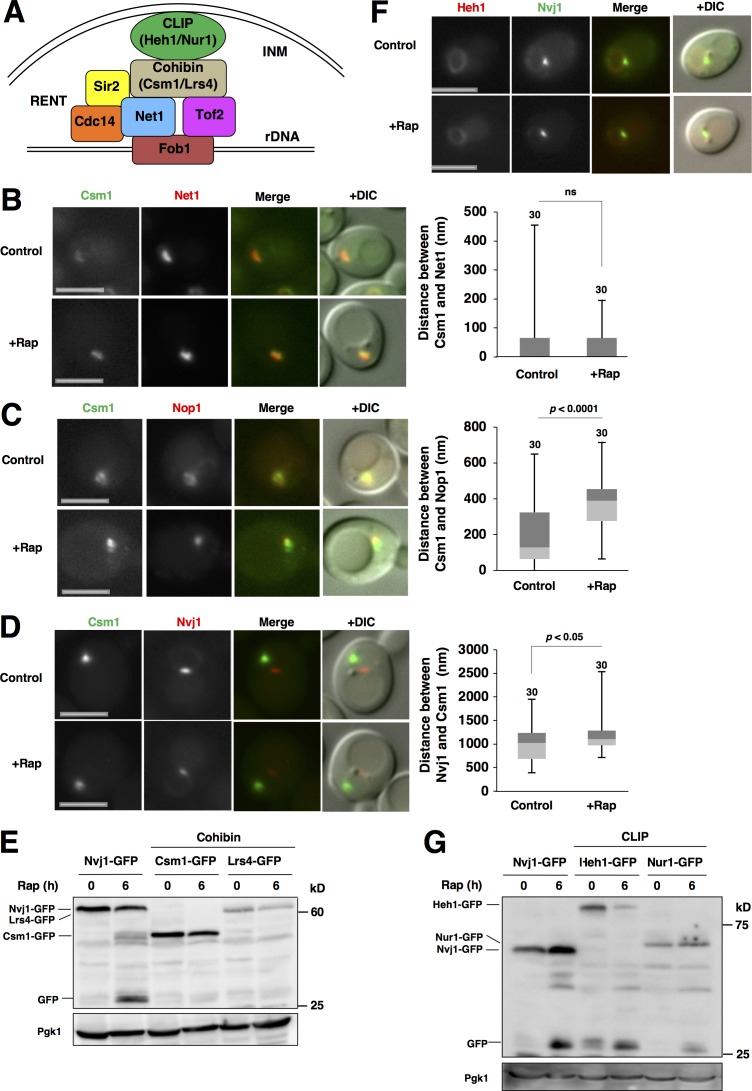
Cohibin is colocalized with rDNA even after TORC1 inactivation
Two complexes, cohibin (Lrs4 and Csm1) and chromosome linkage inner nuclear membrane (INM) protein (CLIP; Heh1 and Nur1), anchor rDNA to the INM via the rDNA-binding protein Fob1, RENT complex (Net1–Cdc14–Sir2), and Tof2 (Mekhail et al., 2008; Fig. 2 A). In this study, we focused on the rDNA-tethering factors cohibin and CLIP. Because rDNA was relocated to the NVJ-distal region after TORC1 inactivation, we suspected that cohibin and CLIP might be involved in these movements. Cohibin (monitored using Csm1-GFP) colocalized with rDNA (monitored using Net1-RFP) in normal conditions, and this colocalization was still maintained after rapamycin treatment (Fig. 2 B). Conversely, cohibin dissociated from nucleolar protein after rapamycin treatment (Fig. 2 C), and it located farther away from the NVJ (Fig. 2 D), like rDNA. Thus, cohibin seemingly tethered rDNA not only in normal conditions but also in TORC1-inactive conditions. Based on these findings, it is not surprising that nucleophagic degradation of cohibin (Csm1-GFP and Lrs4-GFP) after rapamycin treatment was not detectable (Fig. 2 E).
Figure 2.
Cohibin is colocalized with rDNA even after TORC1 inactivation. (A) A model of the CLIP–cohibin rDNA tethering system to the INM. The CLIP complex consists of Heh1 and Nur1, and the cohibin complex is composed of Csm1 and Lrs4. The RENT complex contains Net1, Cdc14, and Sir2. See the text for details. (B) Cells of strain SCU4922 (CSM1-GFP NET1-mRuby2) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between Csm1-GFP and Net1-RFP peaks is shown in the box plot. (C) Cells of strain SCU1502 (CSM1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Csm1-GFP and RFP-Nop1 peaks is shown in the box plot. (D) Exponentially growing cells of strain SCU4794 (CSM1-GFP NVJ1-mRuby2) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Csm1-GFP and Nvj1-RFP peaks is shown in the box plot. (E) Cells of strains SCU1502 (CSM1-GFP) and SCU1555 (LRS4-GFP) were treated with 200 ng/ml rapamycin for 6 h. Cells of strain SCU3287 (NVJ1-GFP) were used as a positive control for free GFP production from fusion proteins, the indicator of autophagic degradation. Whole-cell extracts were subjected to immunoblotting using an anti-GFP antibody. Pgk1 was detected as the loading control using an anti-Pgk1 antibody. (F) Cells of strain SCU4780 (NVJ1-GFP HEH1-mRuby2) were treated with rapamycin for 1 h. Bars, 2.5 µm. (G) Cells of strains SCU3287 (NVJ1-GFP; the positive control), SCU4205 (HEH1-GFP), and SCU4222 (NUR1-GFP) were treated with rapamycin for 6 h. Whole-cell extracts were subjected to Western blotting.
In contrast, CLIP (monitored using Heh1-GFP) was uniformly distributed throughout the nuclear membrane, but did not accumulate with rDNA, in the nucleolus, or at the NVJ in normal and TORC1-inactive conditions (Fig. 2 F and not depicted). This might result from the fact that Heh1 also mediates another events, e.g., nuclear pore complex quality control (Webster et al., 2016). Because nuclear membranes are also degraded by micro- and macronucleophagy, CLIP that was uniformly distributed in the INM should be degraded by nucleophagy. This was indeed the case: CLIP (monitored using Heh1-GFP and Nur1-GFP) was degraded by nucleophagy, whereas cohibin was not (Fig. 2 G).
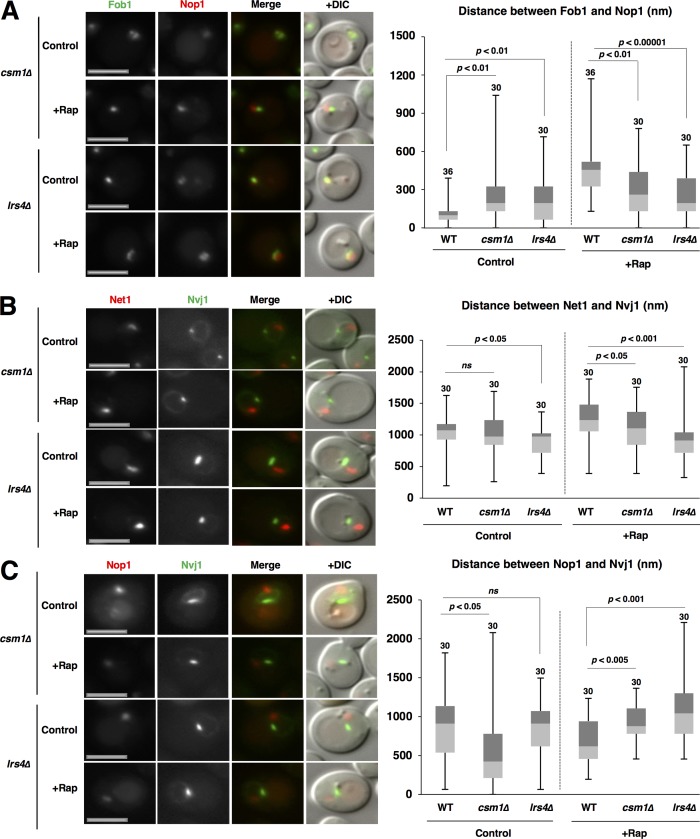
Cohibin is required for repositioning of rDNA and nucleolar proteins
Because cohibin still colocalized with rDNA even after TORC1 inactivation, we assumed that cohibin is critical for the intranuclear repositioning of rDNA after TORC1 inactivation. The separation of rDNA from nucleolar proteins after rapamycin treatment was impeded by the loss of cohibin (Csm1, Lrs4, or both; Figs. 3 A and S6 A). Thus, cohibin is required for rDNA separation from nucleolar proteins. Furthermore, the displacement of rDNA from the NVJ was repressed in cohibin-deficient mutants (Figs. 3 B and S6 B). This indicated that cohibin mediates TORC1 inactivation-induced rDNA repositioning. Surprisingly, loss of cohibin also hindered access of the nucleolar proteins to the NVJ1 after TORC1 inactivation (Figs. 3 C and S6 C). This indicated that cohibin is involved in repositioning of nucleolar protein after TORC1 activation.
