CCL25, CXCL10, and Mn2+ induce three distinct active conformations of integrin α4β7, which have selective high affinity for either MAdCAM-1, VCAM-1, or nonselective high affinity for both ligands. Via this mechanism, integrin α4β7 adopts different active conformations to switch its ligand-binding specificity.
Abstract
Chemokine (C-C motif) ligand 25 (CCL25) and C-X-C motif chemokine 10 (CXCL10) induce the ligand-specific activation of integrin α4β7 to mediate the selective adhesion of lymphocytes to mucosal vascular addressin cell adhesion molecule-1 (MAdCAM-1) or vascular cell adhesion molecule-1 (VCAM-1). However, the mechanism underlying the selective binding of different ligands by α4β7 remains obscure. In this study, we demonstrate that CCL25 and CXCL10 induce distinct active conformers of α4β7 with a high affinity for either MAdCAM-1 or VCAM-1. Single-cell force measurements show that CCL25 increases the affinity of α4β7 for MAdCAM-1 but decreases its affinity for VCAM-1, whereas CXCL10 has the opposite effect. Structurally, CCL25 induces a more extended active conformation of α4β7 compared with CXCL10-activated integrin. These two distinct intermediate open α4β7 conformers selectively bind to MAdCAM-1 or VCAM-1 by distinguishing their immunoglobulin domain 2. Notably, Mn2+ fully opens α4β7 with a high affinity for both ligands. Thus, integrin α4β7 adopts different active conformations to switch its ligand-binding specificity.
Introduction
The recruitment of lymphocytes from blood circulation to different tissues is essential for immune surveillance and host defense (Butcher and Picker, 1996). This recruitment process consists of a highly ordered adhesion cascade that includes the tethering and rolling of lymphocytes along the vessel walls of high endothelial venules, chemokine-induced activation, firm arrest, and extravasation. The initial tethering and rolling of lymphocytes on the endothelium are mediated by the adhesion of selectins and inactive α4 and β2 integrins with their ligands. Then, lymphocytes are stimulated by chemokines, triggering the activation of integrins to mediate cell firm arrest. Chemokines activate integrins by triggering an “inside-out signaling” that converts the inactive integrin (in a low-affinity bent conformation) into its active form, characterized by a high-affinity extended conformation (Takagi and Springer, 2002; Carman and Springer, 2003). EM and atomic structures of integrins have shown that the integrin extracellular domain exists in at least three distinct global conformational states: bent with a closed headpiece, extended with a closed headpiece, and extended with an open headpiece. The closed and open headpieces have a low and high affinity for the ligand, respectively. The equilibrium among these different states is regulated by integrin inside-out signaling (Beglova et al., 2002; Springer and Dustin, 2012). The transition from low-affinity to high-affinity integrin is accompanied by a series of conformational rearrangements including extension of the extracellular domain, a swing-out of the β-subunit hybrid domain and the attached plexin/semaphorin/integrin (PSI) domain, causing a 62° reorientation between the βI (βA) and hybrid domains, a 7-nm separation between the knees of the α and β legs (Kim et al., 2003; Xiao et al., 2004), and a rearrangement of the ligand-binding metal ion-dependent adhesion site (MIDAS) in the βI domain (Springer and Dustin, 2012).
The tissue specificity of lymphocyte homing is tightly controlled by adhesion between the homing molecules on lymphocytes and their specific ligands on the vascular endothelial cells of various tissues (Mora and von Andrian, 2006). However, most integrins on lymphocytes can recognize multiple ligands (Humphries et al., 2006), which may hinder lymphocyte trafficking to specific tissues. For example, integrin α4β7 is a lymphocyte homing receptor that can bind to two ligands, mucosal vascular addressin cell adhesion molecule-1 (MAdCAM-1) and vascular cell adhesion molecule-1 (VCAM-1), which are expressed in different tissues. The primary ligand for α4β7 is MAdCAM-1, which is specifically expressed on the endothelium of high endothelial venules in the gut and gut-associated lymphoid tissues such as Peyer’s patches (Springer, 1994; Berlin et al., 1995; Cox et al., 2010), whereas VCAM-1 is widely expressed on stimulated endothelial cells of blood vessels, peripheral lymph nodes, and bone marrow (Berlin-Rufenach et al., 1999). MAdCAM-1 and VCAM-1 both belong to the Ig superfamily. MAdCAM-1 contains two Ig domains and a mucin-like domain, whereas VCAM-1 is formed by seven Ig domains. They have been reported to bind α4β7 through their N-terminal two Ig domains (Pepinsky et al., 1992; Tan et al., 1998). The Ig domain 1 (D1) of MAdCAM-1 and VCAM-1 has a similar compact structure containing the key integrin-binding residue (Asp42 in MAdCAM-1 and Asp40 in VCAM-1) located on the protruding CD loop. However, Ig domain 2 (D2) of MAdCAM-1 and VCAM-1 is elongated by inserts in several interstrand loops. D2 of MAdCAM-1 contains a D strand and belongs to the I1 set. In contrast, VCAM-1 D2 lacks a D strand but contains an A′ strand and has been classified as a member of the I2 set. It is reported that D2 in MAdCAM-1 and VCAM-1 plays a role in determining integrin binding specificity (Newham et al., 1997).
Our previous study has revealed that chemokine (C-C motif) ligand 25 (CCL25) stimulation promotes α4β7-mediated lymphocyte adhesion to MAdCAM-1 but suppresses adhesion to VCAM-1, whereas C-X-C motif chemokine 10 (CXCL10) stimulation has the opposite effect (Sun et al., 2014). Mechanistically, CCL25 and CXCL10 activate the p38α MAPK–PKCα and c-Src–Syk pathways, respectively, which leads to different phosphorylation states of the β7 tail and distinct talin and kindlin-3 binding patterns, resulting in unique affinities of α4β7 for different ligands. Notably, CXCL10-induced activation of c-Src–Syk pathway is similar to the signaling mediated by P-selectin glycoprotein ligand-1, which induces an intermediate state of β2 integrins in neutrophils (Ma et al., 2004; Xu et al., 2007). Although the intracellular signaling that induces the ligand-specific activation of integrin α4β7 has been illustrated, the mechanism underlying the selective binding of the activated α4β7 integrins to different ligands is unclear.
In this study, using atomic force microcopy (AFM)–based single-cell force spectroscopy (SCFS; Benoit et al., 2000; Helenius et al., 2008; Zhang et al., 2009), we show that CCL25 stimulation of RPMI 8866–CXCR3 cells increases the single-molecule affinity of α4β7 for MAdCAM-1 but decreases its affinity for VCAM-1, whereas CXCL10 produces the opposite effect. In contrast, Mn2+-activated α4β7 shows maximum affinity for both ligands. Structurally, integrin intramolecular fluorescence lifetime imaging microscopy (FLIM)–fluorescence resonance energy transfer (FRET) studies reveal that CCL25 and CXCL10 stimulation induces two intermediate open active conformations of α4β7, whereas Mn2+ induces a fully open conformation of α4β7. Notably, CCL25-activated α4β7 has a more extended structure than CXCL10-activated integrin. Computational molecular dynamics (MD) simulation consistently identifies two distinct intermediate open conformers of the α4β7 headpiece with inverse binding free energy to the same ligand. Although the primary binding site for α4β7 is in D1 of MAdCAM-1 and VCAM-1, a swap of D2 in these two ligands reverses the ligand preference of CCL25- and CXCL10-activated α4β7 integrins, suggesting that D2 in MAdCAM-1 and VCAM-1 serves as the identity element distinguished by the two distinct intermediate open conformers of α4β7. Thus, CCL25, CXCL10, and Mn2+ induce three distinct active conformations of integrin α4β7, which selectively bind to either MAdCAM-1 or VCAM-1 or nonselectively bind to both ligands.
Results
CCL25 and CXCL10 trigger the ligand-specific regulation of α4β7 single-molecule affinity
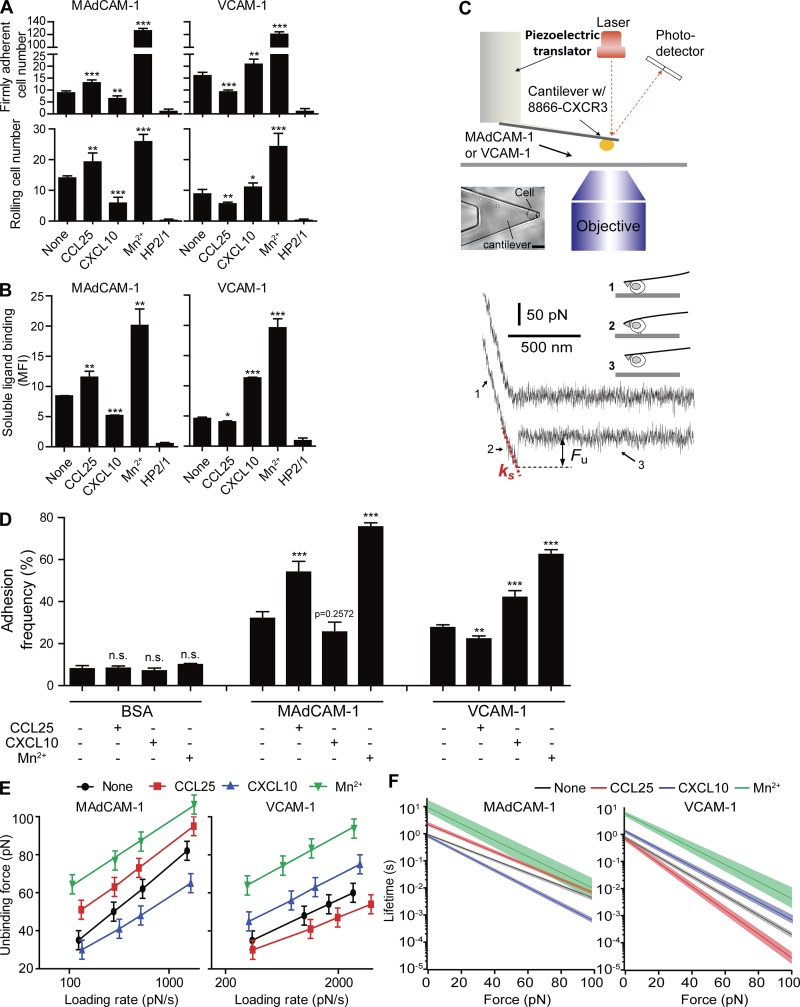
Our previous study has demonstrated that integrin α4β7 is activated by chemokines CCL25 and CXCL10 in a ligand-specific manner to mediate selective adhesion of lymphocytes to either MAdCAM-1 or VCAM-1 (Sun et al., 2014). Specifically, CCL25 stimulation significantly increased adhesion of RPMI 8866–CXCR3 cells, an integrin α4β7+/α4β1− human B lymphocyte cell line that expresses the CCL25 receptor CCR9 and the CXCL10 receptor CXCR3 (Fig. S1 A), to immobilized MAdCAM-1 substrates at a wall shear stress of 1 dyn/cm2 but suppressed cell adhesion to VCAM-1 substrates (Fig. 1 A). In contrast, CXCL10 increased cell adhesion to VCAM-1 but suppressed cell adhesion to MAdCAM-1. However, activation of α4β7 with 0.5 mM Mn2+ greatly increased the number of cells that adhered to both ligands. Cells treated with the α4 blocking antibody HP2/1, which recognizes epitope within residues 195–268 in the β-propeller domain of the α4 subunit, did not adhere to either ligand, indicating that this cell adhesion is mediated by integrin α4β7 (Fig. 1 A; Kamata et al., 1995). The binding of soluble MAdCAM-1 or VCAM-1 to α4β7 in response to chemokine stimulation showed consistent results (Fig. 1 B). These results indicate that CCL25 and CXCL10 induce distinct activation of α4β7 integrin, leading to the switch in α4β7 ligand specificity for MAdCAM-1 and VCAM-1. Notably, the opposite regulation of α4β7 adhesion by CCL25 and CXCL10 was not applicable to another α4β7 ligand fibronectin because both chemokines promoted cell adhesion to fibronectin splice variants (Fig. S2 A; Pankov and Yamada, 2002).
