Abstract
The formation of heparan sulfate occurs within the lumen of the endoplasmic reticulum–Golgi complex–trans-Golgi network by the concerted action of several glycosyltransferases, an epimerase, and multiple sulfotransferases. In this report, we have examined the location and interaction of tagged forms of five of the biosynthetic enzymes: galactosyltransferase I and glucuronosyltransferase I, required for the formation of the linkage region, and GlcNAc N-deacetylase/N-sulfotransferase 1, uronosyl 5-epimerase, and uronosyl 2-O-sulfotransferase, the first three enzymes involved in the modification of the chains. All of the enzymes colocalized with the medial-Golgi marker α-mannosidase II. To study whether any of these enzymes interacted with each other, they were relocated to the endoplasmic reticulum (ER) by replacing their cytoplasmic N-terminal tails with an ER retention signal derived from the cytoplasmic domain of human invariant chain (p33). Relocating either galactosyltransferase I or glucuronosyltransferase I had no effect on the other's location or activity. However, relocating the epimerase to the ER caused a parallel redistribution of the 2-O-sulfotransferase. Transfected epimerase was also located in the ER in a cell mutant lacking the 2-O-sulfotransferase, but moved to the Golgi when the cells were transfected with 2-O-sulfotransferase cDNA. Epimerase activity was depressed in the mutant, but increased upon restoration of 2-O-sulfotransferase, suggesting that their physical association was required for both epimerase stability and translocation to the Golgi. These findings provide in vivo evidence for the formation of complexes among enzymes involved in heparan sulfate biosynthesis. The functional significance of these complexes may relate to the rapidity of heparan sulfate formation.
Keywords: glycosaminoglycans‖sulfation‖epimerization‖enzyme localization‖Golgi complex
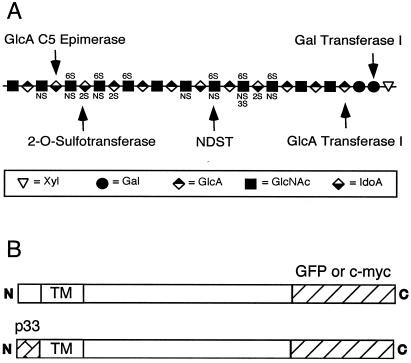
The biosynthesis of heparan sulfate initiates by the translation of a proteoglycan core protein and the assembly of the so-called linkage region tetrasaccharide on specific serine residues (–GlcAβ1,3Galβ1,3Galβ1,4Xylβ1-O-Ser). The chain then polymerizes by the addition of alternating N-acetylglucosamine (GlcNAcα1,4) and glucuronic acid (GlcAβ1,4) residues. A series of modification reactions takes place simultaneously that involves at least six enzymatic activities: (i) N-deacetylation of a portion of GlcNAc residues, (ii) N-sulfation of the resulting unsubstituted amino groups to form GlcNS units, (iii) 5-epimerization of adjacent d-GlcA residues to form l-iduronic acid (IdoA), (iv) 2-O-sulfation of l-IdoA and more rarely d-GlcA residues, (v) 6-O-sulfation of glucosamine units, and (vi) occasional 3-O-sulfation of glucosamine residues (Fig. 1A). A major question concerns how these enzymes orchestrate the formation of specific oligosaccharide sequences with unique binding properties for ligands (1, 2). Multiple isozymes exist for several of the transferases that differ in substrate specificity and temporal/spatial expression during development (3–5). The physical arrangement of the enzymes in the Golgi complex could also play a role.
Figure 1.
Heparan sulfate and chimeric enzymes. (A) Schematic representation of heparan sulfate. The arrows indicate the sites of action for the enzymes examined in this study. NDST, N-deacetylase/N-sulfotransferase 1. (B) To localize the enzymes, green fluorescent protein (GFP) was spliced on the C termini of the enzymes. To relocate the enzymes, the cytoplasmic tail of the enzymes was replaced with the endoplasmic reticulum (ER)-retention domain from human invariant chain (p33). TM, transmembrane domain.
With the exception of one glucosaminyl 3-O-sulfotransferase isozyme (6), all of enzymes involved in heparan sulfate synthesis are type II membrane proteins, with relatively short cytoplasmic tails, a single hydrophobic transmembrane segment, and a stem region that is thought to support a luminally oriented globular catalytic domain. Like most enzymes involved in glycosylation, all of the enzymes of heparan sulfate synthesis are thought to reside in elements of the ER–Golgi–trans-Golgi network (TGN) membrane system (7, 8). Evidence suggests that the transmembrane domains and/or the stem sequences play important roles in sorting and retention of the enzymes in their respective locations (9–11). Differential location of the enzymes and the potential for supramolecular complexes of multiple enzymes could help channel substrates in a selective manner and affect the structure of the chains (12).