Figure 3.
Cohibin is required for repositioning of rDNA and nucleolar proteins. (A) Cells of strains SCU359 (FOB1-GFP), SCU4431 (csm1Δ FOB1-GFP), and SCU4798 (lrs4Δ FOB1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Fob1-GFP and RFP-Nop1 peaks is shown in the box plot. (B) Cells of strains SCU4433 (NVJ1-GFP NET1-mRuby2), SCU4782 (csm1Δ NVJ1-GFP NET1-mRuby2), and SCU4841 (lrs4Δ NVJ1-GFP NET1-mRuby2) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Nvj1-GFP and Net1-RFP peaks is shown in the box plot. (C) Cells of strains SCU3287 (NVJ1-GFP), SCU4790 (csm1Δ NVJ1-GFP), and SCU4800 (lrs4Δ NVJ1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Nvj1-GFP and RFP-Nop1 peaks is shown in the box plot.
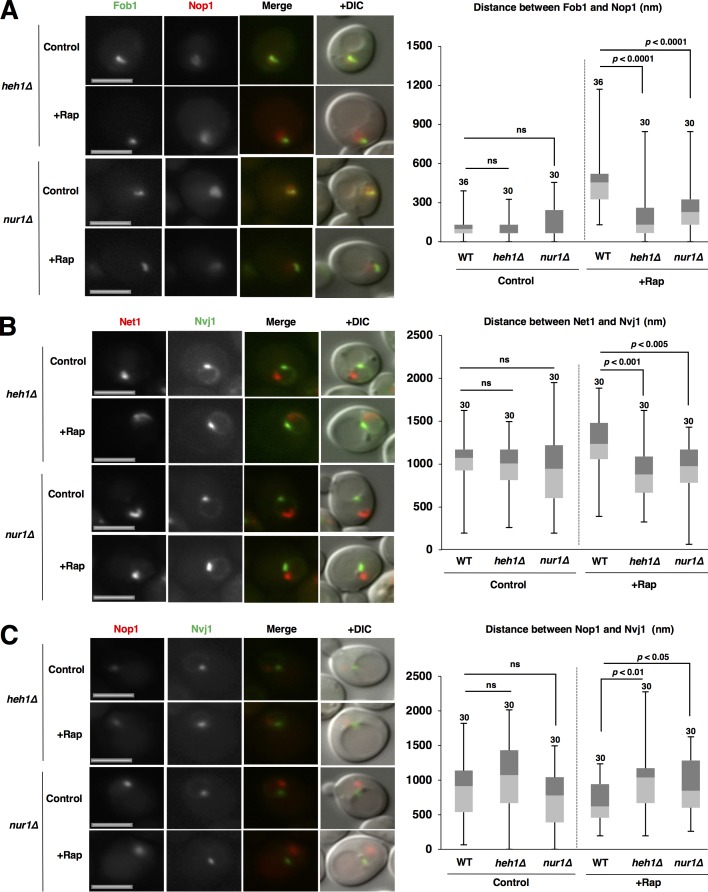
CLIP is required for repositioning of rDNA and nucleolar proteins
We also assessed the involvement of CLIP in repositioning of rDNA and nucleolar proteins. Loss of CLIP (Heh1, Nur1, or both) compromised colocalization of rDNA and nucleolar proteins in normal conditions and the separation of rDNA from nucleolar proteins after rapamycin treatment, similar to the loss of cohibin (Figs. 4 A and S6 A). Thus, CLIP is also required for rDNA separation from nucleolar proteins after TORC1 inactivation. Furthermore, rDNA escape from the NVJ was repressed in these CLIP mutants (Figs. 4 B and S6 B). Thus, CLIP was also involved in TORC1 inactivation-induced rDNA repositioning, like cohibin. In addition, loss of CLIP hampered access of the nucleolar proteins to the NVJ1 after rapamycin treatment (Figs. 4 C and S6 C). Together, the cohibin–CLIP axis is critical for the repositioning of rDNA and nucleolar proteins after TORC1 inactivation.
Figure 4.
CLIP is required for repositioning of rDNA and nucleolar proteins. (A) Cells of strains SCU359 (FOB1-GFP), SCU4768 (heh1Δ FOB1-GFP), and SCU4429 (nur1Δ FOB1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Fob1-GFP and RFP-Nop1 peaks is shown in the box plot. (B) Cells of strains SCU4433 (NVJ1-GFP NET1-mRuby2), SCU4766 (heh1Δ NVJ1-GFP NET1-mRuby2), and SCU4758 (nur1Δ NVJ1-GFP NET1-mRuby2) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between Nvj1-GFP and Net1-RFP peaks is shown in the box plot. (C) Cells of strains SCU3287 (NVJ1-GFP), SCU4764 (heh1Δ NVJ1-GFP), and SCU4756 (nur1Δ NVJ1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between the Nvj1-GFP and RFP-Nop1 peaks is shown in the box plot.
Cohibin is required for nucleophagic degradation of nucleolar proteins
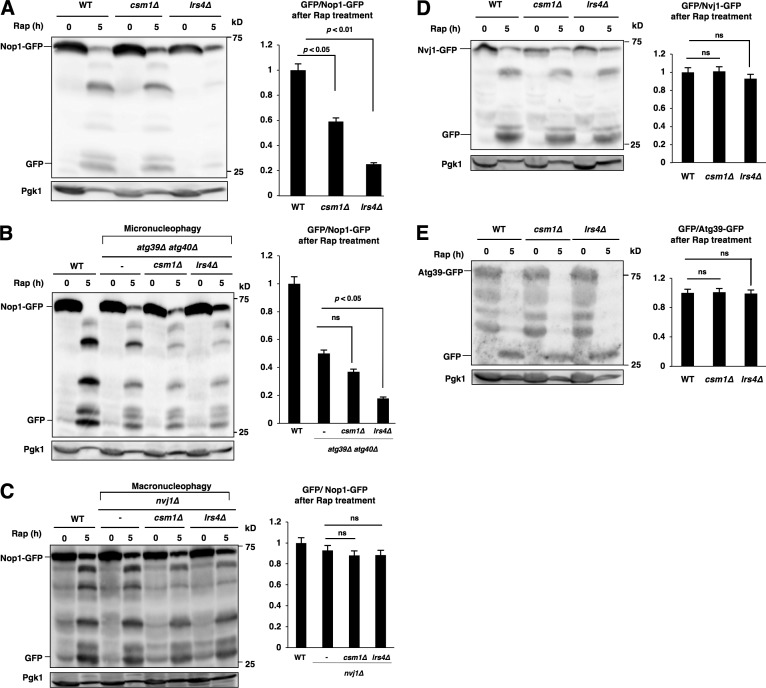
Loss of cohibin impaired the access of nucleolar proteins to the NVJ after TORC1 inactivation; therefore, we suspected that nucleophagic degradation of nucleolar proteins should be compromised by the loss of cohibin. Bulk autophagy (monitored using GFP-Atg8; Shintani and Klionsky, 2004) was not compromised by the loss of cohibin (not depicted), whereas autophagic degradation of the nucleolar proteins (monitored using Nop1-GFP) was substantially reduced by the loss of Lrs4 and, to a lesser extent, by loss of Csm1 (Fig. 5 A). Thus, cohibin was required for nucleophagic degradation of the nucleolar proteins after TORC1 inactivation, which was most likely caused by insufficient relocation of nucleolar proteins to the NVJ.