Figure 1.
Regulation of integrin α4β7–ligand binding affinity by chemokines and Mn2+. (A) Adhesion of RPMI 8866–CXCR3 cells to immobilized MAdCAM-1 and VCAM-1 substrates at 1 dyn/cm2 before and after stimulation with chemokines or 0.5 mM Mn2+. Data are represented as mean ± SD of technical quintuplicate samples (n = 5). (B) Binding of soluble MAdCAM-1 and VCAM-1 to RPMI 8866–CXCR3 cells before and after stimulation with chemokines or 0.5 mM Mn2+. Data are represented as mean ± SD of technical triplicate samples (n = 3). 2 µg/ml mAb HP2/1 was used to block the function of α4 integrin in A and B. MFI, mean fluorescence intensity. (C) AFM schematic. Top: Diagram illustrating key components of the custom-built setup. Bar, 20 µm. Bottom: Representative force-displacement retraction trace without (upper trace) or with (lower trace) adhesion. Fu, unbinding force of the α4β7–ligand complex. ks, system spring constant derived from the slope of the force-displacement trace. The cantilever retraction rate of the measurements was 3.7 µm/s. Bottom right: The three stages of stretching and detaching a single cell from the substrate. (D) Adhesion frequency of the AFM measurements for RPMI 8866–CXCR3 cells. Data are represented as mean ± SEM of >500 repeated force scans conducted using multiple cell-probe pairs. (E) Dynamic force spectra of single-bond MAdCAM-1 and VCAM-1 interactions with and without stimulation. Uncertainty in force is shown as half of the bin width of unbinding force histograms (Fig. S4, A and B). The linear fits of the DFS were obtained using Eq. 2. The fitted lines are superimposed on the respective DFS. (F) The force-dependent lifetimes of single-bond MAdCAM-1 and VCAM-1 interactions are given by Eq. 1. Shaded areas indicate an uncertainty of one SD. Two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., P > 0.05.
To gain insight into the molecular mechanism underlying the chemokine-induced switch in integrin α4β7 ligand specificity, we applied AFM-based SCFS technology to study the biophysical dynamics of chemokine-induced interaction between α4β7 and MAdCAM-1 or VCAM-1 at the single-molecule level. A single RPMI 8866–CXCR3 cell before or after chemokine stimulation was captured by a poly l-lysine–functionalized AFM cantilever (Fig. 1 C; Zhang et al., 2002; Helenius et al., 2008). This converted the live cell into a probe, which was brought into contact with MAdCAM-1− or VCAM-1−adsorbed surfaces. The cantilever was withdrawn at a constant speed, detaching the cell from its binding place, exhibiting two typical force–distance curves (Fig. 1 C). The lower curve revealed a linear increase in force followed by a single sharp transition that signaled the breakage of a single α4β7–ligand complex. Detachment forces may also stem from nonspecific interactions between cell and surface, or multiple α4β7–ligand bonds. To ensure specific interactions can be distinguished from nonspecific interactions, SCFS on a ligand-coated surface was compared with a surface blocked by BSA. A significant decrease in adhesion frequency to 10% or less occurred when α4β7 ligand was absent (BSA-coated surface), suggesting that the vast majority of the recorded unbinding forces were from specific α4β7–ligand interactions (Fig. 1 D). Nonspecific interactions can be clearly distinguished from specific interactions from their smaller magnitudes (Fig. S3). Moreover, when the loading rate increases, the magnitude of nonspecific interactions did not change significantly, whereas specific α4β7–ligand–unbinding force increased with loading rates (Fig. S4, A and B). Similarly, multiple interactions can be distinguished from single bonds from their multiple sequential unbinding events (Fig. S3 B). In the rare case that multiple bonds ruptured at the same time, unbinding of multiple bonds was reflected as a very small population of forces with greater magnitudes on the unbinding force histograms (Fig. S4, A and B).
To ensure measurement of a single-molecule interaction, the contact force and contact time between the cell and ligand-coated surface was optimized to reach an adhesion frequency of 30–40% in the force measurements (Fig. 1 D). Assuming that the adhesion bond formation obeyed Poisson statistics, an adhesion frequency of ∼33% in the force measurements implies that among the observed unbinding events, the probabilities of forming single, double, and triple adhesion bonds between the cell and surface were 81%, 16%, and 2%, respectively (Chesla et al., 1998). Therefore, our experimental conditions ensured a >80% probability that the adhesion event was mediated by a single bond (Evans, 2001), and only the single-bond unbinding force was used for further data analysis. Under the same contact force and time, the frequency of α4β7−MAdCAM-1 adhesion increased from 31% to 55% after CCL25 stimulation but decreased to 25% after CXCL10 stimulation (Fig. 1 D). Conversely, the frequency of α4β7−VCAM-1 adhesion decreased from 28% to 22% after CCL25 treatment but increased to 43% after CXCL10 treatment under the same conditions. Activation of α4β7 with 0.5 mM Mn2+ greatly increased cell adhesion frequency to both ligands (Fig. 1 D). When adhesion frequency was >40%, we further decreased the contact force and time to lower the adhesion frequency to ∼33%.
First, the force distribution for unbinding of the single-bond α4β7–ligand complexes before or after chemokine stimulation under a series of increasing loading rates was obtained (Fig. S4). In general, the force distribution was shifted toward higher values with increasing loading rates. The unbinding forces of the single-bond α4β7−MAdCAM-1 complexes increased linearly with the logarithm of the loading rate, ranging from 35 to 80 pN over a loading rate of ∼100 to ∼1,500 pN/s (Fig. 1 E). Upon stimulation with CCL25, α4β7−MAdCAM-1 complexes exhibited significantly enhanced unbinding forces compared with those without chemokine treatment. In contrast, CXCL10 treatment significantly lowered the unbinding forces under similar loading rates, indicating that CCL25 and CXCL10 influence the mechanical strength of α4β7−MAdCAM-1 complexes in opposite ways. Compared with MAdCAM-1, CCL25 and CXCL10 had opposite effects on the unbinding forces of α4β7−VCAM-1 interactions. Activation of α4β7 with 0.5 mM Mn2+ greatly increased the unbinding forces of both α4β7−MAdCAM-1 and α4β7−VCAM-1 complexes.
A more detailed analysis of single-bond α4β7–ligand dissociation properties was conducted by fitting the acquired dynamic force spectrum (DFS) data to the Bell-Evans model (Evans and Ritchie, 1997). The force-dependent lifetime of single-bond α4β7−MAdCAM-1 and α4β7−VCAM-1 interactions (Eq. 1 and Fig. 1 F) showed an increased lifetime for CCL25-activated α4β7−MAdCAM-1 complexes but a decreased lifetime for CXCL10-activated α4β7−MAdCAM-1 complexes compared with those without chemokine treatment. In contrast, CCL25 and CXCL10 had opposite effects on the bond lifetime of α4β7−VCAM-1 interactions. Mn2+ stimulation increased the force-dependent lifetime of single-bond α4β7−MAdCAM-1 and α4β7−VCAM-1 interactions. Moreover, the Bell-Evans model (Eq. 2) yielded a dissociation rate in the absence of force (k0) and an activation barrier width (γ; Table S1). Stimulation with CCL25 resulted in a lower α4β7−MAdCAM-1 dissociation rate constant (k0) but a higher k0 for the α4β7−VCAM-1 complex, whereas CXCL10 had the opposite effect.
In addition to using the Bell-Evans model, we also fitted the unbinding forces at different loading rates to the Dudko-Hummer-Szabo model, which supports catch bond (Dudko, 2009). The resulting lifetimes of α4β7–ligand bonds for unstimulated, CCL25-treated, and CXCL10-treated cells were comparable with the Bell-Evans model fit (Figs. 1 F and S4, C and D). However, there is no indication that α4β7–ligand formed catch bonds under the loading rates tested in the current study (Fig. S4, C and D).
Collectively, these data demonstrate that CCL25 and CXCL10 induce distinct ligand-specific alterations of the single-bond affinity of α4β7 for MAdCAM-1 and VCAM-1, consistent with the effects of chemokines on cell adhesion in flow (Fig. 1 A) and soluble ligand binding (Fig. 1 B).
CCL25 and CXCL10 induce distinct global conformational changes of integrin α4β7
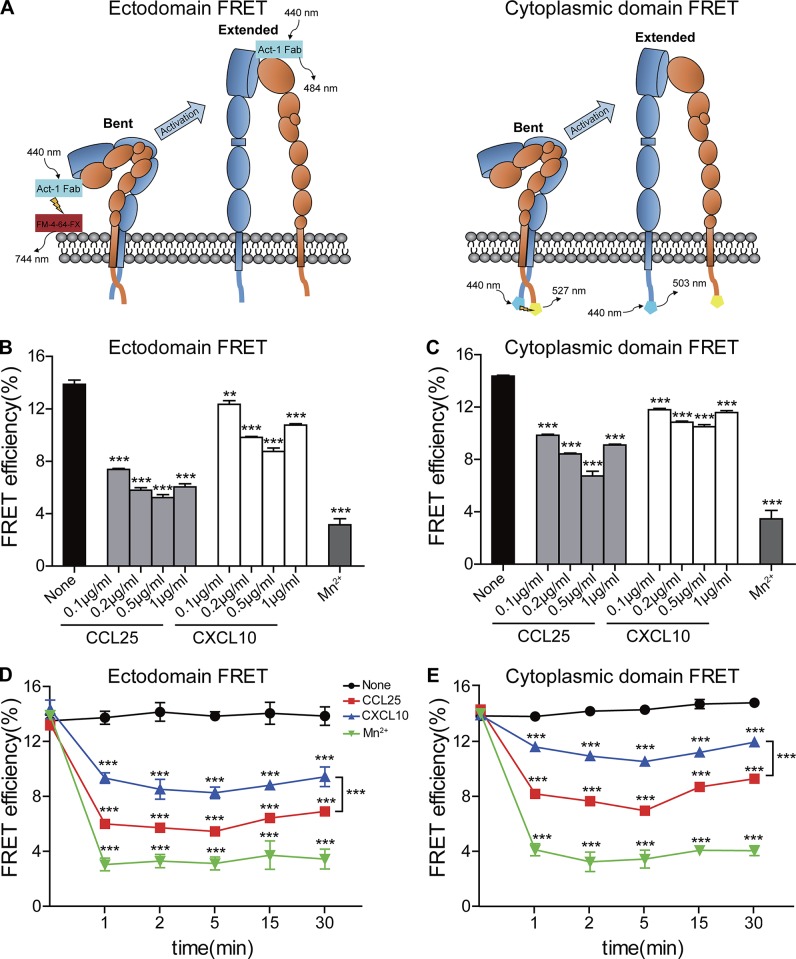
Upon activation, integrins change from a low-affinity to a high-affinity state, which is associated with extension of the ectodomain and separation of the α/β cytoplasmic tails (Adair and Yeager, 2002; Takagi and Springer, 2002; Ye et al., 2008; Lindert et al., 2009; Campbell and Humphries, 2011). To investigate the conformational changes of integrin α4β7 triggered by CCL25 and CXCL10, we used two integrin intramolecular FLIM-FRET systems to examine the influence of chemokines on α4β7 ectodomain extension and tail separation on the plasma membrane of live RPMI 8866–CXCR3 cells, respectively (Fig. 2 A).
Figure 2.
Integrin α4β7 conformational changes induced by chemokines and Mn2+. (A) Experiment setup for measuring FRET efficiency between integrin α4β7 βI domain and the plasma membrane (ectodomain FRET) and FRET efficiency between integrin α4 and β7 cytoplasmic domains (cytoplasmic domain FRET). A composite of all molecules used is depicted. (B and C) Ectodomain FRET efficiency (B) and cytoplasmic domain FRET efficiency (C) in cells before and after treatment with indicated concentration of chemokines or 0.5 mM Mn2+. (D and E) Ectodomain FRET efficiency (D) and cytoplasmic domain FRET efficiency (E) in cells over a 30-min time course of treatment with 0.5 µg/ml chemokines or 0.5 mM Mn2+. Data are represented as mean ± SEM of 6–12 cells per condition (n = 6–12). Two-tailed Student’s t test. **, P < 0.01; ***, P < 0.001.
