Supplemental Digital Content is available in the text
Keywords: differentially expressed gene, kidney-Yin deficiency, Liuwei Dihuang pill, postmenopausal osteoporosis
Abstract
This study aimed to investigate the potential therapeutic targets of Liuwei Dihuang pill (LDP) in the treatment of postmenopausal osteoporosis with kidney-Yin deficiency (PMO-KY).
Gene expression data were downloaded from the GEO database, including 4 PMO-KY samples and 3 healthy postmenopausal controls from GSE56116, as well as 3 PMO-KY samples before LDP treatment and 3 PMO-KY samples after three months of LDP treatment from GSE57273. Limma package was used to identify differentially expressed genes (DEGs). Afterwards, the potential target genes of LDP (namely key DEGs) were identified according to the comparison of DEGs in PMO-KY group and the DEGs in LDP treatment groups. Subsequently, iRegulon plugin in Cytoscape software was used to predict potential transcription factors (TFs) that regulated the key DEGs, and Comparative Toxicogenomics Database was utilized to identify known PMO-related genes among the key DEGs.
Totally, 202 and 2066 DEGs were identified between PMO-KY and controls, as well as after-treatment and before-treatment groups, respectively. Among them, 52 DEGs were up-regulated in PMO-KY but down-regulated after LDP treatment, and 8 TFs were predicted to these DEGs. Furthermore, 34 DEGs were down-regulated in PMO-KY but up-regulated after treatment, and 7 TFs were predicted to regulate these DEGs. Additionally, 43 of the 86 key DEGs were known PMO-related genes.
NCOA3, TCF4, DUSP6, PELI2, and STX7 were predicted to be regulated by HOXA13. In the PMO-KY treatment, NCOA3, TCF4, DUSP6, PELI2, and STX7 might be the potential therapeutic targets of LDP. However, further investigation is required to confirm these genes.
1. Introduction
Postmenopausal osteoporosis (PMO) is characterized by deterioration of bone microarchitecture and chronic decrease of bone mass, and it generally leads to osteoporotic fracture, a major cause of disability and mortality in elderly women.[1] Approximately 40% of postmenopausal women suffer from PMO, and patients with PMO may steadily increase in future due to population ageing.[2] PMO is a chronic and progressive process that involves osteocytes, lining cells, osteoclasts, osteoblasts, T lymphocytes, and so on. From the perspective of traditional Chinese medicine, kidney deficiency is considered as one of the factors causing PMO.[3] According to the “Dialectical reference standards of traditional Chinese medicine deficiency syndrome,”[4] patients with at least 3 symptoms of “aching pain in spinal column,” “limp in shin or pain in heel,” “tinnitus or epicophosis,” “hair loss or gomphiasis,” “dribble of urine or urinary incontinence” and “sexual dysfunction” are diagnosed with kidney deficiency. If the patients with kidney deficiency simultaneously have three of the symptoms “dysphoria in chestpalms-soles,” “mouth and throat dry,” “red tongue or little coating” and “pulse breakdown,” as well as one of the symptoms “hectic fever at afternoon,” “coprostasis or red-yellow urine” and “night sweat,” they will be diagnosed with kidney-Yin deficiency.