Figure 5.
Cohibin is required for nucleophagic degradation of nucleolar proteins. (A) Cells of strains BY4741 (WT), SCU2761 (csm1Δ), and SCU4885 (lrs4Δ) harboring plasmid pSCU740 (NOP1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. Free GFP processed from GFP-tagged protein after rapamycin treatment was measured using ImageJ and quantified by calculating the ratio of cleaved free GFP to uncleaved full-length protein. (B) Cells of strains US356 (WT), SCU4575 (atg39Δ atg40Δ), SCU5037 (atg39Δ atg40Δ csm1Δ), and SCU5039 (atg39Δ atg40Δ lrs4Δ) harboring plasmid pSCU740 (NOP1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. (C) Cells of strains US356 (WT), SCU4514 (nvj1Δ), SCU5041 (nvj1Δ csm1Δ), and SCU5043 (nvj1Δ lrs4Δ) harboring plasmid pSCU740 (NOP1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. (D) Cells of strains SCU3287 (NVJ1-GFP), SCU4790 (csm1Δ NVJ1-GFP), and SCU4800 (lrs4Δ NVJ1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. (E) Cells of strains SCU5143 (ATG39-GFP), SCU5145 (csm1Δ ATG39-GFP), and SCU5147 (lrs4Δ ATG39-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. The mean (± SD) was determined from three independent experiments, and relative values normalized against the value in control cells are shown. The p-values were calculated using two-tailed Student's t test.
The fact that access of nucleolar proteins to the NVJ and nucleophagic degradation of nucleolar proteins were both impeded by the loss of cohibin allowed us to suspect whether “micronucleophagic” degradation of nucleolar proteins might be compromised by the loss of cohibin. We tested this idea using atg39Δ atg40Δ background cells that were defective in macronucleophagy: namely, they were capable of micronucleophagy only. Indeed, micronucleophagic degradation of nucleolar proteins was significantly reduced by the loss of Lrs4, although its reduction in csm1Δ cells was not significant (Fig. 5 B). In contrast, macronucleophagic degradation (in nvj1Δ background cells) of nucleolar proteins was not significantly reduced by the loss of cohibin (Fig. 5 C). Thus, cohibin was important for micronucleophagic (but not macronucleophagic) degradation of nucleolar proteins.
Nvj1 itself is degraded during micronucleophagy, and thereby Nvj1-GFP is used for micronucleophagy assay (Krick et al., 2008). Micronucleophagy (monitored using Nvj1-GFP) after rapamycin treatment was not hindered in cohibin mutants (Fig. 5 D). In addition, macronucleophagy itself (monitored using Atg39-GFP) was not impeded by the loss of cohibin (Fig. 5 E). Thus, cohibin is not required for either micronucleophagy or macronucleophagy.
CLIP is required for nucleophagic degradation of nucleolar proteins
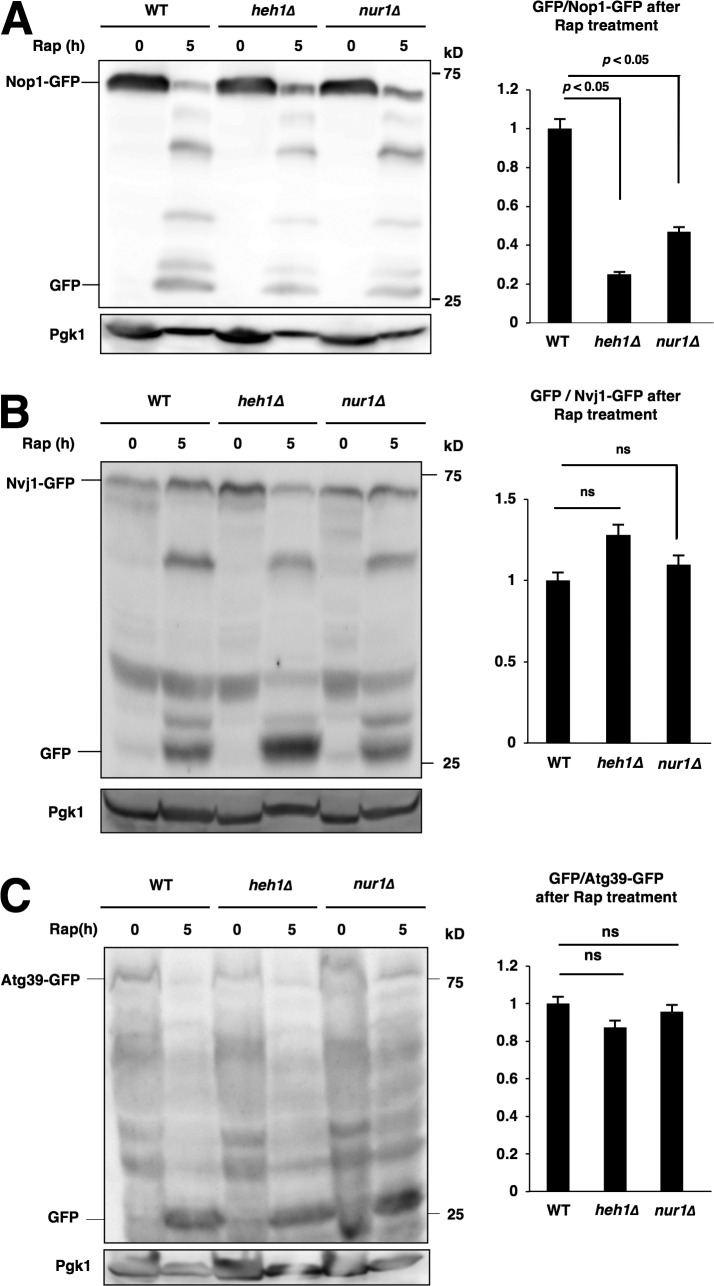
Next, we tested whether CLIP is also involved in nucleophagic degradation of nucleolar proteins. Again, we found that loss of CLIP compromised the nucleophagic degradation of nucleolar proteins (Nop1-GFP), but not Nvj1-GFP, Atg39-GFP, or GFP-Atg8 (Fig. 6, A–C; and not depicted). Thus, CLIP was required for the nucleophagic degradation of nucleolar proteins, but not nucleophagy (macro- and micronucleophagy) itself or bulk autophagy. Collectively, the CLIP–cohibin axis was required for the nucleophagic degradation of nucleolar proteins.
Figure 6.