In this report, we have examined the location and potential interaction of enzymes involved in heparan sulfate synthesis by using an in vivo approach pioneered by Nilsson and coworkers in their study of glycoprotein processing enzymes in the Golgi (13, 14). These investigators grafted an ER retention sequence on the N terminus of Golgi glucosaminyltransferase I and discovered that retention of the enzyme in the ER simultaneously relocated α-mannosidase II. This finding led to the kin-recognition hypothesis as a means for colocalization of Golgi enzymes involved in a common pathway (10, 13, 14). Using this technique, we now show that two of the enzymes that act early in heparan sulfate biosynthesis localize independently (galactosyltransferase I and glucuronosyltransferase I), whereas two of the modification enzymes that act later in the pathway physically interact [uronosyl 2-O-sulfotransferase (2OST) and 5-epimerase (Epi)]. Complex formation affects the location and activity of the epimerase and may play a functional role in chain modification by ensuring rapid and complete 2-O-sulfation of iduronic acid (IdoA) units generated by the epimerase.
Materials and Methods
Cell Culture.
Chinese hamster ovary cells (CHO-K1) were obtained from the American Type Culture Collection (ATCC CCL61). CHO mutants pgsF17, deficient in 2OST, pgsG-224, deficient in glucuronosyltransferase I, and pgsB-761, deficient in galactosyltransferase, I were described previously (15–17). The cells were grown under an atmosphere of 5% CO2 in air and 100% relative humidity in Ham's F-12 growth medium or F12/DMEM (1:1, vol/vol) (Life Technologies) supplemented with 10% (vol/vol) FBS (HyClone), 100 μg/ml streptomycin sulfate, and 100 units/ml penicillin G.
Recombinant DNA.
Full-length cDNA encoding human invariant chain (hIip33) was obtained from Michael R. Jackson (Johnson Research Institute, La Jolla, CA). PCR fragments encoding the first 282 bp of hIip33 cDNA were generated with an NheI restriction site at the 5′ end and EcoRI or XhoI restriction sites at the 3′ end. After digestion with the appropriate restriction enzymes, the fragment was ligated to the N terminus of each specific enzyme, just before the first amino acid of the predicted transmembrane domain. cDNAs encoding murine GlcNAc N-deacetylase/N-sulfotransferase (NDST1) and galactosyltransferase I (GalTI) and CHO glucuronosyltransferase I (GlcATI), 2OST, and Epi were inserted in-frame with GFP (pEGFP-N1) or with a c-Myc-tag (pcDNA3.1) at the C terminus. Cells were transfected by using Lipofectamine (Life Technologies) in accordance with the manufacturer's instructions. In transient expression experiments, the cells were analyzed after 2 days. Stable cell clones were selected in 400 μg/ml G418 and screened by flow cytometry to obtain strains with comparable levels of GFP or Myc-tag expression.
Fluorescence Microscopy.
Cells grown on 24-well glass microscope slides were rinsed with PBS (18), fixed for 1 h with 2% paraformaldehyde in 75 mM phosphate buffer, and permeabilized with 0.1% Triton X-100 in PBS containing 0.1% BSA. Primary antibodies against α-mannosidase II (medial Golgi marker, rabbit polyclonal antibody, a gift from Marilyn G. Farquhar, Univ. of California at San Diego), calreticulin (ER marker, rabbit polyclonal antibody, StressGen Biotechnologies, Victoria, Canada), and mouse monoclonal anti-Myc (Invitrogen) were diluted 1:400 in PBS containing 1% BSA and incubated with fixed cells in a humid chamber for 1 h. After several washes the cells were incubated with 4′,6-diamidino-2-phenylindole (DAPI) to stain nuclei and a secondary antibody (anti-rabbit IgG coupled with CY5 from Accurate Chemicals, or anti-rabbit IgG coupled with Alexa from Molecular Probes) diluted 1:200 in PBS containing 1% BSA. Myc-tagged proteins were visualized with tetramethylrhodamine isothiocyanate (TRITC)-labeled anti-mouse IgG (Sigma). The coverslips were washed several times with PBS containing 0.1% BSA and mounted with Vectashield mounting medium (Vector Laboratories). Digital images were captured with a Hamamatsu C5810 three-color chilled charge-coupled device (CCD) camera mounted on a Zeiss Axiophot (100× lens) microscope. For deconvolution, images were captured with a Photometrics CCD mounted on a Nikon microscope adapted to a DeltaVision (Applied Precision, Issaquah, WA) deconvolution imaging system. The data sets were deconvoluted and analyzed by using SoftWorx software (Applied Precision) on a Silicon Graphics Octane workstation.
Detergent and Salt Extraction.