Currently, worldwide approved therapeutics for PMO are vitamin D, bisphosphonates (eg, alendronate, zoledronate, ibandronate, and risedronate), selective estrogen-receptor modulators, parathyroid hormone, anti-RANKL, calcitonin, parathyroid hormone (PTH1-84), teriparatide (PTH1-34), denosumab, menatetrenone, alfacalcidol, and strontium ranelate.[5–8] However, drug intolerance, adverse effect, or price of these drugs limit their application and effectivity.[8] Liuwei Dihuang pill (LDP), a traditional Chinese medicine formula with a history of thousands of years, is an effective but lower-cost therapeutic for PMO, especially the PMO with kidney-Yin deficiency (PMO-KY).[9–12] After 12 weeks of treatment with LDP, serum levels of osteocalcin and alkaline phosphatase were decreased, and bone mineral density (BMD) of femurs and biomechanics of lumbar vertebra were improved in a PMO rat model.[10] With a 95% curative rate, LDP can increase BMD, inhibit bone absorption, and promote bone formation in clinic.[9] As LDP is a complex drug that is composed of 6 Chinese crude herbs, studies have been conducted to investigate its therapeutic mechanisms. Xia et al[10] suggested that LDP might alleviate PMO via up-regulating β-catenin, Runx2, Lrp-5, and Osx that were involved in canonical Wnt/β-catenin signaling pathway in osteoblast. Based on microarray analysis, Xie et al identified that PRLR, INSR, ASB1, CLCF1, PROK2, GPR27, C3orf35, JUN, and JUNB were down-regulated in patients with PMO-KY compared with healthy postmenopausal women, while ASB1, CLCF1, JUN, and JUNB were significantly up-regulated after LDP treatment for 3 months.[13,14] In addition, 25 genes were differentially expressed in monocytes in peripheral blood of PMO-KY patients after LDP treatment, including the genes associated with immune response (eg, PRL, PRLR, OSM, ISG15, and IL4R), cell growth (eg, F2 and SOCS3), JAK/STAT pathway (eg, PIAS1 and SOCS4), and SH3/SH2 adaptor (eg, SRC and STAM), as well as regulator genes that interact with STAT proteins (eg, JUN, JUNB, SMAD3, SMAD5, SMAD4, SP1, and YY1).[13] However, these studies mainly focus on several genes, and regulatory mechanism of gene expression like transcription factor (TF) has not been studied. Thus, the precise therapeutic mechanisms of LDP in treatment of PMO-KY have not been fully understood.
In the present study, comprehensive bioinformatics methods were utilized to re-analyze 2 microarray datasets uploaded to the Gene Expression Omnibus (GEO) by Xie et al[10] (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56116; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57273) As a consequence, potential target genes of LDP in the treatment of PMO-KY were identified, providing a novel direction for drug design in future.
2. Materials and methods
2.1. Microarray data
Gene expression data about PMO-KY were obtained from the GEO database, including datasets GSE56116 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56116) and GSE57273 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57273). The dataset GSE56116 contained 4 peripheral blood samples from PMO-KY patients, 3 samples from PMO patients with kidney Yang deficiency, 3 samples from PMO patients without kidney deficiency, and 3 healthy postmenopausal controls. Only the 4 PMO-KY samples (PMO-KY group) and 3 healthy postmenopausal controls (control group) were utilized for this analysis as our study focused on only PMO-KY. Their average age were 61.15 ± 4.381 years old, GSE57273 contained 6 peripheral blood samples from 3 PMO-KY patients before LDP treatment (n = 3, before-treatment group) and after 3 months of LDP treatment (n = 3, after-treatment group). Both GSE56116 and GSE57273 data were produced by the platform of Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Feature Number version). They were approved by ethics committee Fujian Academy of Traditional Chinese Medicine.
2.2. Data preprocessing
As GSE56116 and GSE57273 included only Agilent chips with single-channel, the Agilent one-color method in limma package (version 3.26.3, http://www.bioconductor.org/packages/3.0/bioc/html/limma.html)[15] was utilized to preprocess microarray data, including background correction, normalization between arrays, condensation of microarray data, transformation of probes into gene symbol, and expression calculation.
2.3. Identification of DEGs
Unpaired t-test method in limma package[15] was utilized to obtain the DEGs between PMO-KY and control groups, as well as after-treatment and before-treatment groups. Only DEGs with |log2 fold change| ≥ 0.5 and P < .05 were considered as significant DEGs, and they were clustered using g-plots package (version 2.4)[16] based on their expression levels.
2.4. Prediction of potential target genes of LDP
Based on the DEGs between PMO-KY and control groups, as well as DEGs between after-treatment and before-treatment groups, the key DEGs with opposite change patterns were identified using online software Venn-Diagram-online (http://www.bioinformatics.lu/venn.php#userconsent#). Expression change patterns of these DEGs between PMO-KY and control groups were different from those between after-treatment and before-treatment groups, indicating that they might be the target genes of LDP in the treatment of PMO-KY. In order to verify the accuracy of these key DEGs, Pvclust package (version 1.2-1, https://cran.r-project.org/)[17] was utilized to cluster these DEGs. This package provides approximately unbiased (AU) P value, as well as bootstrap probability (BP) value. AU P value is calculated using multi-scale bootstrap resampling, whereas BP value is calculated using normal bootstrap resampling. Results based on AU p-value are more accurate than those based on BP value. In this study, Pvclust-clusters with AU P value > 95% were considered as significant clusters.