CLIP is also required for nucleophagic degradation of nucleolar proteins. (A) Cells of strains BY4741 (WT), SCU4419 (heh1Δ), and SCU4400 (nur1Δ) harboring plasmid pSCU740 (NOP1-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting. (B) Cells of strains SCU3287 (NVJ1-GFP), SCU4764 (heh1Δ NVJ1-GFP), and SCU4756 (nur1Δ NVJ1-GFP) were treated with rapamycin for 5 h. (C) Cells of strains SCU5143 (ATG39-GFP), SCU5149 (heh1Δ ATG39-GFP), and SCU5151 (nur1Δ ATG39-GFP) were treated with rapamycin for 5 h. Whole-cell extracts were subjected to Western blotting.
rDNA escapes from nucleophagic degradation in CLIP- or cohibin-deficient cells
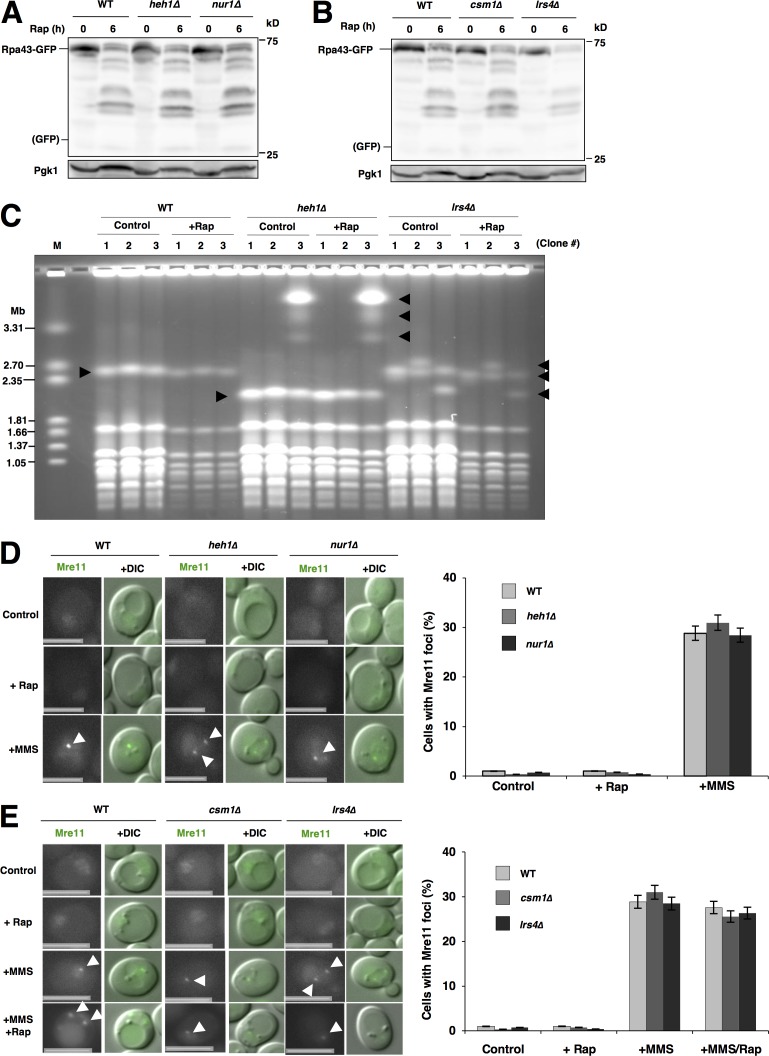
The rDNA-binding protein Rpa43 was not degraded by autophagy/nucleophagy after TORC1 inactivation in the WT cells, similar to cohibin, because no free GFP was detected after rapamycin treatment (Fig. 7, A and B). This supported the fact that rDNA can escape from nucleophagy. We noted that the protein levels of Rpa43-GFP decreased after TORC1 inactivation, even though no free GFP was detected. This suggested that another degradative pathway (probably the ubiquitin–proteasome system) degrades Rpa43. This idea was confirmed by a finding that the proteasome inhibitor MG132 in combination with 0.003% SDS (in pdr5Δ background cells; Liu et al., 2007) repressed the decrease in Rpa43-GFP after rapamycin treatment (Fig. S6 D). This indicated that the ubiquitin–proteasome system promotes Rpa43 degradation after rapamycin treatment.
Figure 7.
rDNA escapes from nucleophagic degradation in CLIP- or cohibin-deficient cells. (A) Cells of strains SCU366 (RPA43-GFP), SCU4770 (heh1Δ RPA43-GFP), and SCU4916 (nur1Δ RPA43-GFP) were treated with rapamycin for 6 h. Whole-cell extracts were subjected to Western blotting. (B) Cells of strains SCU366 (RPA43-GFP), SCU4918 (csm1Δ RPA43-GFP), and SCU4920 (lrs4Δ RPA43-GFP) were treated with rapamycin for 6 h. Whole-cell extracts were subjected to Western blotting. (C) PFGE analysis to monitor rDNA stability in the rapamycin-treated cells. Three independent colonies of strains the WT (BY4741), SCU4419 (heh1Δ), and SCU4885 (lrs4Δ) were used. Exponentially growing cells were treated with rapamycin for 6 h. After PFGE, the gel was stained with ethidium bromide. Positions of chromosome XII that has the rDNA are indicated by arrowheads. M, size marker (H. wingei). (D) Cells of strains SCU2139 (MRE11-GFP), SCU4762 (heh1Δ MRE11-GFP), and SCU4926 (nur1Δ MRE11-GFP) were treated with rapamycin for 3 h or 0.1% MMS for 1 h. Arrowheads indicate Mre11 foci. Bars, 2.5 µm. Cells with Mre11-GFP foci were counted in each experiment (>200 cells) and the mean value (± SD) obtained from three independent experiments is shown as a percentage. (E) Cells of strains SCU2139 (MRE11-GFP), SCU4786 (csm1Δ MRE11-GFP), and SCU4924 (lrs4Δ MRE11-GFP) were treated with 200 ng/ml rapamycin for 3 h, 0.1% MMS for 1 h, or MMS together with rapamycin for 1 h. Arrowheads indicate Mre11 foci. Bars, 2.5 µm. Cells with Mre11-GFP foci were counted in each experiment (>200 cells) and the mean value (± SD) obtained from three independent experiments is shown as a percentage.
Because rDNA–nucleolar protein dissociation was impaired in cohibin and CLIP mutant cells after TORC1 inactivation, we wondered whether rDNA was accidentally degraded by nucleophagy in these mutant cells. However, no nucleophagic degradation of the rDNA marker Rpa43-GFP was found even in cells lacking cohibin or CLIP (Fig. 7, A and B). This suggested that rDNA escapes from nucleophagic degradation even if rDNA is not properly dissociated from nucleolar proteins by the loss of the CLIP–cohibin axis. To assess this, chromosome stability after rapamycin treatment was determined by pulsed-field gel electrophoresis (PFGE) analysis (Kobayashi et al., 2004). In S. cerevisiae, ∼150 rDNA repeats are tandemly arranged on chromosome XII (Chr. XII), which is the longest chromosome in yeast. Loss of cohibin or CLIP resulted in marked changes in rDNA copy number, leading to fluctuations in Chr. XII length (Mekhail et al., 2008; Fig. 7 C). However, Chr. XII showed no further instability after rapamycin treatment in the WT, heh1, and lrs4Δ cells. This confirmed that Chr. XII, including rDNA repeats in cells lacking the CLIP–cohibin axis, was not injured by nucleophagy after TORC1 inactivation. In addition, no DNA damage (monitored using the formation of Mre11 foci) occurred after rapamycin treatment, not only in the WT cells but also in cells lacking CLIP or cohibin, although Mre11 foci were generated in cells treated with the DNA-damaging agent methyl methanesulfonate (MMS), which was used as the positive control (Fig. 7, D and E). It could be suspected that rapamycin represses the DNA damage response, even if DNA damage occurs. We demonstrated that this was not the case because MMS-induced Mre11 focus formation was not impaired by rapamycin addition (Fig. 7, D and E). Thus, rDNA dissociation defects in CLIP- or cohibin-deficient cells caused impaired nucleophagic degradation of nucleolar proteins but no rDNA damage/degradation.