Act-1 mAb recognizes human integrin α4β7 heterodimer and binds to the top of the interface between α4-subunit β-propeller and β7-subunit βI domain (Yu et al., 2012), which makes it an ideal antibody for labeling the top head of α4β7. To assess the orientation of the α4β7 ectodomain relative to the plasma membrane, the top head domain of α4β7 was labeled with Atto 425–Act-1 Fab fragment as the FRET donor, and the cell membrane was labeled with a lipophilic probe, FM4-64 FX (FM), as the FRET acceptor (Fig. 2 A; Chigaev et al., 2007; Xiong et al., 2009; Pan et al., 2010). The fluorescence lifetime of Atto 425–Act-1 Fab was monoexponential with a time constant of ∼3.493 ± 0.017 ns (mean ± SD; Table S2), and it fitted well with a biexponential function in the presence of acceptor. RPMI 8866–CXCR3 cells were pretreated with increasing concentration of CCL25 or CXCL10 or 0.5 mM Mn2+ for 5 min. Compared with unstimulated RPMI 8866–CXCR3 cells, CCL25 or CXCL10 treatment induced a significantly lower FRET signal (Fig. 2 B), suggesting that the α4β7 head domain moves away from the cell membrane after chemokine stimulation. Notably, cells treated with CCL25 showed a lower FRET efficiency than CXCL10-treated cells regardless of the chemokine concentration used, suggesting that CCL25 stimulation induces a more extended conformation of the α4β7 ectodomain than CXCL10 treatment independent of chemokine dose. Cells treated with 0.5 mM Mn2+ showed the lowest FRET signal. These data suggest that CCL25 stimulation induces a more extended conformation of the α4β7 ectodomain than CXCL10 treatment, whereas Mn2+ induces the most extended conformation. In the range of 0.1–1 µg/ml chemokines, the middle concentration of 0.5 µg/ml CCL25 or CXCL10 induced the most extension of α4β7 (Fig. 2 B), which is consistent with previous studies reporting that a high level of chemokines may attenuate its chemotactic activity (Grimm et al., 1998; Sordi et al., 2005).
To examine the effects of chemokines on the separation of integrin α4β7 cytoplasmic tails, we established α4β7 knockout (KO) RPMI 8866–CXCR3 cells and stably reexpressed the α4 and β7 subunits with mTurquoise2 and mCitrine fused at the C termini of their cytoplasmic domains, respectively (Figs. 2 A and S1 B; Kim et al., 2003; Hyun et al., 2009; Pan et al., 2010; Bajar et al., 2016). The fluorescence lifetime of mTurquoise2-α4 was monoexponential with a time constant of ∼3.988 ± 0.022 ns (mean ± SD; Table S2), which fitted well with a biexponential function in the presence of acceptor. CCL25 or CXCL10 treatment significantly decreased the FRET signals in those cells (Fig. 2 C), suggesting separation of the α4 and β7 cytoplasmic domains. In the range of 0.1–1 µg/ml chemokines, CCL25 induced a lower FRET efficiency than CXCL10 (Fig. 2 C), suggesting that CCL25 stimulation induces more separation of α4β7 cytoplasmic domains than CXCL10 treatment independent of chemokine dose. In addition, Mn2+ induces the lowest FRET signal (Fig. 2 C). These data suggest that CCL25 stimulation induces more separation of α4β7 cytoplasmic domains than CXCL10 treatment, whereas Mn2+ induces the most separation of integrin tails. Interestingly, αIIbβ3 has been reported to exist in partially extended intermediate open conformations without separation of the lower legs (Xu et al., 2016). The difference in cytoplasmic domain separation of the intermediate open α4β7 and αIIbβ3 may be a result of the intrinsic difference between the two integrins, including stronger interactions between αIIbβ3 leg domains (Kamata et al., 2005) and dissimilar energy barrier between different conformational states among different integrins (Askari et al., 2009).
To investigate whether the different conformational changes induced by CCL25 and CXCL10 are affected by chemokine treatment time, cells were pretreated with 0.5 µg/ml chemokines for 1–30 min (Fig. 2, D and E; and Table S2). Consistently with the results above, CCL25 stimulation induces more extension of ectodomain and separation of cytoplasmic domains in integrin α4β7 than CXCL10 treatment regardless of chemokine treatment time, whereas Mn2+ induces the most conformational changes. It is noteworthy that chemokine-induced conformer-specific activation of α4β7 can last for at least 30 min, which is consistent with the previous study reporting that integrin α4β7 exhibits sustained activation upon chemokine treatment (Sun et al., 2014).
Collectively, CCL25 and CXCL10 induce two distinct intermediate open integrin α4β7 conformers with a high affinity specifically for either MAdCAM-1 or VCAM-1. Mn2+ induces a fully open conformation with a nonselective high affinity for both ligands.
Two intermediate conformers of the α4β7 headpiece show inverse binding free energy to the same ligand
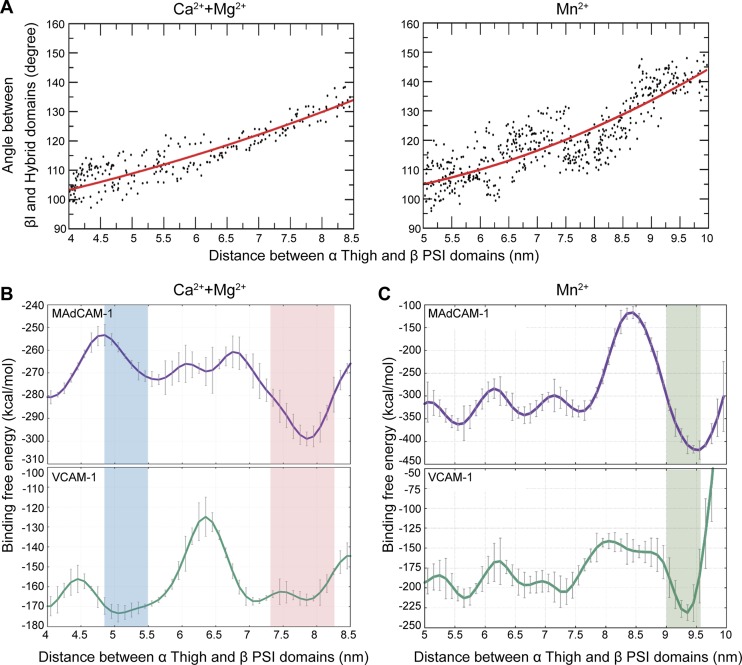
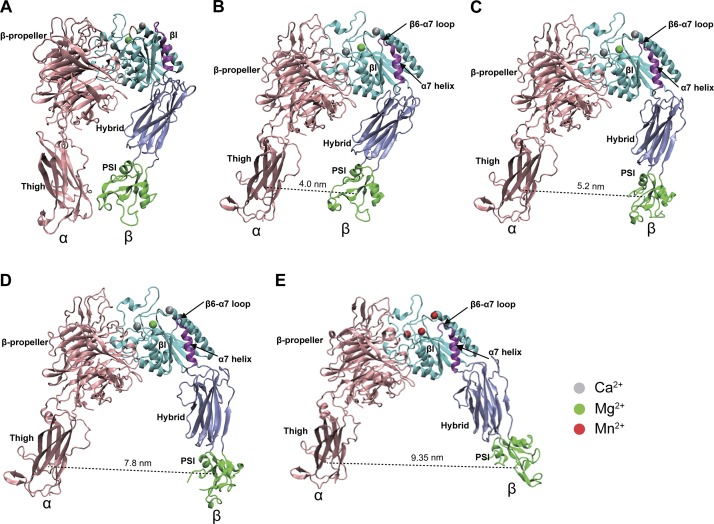
To further investigate the correlation between different active conformations of integrin α4β7 and the ligand-specific affinity regulation by chemokines or Mn2+, we applied MD simulations to identify the binding free energy of MAdCAM-1 and VCAM-1 for the α4β7 headpiece during its transition from the closed to open conformation. Because the α4β7 headpiece structure (PDB ID 3V4P; Yu et al., 2012) does not contain a PSI domain, we first used the MODELLER (Sali et al., 1995) package with the α4β7 headpiece and PSI-containing αIIbβ3 headpiece (PDB ID 3NID; Zhu et al., 2010) crystal structures as a template to construct the homology model of the five-domain headpiece of α4β7 containing the β-propeller and Thigh domains (residues 1–586) of the α4 subunit and the βI, hybrid, and PSI domains (residues 41–503) of the β7 subunit. Superposition of the initial five-domain α4β7 headpiece structure and αIIbβ3 bent, closed ectodomain structure (PDB ID 3FCS; Zhu et al., 2008) indicated that the initial α4β7 headpiece structure for MD simulation to begin with was completely bent (Fig. S5, A–C). MAdCAM-1 and VCAM-1 D1-D2 structures were used as the ligands in MD simulations because D1-D2 contains essential α4β7 binding interfaces that mediate efficient α4β7 binding (Newham et al., 1997). The MAdCAM-1 and VCAM-1 binding modes were determined by the ZDOCK (Pierce et al., 2014) program and were further equilibrated using a two-domain α4β7 headpiece fragment containing the β-propeller domain (residues 1–428) of the α4 subunit and the βI domain (residues 152–391) of the β7 subunit in MD simulations. Next, the ligand-bound five-domain α4β7 headpieces were obtained by superimposing the equilibrated two-domain α4β7 headpieces to the five-domain model of α4β7 in MD simulations. Because the distance between the α Thigh and β PSI/hybrid domains has been generally used to define the conformational changes during integrin activation (Springer and Dustin, 2012; Zhu et al., 2013), the distance between the centers of mass of the α Thigh domain and β PSI domain was selected as a collective variable (CV) for biasing the conformational changes of the ligand-bound α4β7 headpieces, and adiabatic bias molecular dynamic (ABMD; Marchi and Ballone, 1999) simulations were used to drive the conformational transition of α4β7 from the closed to open state. The virtual atoms used to define the distance between the Thigh and PSI domains are shown in Table S7. The secondary structure elements of the α4β7 headpiece during MD simulations showed no significant changes (Fig. S5 D). During the transition of the ligand-bound α4β7 headpieces from the closed to open state in Ca2+ + Mg2+ condition, which are associated with a 4.0–8.5–nm separation between the Thigh and PSI domains and a swing-out of hybrid domain with the angle between the βI and hybrid domains changing from acute to obtuse (Fig. 3 A), the binding free energy profiles showed an inverse binding free energy of the α4β7 headpiece to MAdCAM-1 and VCAM-1 along with conformational transition (Fig. 3 B). Compared with the closed α4β7 headpiece, the first intermediate open conformer of α4β7, with a distance between the Thigh and PSI domains from 4.85 to 5.5 nm, showed lower binding free energy to VCAM-1 but higher binding free energy to MAdCAM-1 (Fig. 3 B), suggesting that this intermediate open conformer of α4β7 has increased ligand-binding affinity for VCAM-1 and decreased affinity for MAdCAM-1. In contrast with the first intermediate open conformer of α4β7, a second intermediate open α4β7 conformer with a Thigh and PSI distance from 7.3 to 8.25 nm showed inverse binding free energy to MAdCAM-1 and VCAM-1, suggesting decreased ligand-binding affinity for VCAM-1 and increased affinity for MAdCAM-1. During the transition of the ligand-bound α4β7 headpieces from the closed to open state in Mn2+ condition, an open α4β7 conformer with a distance between the Thigh and PSI domains from 9.0 to 9.57 nm showed significantly decreased binding free energy to MAdCAM-1 and VCAM-1 (Fig. 3 C), suggesting that Mn2+ induces a fully open conformation with a nonselective high affinity for both ligands. Moreover, this open α4β7 conformer exhibited lower binding free energy compared with the chemokine-induced intermediate open conformers (Fig. 3, B and C). These results are consistent with the chemokine- and Mn2+-induced changes in ligand-binding affinity and conformation of α4β7. Compared with the closed integrin α4β7 headpiece structure from the MD simulation, two intermediate open α4β7 conformers with inverse binding free energy to MAdCAM-1 and VCAM-1 and a fully open α4β7 conformer in Mn2+ condition with lowest binding free energy for both ligands showed the downward movement of the α7 helix and reshaping of the β6–α7 loop in the β7I domain and the change of the angle between the βI and hybrid domains from acute to obtuse (Fig. 4), which are the major conformational changes in integrin β head domain associated with integrin activation (Yang et al., 2004; Springer and Dustin, 2012).