2.5. Functional analysis of key DEGs based on literature mining
For the key DEGs identified above, literature mining was performed using “Gene Cluster With Literature Profiles” module in the online software GenCLiP 2.0 (http://ci.smu.edu.cn/)[18] to investigate their bio-functions. The corresponding criteria were Hit ≥ 3 and P value < .05. “Gene Cluster With Literature Profiles” module can use a fuzzy cluster algorithm to generate statistically over-represented keywords and thus to annotate the input genes, and Medline abstracts are utilized to link genes and keywords.
2.6. Construction of TF-DEG regulatory network
iRegulon uses a genome-wide ranking-and-recovery approach to enrich TFs and their optimal targets based on TRANSFAC, ENCODE, HOMER, JASPAR, and SwissRegulon databases, and it is generally utilized to construct transcriptional regulatory network. Therefore, iRegulon plugin[19] in Cytoscape (version 2.8; http://cytoscape.org)[20] was applied to predict the potential TFs targeting the key DEGs identified above. The criteria for this analysis were false discovery rate on motif similarity ≤ 0.001, identity between orthologous genes ≥ 0.05, and normalized enrichment score (NES) > 4.
2.7. Known PMO-related genes
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org/)[21] records various disease-related genes based on precise document processing and drug-target prediction. To examine whether the identified key genes were known PMO-related genes, CTD database was used to search genes related to “postmenopausal osteoporosis” at November 18, 2015.
3. Results
3.1. Identified DEGs
After data preprocessing, an expression matrix containing 12132 genes was generated from both GSE56116 and GSE57273. A total of 202 DEGs were identified between PMO-KY and control groups, including 101 up- and 101 down-regulated DEGs. Furthermore, 2066 DEGs were identified between After-treatment and Before-treatment groups, including 1157 up- and 909 down-regulated DEGs. After clustering analysis, the 202 DEGs clearly classified the PMO-KY and control samples (Fig. 1A), whereas the 2066 DEGs clearly distinguished the After-treatment and Before-treatment samples (Fig. 1B).
Figure 1.
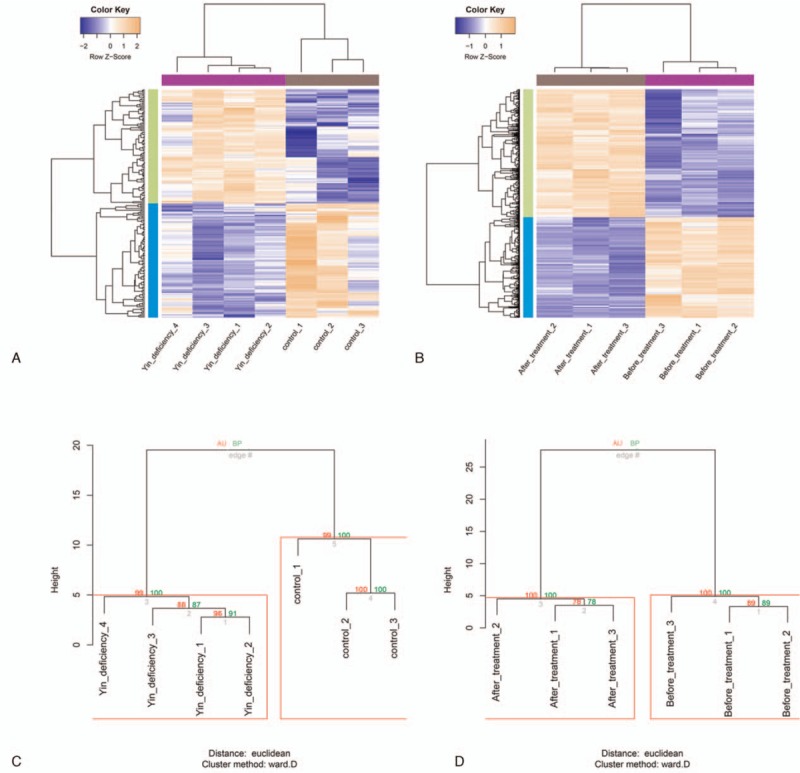
DEGs and key DEGs. A, Heatmaps of the 202 DEGs between PMO-KY and control groups. B, Heat maps of the 2066 DEGs between after-treatment and before-treatment groups. Blue stands for low expression level, whereas sandy brown represents high expression level. C, Clustering of the 86 key DEGs in PMO-KY and control samples. D, Clustering of the 86 key DEGs in after-treatment and before-treatment samples. Red letters on the edge stand for AU P-value (%), and green letters on the edge represent BP value (%). AU P-value > 95% are highlighted by red rectangles. AU P-value = approximately unbiased P-value, BP value = bootstrap probability value, DEGs = differentially expressed genes, LDP = Liuwei Dihuang pill, PMO-KY = postmenopausal osteoporosis with kidney-Yin deficiency.