Nucleophagy is required for the repositioning of rDNA and nucleolar proteins
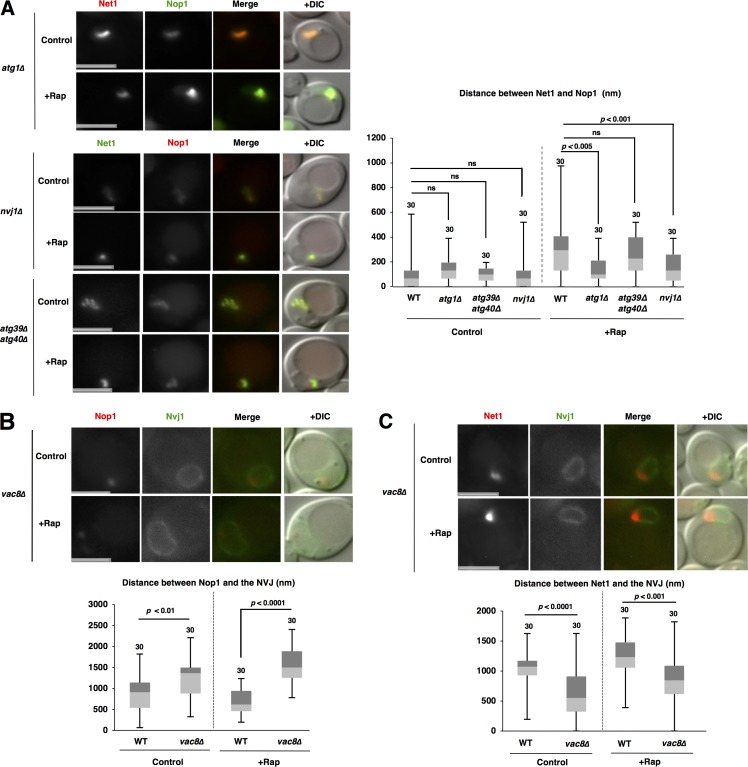
What produces the force for repositioning of rDNA and nucleolar proteins after TORC1 inactivation? We doubted whether nuclear deformation induced by nucleophagy at the NVJ promoted repositioning. To test this idea, we first estimated rDNA–nucleolar protein dissociation in atg1Δ cells in which no autophagic activity is detected. Loss of Atg1 compromised rDNA–nucleolar protein dissociation (Fig. 8 A). Furthermore, the dissociation was significantly repressed in micronucleophagy-deficient nvj1Δ cells, but not in macronucleophagy-deficient atg39Δ atg40Δ cells. These findings indicate that micronucleophagy is required for rDNA–nucleolar protein dissociation.
Figure 8.
Nucleophagy is required for the repositioning of nucleolar proteins and rDNA. (A) Cells of strains SCU5056 (NET1-GFP), SCU5019 (atg1Δ NET1-mRuby2), SCU5021 (atg39Δ atg40Δ NET1-GFP), and SCU5023 (nvj1Δ NET1-GFP) harboring plasmid pSCU740 (pNOP1-GFP) or pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between Net1 and Nop1 fluorescent peaks is shown in the box plot. (B) Cells of strains SCU3287 (NVJ1-GFP) and SCU5153 (vac8Δ NVJ1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between Nvj1-GFP and RFP-Nop1 peaks measured as described in Materials and methods (also see Fig. S1 B) is shown in the box plot. (C) Cells of strains SCU4433 (NVJ1-GFP NET1-mRuby2) and SCU4435 (vac8Δ NVJ1-GFP NET1-mRuby2) were treated with rapamycin for 1 h. Bars, 2.5 µm. The distance between Nvj1-GFP and Net1-RFP peaks is shown in the box plot.
We next assessed whether micronucleophagy is also required for the opposing movements of nucleolar proteins and rDNA after TORC1 inactivation. Micronucleophagy defects caused by the loss of Vac8 hindered the access of nucleolar proteins to the NVJ after rapamycin treatment (Figs. 8 B and S1 B). Of note, Nvj1–Vac8 interaction is essential for micronucleophagy, but not for NVJ formation: the nucleus still contacted to the vacuole even in the absence of Vac8, although accumulation of Nvj1 at the NVJ was abolished, as described previously (Kvam and Goldfarb, 2004). Additionally, deficiency of micronucleophagy abrogated the displacement of rDNA from the NVJ after rapamycin treatment (Fig. 8 C). Collectively, these results show that micronucleophagy is required for the repositioning of nucleolar proteins and rDNA after TORC1 inactivation.
Spatiotemporal dynamics of the repositioning of nucleolar proteins and rDNA after TORC1 inactivation
First, we suspected that the repositioning of nucleolar proteins and rDNA after TORC1 inactivation should be a prerequisite for nucleophagic degradation of nucleolar proteins and protection of rDNA against nucleophagic degradation. It was indeed the case: the repositioning of nucleolar proteins and rDNA already occurred after 1 h of TORC1 inactivation (as mentioned above), although autophagy is not significantly induced after 1 h of nutrient starvation and TORC1 inactivation (Takeshige et al., 1992). We further monitored temporal and spatial dynamics of the separation of rDNA and nucleolar proteins and the access of nucleolar proteins to the NVJ up to 6 h after rapamycin treatment (Fig. 9, A and B), because nucleophagic degradation of nucleolar proteins (monitored by Nop1-GFP processing) was almost completed after 5 h of rapamycin treatment (Fig. 5, A and B). Nop1-GFP was markedly degraded after 5 h of rapamycin treatment (Fig. 5, A and B), and nucleolar Nop1 signals were markedly decreased in cells after 6 h of rapamycin treatment (Fig. 9, C and D). The budding yeast S. cerevisiae possesses a single nucleolus in the cell, and the single nucleolar protein signal in the cell did not separate into smaller signals with up to 6 h of rapamycin treatment (Fig. 9, C and D; and not depicted). This suggested that a portion of the single cluster of nucleolar proteins might be gradually and partially degraded by nucleophagy after TORC1 inactivation.
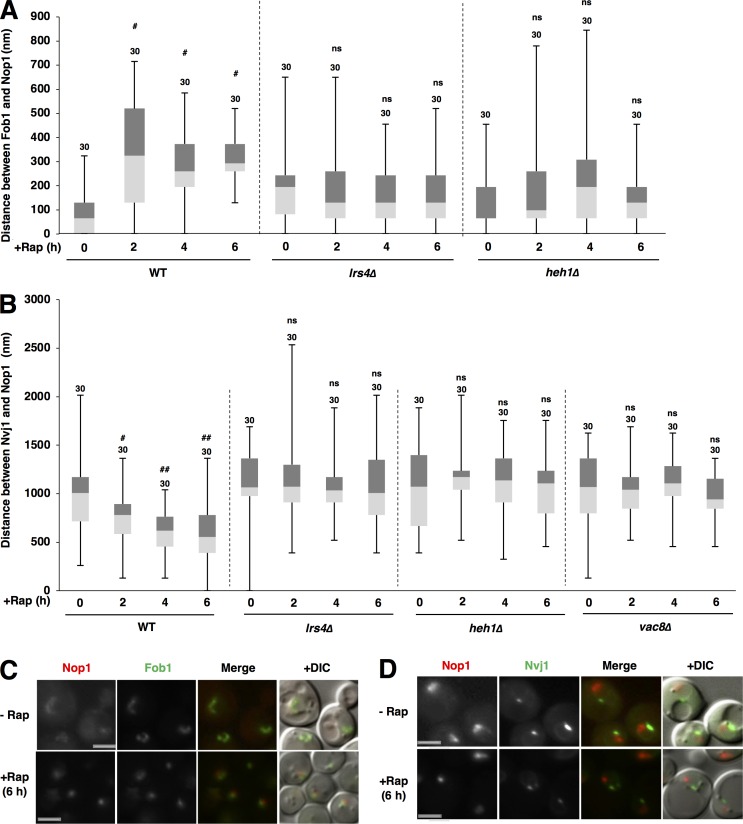
Figure 9.