As described in ref. 19, cells were harvested by scraping in 50 mM Mes buffer (2-N-morpholinoethane-sulfonic acid), pH 6.5, containing 0.5% Triton X-100 and protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride, 10 μM pepstatin A, 10 μM leupeptin, and 25 μg/ml aprotinin). The samples were incubated with 0–300 mM NaCl for 1 h with gentle rotation on ice, and then centrifuged for 15 min at 15,000 × g. The supernatant was then centrifuged for 1 h at 100,000 × g. The final supernatants were adjusted to the same volume in SDS sample buffer, separated by SDS/PAGE, and transferred electrophoretically to nitrocellulose membranes. Myc-tagged proteins were detected with anti-Myc mAb (Invitrogen), and chimeras containing GFP were detected with an anti-GFP mAb (JL-8; CLONTECH). The secondary antibody, goat anti-mouse IgG (H + L) conjugated to horseradish peroxidase (Bio-Rad) was detected by chemiluminescence.
Immunoprecipitation.
pgF17 cells stably transfected with 2OST-GFP were transiently transfected with Epi-Myc. After 2 days, the cells were lysed in 50 mM Tris⋅HCl pH 7.4/0.5% Triton X-100/50 mM NaCl/5 mM CaCl2/5 mM MgCl2/20 mM benzamidine/1 mM PMSF/50 mM aminocaproic acid/10 mM iodoacetamide. The lysates were centrifuged at 12,000 × g for 20 min, precleared for 30 min with 30 μl of staphylococcal protein A-Sepharose (Amersham Pharmacia) at 4°C, and then incubated with 0.5 mg of mouse anti-GFP mAb for 4 h, followed by incubation with 50 μl of Protein A-Sepharose overnight. The beads were washed three times with PBS, boiled for 5 min in 50 μl of SDS-PAGE sample buffer and analyzed by SDS-PAGE electrophoresis. Proteins were transferred to nitrocellulose membrane (Millipore), blocked with 4% albumin in PBS, containing 0.05% Tween-20 and developed with mouse anti-GFP mAb or anti-Myc mAb. Epi-Myc and 2OST-GFP were detected with calf anti-mouse IgG secondary antibody conjugated to horseradish peroxidase. The specific proteins were detected by using ECL Western blotting detection kit (Amersham Pharmacia) and exposed to x-ray film (Kodak) for 1 min.
Enzymatic Assays.
Enzyme activities were measured in cell homogenates as described previously (4, 15–17, 20). Cells were grown to confluence in six-well plates, rinsed three times with cold PBS, and detached with a rubber policeman in 40 μl of buffer containing 0.25 M sucrose, 50 mM Tris⋅HCl (pH 7.5), 1% (wt/vol) Triton X-100, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1 mM PMSF. Aliquots of the cell extracts were stored at −20°C and sonicated before assay.
Results
Heparan sulfate synthesis occurs in a sequential fashion by the transfer of single monosaccharides to a nascent chain attached to a proteoglycan protein core (Fig. 1A). The initial reactions consist of xylosylation at specific serine residues, followed by attachment of two galactose units and GlcA. The alternating addition of GlcNAc and GlcA units forms the backbone of the chain, and a series of modification reactions then ensues, in which subsets of GlcNAc residues undergo N-deacetylation and N-sulfation, adjacent d-GlcA units epimerize to l-IdoA, and the uronic acids and glucosaminyl residues undergo sulfation at various positions (Fig. 1A). Prior studies have shown that the assembly process is quite rapid (21), suggesting that the enzymes may be located in the same subcellular compartment, possibly in complexes, to facilitate channeling of reaction products to downstream enzymes in the pathway. Two of the enzymes actually exist as bifunctional proteins containing dual activities: NDST isozymes catalyze GlcNAc N-deacetylation and N-sulfation, and EXTs catalyze GlcNAc and GlcA copolymerization (2). Here, we have examined the location and interaction of the enzymes in greater detail by expressing recombinant forms of GalTI and GlcATI, which catalyze early reactions involved in the formation of the linkage region tetrasaccharide, and Epi and 2OST, which modify the GlcA residues after chain polymerization.
Location and Relocation of GalTI and GlcATI.
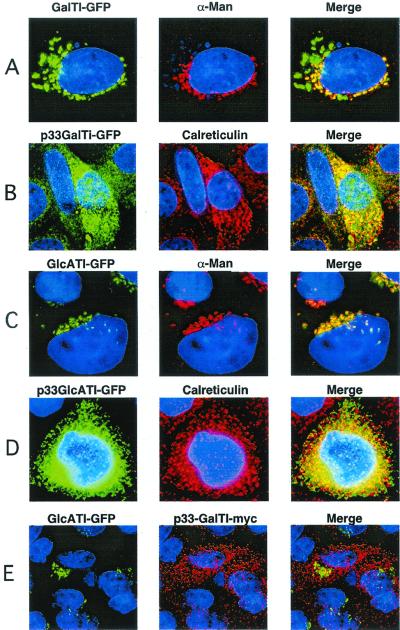
Chimeric forms of GalTI and GlcATI containing C-terminal GFP were introduced into CHO cells (Fig. 1B), where they colocalized with the medial Golgi marker, α-mannosidase II (22) (Fig. 2 A and C, respectively). Both of these chimeric enzymes were enzymatically active as judged by activity assays and their ability to correct heparan sulfate biosynthesis in corresponding mutants of CHO cells (16, 17). By deconvolution fluorescence microscopy, GlcATI-GFP completely colocalized with α-mannosidase II, whereas GalTI-GFP was located in compartments shared by a α-mannosidase II as well as a nearby compartment that does not contain the marker. Because GalTI action precedes GlcATI biosynthetically, this compartment may represent the cis Golgi network or the ER–Golgi intermediate compartment (23).