3.2. Potential target genes of LDP
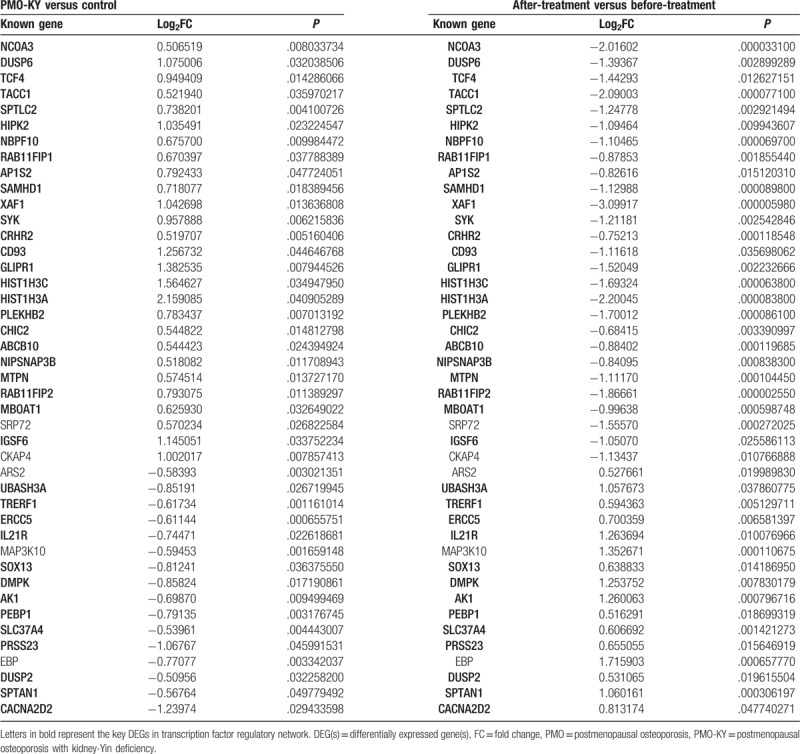
After comparing the 202 DEGs with the 2066 DEGs, a total of 86 genes were identified as key DEGs. Among these genes, 52 DEGs were up-regulated in PMO-KY but down-regulated after LDP treatment, and 34 DEGs were down-regulated in PMO-KY but up-regulated after LDP treatment. These DEGs might be the target genes of LDP. After clustering analysis, PMO-KY and control samples (Fig. 1C), as well as after-treatment and before-treatment samples (Fig. 1D), were clearly distinguished based on the 86 key DEGs, indicating the specificity of these DEGs. The total key DEGs are shown in Supplementary Table 1.