Spatiotemporal dynamics of the repositioning of nucleolar proteins and rDNA after TORC1 inactivation. (A) Cells of strains SCU359 (FOB1-GFP), SCU4798 (lrs4Δ FOB1-GFP), and SCU4768 (heh1Δ FOB1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for up to 6 h. Cell images with GFP and RFP signals were captured using a fluorescence microscope. The distance between Fob1-GFP and RFP-Nop1 peaks, measured as described in Materials and methods (also see Fig. S1 C) is shown in the box plot. Numbers above the bars are sample sizes. Significant differences (#, P < 0.0001) between rapamycin-treated cells and nontreated control cells (time 0 h) were calculated using two-tailed Mann–Whitney U test. (B) Cells of strains SCU3287 (NVJ1-GFP), SCU4800 (lrs4Δ NVJ1-GFP), SCU4764 (heh1Δ NVJ1-GFP), and SCU5153 (vac8Δ NVJ1-GFP) harboring plasmid pSCU618 (pRFP-NOP1) were treated with rapamycin for up to 6 h. Cell images with GFP and RFP signals were captured using a fluorescence microscope. The distance between Nvj1-GFP and RFP-Nop1 peaks, measured as described in Materials and methods (also see Fig. S1 A) is shown in the box plot. Numbers above the bars are sample sizes. Significant differences (#, P < 0.01; ##, P < 0.0001) between rapamycin-treated cells and nontreated control cells (time 0 h) were calculated using two-tailed Mann–Whitney U test. (C and D) Representative cell images with GFP and RFP signals in the WT cells treated with rapamycin for 6 h and without rapamycin. Bars, 2.5 µm.
In the WT cells, the separation of rDNA and nucleolar proteins already occurred after 1 h of rapamycin treatment (Fig. 1 C), but the separation did not gradually increase after rapamycin treatment (Fig. 9 A). Conversely, the access of nucleolar proteins to the NVJ also started after 1 h of rapamycin treatment (Fig. 1 A), and the access gradually increased after rapamycin treatment (Fig. 9 B, WT). However, the repositioning was abolished in heh1Δ or lrs4Δ cells even after 6 h of rapamycin treatment (Fig. 9, A and B). This suggested that loss of CLIP or cohibin abolishes, and not merely inhibits, the repositioning of nucleolar proteins and rDNA after TORC1 inactivation. In addition, the repositioning of nucleolar proteins was completely abolished in vac8Δ cells even after 6 h of rapamycin treatment (Fig. 9 B). This suggested that loss of micronucleophagy abolishes the repositioning of nucleolar proteins and rDNA after TORC1 inactivation.
The CLIP–cohibin axis is required for survival during nutrient starvation
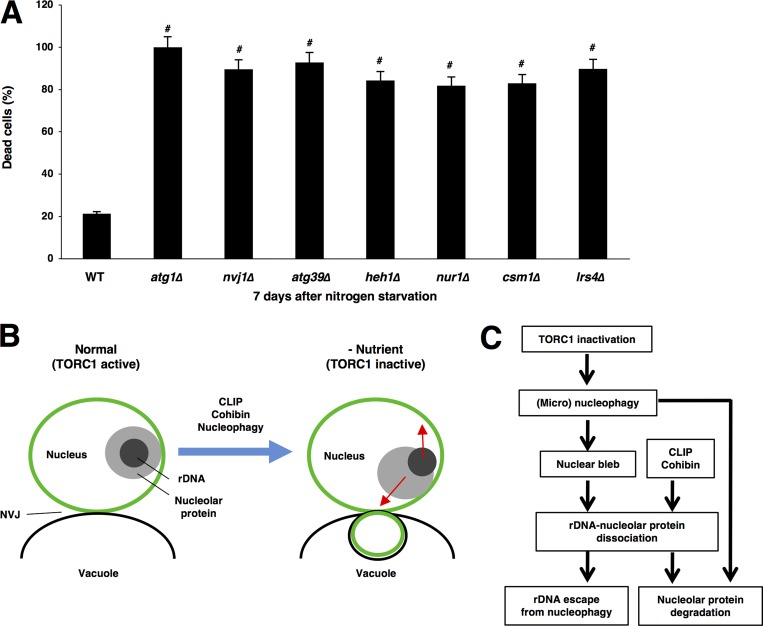
Loss of the core autophagy component Atg1 rapidly reduced cell survivability during nitrogen starvation (Tsukada and Ohsumi, 1993), indicating that autophagy induction after nutrient starvation is critical for cell survival. Cells lacking Atg39 were susceptible to long-term nitrogen deprivation, suggesting that Atg39-dependent macronucleophagy is critical for survival in nutrient starvation conditions (Mochida et al., 2015; Fig. 10 A). We found that loss of Nvj1 also decreased cell survivability during nitrogen starvation, suggesting that Nvj1-dependent micronucleophagy is also important for survival in nutrient-deprived conditions (Fig. 10 A). Thus, both micro- and macronucleophagy should be required for cell viability. Because the CLIP–cohibin axis is important for nucleophagic degradation of nucleolar proteins, we suspected that this axis is critical for cell survival during starvation. Indeed, cells lacking cohibin or CLIP also lost cell viability after nitrogen starvation (Fig. 10 A). This indicated that the CLIP–cohibin axis is critical for cell survival during starvation and suggested that CLIP–cohibin-dependent nucleophagic degradation of nucleolar proteins is critical for adaptation to nutrient starvation.
Figure 10.
The CLIP–cohibin axis is required for survival during nitrogen starvation. (A) Strains BY4741 (WT), SCU3365 (atg1Δ), SCU3368 (nvj1Δ), SCU4325 (atg39Δ), SCU4419 (heh1Δ), SCU4400 (nur1Δ), SCU2761 (csm1Δ), and SCU4885 (lrs4Δ) were used. Exponentially growing cells were subjected to nitrogen starvation for 7 d. Dead cells were stained with 2 µg/ml phloxine B. Cells stained with phloxine B counted as dead cells in each experiment (>100 cells), and the mean value (± SD) obtained from three independent experiments is shown as a percentage. Significant differences (#, P < 0.001) between mutants and the WT were calculated using two-tailed Student's t test. (B) A cartoon for the movements of nucleolar proteins (nucleoli) and rDNA after TORC1 inactivation. CLIP, cohibin, and nucleophagy are required for this repositioning. (C) A hypothetical model for rDNA and nucleolar protein repositioning after TORC1 inactivation in the context of nucleophagy. The CLIP–cohibin axis is required for the repositioning. Micronucleophagy is necessary not only for nucleolar protein degradation but also for the preceding repositioning. See the text for details.
Discussion
In this study, we showed several important findings: (a) TORC1 inactivation promoted opposing movements of nucleolar proteins and rDNA, namely, access of nucleolar proteins to the NVJ and displacement of rDNA from the NVJ, resulting in rDNA–nucleolar protein dissociation; (b) the rDNA-tethering CLIP–cohibin system mediated these movements; and (c) micronucleophagy also promoted this repositioning (Fig. 10, B and C).
We assumed that after TORC1 inactivation, rDNA is probably relocalized to the NVJ-distal region along the INM via the CLIP–cohibin system. It is unknown how rDNA is directed toward the NVJ-distal region. What kind of factor determines this direction of movement? Such a factor might be controlled in the context of the vacuolar position, namely the NVJ. Here, we also found that loss of CLIP or cohibin impaired the access of nucleolar proteins to the NVJ after TORC1 inactivation, suggesting that the CLIP–cohibin axis is involved in nucleolar protein repositioning after TORC1 inactivation, albeit with no information as to a tethering system for nucleolar proteins to the INM. Alternatively, nucleolar protein repositioning after TORC1 inactivation may be indirectly dependent on nucleoplasm/chromosome repositioning. If dynamic changes in nuclear shape and size occurred after TORC1 inactivation, they also might influence the repositioning of rDNA and nucleolar proteins. We measured nuclear shape and size before and after rapamycin treatment and could not detect significant changes in them after rapamycin treatment (Fig. S7, A–D). This suggested that the repositioning of rDNA and nucleolar proteins was not caused by dynamic changes in nuclear shape and size.