Figure 3.
MD simulation of the binding free energy of MAdCAM-1 and VCAM-1 for the α4β7 headpiece during its transition from the closed to open conformation. (A) Relationship between the Thigh and PSI distance and βI and hybrid domain–domain angle in Ca2+ + Mg2+ and Mn2+ conditions. (B and C) Binding free energy profiles of the α4β7 headpiece to MD1-MD2 (top) and VD1-VD2 (bottom) in Ca2+ + Mg2+ condition (B) and Mn2+ condition (C). Twenty intermediate states with 500 snapshots each (a total of 10,000 conformations) were used to evaluate the binding free energy using the MM/GBSA method to obtain statistically meaningful results. Data are represented as mean ± SD per condition.
Figure 4.
Integrin α4β7 headpiece structures from the MD simulation. (A) The initial structure of integrin α4β7 headpiece for MD simulation. (B–D) Snapshots of integrin α4β7 headpiece structures in closed, low-affinity state (B), intermediate state with a distance of 5.2 nm between the Thigh and PSI domains (C), and intermediate state with a distance of 7.8 nm between the Thigh and PSI domains (D). MIDAS was occupied by Mg2+, and ADMIDAS and SyMBS were occupied by Ca2+. (E) Snapshot of integrin α4β7 headpiece structure in fully open active state with a distance of 9.35 nm between the Thigh and PSI domains. MIDAS, ADMIDAS, and SyMBS were occupied by Mn2+. The three metal ion binding sites are SyMBS, MIDAS, and ADMIDAS from left to right.
CCL25- and CXCL10-activated α4β7 integrins distinguish MAdCAM-1 and VCAM-1 by recognizing their D2
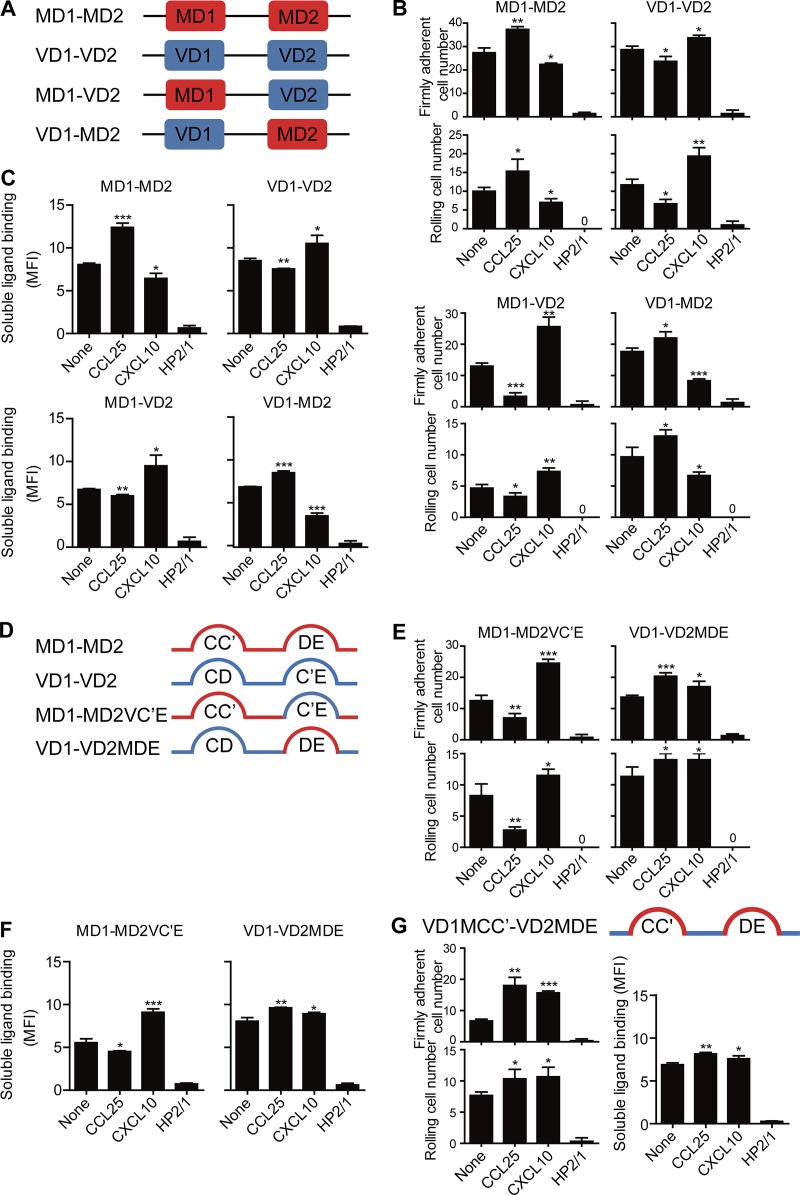
Next, we aimed to determine the structural elements in MAdCAM-1 and VCAM-1 responsible for the opposite ligand preference of CCL25- and CXCL10-activated α4β7 integrins. Although the essential integrin-binding motif locates in D1 of MAdCAM-1 and VCAM-1, D2 is also required for efficient integrin binding (Newham et al., 1997; Green et al., 1999). Therefore, we generated recombinant D1-D2 of human MAdCAM-1 (MD1-MD2; Val1 to Ser204 numbering without signal peptide) and VCAM-1 (VD1-VD2; Phe1 to Tyr196 numbering without signal peptide) proteins with a C-terminal–fused Fc region (CH2 and CH3 domains) of human IgG1 (Fig. 5 A). Consistent with the results of chemokine-induced cell adhesion to full-length MAdCAM-1 and VCAM-1 in flow (Fig. 1 A), CCL25 significantly increased adhesion of RPMI 8866–CXCR3 cells to immobilized MD1-MD2 at a wall shear stress of 1 dyn/cm2 but suppressed cell adhesion to VD1-VD2 substrates, whereas CXCL10 showed the opposite effect (Fig. 5 B). The binding of soluble MD1-MD2 or VD1-VD2 to α4β7 in response to chemokine stimulation showed consistent results (Fig. 5 C). Thus, D1-D2 of MAdCAM-1 and VCAM-1 is sufficient to mediate the ligand-specific adhesion to CCL25- and CXCL10-activated α4β7 integrins.
Figure 5.
Effect of the domain swap and loop swap mutations in MAdCAM-1 and VCAM-1 on α4β7 adhesion. (A) Schematic diagram of MAdCAM-1 and VCAM-1 constructs. MD1-MD2, VD1-VD2, and chimeric proteins containing MD1-VD2 or VD1-MD2. (B) Adhesion of RPMI 8866–CXCR3 cells to immobilized MAdCAM-1/VCAM-1 D1-D2 proteins and MAdCAM-1/VCAM-1 D1-D2 proteins with swapped D2 at 1 dyn/cm2 before and after chemokine stimulation. (C) Binding of soluble MAdCAM-1/VCAM-1 D1-D2 proteins and MAdCAM-1/VCAM-1 D1-D2 proteins with swapped D2 to RPMI 8866–CXCR3 cells before and after stimulation with chemokines. (D) Schematic diagram of MAdCAM-1 and VCAM-1 loop swap constructs. The EEEPQGDED motif in the MAdCAM-1 DE loop and the DADRKSLET motif in the VCAM-1 C′E loop were swapped to generate chimeric MAdCAM-1 (MD1-MD2VC′E) and VCAM-1 (VD1-VD2MDE) proteins with swapped C′E and DE loops. (E) Adhesion of RPMI 8866–CXCR3 cells to the loop-swapped MAdCAM-1 and VCAM-1 substrates at 1 dyn/cm2 before and after chemokine stimulation. (F) Binding of soluble loop-swapped MAdCAM-1/VCAM-1 D1-D2 proteins to RPMI 8866–CXCR3 cells before and after stimulation with chemokines. (G) Adhesion of RPMI 8866–CXCR3 cells to the CD and C′E loop-swapped VCAM-1 substrate at 1 dyn/cm2 before and after chemokine stimulation (left). Binding of soluble loop-swapped VCAM-1 D1-D2 protein to RPMI 8866–CXCR3 cells before and after stimulation with chemokines (right). 2 µg/ml mAb HP2/1 was used to block the function of α4 integrin. MFI, mean fluorescence intensity. Data are represented as mean ± SD of technical triplicate samples (n = 3). Two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further investigate the roles of D1 and D2 in mediating the ligand-specific adhesion to CCL25- and CXCL10-activated α4β7 integrins, we swapped the D2 of MAdCAM-1 and VCAM-1 to generate chimeric proteins containing MAdCAM-1 D1 and VCAM-1 D2 (MD1-VD2) or VCAM-1 D1 and MAdCAM-1 D2 (VD1-MD2; Fig. 5 A). Strikingly, CCL25 and CXCL10 induced cell adhesion patterns on MD1-VD2 substrates similar to those observed on VD1-VD2 substrates (Fig. 5 B), suggesting that replacement of MD2 with VD2 changed MAdCAM-1 to VCAM-1 in terms of the ligand-binding preference of CCL25- and CXCL10-activated α4β7. On VD1-MD2 substrates, chemokines induced cell adhesion patterns similar to those observed on MD1-MD2 (Fig. 5 B), suggesting that replacing VD2 with MD2 converted the identity of VCAM-1 to MAdCAM-1 for chemokine-activated α4β7. Also, the binding of soluble MD1-VD2 or VD1-MD2 to α4β7 in response to chemokine stimulation showed consistent results (Fig. 5 C). Thus, a swap of D2 in MAdCAM-1 and VCAM-1 reverses the ligand preference by CCL25- and CXCL10-activated α4β7. These data indicate that D2 in MAdCAM-1 and VCAM-1 is the structural element for the opposite ligand preference of CCL25- and CXCL10-activated α4β7 integrins.