Figure 2.
Localization of chimeric enzymes involved in the formation of linkage region. Wild-type CHO cells were transiently transfected with the indicated chimeric enzymes and imaged by deconvolution microscopy (Materials and Methods). (A and C) Golgi localization of GalTI-GFP and GlcATI-GFP, respectively. (B and D) ER localization of p33-GalTI-GFP and p33-GlcATI-GFP, respectively. (E) Coexpression of GlcATI-GFP (green) and p33-GalTI-Myc (red). α-Man, α-mannosidase II, the medial Golgi marker; Calreticulin, ER marker; Merge, colocalization (yellow) of the chimeric enzymes with each other or the markers.
Since GalTI and GlcATI overlap in the same compartment, we next determined whether they could associate physically. In these experiments, we used a retargeting method described by Nilsson et al. (14), in which an ER retention peptide from the human invariant chain (hIip33) is spliced onto the N terminus of one of the transferases (Fig. 1B). The recombinant proteins, designated p33-GalTI-GFP and p33-GlcATI-GFP, were enzymatically active, but were located in the ER, colocalizing with the ER marker calreticulin (Fig. 2 B and D, respectively). To test for interactions, a Myc-tagged form of p33-GalTI was prepared and cotransfected with GlcATI-GFP. These recombinant proteins migrated independently to the ER and Golgi compartments, respectively (Fig. 2E). Similar results were obtained when p33-GlcATI-Myc was cotransfected with GalTI-GFP (data not shown). Furthermore, the location of each chimeric enzyme was unaffected when expressed in mutant cell lines lacking endogenous GalTI (17) or GlcATI (16) (data not shown), suggesting that the native forms of the enzymes do not interact with the chimeras. Thus, GalTI and GlcATI overlap in location, but do not appear to physically interact.
Location and Relocation of 2OST and Epi.
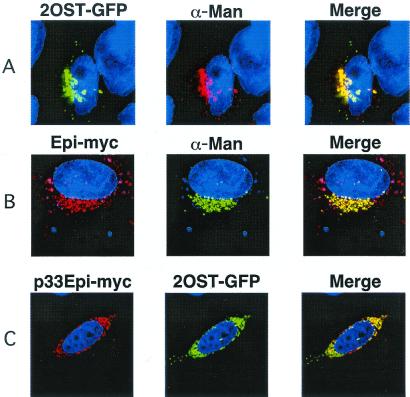
We next examined the location and interaction of Epi, which converts GlcA units in heparan sulfate to IdoA, and 2OST, which adds sulfate preferentially to the C2 position of the resulting IdoA units, preventing reversal of the epimerization reaction (24). Like GalTI and GlcATI, tagged forms of Epi and 2OST showed a typical juxtanuclear Golgi position, colocalizing with α-mannosidase II (Fig. 3 A and B). The p33 tagged forms of these enzymes also relocated to the ER, colocalizing with calreticulin (Fig. 3C shows p33-Epi-Myc). However, in contrast to the behavior of GalTI and GlcATI, 2OST-GFP in cells containing p33-Epi-Myc was found in the ER (Fig. 3C). This relocation of 2OST was selective because GFP-tagged forms of NDST1, GalTI, and GlcATI maintained their typical Golgi localization in cells containing p33-Epi-Myc (data not shown).
Figure 3.
ER-localized Epi relocates 2OST. (A and B) Wild-type CHO cells were transiently transfected with chimeric 2OST-GFP or Epi-Myc and their location was determined by deconvolution microscopy (Materials and Methods). (C) Cells were cotransfected with p33-Epi-Myc and 2OST-GFP. The two enzymes colocalized in the ER, in the compartments containing the calreticulin marker. α-Man, α-mannosidase II, the medial Golgi marker; Calreticulin, the ER marker; Merge, colocalization (yellow) of the chimeric enzymes with each other or the markers.