It is not surprising that when the access of nucleolar proteins to the NVJ is not properly achieved by CLIP or cohibin deficiency, nucleophagic (especially, micronucleophagic) degradation of nucleolar proteins is repressed, although nucleophagy itself seems to occur efficiently (Fig. 5, A–E; and Fig. 6, A–C). In this case, rDNA was not still degraded by nucleophagy. This suggested that the CLIP–cohibin system is required for rDNA/nucleolar protein repositioning after TORC1 inactivation but that it is not essential for rDNA escape from nucleophagy. We noted that rDNA (and nucleolar proteins) was still resident near the nuclear membrane in cohibin- or CLIP-deficient cells (Fig. 3, B and C; and Fig. 4, B and C). This suggested that another system might tether rDNA to the INM. Nevertheless, the nuclear membrane (monitored using Nvj1-GFP and Atg39-GFP), but not rDNA, was degraded by nucleophagy (Fig. 7, C–E). This suggested that rDNA may still escape from the INM just before the execution of nucleophagy, which guarantees rDNA escape from nucleophagic degradation even if the CLIP–cohibin system suffers dysfunction. In this study, we found no connection between rDNA–nucleolar repositioning and macronucleophagy. It is largely unknown how and where macronucleophagy occurs on the nuclear envelope. Even if macronucleophagy can occur everywhere on the nuclear membrane, rDNA was able to escape from macronucleophagy if it detached from the INM before nuclear blebbing.
We also found that micronucleophagy itself was required for proper repositioning of nucleolar proteins and rDNA after TORC1 inactivation (Fig. 8, A–C). We assumed that micronucleophagy-induced nuclear deformation (invagination of the nuclear envelope into the vacuole) promoted this repositioning. Nuclear projection already started in cells 1 h after rapamycin treatment (Fig. S7 E), when the repositioning of nucleolar proteins and rDNA occurred. Alternatively, micronucleophagy may promote the movements of nucleolar proteins and rDNA independently of nuclear deformation. These distinct possibilities remain to be examined.
In mammalian cells, macronucleophagy and micronucleophagy similar to the processes found in budding yeast have not been found. In addition, there is no information about mammalian counterparts of Nvj1 and Atg39. However, macroautophagy eliminated aberrant micronuclei (Rello-Varona et al., 2012), suggesting that the activity of autophagic elimination of the nucleus is also conserved in mammals. Additionally, several mammalian autophagy proteins are present in the nucleus, including Atg8/LC3, Atg5, and Atg7 (Drake et al., 2010; Shoji et al., 2010; Lee et al., 2012; Simon et al., 2014; Dou et al., 2015; Huang et al., 2015), although whether they are involved in nucleophagy is unclear. Upon starvation, mammalian cells no longer need nucleolar proteins (not only Ribi proteins but also preribosomes under construction), like yeast cells. The degradation of these proteins and subsequent nutrient recycling should be beneficial for survival in starvation conditions. We anticipated that mammalian nucleolar proteins are similarly degraded by nucleophagy upon nutrient starvation and TORC1 inactivation. This study might establish new perspectives in mammalian nucleophagy.
Materials and methods
Strains, plasmids, and media
S. cerevisiae strains and plasmids used are listed in Tables S1 and S2, respectively. Glucose-containing yeast-peptone-dextrose medium with 0.01% adenine (YPAD) and synthetic minimal medium complemented with the appropriate nutrients for plasmid maintenance were prepared using standard methods. For assessment of autophagy, when cells harbored plasmids, cells were precultured in synthetic minimal medium with the appropriate nutrients and then cultured in YPAD. For nitrogen-starvation experiments, cells were transferred into synthetic minimal medium without ammonium sulfate (SD-N medium).
Western blotting analysis
Exponentially growing cells were used for experiments. Proteins were extracted using a postalkaline extraction method in accordance with a previous study (Kushnirov, 2000). In brief, cells (10 ml culture, OD600 = 0.2–0.8) were treated with 200 µl of 0.1 M NaOH for 5 min, and the pellet was collected by centrifugation. The pellet was resuspended in sample buffer (60 mM Tris-HCl, pH 6.8, 5% glycerol, 2% SDS, 4% 2-mercaptoethanol, and 0.0025% bromophenol blue) at 95°C for 5 min. Crude extracts were cleared by centrifugation, and the supernatant was used for Western blotting analysis. We used anti-GFP mouse monoclonal antibody (sc-9996; Santa Cruz) and an anti-Pgk1 mouse monoclonal antibody (A-6457; Thermo Fisher Scientific). Chemiluminescence signals from Western BLoT Quant HRP Substrate (DS-T7102; Takara Bio) for HRP and Immuno Shot (IS-012-250; Cosmo Bio) as an immunoreaction enhancer solution were detected using an image analyzer (LAS3000mini; Fuji). All Western blotting experiments were performed independently at least three times to confirm the reproducibility of the results. Relative protein amounts were measured using ImageJ (National Institutes of Health). The mean and SD were determined for each sample from three independent experiments, and relative values normalized against the value in control cells are shown. For statistical analysis, p-values were calculated using two-tailed Student’s t test.
Microscope observations
Exponentially growing cells were used for experiments. Cell, GFP, and RFP images were captured using a Axio Imager M1 microscope with a cooled CCD camera (AxioCam MRm; Carl Zeiss). All microscope observations were performed independently at least three times to confirm the reproducibility of the results.
For the determination of distances between rDNA and the NVJ, we used cells expressing Net1-RFP and Nvj1-GFP. We performed line scanning along a straight line passing through the middle point of the Nvj1-GFP signals and the brightest point of Net1-RFP signals (Fig. S1 A). We measured the distance between the fluorescence peaks of Nvj1-GFP and Net1-RFP by line scanning using a microscope and imaging software (Fig. S1 A, right). When we determined the distance between nucleolar proteins and the NVJ, we similarly measured distances between the fluorescence peaks of Nvj1-GFP and RFP-Nop1. In the case of vac8Δ cells, Nvj1 did not accumulate in the NVJ, but instead the nuclear membrane and the vacuolar membrane still contacted each other. Considering Nvj1-GFP signals and bright-field images of vacuolar membranes, we determined the nuclear–vacuolar contact site. We drew a straight line passing through the middle point of the contact site and the brightest point of Net1-RFP signals (Fig. S1 B). For determination of the distance between nucleolar proteins and rDNA, we used cells expressing RFP-Nop1 and Fob1-GFP. We drew a straight line passing through the brightest points of RFP-Nop1 and Fob1-GFP signals and measured the distance between the fluorescence peaks (Fig. S1 C). All data from three independent experiments are shown as box plots. For statistical analysis, p-values were calculated using a two-tailed Mann–Whitney U test.
3D analysis of GFP and RFP signals
Exponentially growing cells were used for experiments. GFP and RFP images were collected using a DeltaVision Core microscope (Applied Precision) with CoolSNAP HQ2 CCD camera (Photometrics) as a stack of pictures, which were deconvolved. The distance between GFP and RFP signals was measured using Imaris software (Bitplane). All data from three independent experiments are shown as box plots. For statistical analysis, p-values were calculated using two-tailed Mann–Whitney U test.
Chromatin immunoprecipitation (ChIP) assay
Exponentially growing cells of the tandem affinity purification (TAP)–tagged Fob1 (FOB1-TAP) and Net1 (NET1-TAP) strains were used for the ChIP assay. Cells were cultured in YPAD medium and treated with rapamycin for 1 h. ChIP assay was performed as described previously (Johzuka and Horiuchi, 2007). rDNA region containing Fob1-binding site was amplified using the following primers: forward, 5′-GTGAAAGGATTTGCCCGGACAGTTTG-3′, and reverse, 5′-AGTAGCCTCATCCTTTTACGCTGCCT-3′. DNA contents in the input and immunoprecipitation samples were analyzed by real-time PCR using a Thermal Cycler Dice (Takara Bio) and SYBR premix EX Taq (Takara Bio). WT strain without TAP tag (BY4741) was used as a control. Experiments were performed in triplicate to achieve reproducibility. DNA content in each immunoprecipitation sample relative to that of its input sample is shown as mean ± SD.
Cell viability assay
Cell viability was assessed as described previously (Suzuki et al., 2011). In brief, exponentially growing cells were shifted to SD-N medium and incubated for 7 d. Dead cells were stained with 2 µg/ml phloxine B (Sigma-Aldrich). Fluorescence microscopy was performed with a FITC filter. More than 100 cells were examined for each strain, and the proportion of phloxine-positive (dead) cells to total cells observed per bright-field image was calculated. All data (mean ± SD) from three independent experiments are shown. For statistical analysis, p-values were calculated using two-tailed Student’s t test.