The VCAM-1 C′E loop and the MAdCAM-1 DE loop are critical structural elements for ligand preference of chemokine-activated α4β7
The C′E loop in VD2 and the DE loop in MD2 contain regulatory residues adjacent to integrin contact sites and play a role in regulating α4β7 binding (Newham et al., 1997; Green et al., 1999; Sun et al., 2011). To investigate the contribution of these loops in the ligand-binding preference of CCL25- and CXCL10-activated α4β7 integrins, we swapped the EEEPQGDED motif of the MAdCAM-1 DE loop and the DADRKSLET motif of the VCAM-1 C′E loop to generate chimeric MAdCAM-1 (MD1-MD2VC′E) and VCAM-1 (VD1-VD2MDE) proteins with swapped C′E and DE loops (Fig. 5 D). Interestingly, a flow chamber assay showed similar adhesion patterns of RPMI 8866–CXCR3 cells on MD1-MD2VC′E and VD1-VD2 substrates at the wall shear stress of 1 dyn/cm2 after chemokine treatment (Fig. 5, B and E). The binding of soluble MD1-MD2VC′E to α4β7 in response to chemokine stimulation showed consistent results (Fig. 5 F). These data indicate that replacement of the DE loop in MAdCAM-1 with the VCAM-1 C′E loop changed MAdCAM-1 to VCAM-1 in terms of the ligand-binding preference of CCL25- and CXCL10-activated α4β7. For VCAM-1 D1-D2 with the C′E loop replaced by the MAdCAM-1 DE loop, CCL25-treated RPMI 8866–CXCR3 cells showed increased cell adhesion to VD1-VD2MDE substrates in flow, similar to cell adhesion to MAdCAM-1 (Fig. 5, B and E), suggesting that CCL25-activated α4β7 recognizes VCAM-1 as MAdCAM-1 after replacing the C′E loop in VCAM-1 with the MAdCAM-1 DE loop. However, CXCL10-treated cells still showed increased adhesion to VD1-VD2MDE, which was different from the decreased cell adhesion to MAdCAM-1 after CXCL10 treatment. The binding of soluble VD1-VD2MDE to α4β7 in response to chemokine stimulation showed consistent results (Fig. 5 F). Thus, replacement of the C′E loop in VCAM-1 with the MAdCAM-1 DE loop converted the identity of VCAM-1 to MAdCAM-1 only for CCL25-activated α4β7, suggesting that structural elements in addition to the DE loop in MD2 are required for the ligand preference of CXCL10-activated α4β7. Next, we replaced both the CD loop in VD1 and the C′E loop in VD2 with the CC′ loop and DE loop in MAdCAM-1 to generate VD1MCC′-VD2MDE (Fig. 5 G). VD1MCC′-VD2MDE showed similar cell adhesion and soluble ligand binding results as VD1-VD2MDE, suggesting that structural elements in addition to the CC′ and DE loops in MAdCAM-1 are required for the ligand preference of CXCL10-activated α4β7 (Fig. 5, E–G). Collectively, these results demonstrate that the C′E and DE loops are major “fingerprint” structural elements in VCAM-1 and MAdCAM-1 that are recognized by CCL25- or CXCL10-activated α4β7 integrins to support distinct ligand-specific adhesion.
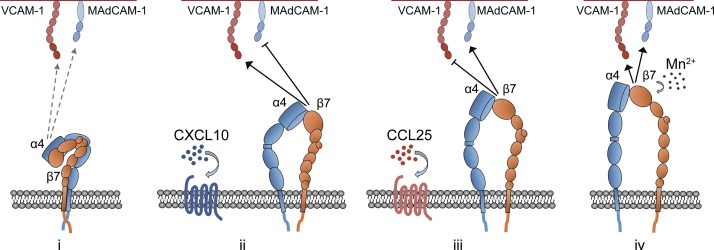
Collectively, CCL25 and CXCL10 trigger the switch in ligand specificity of integrin α4β7 by inducing two unique intermediate open conformers of α4β7 integrins, which have opposite ligand preference by distinguishing D2, especially the DE and C′E loops, in MAdCAM-1 and VCAM-1. Moreover, Mn2+ induces maximal activation of α4β7, which shows nonpreferable high-affinity binding to both ligands (Fig. 6). Thus, our findings demonstrate that CCL25, CXCL10, and Mn2+ induce three distinct active conformations of integrin α4β7, which have selective high affinity for either MAdCAM-1, VCAM-1, or nonselective high affinity for both ligands.
Figure 6.
Schematic representation of chemokine- or Mn2+-induced integrin α4β7 conformers with distinct ligand-binding specificity. (i) The resting integrin α4β7 with closed conformation binds to MAdCAM-1 and VCAM-1 in low affinity. CXCL10 (ii) and CCL25 (iii) induce two different intermediate open conformers of α4β7, which show selective binding to VCAM-1 or MAdCAM-1, respectively. (iv) Mn2+-induced fully open conformer of α4β7 shows nonselective high-affinity binding to both ligands.
Discussion
In this study, we demonstrate that CCL25, CXCL10, and Mn2+ induce three distinct active conformations of integrin α4β7, each of which has a unique ligand-binding preference. Our previous study reports that CCL25 and CXCL10 activate the p38α MAPK–PKCα and c-Src–Syk pathways, respectively, which leads to ligand-specific activation of α4β7. Interestingly, P-selectin glycoprotein ligand-1 signaling via selectins can activate Src family kinases and Syk in neutrophils, which is similar to CXCL10 signaling in our study, and induces an intermediate active state of β2 integrins (Ma et al., 2004; Stadtmann et al., 2013). Moreover, integrin αLβ2 (lymphocyte function-associated antigen-1) has been reported to show conformer-specific activation regulated by a chemokine-triggered Rho signaling module (Bolomini-Vittori et al., 2009). Inhibition of small GTPase Rac1 converts CXCL12-induced high-affinity αLβ2 to low-intermediate open conformation, whereas inhibition of Cdc42 activity induces a more open conformation of αLβ2 than CXCL12-stimulated αLβ2. These data demonstrate that β2 integrins may also be differentially activated and have different activate states.
Using AFM-based SCFS, we quantified the mechanical strength between α4β7 and MAdCAM-1 or VCAM-1 at the single-bond level. The force spectra of the α4β7−MAdCAM-1/VCAM-1 interactions provided insight into the dissociation pathway of the complexes. We elected to use SCFS (Benoit, 2002; Helenius et al., 2008) because this assay can closely resemble the flow chamber assay, in which live lymphocytes interact with a surface coated with either MAdCAM-1 or VCAM-1. Taking advantage of the AFM’s high force sensitivity, the SCFS may reveal the biophysical and molecular insights that can hardly be detected by ensemble measurements. The single-bond unbinding forces ranged from 32 to 80 pN for the α4β7−MAdCAM-1 complexes and from 30 to 60 pN for the α4β7−VCAM-1 complexes under loading rates ranging from ∼100 to ∼2,700 pN/s. The loading rate range was chosen to cover the estimated physiological loading rates (i.e., 125–2,500 pN/s) of cellular tethered bonds in the vasculature (Rinko et al., 2004). Our analysis of the unbinding of the α4β7–ligand complex using the Bell-Evans model (Eq. 2) clearly suggested distinct biophysical characters of the α4β7−MAdCAM-1 and α4β7−VCAM-1 complexes reflected by changes in the dissociation rate constant (k0) of the bonds (Table S1). For the α4β7−MAdCAM-1 complex, the k0 was 1.03 s−1 for the unstimulated complex and 0.40 s−1 for the high-affinity complex stimulated by CCL25, whereas their transition state positions (γ) showed a slight increase. These results indicate that CCL25 stimulation resulted in a significant decrease in the dissociation rate by 2.5-fold. Upon CXCL10 treatment, although the k0 value of 1.17 s−1 increased slightly, the γ value increased significantly from 2.19 Å to 2.93 Å. The longer barrier width suggests that the α4β7−MAdCAM-1 complex is less resistant to mechanical pulling (Fig. 1 F). In contrast, we obtained opposite results for the α4β7−VCAM-1 complex; the k0 was 1.58 s−1 for the unstimulated complex and 0.83 s−1 for the high-affinity complex stimulated by CXCL10, whereas their positions of the transition state (γ) showed little difference. However, CCL25 resulted in a small increase in k0 and a greater increase in γ. These results indicate that the unstressed dissociation rates of α4β7−MAdCAM-1 and α4β7−VCAM-1 bonds that allow cell rolling are 0.40 to 1.17 s−1 and 0.83 to 1.75 s−1, respectively, which share similar unstressed dissociation rates of selectin–ligand bonds (0.22–1.4 s−1; Hanley et al., 2004; Klopocki et al., 2008). Moreover, Mn2+ induced a maximal decrease in k0 values for both the α4β7−MAdCAM-1 and α4β7−VCAM-1 bonds, suggesting indiscriminate high-affinity binding to both ligands.
Integrin activation is associated with integrin molecule extension coupled with hybrid domain swing-out and separation of the α/β leg domains (Takagi et al., 2002; Kim et al., 2003). The distance between the α Thigh and β I-EGF1 domains ranges from 4.5 to 12 nm along with the integrin headpiece changing from a closed to a fully open conformation (Springer and Dustin, 2012). In our MD simulation system, we defined a reference point of the binding free energy at the position with a distance of 4.0 nm between the Thigh and PSI domains, which represents the closed state of the ligand-bound integrin headpiece. The distance between α Thigh domain and β PSI domain from 4.0 nm to 10.0 nm represents the conformational transition from a closed headpiece to an open headpiece.
An interesting experiment using MAdCAM-1 and VCAM-1 with swapped D1 has shown that the chimeric protein VD1–MD2–mucin–α4β7 interaction is abolished by a α4β7−MAdCAM-1−specific blocking antibody, Act-1, whereas Act-1 shows no inhibitory function on MD1–VD2,3,7–α4β7 binding (Green et al., 1999), suggesting that α4β7 has a distinct binding interface with MAdCAM-1 and VCAM-1 D2. Furthermore, another study has shown that α4β7 has an accessory binding site located in VCAM-1 D2, and the analogous site in MAdCAM-1 D2 is markedly different in size and sequence, which has a role in the determination of integrin binding specificity (Newham et al., 1997). In our study, we showed that D2 of MAdCAM-1 and VCAM-1 is the crucial structural element distinguished by CCL25- and CXCL10-activated α4β7, thus mediating selective adhesion to different ligands (Fig. 5, B and C).
It has been reported that DE loop in MAdCAM-1 D2 and the C′E loop in VCAM-1 D2 contribute to the integrin binding specificity (Newham et al., 1997). The DE loop and the C′E loop contain negatively charged motifs, which are prominent and contribute to the concentration of the electrostatic potential surface of these proteins (Jones et al., 1995; Tan et al., 1998). Moreover, residues from these loops are located on the same face as the primary integrin-binding Asp residue in CC′ and CD loop in D1 of MAdCAM-1 and VCAM-1, respectively, and are reported to be involved in α4β7 binding (Tan et al., 1998; Wang and Springer, 1998). Our results showed that swap of DE and C′E loop in MAdCAM-1 and VCAM-1 completely switched the identity of both ligands for CCL25-activated α4β7. However, for CXCL10-activated α4β7, it only switched the identity of MAdCAM-1 to VCAM-1 but failed to switch the identity of VCAM-1 to MAdCAM-1, suggesting that structural elements in addition to the DE loop in MAdCAM-1 are required for the ligand-binding preference of CXCL10-activated α4β7. It is noteworthy that most key regulatory residues responsible for ligand specificity in VCAM-1 D2 belong to the C′E loop, whereas a region in MAdCAM-1 D2 (residues 143–150) beyond the DE loop also contributes to integrin binding (Newham et al., 1997; Green et al., 1999). Thus, other residues besides the DE loop in MAdCAM-1 D2 may contribute to the ligand-binding preference of CXCL10-activated α4β7.
Although D1-D2 of MAdCAM-1 and VCAM-1 is sufficient to bind α4β7 (Green et al., 1999), the mucin-like domain of MAdCAM-1 and the remaining five Ig domains in VCAM-1 are believed to extend the integrin-binding domains well above the cell surface for efficient integrin binding (Tan et al., 1998). Indeed, compared with cell adhesion to full-length MAdCAM-1 and VCAM-1 substrates (Fig. 1 A), our results showed that RPMI 8866–CXCR3 cells displayed similar adhesive behaviors but a lower number of adherent cells when the same concentration of MAdCAM-1 and VCAM-1 D1-D2 proteins was used (Fig. S2 B). Thus, a higher concentration of MAdCAM-1 and VCAM-1 D1-D2 proteins was used to achieve a comparable number of adherent cells in the flow assay (Fig. 5 B).
Our study demonstrates that integrin α4β7 can undergo conformer-specific activation to adopt different active conformational states physiologically. More importantly, each of the active conformers has a unique ligand-binding preference, leading to the switch in integrin ligand specificity to precisely regulate the tissue specificity of lymphocyte homing. Thus, different integrin intermediate open states can be physiologically induced and stably exist, which have diverse biological functions other than simply lower ligand-binding affinities compared with fully activated integrin.