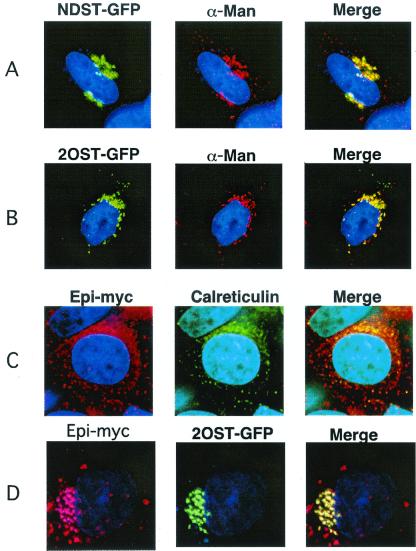
To exclude the possibility that the relocation of 2OST by p33-Epi was an artifact caused by protein overexpression or the introduction of the C-terminal tags, we examined the location of Epi-Myc in pgsF17 mutant CHO cells. This mutant lacks endogenous 2OST activity and mRNA, suggesting that it also does not contain 2OST protein (15). In these cells, 2OST-GFP and NDST1-GFP colocalized with α-mannosidase II (Fig. 4 A and B), but transfected Epi-Myc localized to the ER, coinciding with calreticulin (Fig. 4C). The relocation of Epi to the ER in these cells suggested that the movement of Epi to the Golgi may depend on the presence of 2OST. To confirm this idea, pgsF17 cells were transfected with a cDNA encoding wild-type 2OST. Reexpression of the enzyme reverted the location of Epi to the Golgi, and its new location completely coincided with 2OST-GFP (Fig. 4D). Introduction of p33–2OST in the mutant did not affect the location of Epi (data not shown). Together with the results shown in Fig. 3, these findings provided strong evidence that 2OST and Epi can form a complex and that the interaction is required for translocation of both Epi and 2OST to the Golgi.
Figure 4.
Golgi localization of Epi depends on the presence of 2OST. pgsF17 cells lacking endogenous 2OST were transiently transfected with NDST1-GFP (A), 2OST-GFP (B), or Epi-Myc (C). NDST1 and 2OST colocalized with the α-mannosidase marker, whereas Epi-Myc was found in the ER. In D, the mutant was stably transfected with 2OST cDNA to the correct the enzyme deficiency, and then transiently transfected with 2OST-GFP or Epi-Myc.
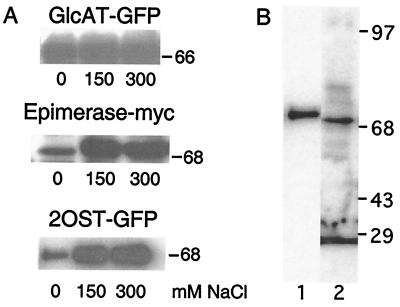
Complexes of Golgi enzymes have been inferred to exist by a requirement for high salt for solubilization from detergent extracts of cell membranes (19). For example, glucosaminyltransferase I and α-mannosidase require high salt for solubilization and sediment as complexes (19). These enzymes also exhibit mutual relocation when one of the proteins is retargeted by using the p33 grafting method (14). In contrast, late-acting Golgi enzymes involved in glycoprotein assembly (β1,4-galactosyltransferase and α1,2-fucosyltransferase) readily solubilize in low salt and migrate as monomers in sucrose density gradient centrifugation. When the solubilization scheme was applied to Epi and 2OST, we found that both enzymes required high salt for solubilization, consistent with the idea that they resided in the Golgi as a complex. In contrast, GlcATI-GFP solubilized readily in salt-free buffer (Fig. 5A).
Figure 5.
Physical interaction of Epi and 2OST. (A) CHO cells expressing GlcATI-GFP, Epi-Myc, or 2OST-GFP were extracted in buffer containing 50 mM Mes (pH 6.5), 0.5% Triton X-100, and 0, 150, or 300 mM NaCl as indicated. The cell extracts were centrifuged, separated by SDS/gel electrophoresis, and analyzed by immunoblotting (Materials and Methods). (B) Samples of cells containing Epi-Myc and 2OST-GFP were immunoprecipitated with antibody to GFP and analyzed by SDS/PAGE. Lane 1, Western blot using mouse anti-Myc mAb. Lane 2, Western blot using mouse anti-GFP mAb.
To confirm the interaction, samples of pgsF17 cells, stably transfected with 2OST-GFP, were transiently transfected with Epi-Myc and reacted with an anti-GFP mAb. The immunoprecipitates were then analyzed by SDS/PAGE and Western blotting with anti-GFP mAb or anti-Myc mAb (Fig. 5B). The anti-Myc mAb reacted with single band that migrated at the expected molecular mass of the epimerase (20). The mAb against GFP reacted with a somewhat larger band at the expected mass of the chimera (≈70 kDa). The anti-GFP mAb also reacted with proteins that varied in position and intensity in independent experiments, both in transfected and nontransfected cells, suggesting that these bands were nonspecific.
Location of Epi Alters Enzyme Activity.