PFGE
Cells of strains WT (BY4741), SCU4419 (heh1Δ), and SCU4885 (lrs4Δ) were inoculated to YPAD plates and incubated for 2 d. Three independent colonies were picked up and incubated in YPAD medium. Exponentially growing cells (∼108 cells) were treated with rapamycin for 6 h. PFGE was performed as previously described (Kobayashi et al., 2004). Genomic DNA was prepared in low-melting-temperature agarose plugs. Conditions of electrophoresis were 300–900-s pulse time, 100 V for 68 h at 14°C, in 1% agarose gel in 0.5× TBE (CHEF Mapper XA; Bio-Rad). The angle was 120°. Hansenula wingei chromosomal DNA size marker (Bio-Rad) was also loaded. After electrophoresis, the gel was stained with 0.5 µg/ml ethidium bromide and photographed.
Online supplemental material
Fig. S1 shows the procedures to measure distances between rDNA, nucleolar proteins, and the NVJ. Fig. S2 demonstrates that Rpa43 moves far from the NVJ and dissociates from nucleolar proteins after TORC1 inactivation. Fig. S3 shows profiling of several rDNA markers after TORC1 inactivation. Fig. S4 shows that nitrogen starvation promotes intranuclear repositioning of nucleolar proteins and rDNA. Fig. S5 demonstrates 3D analysis of the relocation of nucleolar proteins to the NVJ and rDNA escape from the NVJ after TORC1 inactivation. Fig. S6 shows that the CLIP–cohibin axis is required for the relocation of rDNA and nucleolar proteins after TORC1 inactivation and that Rpa43 is degraded in a proteasome-dependent manner. Fig. S7 shows an assessment of nuclear shape change and size after TORC1 inactivation and nuclear invagination during micronucleophagy. Table S1 shows strains used in this study. Table S2 shows plasmids used in this study.
Supplementary Material
Acknowledgments
We thank Fumiyoshi Abe (Aoyama Gakuin University, Sagamihara, Japan), Ed Hurt, Danesh Moazed (Harvard Medical School, Boston, MA), Yoshinori Ohsumi, and Uttam Surana for gifts of materials; Katsuki Johzuka for technical advice; and members of the T. Ushimaru laboratory, Ayumu Yamamoto and Chihiro Horigome, for helpful discussions. We also thank the Imaging Core Facility at the Biozentrum of University of Basel for help and advice with image acquisition and analysis.
This work was supported in part by the Japan Society for the Promotion of Science (grants 19370082 and 23570225 to T. Ushimaru).
The authors declare no competing financial interests.
Author contributions: T. Ushimaru designed research. M.G. Mostofa mainly conducted the experiments. M.A. Rahman, N. Koike, A.M. Yeasmin, N. Islam, S. Hosoyamada, and M. Shimobayashi partially performed the experiments. T.M. Waliullah technically supported the experiments. T. Kobayashi and M.N. Hall supervised some experiments. T. Ushimaru mainly wrote the paper. M.G. Mostofa helped to write the paper.
References
- Dawaliby R., and Mayer A.. 2010. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol. Biol. Cell. 21:4173–4183. 10.1091/mbc.e09-09-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z., Xu C., Donahue G., Shimi T., Pan J.A., Zhu J., Ivanov A., Capell B.C., Drake A.M., Shah P.P., et al. 2015. Autophagy mediates degradation of nuclear lamina. Nature. 527:105–109. 10.1038/nature15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake K.R., Kang M., and Kenworthy A.K.. 2010. Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3. PLoS One. 5:e9806 10.1371/journal.pone.0009806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Xu Y., Wan W., Shou X., Qian J., You Z., Liu B., Chang C., Zhou T., Lippincott-Schwartz J., and Liu W.. 2015. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell. 57:456–466. 10.1016/j.molcel.2014.12.013 [DOI] [PubMed] [Google Scholar]
- Johzuka K., and Horiuchi T.. 2007. RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells. 12:759–771. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., and Nomura M.. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 117:441–453. 10.1016/S0092-8674(04)00414-3 [DOI] [PubMed] [Google Scholar]
- Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.L., Millen J., Goldfarb D.S., and Thumm M.. 2008. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 19:4492–4505. 10.1091/mbc.e08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J.B., Schwarz H., and Mayer A.. 2004. Determination of four sequential stages during microautophagy in vitro. J. Biol. Chem. 279:9987–9996. 10.1074/jbc.M307905200 [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. 2000. Rapid and reliable protein extraction from yeast. Yeast. 16:857–860. [DOI] [PubMed] [Google Scholar]
- Kvam E., and Goldfarb D.S.. 2004. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 117:4959–4968. 10.1242/jcs.01372 [DOI] [PubMed] [Google Scholar]
- Kvam E., and Goldfarb D.S.. 2007. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 3:85–92. 10.4161/auto.3586 [DOI] [PubMed] [Google Scholar]
- Lee I.H., Kawai Y., Fergusson M.M., Rovira I.I., Bishop A.J., Motoyama N., Cao L., and Finkel T.. 2012. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 336:225–228. 10.1126/science.1218395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Apodaca J., Davis L.E., and Rao H.. 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques. 42:158–162. 10.2144/000112389 [DOI] [PubMed] [Google Scholar]
- Loewith R., and Hall M.N.. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 189:1177–1201. 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K., Seebacher J., Gygi S.P., and Moazed D.. 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 456:667–670. 10.1038/nature07460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen J.I., Krick R., Prick T., Thumm M., and Goldfarb D.S.. 2009. Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy. 5:75–81. 10.4161/auto.5.1.7181 [DOI] [PubMed] [Google Scholar]
- Miyazaki T., and Kobayashi T.. 2011. Visualization of the dynamic behavior of ribosomal RNA gene repeats in living yeast cells. Genes Cells. 16:491–502. 10.1111/j.1365-2443.2011.01506.x [DOI] [PubMed] [Google Scholar]
- Mochida K., Oikawa Y., Kimura Y., Kirisako H., Hirano H., Ohsumi Y., and Nakatogawa H.. 2015. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 522:359–362. 10.1038/nature14506 [DOI] [PubMed] [Google Scholar]
- Müller O., Sattler T., Flötenmeyer M., Schwarz H., Plattner H., and Mayer A.. 2000. Autophagic tubes. J. Cell Biol. 151:519–528. 10.1083/jcb.151.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., and Ohsumi Y.. 2009. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D.S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., and Klionsky D.J.. 2013. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics. 194:341–361. 10.1534/genetics.112.149013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rello-Varona S., Lissa D., Shen S., Niso-Santano M., Senovilla L., Mariño G., Vitale I., Jemaá M., Harper F., Pierron G., et al. 2012. Autophagic removal of micronuclei. Cell Cycle. 11:170–176. 10.4161/cc.11.1.18564 [DOI] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., and Goldfarb D.S.. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:129–141. 10.1091/mbc.e02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler T., and Mayer A.. 2000. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J. Cell Biol. 151:529–538. 10.1083/jcb.151.3.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., and Klionsky D.J.. 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279:29889–29894. 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji J.Y., Kikuma T., Arioka M., and Kitamoto K.. 2010. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS One. 5:e15650 10.1371/journal.pone.0015650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H.U., Yousefi S., Schmid I., and Friis R.. 2014. ATG5 can regulate p53 expression and activation. Cell Death Dis. 5:e1339 10.1038/cddis.2014.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S.W., Onodera J., and Ohsumi Y.. 2011. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One. 6:e17412 10.1371/journal.pone.0017412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., and Ohsumi Y.. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301–311. 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M., and Ohsumi Y.. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174. 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- Webster B.M., Thaller D.J., Jäger J., Ochmann S.E., Borah S., and Lusk C.P.. 2016. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 35:2447–2467. 10.15252/embj.201694574 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.