Materials and methods
Antibodies and reagents
Human CCL25 and CXCL10 were from R&D Systems. The antibodies used were to human α4 (HP2/1; Abcam), human CCR9 (557975; BD), and human CXCR3 (CD183; 550633; BD). Mouse mAb 9F10 against human α4, rat mAb FIB504 against human β7, and rat mAb AIIB2 against human β1 were prepared by using hybridomas (Developmental Studies Hybridoma Bank). Alexa Fluor 647–conjugated goat anti–mouse IgG (A21236), FITC-conjugated goat anti–rat IgG (629511), and FITC-conjugated goat anti–mouse IgG (626511) were from Invitrogen. Natural plasma fibronectin (F2006) was from Sigma-Aldrich. Fibronectin CS1 peptide (3624-FN-050) was from R&D Systems. Fibronectin alternatively spliced domain A (EDA) segment (ab187877) was from Abcam. Act-1 against human α4β7 was as previously described (Sun et al., 2014). Atto 425 N-hydroxysuccinimide (16805) was from Sigma-Aldrich.
cDNAs and cell lines
cDNAs of human α4 and β7 subunits were constructed in vector pcDNA3.1/Hygro(–; Invitrogen). cDNAs encoding human α4 and β7 subunits with C-terminal–fused mTurquoise2 and mCitrine, respectively, were constructed in vector pCDH-puro (Invitrogen). Human integrin α4 and β7 single-guide RNA (sgRNA) designed by GN20GG rule (Ran et al., 2013) were constructed into vector lentiCRISPR v2 (Addgene). The 20-nucleotide sgRNA sequence was 5′-GAGCTGTTCGCACGTCTGGC-3′ for the ITGA4 gene and 5′-GCGGCGCTGCGCCCGACGAG-3′ for the ITGB7 gene. The sgRNA-resistant point mutations were generated using QuikChange (Agilent Technologies) in WT α4β7 with or without C-terminal–fused mTurquoise2 and mCitrine. An RPMI 8866 cell line stably expressing CXCR3 (RPMI 8866–CXCR3) was generated by electroporation of CXCR3 cDNA followed by selection using puromycin (2 µg/ml).
AFM-based SCFS
Single-cell force measurements on integrin α4β7–ligand interactions were conducted using a custom-built AFM as described earlier (Fu et al., 2015). The custom-built setup was used to measure rupture forces between the MAdCAM-1 or VCAM-1 (20 µl of 10 µg/ml) coated surfaces and a single RPMI 8866–CXCR3 cell picked up via interaction with the triangular area of the C-cantilever (MLCT microlever probes; Bruker Nano). For cells stimulated with chemokines or Mn2+, the MAdCAM-1 concentration was decreased to 5 µg/ml and 2.5 µg/ml, respectively. The cantilevers were calibrated using a thermal fluctuation method (Hutter and Bechhoefer, 1993). The spring constants (12 ± 3 pN/nm) of the calibrated cantilevers agreed with the values specified by the manufacturer. For cells without stimulation, a contact force of 200 pN and contact time of 0.2 s were used. For cells pretreated with chemokines or Mn2+, contact force and duration were lowered to 100 pN and 0.08 s to ensure the detection of single-molecule interactions. All measurements for the chemokine-stimulated cell were recorded within 1 h after chemokine stimulation. Force–distance curves were recorded and analyzed using IGOR Pro software (Wave Metrics).
Fitting the acquired DFS data to the Bell-Evans model
According to this model, a pulling force f distorts the intermolecular potential of a ligand–receptor complex, leading to a lowering of the activation energy and an increase in the dissociation rate k(f),
| (1) |
where k0 is the dissociation rate constant in the absence of a pulling force, γ is the position of the transition rate, T is the absolute temperature, and kb is the Boltzmann’s constant. For a constant loading rate rf, the model can be described as
| (2) |
Fitting the acquired DFS data to the Dudko-Hummer-Szabo model
Lifetimes (τ) for RPMI 8866–CXCR3 cell−MAdCAM-1 or RPMI 8866–CXCR3 cell−VCAM-1 interactions as a function of the applied force F were obtained by transforming the histograms of unbinding force under different loading rates. For each histogram, N is the total number of bins of width Let the number of counts in the bin be and then the total of counts is , resulting in the probability and the density
Thus, the force-dependent lifetime is
where and is the loading rate (Kim et al., 2010; Zhang et al., 2015).
MD simulation
The first two domains (D1-D2) of MAdCAM-1 and VCAM-1 contain essential α4β7 binding interfaces to mediate efficient integrin α4β7 binding. The primary interaction between α4β7 and ligands forms between Mg2+ or Mn2+ at the MIDAS site of the β7I domain and Asp42 in MAdCAM-1 D1 or Asp40 in VCAM-1 D1 (Newham et al., 1997; Zhang and Chen, 2012). The MAdCAM-1 and VCAM-1 binding modes were identified using the two-domain α4β7 headpiece fragment containing the α4 β-propeller domain (residues 1–428) and the β7I domain (residues 152–391, with Mg2+ at MIDAS and Ca2+ at adjacent to MIDAS (ADMIDAS) and synergistic metal ion-binding site (SyMBS) in Ca2+ + Mg2+ condition; and with Mn2+ at MIDAS, ADMIDAS, and SyMBS in Mn2+ condition) extracted from the integrin α4β7 closed headpiece structure (PDB ID 3V4P; Yu et al., 2012). The MAdCAM-1 D1-D2 structure (PDB ID 1GSM; Dando et al., 2002) was used as a ligand to perform rigid-body docking around the two-domain α4β7 headpiece fragment. A total of 2,000 predictions were generated using the ZDOCK program, and the possible binding modes were determined by measuring the distance between MAdCAM-1 Asp42 (CG atom) and the Mg2+ or Mn2+ at MIDAS followed by equilibration using MD simulations. The initial VCAM-1 binding mode was obtained by superimposing VCAM-1 (PDB ID 1IJ9; Taylor et al., 2001) D2 to MAdCAM-1 D2 after 100 ns MD simulations of the MAdCAM-1–bound complex, and then 100 ns MD equilibrations of the VCAM-1–bound complex were performed with an upper boundary wall potential (restricted within 3 Å) between VCAM-1 Asp40 (CG atom) and the Mg2+ or Mn2+ at MIDAS. Finally, the MAdCAM-1– and VCAM-1–bound five-domain α4β7 headpieces were obtained by superimposing these equilibrated two-domain α4β7 headpiece fragments in aforesaid MD simulations to the five-domain α4β7 model structure containing the β-propeller and Thigh domains (residues 1–586) of the α4 subunit and the βI, hybrid, and PSI domains (residues 41–503) of the β7 subunit.
The initial complexes for MD simulations were first handled using the pdb2gmx module in the Gromacs (Van Der Spoel et al., 2005) package to add missing hydrogens and detect disulfide bridges and protonation states of titratable residues. For the two-domain α4β7 headpiece fragments, the conformations were centered into a 12.5 × 11.0 × 8.0 nm rectangle box, and dissolved with 32,347 TIP3P water molecules. The five-domain α4β7 headpiece fragments were centered into a rectangle box with the size of 11.9 × 17.8 × 15.9 nm, and dissolved with 109,023 TIP3P water molecules. Subsequently, 0.1 M NaCl ions were added to neutralize the net charge of the whole system, which yields the final system containing a total of 110,208 atoms for the two-domain complex and 346,103 atoms for the five-domain complex, respectively.
The MD simulations were performed using Gromacs (5.0.4; Abraham et al., 2015) with the CHARMM36 force field (MacKerell et al., 1998). The steepest descent algorithm was used to minimize the whole system before it was gradually heated to 300 K with a position restraint potential to the protein heavy atoms. The leapfrog integrator was used with an integration time-step of 2 fs under substance/volume/temperature conditions. The modified Berendsen (V-rescale; Berendsen et al., 1984) thermostat was used to control the temperature of the systems at 300 K with a time constant of 1 ps. The Particle Mesh Ewald method (Darden et al., 1993) was used to compute the electrostatic interactions with a real-space cutoff distance of 1 nm. The same cutoff value was chosen for treating the van der Waals interactions. The SETTLE algorithm (Miyamoto and Kollman, 1992) was used to constrain water molecules, and all nonwater bonds were constrained using the LINCS algorithm (Hess et al., 1997).
To guarantee that the intermediate states were sufficiently sampled during the conformational changes, five rounds of ABMD simulations were performed by driving the distance CV from the closed state to open state. 20 MAdCAM-1– and VCAM-1–bound intermediates, respectively, were selected from ABMD simulations to evaluate the ligand-binding affinity differences during the conformational changes. These 40 intermediates were used as initial conformations to conduct MD simulations. A harmonic restraint (75 kJ/mol/nm2) was exerted on each intermediate state to maintain the distance CV. Each MD simulation lasts for 150 ns, and a total of 3 µs MD trajectories were aggregated for each ligand. The last 50 ns of 20 intermediate states with 500 snapshots each (a total of 10,000 conformations) were used to evaluate binding free energy using the molecular mechanics/generalized born surface area (MM/GBSA) method (Genheden and Ryde, 2015). The dielectric constant ε = 4 is used for the protein in MM/GBSA calculations.
Flow chamber assay
The flow chamber assay was performed as described (Chen et al., 2004). A polystyrene Petri dish was coated with 20 μl of MAdCAM-1/Fc, VCAM-1/Fc (20 µg/ml) or MAdCAM-1, VCAM-1 D1-D2/Fc (80 µg/ml) alone, or chemokines (2 µg/ml) in coating buffer (PBS and10 mM NaHCO3, pH 9.0) for 1 h at 37°C followed by blocking with 2% BSA in coating buffer for 1 h at 37°C. Cells were diluted to 1 × 106 cells/ml in HBSS (10 mM Hepes) containing different divalent cations (1 mM Ca2+ + Mg2+ for unstimulated and chemokine-treated conditions, and 0.5 mM Mn2+ for Mn2+-treated conditions) and immediately perfused through the flow chamber at a constant flow of 1 dyn/cm2. For the MAdCAM-1/VCAM-1 domain swap or loop swap mutants, the polystyrene Petri dish was coated with 20 μl of 80 µg/ml chimeric ligand. For the fibronectin splice variants, the polystyrene Petri dish was coated with 20 μl of 40 µg/ml plasma fibronectin or 80 µg/ml CS1 peptide and EDA fragment. All adhesive interactions between the flowing cells and the coated substrates were determined by manually tracking the motions of individual cells for 1 min as previously described (Sun et al., 2014). The motion of each adherent cell was monitored for 10 s after the initial adhesion point, and two categories of cell adhesion were defined. Adhesion was defined as rolling if the adherent cells were followed by rolling motions ≥5 s with a velocity of at least 1 µm/s, whereas a firmly adherent cell was defined as a cell that remained adherent and stationary for at least 10 s.
Soluble ligand binding assay
Soluble ligand binding assay was performed as described (Sun et al., 2014). RPMI 8866–CXCR3 cells were diluted in HBSS (10 mM Hepes) containing different divalent cations (1 mM Ca2+ + Mg2+ for unstimulated and chemokine-treated conditions and 0.5 mM Mn2+ for Mn2+-treated condition). Cells before and after chemokine (0.5 µg/ml) or Mn2+ stimulation were fixed with paraformaldehyde (3.7%). Then, 50 µg/ml MAdCAM-1−his−Alexa Fluor 647 fusion protein or VCAM-1−his−Alexa Fluor 647 fusion protein was added to the mixture and incubated for 30 min at RT. For the MAdCAM-1/VCAM-1 domain swap or loop swap mutants, 200 µg/ml ligand-his-FITC fusion proteins were used. Next, cells were washed twice and measured using a FACSCalibur (BD) and analyzed using FlowJo software (TreeStar).