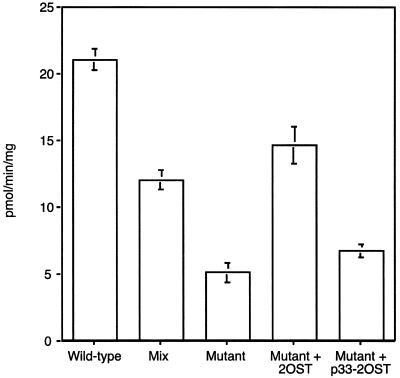
To determine whether the interaction of Epi and 2OST might affect enzyme activity, we measured epimerase in extracts prepared from pgsF17 and wild-type cells. As shown in Fig. 6, extracts of pgsF17 cells contained less Epi activity compared with the wild-type (5 ± 1 pmol/min per mg of protein versus 22 ± 2 pmol/min per mg, respectively). This difference was not due to altered kinetic properties of the enzyme, because the substrate was saturating in the in vitro reactions. Mixing experiments, in which equal amounts of pgsF17 and wild-type extracts were combined and assayed showed that the resultant activity was the arithmetic mean of the individual samples (Fig. 6). This finding suggested that the lower activity in pgsF17 cells was not due to the presence of an inhibitor or the absence of a soluble activator in the wild type. However, transfection of pgsF17 with 2OST restored Epi activity to wild-type levels (15 ± 3 pmol/min per mg). In contrast, introduction of p33–2OST did not (7 ± 1 pmol/min per mg). Thus, the restorative effect of 2OST was likely due to relocation of Epi to the Golgi (Fig. 4D), which may serve to stabilize the enzyme against degradative processes active in the ER (25).
Figure 6.
Decreased epimerase activity in 2OST-deficient cells. Wild-type, pgsF17, and pgsF17 cells stably transfected with 2OST or p33–2OST were assayed for Epi activity. “Mix” refers to a sample containing equal amounts of wild-type and mutant extracts at half the level in samples assayed alone.
Discussion
Nearly all of the enzymes involved in heparan sulfate biosynthesis have now been molecularly cloned and partially characterized, but very little is known about their organization and distribution in the ER-Golgi-TGN, where assembly of the chains takes place. Like glycosyltransferases required for glycolipid and glycoprotein assembly, the biosynthetic enzymes for heparan sulfate are likely to be arrayed across the secretory apparatus in the order in which the reactions occur (8). However, significant overlap exists, dependent in part on the size and number of cisternae, which can vary in different cell types. CHO cells have relatively simple Golgi, and as shown here all five of the enzymes appeared to locate in the compartment containing the α-mannosidase II marker. Prior studies have suggested that NDST1 may be located in the TGN in mouse LTA cells, probably reflecting differences in the arrangement of the Golgi in these cells (26).
Analysis of two early reactions in the pathway catalyzed by GalTI and GlcATI showed that these enzymes colocalize with the medial-Golgi marker, α-mannosidase II, but GalTI also appears in a compartment lacking α-mannosidase II. Although the identity of this compartment is not yet known, it may represent the transitional elements between the ER and Golgi. Xylosyltransferase (XylT) is thought to be located in the ER and transitional elements possibly related to the ER–Golgi intermediate compartment based on electron microscopic autoradiography, immunocytochemistry, and localization of the decarboxylase that generates UDP-xylose (27–29). In addition, GalTI can physically interact with XylT in vitro (30–32), suggesting that they may reside in the same compartment in vivo. Because XylT precedes galactosyltransferase in the pathway, it is reasonable to assume that the association may occur in a more proximal biosynthetic compartment. The recent purification and cloning of XylT should help clarify this issue (33, 34).
The formation of the linkage region is completed by the action of galactosyltransferase II (GalTII) and GlcATI. GalTII has been recently cloned and shown to also overlap with α-mannosidase II as wells as CALNUC, a marker of the cis Golgi (35). The physical interaction of GalTII with GlcATI or GalTI has not yet been explored. GalTI apparently does not interact with GlcATI by the criteria of relocation in the p33 grafting technique and ease of extraction (Fig. 2). The ability to readily solubilize GlcATI from microsomal membranes suggests that complexes may not exist (Fig. 5), implying that none of these enzymes may be associated with one another.
The data presented here represent in vivo evidence for the formation of a complex between sequential enzymes in the pathway of heparan sulfate biosynthesis, Epi and 2OST. This conclusion is based on their mutual relocation when one of the proteins is relocated to the ER, on the mislocalization of a tagged form of Epi in a mutant cell line lacking 2OST, on the requirement for high salt to solubilize both enzymes from microsomal membranes, and coimmunoprecipitation. An important question concerns the functional significance of the Epi–2OST interaction. Substrate channeling through this enzyme complex would ensure a high degree of sulfation after epimerization has occurred, which is borne out by structural studies of heparan sulfate in CHO cells and other cell types (15, 36). A second function concerns the proper placement of the enzymes in the Golgi, either in the same compartment or distal to the site of chain polymerization. Chain polymerization occurs by the alternating addition of GlcNAc and GlcA residues, and these reactions are catalyzed by one or more enzymes (EXT1 and EXT2) having both transferase activities (37–41). It is interesting to note that EXT1 and EXT2 can form heterooligomers apparently required for maximal activity in vitro and translocation to the Golgi in vivo (42), reminiscent of the behavior of epimerase and 2OST. Whether a relationship exists between these two systems is unknown.