FLIM-FRET assay
FLIM-FRET assay was performed as described (Askari et al., 2010). FLIM utilizes only the donor fluorescence, thus avoiding the problem of misexcitation of the acceptor, and can determinate the FRET efficiency and the binding fraction of the FRET pairs independent of the fluorophores’ concentration (Xiong et al., 2009; Takahashi et al., 2015). For detecting the orientation of integrin ectodomain relative to cell membrane, cells were stained with 20 µg/ml Atto 425–conjugated Act-1 Fab for 40 min at 37°C. After two washes, cells were labeled with 10 µM FM4-64 FX (Invitrogen) for 4 min on ice, and washed once. To estimate whether the chemokine induced ligand-specific activation of α4β7 depends on distinct chemokine dose, we performed a chemokine dose–response FRET assay with Hepes-buffered saline (1 mM Ca2+ + Mg2+) in the plate containing varying levels of chemokines (0.1–1 µg/ml) within 5 min. To analyze the time-response of chemokine and Mn2+-induced integrin α4β7 global conformational changes, cells were incubated with or without 0.5 µg/ml soluble chemokines or 0.5 mM Mn2+ for 1–30 min at 37°C. For detecting the association of integrin cytoplasmic tails, α4-mTurquoise2/β7-Citrine RPMI 8866–CXCR3 cells were treated as above. FRET was detected and quantified by FLIM using a (time domain) time correlated single photon counting approach. The inverted laser-scanning microscope Nikon A1 with a 60× oil immersion 1.4 NA Plan Apochromat objective equipped with a 440-nm pulsed laser (Picoquant) tuned at 20 MHz and single-photon counting electronics (PicoHarp 300) were used to excite the donor alone (Atto 425–Act-1 Fab/α4-mTurquoise2) and the donor in the presence of acceptor (Atto 425–Act-1 Fab + FM4-64 FX/α4-mTurquoise2 + β7-mCitrine). The emitted photons passed through a 482/35-nm band-pass filter and were detected with a PMA hybrid detector (Picoquant). FLIM data were measured until 200 photons per pixel were collected. The acquired fluorescence decays coming from regions of interest comprising the cell membrane were tail-fitted using Symphotime 64 software (Picoquant) with one- and two-exponential theoretical models in both donor alone (Atto 425–Act-1 Fab/α4-mTurquoise2) and the donor in the presence of acceptor (Atto 425–Act-1 Fab + FM4-64 FX/α4-mTurquoise2 + β7-mCitrine) assays. The reduced χ2 parameter was used to judge the goodness of fit, which was deemed acceptable for 0.8 < χ2 < 1.2 (Tables S3, S4, S5, and S6). Fluorescent intensity decays were suitably fitted to a one-exponential decay model where acceptor was absent and a two-exponential model when both donor and acceptor were present to extract mean lifetimes. For two-exponential fits, the lifetimes were the weighted mean of the two fitted lifetime components. The donor lifetime obtained from a single exponential fit from cells (∼3.493 ns of Atto 425–Act-1 Fab and ∼3.988 ns of α4-mTurquoise2) expressing the donor alone was used for the noninteracting fraction of the double exponential model in the corresponding cotransfected cell. The relative FRET efficiency was calculated as
| (3) |
where τ is the mean lifetime obtained from the exponential fit of the decay curve of the donor (Atto 425/mTurquoise2) alone (τD) or of the donor in the presence of the acceptor (FM4-64 FX/mCitrine; τDA).
Statistical analysis
Statistical significance was determined by two-tailed Student’s t test using PRISM software (5.00, GraphPad Software; Figs. 1, 2, 5, and S2; and Tables S1 and S2). The resulting p-values are indicated as follows: n.s., P > 0.05; *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; and ***, P < 0.001. To judge the goodness of one- and two-exponential theoretical fits of fluorescent intensity decays in FLIM-FRET, χ2 analyses were done using Symphotime 64 software (Picoquant; Tables S3 and S4). To compare the fits, the extra sum-of-squares F test was applied using SPSS software (version 19; IBM SPSS for Windows; Tables S3, S4, S5, and S6). For parametric tests, data distribution was assumed to be normal but was not formally tested.
Online supplemental material
Fig. S1 shows the expression of integrins and chemokine receptors on the surfaces of different RPMI 8866–CXCR3 cell lines. Fig. S2 shows adhesion of RPMI 8866–CXCR3 cells to immobilized fibronectin splice variants and MAdCAM-1/VCAM-1 (D1-D2; 20 µg/ml) in flow before and after treatment with chemokines or Mn2+. Fig. S3 shows representative single force–distance (retraction) curves and force histograms of forces between a RPMI 8866–CXCR3 cell and a BSA-coated or MAdCAM-1−coated surface. Fig. S4 presents unbinding force histograms of RPMI 8866–CXCR3 cell−MAdCAM-1 and VCAM-1 interactions and the analysis of specific unbinding forces with the Dudko-Hummer-Szabo model. Fig. S5 shows the structures of the α4β7 headpiece. Table S1 shows Bell-Evans model parameters of α4β7−MAdCAM-1 and α4β7−VCAM-1 complexes. Table S2 shows the time-response lifetimes of FRET donor in absence and presence of acceptor. Tables S3 and S4 show the χ2 values for one- and two-exponential fits of fluorescent intensity decays in ectodomain FRET and cytoplasmic domain FRET, respectively. Tables S5 and S6 show the maximum studentized residuals for one- and two-exponential fits of fluorescent intensity decays in ectodomain FRET and cytoplasmic domain FRET, respectively. Table S7 shows the virtual atoms used for defining the distance between Thigh and PSI domains and the angle between βI and hybrid domains in integrin α4β7 headpiece.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31525016, 21625302, and 31471309), the National Basic Research Program of China (2014CB541905), the American Heart Association (11SDG5420008), Personalized Medicines-Molecular Signature-Based Drug Discovery and Development, the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA12010101), and the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs International Partnership Program for Creative Research Teams. The authors acknowledge the support of the Sanofi-Aventis Shanghai Institutes for Biological Science scholarship program.
The authors declare no competing financial interests.
Author contributions: S. Wang, G. Ge, G. Li, X.F. Zhang, and J. Chen designed experiments; S. Wang, C. Wu, Y. Zhang, Q. Zhong, H. Sun, and W. Cao performed experiments and analyzed data; S. Wang, G. Li, X.F. Zhang, and J. Chen interpreted results; and the manuscript was drafted by S. Wang, X.F. Zhang, and Y. Zhang and edited by J. Chen.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19-25. http://www.sciencedirect.com/science/article/pii/S2352711015000059.
- Adair B.D., and Yeager M.. 2002. Three-dimensional model of the human platelet integrin alpha IIbbeta 3 based on electron cryomicroscopy and x-ray crystallography. Proc. Natl. Acad. Sci. USA. 99:14059–14064. 10.1073/pnas.212498199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askari J.A., Buckley P.A., Mould A.P., and Humphries M.J.. 2009. Linking integrin conformation to function. J. Cell Sci. 122:165–170. 10.1242/jcs.018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askari J.A., Tynan C.J., Webb S.E., Martin-Fernandez M.L., Ballestrem C., and Humphries M.J.. 2010. Focal adhesions are sites of integrin extension. J. Cell Biol. 188:891–903. 10.1083/jcb.200907174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajar B.T., Wang E.S., Zhang S., Lin M.Z., and Chu J.. 2016. A Guide to Fluorescent Protein FRET Pairs. Sensors (Basel). 16:E1488 10.3390/s16091488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglova N., Blacklow S.C., Takagi J., and Springer T.A.. 2002. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nat. Struct. Biol. 9:282–287. 10.1038/nsb779 [DOI] [PubMed] [Google Scholar]
- Benoit M. 2002. Cell adhesion measured by force spectroscopy on living cells. Methods Cell Biol. 68:91–114. 10.1016/S0091-679X(02)68006-9 [DOI] [PubMed] [Google Scholar]
- Benoit M., Gabriel D., Gerisch G., and Gaub H.E.. 2000. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2:313–317. 10.1038/35014000 [DOI] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., and Haak J.R.. 1984. Molecular-Dynamics with Coupling To an External Bath. J. Chem. Phys. 81:3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Berlin C., Bargatze R.F., Campbell J.J., von Andrian U.H., Szabo M.C., Hasslen S.R., Nelson R.D., Berg E.L., Erlandsen S.L., and Butcher E.C.. 1995. alpha 4 integrins mediate lymphocyte attachment and rolling under physiologic flow. Cell. 80:413–422. 10.1016/0092-8674(95)90491-3 [DOI] [PubMed] [Google Scholar]
- Berlin-Rufenach C., Otto F., Mathies M., Westermann J., Owen M.J., Hamann A., and Hogg N.. 1999. Lymphocyte migration in lymphocyte function-associated antigen (LFA)-1-deficient mice. J. Exp. Med. 189:1467–1478. 10.1084/jem.189.9.1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolomini-Vittori M., Montresor A., Giagulli C., Staunton D., Rossi B., Martinello M., Constantin G., and Laudanna C.. 2009. Regulation of conformer-specific activation of the integrin LFA-1 by a chemokine-triggered Rho signaling module. Nat. Immunol. 10:185–194. 10.1038/ni.1691 [DOI] [PubMed] [Google Scholar]
- Butcher E.C., and Picker L.J.. 1996. Lymphocyte homing and homeostasis. Science. 272:60–66. 10.1126/science.272.5258.60 [DOI] [PubMed] [Google Scholar]
- Campbell I.D., and Humphries M.J.. 2011. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3:a004994 10.1101/cshperspect.a004994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman C.V., and Springer T.A.. 2003. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 15:547–556. 10.1016/j.ceb.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Chen J., Takagi J., Xie C., Xiao T., Luo B.H., and Springer T.A.. 2004. The relative influence of metal ion binding sites in the I-like domain and the interface with the hybrid domain on rolling and firm adhesion by integrin alpha4beta7. J. Biol. Chem. 279:55556–55561. 10.1074/jbc.M407773200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesla S.E., Selvaraj P., and Zhu C.. 1998. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys. J. 75:1553–1572. 10.1016/S0006-3495(98)74074-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A., Waller A., Zwartz G.J., Buranda T., and Sklar L.A.. 2007. Regulation of cell adhesion by affinity and conformational unbending of alpha4beta1 integrin. J. Immunol. 178:6828–6839. 10.4049/jimmunol.178.11.6828 [DOI] [PubMed] [Google Scholar]
- Cox D., Brennan M., and Moran N.. 2010. Integrins as therapeutic targets: lessons and opportunities. Nat. Rev. Drug Discov. 9:804–820. 10.1038/nrd3266 [DOI] [PubMed] [Google Scholar]
- Dando J., Wilkinson K.W., Ortlepp S., King D.J., and Brady R.L.. 2002. A reassessment of the MAdCAM-1 structure and its role in integrin recognition. Acta Crystallogr. D Biol. Crystallogr. 58:233–241. 10.1107/S0907444901020522 [DOI] [PubMed] [Google Scholar]
- Darden T., York D., and Pedersen L.. 1993. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums In Large Systems. J. Chem. Phys. 98:10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Dudko O.K. 2009. Single-molecule mechanics: new insights from the escape-over-a-barrier problem. Proc. Natl. Acad. Sci. USA. 106:8795–8796. 10.1073/pnas.0904156106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. 2001. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128. 10.1146/annurev.biophys.30.1.105 [DOI] [PubMed] [Google Scholar]
- Evans E., and Ritchie K.. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541–1555. 10.1016/S0006-3495(97)78802-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Xu Y., Wu C., Moy V.T., and Zhang X.F.. 2015. Anchorage-dependent binding of integrin I-domain to adhesion ligands. J. Mol. Recognit. 28:385–392. 10.1002/jmr.2453 [DOI] [PubMed] [Google Scholar]
- Genheden S., and Ryde U.. 2015. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10:449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N., Rosebrook J., Cochran N., Tan K., Wang J.H., Springer T.A., and Briskin M.J.. 1999. Mutational analysis of MAdCAM-1/alpha4beta7 interactions reveals significant binding determinants in both the first and second immunuglobulin domains. Cell Adhes. Commun. 7:167–181. 10.3109/15419069909010800 [DOI] [PubMed] [Google Scholar]