Unlike other pathways of glycosylation, glycosaminoglycan biosynthesis employs at least two enzymes that have more than one activity. The EXT isozymes (EXT1 and EXT2) each have the capacity to transfer GlcA and GlcNAc to a growing chain, and NDSTs have the capacity to remove acetyl groups and substitute the free amino groups with sulfate. Fused proteins with dual or multiple catalytic functions provide an appealing strategy to effect the rapid assembly of glycans on protein and lipid cores as they pass through the Golgi on their way to the plasma membrane or extracellular matrix. The physical interaction of separate enzymes achieves the same goal, but provides an added degree of control in the system because the abundance of individual subunits can be altered independently.
With the evidence presented here, complexes have now been demonstrated between sequential enzymes involved in the formation of all major glycoconjugates (glycoproteins, glycolipids, and proteoglycans) (13, 43). Apparently enzyme–enzyme interactions can occur in different subcompartments of the Golgi, indicating that “kin-recognition” may be a general mechanism for ensuring that certain protein subsets reside in the appropriate compartments. These complexes may include other components involved in glycosylation, including nucleotide transporters (44) as well as chaperones that participate in quality control. Further studies are needed to determine if such complexes affect the fidelity of the process, the fine structure of the carbohydrate chains, and thus the formation of binding sites for various carbohydrate-binding proteins.
Acknowledgments
We thank Jim Feramisco (Imaging Center, Univ. of California San Diego Cancer Center) for many helpful discussions, Joel Shaper (Johns Hopkins School of Medicine) for providing the cDNA clone for GalTI, Haroko Habuchi (Aichi Medical School) for providing the 2-OST cDNA, and G. Wei (Salk Institute) for providing GlcATI cDNA. A grant from The PEW Latin American Program (to M.A.S.P.) and Grant R37GM33063 from the National Institutes of Health (to J.D.E.) supported this work.
Abbreviations
- CHO
Chinese hamster ovary
- Epi
heparan sulfate uronosyl 5-epimerase
- ER
endoplasmic reticulum
- GalTI
xylose:UDP-Gal galactosyltransferase I
- GFP
green fluorescent protein
- GlcA
glucuronic acid
- GlcATI
galactose:UDP-GlcA glucuronosyltransferase I
- GlcNAc
N-acetylglucosamine
- IdoA
iduronic acid
- NDST
GlcNAc N-deacetylase/N-sulfotransferase
- 2OST
uronosyl:PAPS 2-O-sulfotransferase (PAPS, 3′-phosphoadenosine 5′-phosphosulfate)
- p33
the cytoplasmic domain of human invariant chain (hIip33)
- TGN
trans-Golgi network
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Present address: Universidade Federal De São Paulo, Rua Três De Maio, No 100, 4° Andar—Biologia Molecular, Vila Clementino, São Paulo CEP: 04044020, Brazil.
References
- 1.Lindahl U, Kusche-Gullberg M, Kjellén L. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 2.Esko J D, Lindahl U. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shworak N W, Liu J A, Petros L M, Zhang L J, Kobayashi M, Copeland N G, Jenkins N A, Rosenberg R D. J Biol Chem. 1999;274:5170–5184. doi: 10.1074/jbc.274.8.5170. [DOI] [PubMed] [Google Scholar]
- 4.Aikawa J, Grobe K, Tsujimoto M, Esko J D. J Biol Chem. 2001;276:5876–5882. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- 5.Habuchi H, Tanaka M, Habuchi O, Yoshida K, Suzuki H, Ban K, Kimata K. J Biol Chem. 2000;275:2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 6.Shworak N W, Liu J, Fritze L M S, Schwartz J J, Zhang L J, Logeart D, Rosenberg R D. J Biol Chem. 1997;272:28008–28019. doi: 10.1074/jbc.272.44.28008. [DOI] [PubMed] [Google Scholar]
- 7.Fernández C J, Warren G. J Biol Chem. 1998;273:19030–19039. doi: 10.1074/jbc.273.30.19030. [DOI] [PubMed] [Google Scholar]
- 8.Varki A. Trends Cell Biol. 1998;8:34–40. doi: 10.1016/s0962-8924(97)01198-7. [DOI] [PubMed] [Google Scholar]
- 9.Colley K J. Glycobiology. 1997;7:1–13. doi: 10.1093/glycob/7.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson T, Rabouille C, Hui N, Watson R, Warren G. J Cell Sci. 1996;109:1975–1989. doi: 10.1242/jcs.109.7.1975. [DOI] [PubMed] [Google Scholar]
- 11.Munro S. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salmivirta M, Lidholt K, Lindahl U. FASEB J. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 13.Nilsson T, Hoe M H, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger E G, Warren G. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson T, Slusarewicz P, Hoe M H, Warren G. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- 15.Bai X M, Esko J D. J Biol Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- 16.Bai X M, Wei G, Sinha A, Esko J D. J Biol Chem. 1999;274:13017–13024. doi: 10.1074/jbc.274.19.13017. [DOI] [PubMed] [Google Scholar]
- 17.Esko J D, Weinke J L, Taylor W H, Ekborg G, Rodén L, Anantharamaiah G, Gawish A. J Biol Chem. 1987;262:12189–12195. [PubMed] [Google Scholar]
- 18.Dulbecco R, Vogt M. J Exp Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Opat A S, Houghton F, Gleeson P K. J Biol Chem. 2000;275:11836–11845. doi: 10.1074/jbc.275.16.11836. [DOI] [PubMed] [Google Scholar]
- 20.Crawford B E, Olson S K, Esko J D, Pinhal M A S. J Biol Chem. 2001;276:21538–21543. doi: 10.1074/jbc.M100880200. [DOI] [PubMed] [Google Scholar]
- 21.Höök M, Lindahl U, Hallén A, Bäckström G. J Biol Chem. 1975;250:6065–6071. [PubMed] [Google Scholar]
- 22.Velasco A, Hendricks L, Moremen K W, Tulsiani D R, Touster O, Farquhar M G. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahtinen U, Dahllof B, Saraste J. J Cell Sci. 1992;103:321–333. doi: 10.1242/jcs.103.2.321. [DOI] [PubMed] [Google Scholar]
- 24.Jacobsson I, Lindahl U, Jensen J W, Rodén L, Prihar H, Feingold D S. J Biol Chem. 1984;259:1056–1063. [PubMed] [Google Scholar]
- 25.Lord J M, Davey J, Frigerio L, Roberts L M. Semin Cell Dev Biol. 2000;11:159–164. doi: 10.1006/scdb.2000.0160. [DOI] [PubMed] [Google Scholar]
- 26.Humphries D E, Sullivan B M, Aleixo M D, Stow J L. Biochem J. 1997;325:351–357. doi: 10.1042/bj3250351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann H P, Schwartz N B, Rodén L, Prockop D J. Connect Tissue Res. 1984;12:151–163. doi: 10.3109/03008208408992780. [DOI] [PubMed] [Google Scholar]
- 28.Vertel B M, Walters L M, Flay N, Kearns A E, Schwartz N B. J Biol Chem. 1993;268:11105–11112. [PubMed] [Google Scholar]
- 29.Kearns A E, Vertel B M, Schwartz N B. J Biol Chem. 1993;268:11097–11104. [PubMed] [Google Scholar]
- 30.Schwartz N B, Rodén L. Carbohydr Res. 1974;37:167–180. doi: 10.1016/s0008-6215(00)87072-x. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz N B, Rodén L, Dorfman A. Biochem Biophys Res Commun. 1974;56:717–724. doi: 10.1016/0006-291x(74)90664-0. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz N B. FEBS Lett. 1975;49:342–345. doi: 10.1016/0014-5793(75)80781-2. [DOI] [PubMed] [Google Scholar]
- 33.Götting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K. J Mol Biol. 2000;304:517–528. doi: 10.1006/jmbi.2000.4261. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn J, Götting C, Schnolzer M, Kempf T, Brinkmann T, Kleesiek K. J Biol Chem. 2000;276:4940–4947. doi: 10.1074/jbc.M005111200. [DOI] [PubMed] [Google Scholar]
- 35.Bai, X., Zhou, D., Brown, J. R., Hennet, T. & Esko, J. D. (2001) J. Biol. Chem.276, in press. [DOI] [PubMed]
- 36.Bame K J, Lidholt K, Lindahl U, Esko J D. J Biol Chem. 1991;266:10287–10293. [PubMed] [Google Scholar]
- 37.Lidholt K, Weinke J L, Kiser C S, Lugemwa F N, Bame K J, Cheifetz S, Massagué J, Lindahl U, Esko J D. Proc Natl Acad Sci USA. 1992;89:2267–2271. doi: 10.1073/pnas.89.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 39.McCormick C, Leduc Y, Martindale D, Mattison K, Esford L E, Dyer A P, Tufaro F. Nat Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 40.Wei G, Bai X M, Gabb M M G, Bame K J, Koshy T I, Spear P G, Esko J D. J Biol Chem. 2000;275:27733–27740. doi: 10.1074/jbc.M002990200. [DOI] [PubMed] [Google Scholar]
- 41.Senay C, Lind T, Muguruma K, Tone Y, Kitagawa H, Sugahara K, Lidholt K, Lindahl U, Kusche-Gullberg M. EMBO Rep. 2000;1:282–286. doi: 10.1093/embo-reports/kvd045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCormick C, Duncan G, Goutsos K T, Tufaro F. Proc Natl Acad Sci USA. 2000;97:668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giraudo C G, Daniotti J L, Maccioni H J F. Proc Natl Acad Sci USA. 2001;98:1625–1630. doi: 10.1073/pnas.031458398. . (First Published January 23, 2001; 10.1073/pnas.031458398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirschberg C B, Robbins P W, Abeijon C. Annu Rev Biochem. 1998;67:49–69. doi: 10.1146/annurev.biochem.67.1.49. [DOI] [PubMed] [Google Scholar]