- Grimm M.C., Ben-Baruch A., Taub D.D., Howard O.M., Resau J.H., Wang J.M., Ali H., Richardson R., Snyderman R., and Oppenheim J.J.. 1998. Opiates transdeactivate chemokine receptors: Δand μ opiate receptor-mediated heterologous desensitization. J. Exp. Med. 188:317–325. 10.1084/jem.188.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley W.D., Wirtz D., and Konstantopoulos K.. 2004. Distinct kinetic and mechanical properties govern selectin-leukocyte interactions. J. Cell Sci. 117:2503–2511. 10.1242/jcs.01088 [DOI] [PubMed] [Google Scholar]
- Helenius J., Heisenberg C.P., Gaub H.E., and Muller D.J.. 2008. Single-cell force spectroscopy. J. Cell Sci. 121:1785–1791. 10.1242/jcs.030999 [DOI] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., and Fraaije J.G.E.M.. 1997. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [DOI] [Google Scholar]
- Humphries J.D., Byron A., and Humphries M.J.. 2006. Integrin ligands at a glance. J. Cell Sci. 119:3901–3903. 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter J.L., and Bechhoefer J.. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:1868–1873. 10.1063/1.1143970 [DOI] [Google Scholar]
- Hyun Y.M., Chung H.L., McGrath J.L., Waugh R.E., and Kim M.. 2009. Activated integrin VLA-4 localizes to the lamellipodia and mediates T cell migration on VCAM-1. J. Immunol. 183:359–369. 10.4049/jimmunol.0803388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E.Y., Harlos K., Bottomley M.J., Robinson R.C., Driscoll P.C., Edwards R.M., Clements J.M., Dudgeon T.J., and Stuart D.I.. 1995. Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution. Nature. 373:539–544. 10.1038/373539a0 [DOI] [PubMed] [Google Scholar]
- Kamata T., Puzon W., and Takada Y.. 1995. Identification of putative ligand-binding sites of the integrin alpha 4 beta 1 (VLA-4, CD49d/CD29). Biochem. J. 305:945–951. 10.1042/bj3050945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T., Handa M., Sato Y., Ikeda Y., and Aiso S.. 2005. Membrane-proximal alpha/beta stalk interactions differentially regulate integrin activation. J. Biol. Chem. 280:24775–24783. 10.1074/jbc.M409548200 [DOI] [PubMed] [Google Scholar]
- Kim J., Zhang C.Z., Zhang X., and Springer T.A.. 2010. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 466:992–995. 10.1038/nature09295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Carman C.V., and Springer T.A.. 2003. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 301:1720–1725. 10.1126/science.1084174 [DOI] [PubMed] [Google Scholar]
- Klopocki A.G., Yago T., Mehta P., Yang J., Wu T., Leppänen A., Bovin N.V., Cummings R.D., Zhu C., and McEver R.P.. 2008. Replacing a lectin domain residue in L-selectin enhances binding to P-selectin glycoprotein ligand-1 but not to 6-sulfo-sialyl Lewis x. J. Biol. Chem. 283:11493–11500. 10.1074/jbc.M709785200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindert S., Silvestry M., Mullen T.M., Nemerow G.R., and Stewart P.L.. 2009. Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J. Virol. 83:11491–11501. 10.1128/JVI.01214-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.Q., Plow E.F., and Geng J.G.. 2004. P-selectin binding to P-selectin glycoprotein ligand-1 induces an intermediate state of alphaMbeta2 activation and acts cooperatively with extracellular stimuli to support maximal adhesion of human neutrophils. Blood. 104:2549–2556. 10.1182/blood-2004-03-1108 [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Marchi M., and Ballone P.. 1999. Adiabatic bias molecular dynamics: A method to navigate the conformational space of complex molecular systems. J. Chem. Phys. 110:3697–3702. 10.1063/1.478259 [DOI] [Google Scholar]
- Miyamoto S., and Kollman P.A.. 1992. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13:952–962. 10.1002/jcc.540130805 [DOI] [Google Scholar]
- Mora J.R., and von Andrian U.H.. 2006. T-cell homing specificity and plasticity: new concepts and future challenges. Trends Immunol. 27:235–243. 10.1016/j.it.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Newham P., Craig S.E., Seddon G.N., Schofield N.R., Rees A., Edwards R.M., Jones E.Y., and Humphries M.J.. 1997. Alpha4 integrin binding interfaces on VCAM-1 and MAdCAM-1. Integrin binding footprints identify accessory binding sites that play a role in integrin specificity. J. Biol. Chem. 272:19429–19440. 10.1074/jbc.272.31.19429 [DOI] [PubMed] [Google Scholar]
- Pan Y., Zhang K., Qi J., Yue J., Springer T.A., and Chen J.. 2010. Cation-pi interaction regulates ligand-binding affinity and signaling of integrin alpha4beta7. Proc. Natl. Acad. Sci. USA. 107:21388–21393. 10.1073/pnas.1015487107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R., and Yamada K.M.. 2002. Fibronectin at a glance. J. Cell Sci. 115:3861–3863. 10.1242/jcs.00059 [DOI] [PubMed] [Google Scholar]
- Pepinsky B., Hession C., Chen L.L., Moy P., Burkly L., Jakubowski A., Chow E.P., Benjamin C., Chi-Rosso G., Luhowskyj S., and Lobb R.. 1992. Structure/function studies on vascular cell adhesion molecule-1. J. Biol. Chem. 267:17820–17826. [PubMed] [Google Scholar]
- Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., and Weng Z.. 2014. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 30:1771–1773. 10.1093/bioinformatics/btu097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinko L.J., Lawrence M.B., and Guilford W.H.. 2004. The molecular mechanics of P- and L-selectin lectin domains binding to PSGL-1. Biophys. J. 86:544–554. 10.1016/S0006-3495(04)74133-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Potterton L., Yuan F., van Vlijmen H., and Karplus M.. 1995. Evaluation of comparative protein modeling by MODELLER. Proteins. 23:318–326. 10.1002/prot.340230306 [DOI] [PubMed] [Google Scholar]
- Sordi V., Malosio M.L., Marchesi F., Mercalli A., Melzi R., Giordano T., Belmonte N., Ferrari G., Leone B.E., Bertuzzi F., et al. 2005. Bone marrow mesenchymal stem cells express a restricted set of functionally active chemokine receptors capable of promoting migration to pancreatic islets. Blood. 106:419–427. 10.1182/blood-2004-09-3507 [DOI] [PubMed] [Google Scholar]
- Springer T.A. 1994. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 76:301–314. 10.1016/0092-8674(94)90337-9 [DOI] [PubMed] [Google Scholar]
- Springer T.A., and Dustin M.L.. 2012. Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24:107–115. 10.1016/j.ceb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmann A., Germena G., Block H., Boras M., Rossaint J., Sundd P., Lefort C., Fisher C.I., Buscher K., Gelschefarth B., et al. 2013. The PSGL-1-L-selectin signaling complex regulates neutrophil adhesion under flow. J. Exp. Med. 210:2171–2180. 10.1084/jem.20130664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Wu Y., Qi J., Pan Y., Ge G., and Chen J.. 2011. The CC’ and DE loops in Ig domains 1 and 2 of MAdCAM-1 play different roles in MAdCAM-1 binding to low- and high-affinity integrin alpha4beta7. J. Biol. Chem. 286:12086–12092. 10.1074/jbc.M110.208900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Liu J., Zheng Y., Pan Y., Zhang K., and Chen J.. 2014. Distinct chemokine signaling regulates integrin ligand specificity to dictate tissue-specific lymphocyte homing. Dev. Cell. 30:61–70. 10.1016/j.devcel.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Takagi J., and Springer T.A.. 2002. Integrin activation and structural rearrangement. Immunol. Rev. 186:141–163. 10.1034/j.1600-065X.2002.18613.x [DOI] [PubMed] [Google Scholar]
- Takagi J., Petre B.M., Walz T., and Springer T.A.. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 110:599–611. 10.1016/S0092-8674(02)00935-2 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Sawada W., Noguchi J., Watanabe S., Ucar H., Hayashi-Takagi A., Yagishita S., Ohno M., Tokumaru H., and Kasai H.. 2015. Two-photon fluorescence lifetime imaging of primed SNARE complexes in presynaptic terminals and β cells. Nat. Commun. 6:8531 10.1038/ncomms9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Casasnovas J.M., Liu J.H., Briskin M.J., Springer T.A., and Wang J.H.. 1998. The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition. Structure. 6:793–801. 10.1016/S0969-2126(98)00080-X [DOI] [PubMed] [Google Scholar]
- Taylor P., Bilsland M., and Walkinshaw M.D.. 2001. A new conformation of the integrin-binding fragment of human VCAM-1 crystallizes in a highly hydrated packing arrangement. Acta Crystallogr. D Biol. Crystallogr. 57:1579–1583. 10.1107/S0907444901011209 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., and Berendsen H.J.. 2005. GROMACS: fast, flexible, and free. J. Comput. Chem. 26:1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Wang J., and Springer T.A.. 1998. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol. Rev. 163:197–215. 10.1111/j.1600-065X.1998.tb01198.x [DOI] [PubMed] [Google Scholar]
- Xiao T., Takagi J., Coller B.S., Wang J.H., and Springer T.A.. 2004. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 432:59–67. 10.1038/nature02976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.P., Mahalingham B., Alonso J.L., Borrelli L.A., Rui X., Anand S., Hyman B.T., Rysiok T., Müller-Pompalla D., Goodman S.L., and Arnaout M.A.. 2009. Crystal structure of the complete integrin αVβ3 ectodomain plus an α/β transmembrane fragment. J. Cell Biol. 186:589–600. 10.1083/jcb.200905085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Zhang L., Geng Z.H., Wang H.B., Wang J.T., Chen M., and Geng J.G.. 2007. P-selectin cross-links PSGL-1 and enhances neutrophil adhesion to fibrinogen and ICAM-1 in a Src kinase-dependent, but GPCR-independent mechanism. Cell Adhes. Migr. 1:115–123. 10.4161/cam.1.3.4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.P., Kim E., Swift M., Smith J.W., Volkmann N., and Hanein D.. 2016. Three-Dimensional Structures of Full-Length, Membrane-Embedded Human α(IIb)β(3) Integrin Complexes. Biophys. J. 110:798–809. 10.1016/j.bpj.2016.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Shimaoka M., Chen J., and Springer T.A.. 2004. Activation of integrin beta-subunit I-like domains by one-turn C-terminal alpha-helix deletions. Proc. Natl. Acad. Sci. USA. 101:2333–2338. 10.1073/pnas.0307291101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Liu J., Winkler H., and Taylor K.A.. 2008. Integrin alpha IIb beta 3 in a membrane environment remains the same height after Mn2+ activation when observed by cryoelectron tomography. J. Mol. Biol. 378:976–986. 10.1016/j.jmb.2008.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Zhu J., Mi L.Z., Walz T., Sun H., Chen J., and Springer T.A.. 2012. Structural specializations of α(4)β(7), an integrin that mediates rolling adhesion. J. Cell Biol. 196:131–146. 10.1083/jcb.201110023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., and Chen J.. 2012. The regulation of integrin function by divalent cations. Cell Adhes. Migr. 6:20–29. 10.4161/cam.18702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Deng W., Zhou L., Xu Y., Yang W., Liang X., Wang Y., Kulman J.D., Zhang X.F., and Li R.. 2015. Identification of a juxtamembrane mechanosensitive domain in the platelet mechanosensor glycoprotein Ib-IX complex. Blood. 125:562–569. 10.1182/blood-2014-07-589507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wojcikiewicz E., and Moy V.T.. 2002. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys. J. 83:2270–2279. 10.1016/S0006-3495(02)73987-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Rico F., Xu A.J., and Moy V.. 2009. Atomic force microscopy of protein-protein interactions. In Handbook of Single-Molecule Biophysics. Hinterdorfer P., and van Oijen A., editors. Springer Press; 10.1007/978-0-387-76497-919. [DOI] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., Zhang C., Nishida N., and Springer T.A.. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861. 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu J., Negri A., Provasi D., Filizola M., Coller B.S., and Springer T.A.. 2010. Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood. 116:5050–5059. 10.1182/blood-2010-04-281154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu J., and Springer T.A.. 2013. Complete integrin headpiece opening in eight steps. J. Cell Biol. 201:1053–1068. 10.1083/jcb.201212037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.