3.3. Potential functions of the key DEGs
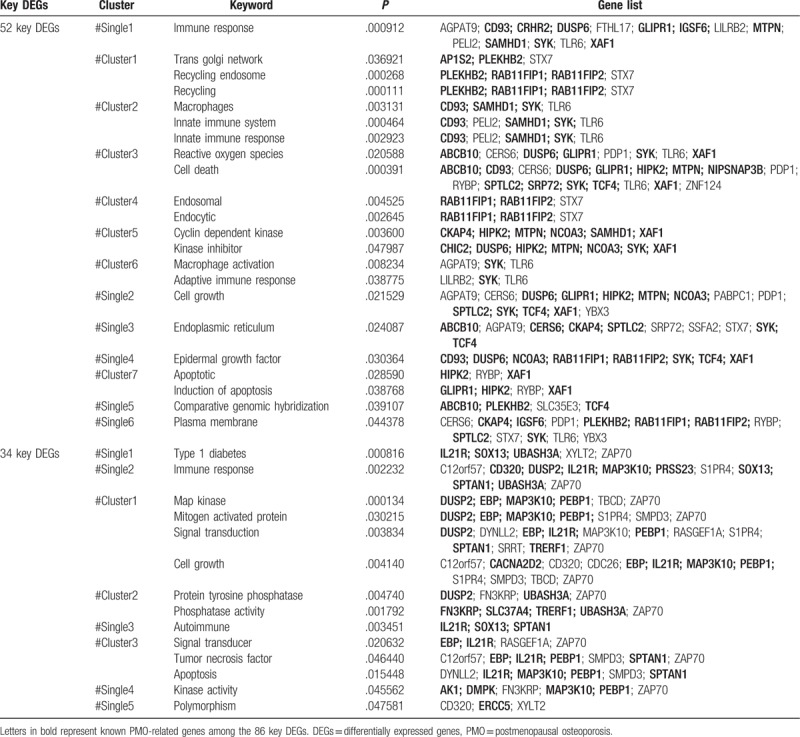
Based on literature mining, a total of 22 functions were enriched by the 52 key DEGs that were up-regulated in PMO-KY but down-regulated after LDP treatment (Fig. 2 A and Table 1). These functions were mainly related to immune response and cell death. Furthermore, 14 functions were enriched by the 34 key DEGs that were down-regulated in PMO-KY but up-regulated after LDP treatment (Fig. 2B and Table 1). These functions were mainly associated with cell growth, apoptosis, and immune response.
Figure 2.
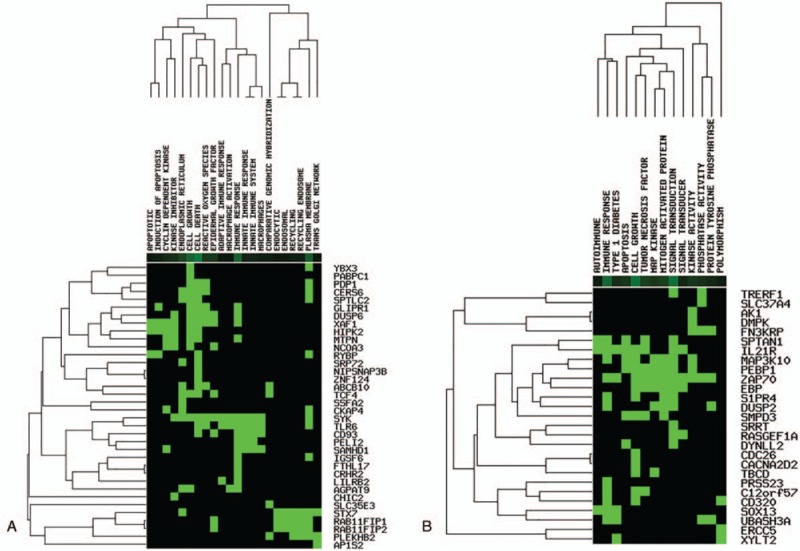
Functions of the 86 key DEGs. A, Functions enriched by the 52 key DEGs. B, Functions enriched by the 34 key DEGs. DEGs = differentially expressed genes.
Table 1.
Functions enriched by the 86 key DEGs.
3.4. TF-DEG regulatory networks
Based on TRANSFAC, ENCODE, HOMER, JASPAR, and SwissRegulon databases, 8 TFs were predicted to regulate 38 of the 52 key DEGs (Fig. 3A), and 7 TFs were predicted to target 27 of the 34 key DEGs (Fig. 3B). Expression levels of the TFs identified here were not significantly changed in PMO-KY or during the LDP treatment. In TF-DEG regulatory network of the 52 key DEGs, 7 DEGs (eg, NCOA3, TCF4, DUSP6, PELI2, and STX7) were predicted to be regulated by at least 5 TFs (Fig. 3A). Among them, all of NCOA3, TCF4, DUSP6, PELI2, and STX7 were predicted to be regulated by HOXA13 (target number = 11). Additionally, in TF-DEG regulatory network of the 34 key DEGs, 2 DEGs were targeted by at least 5 TFs (Fig. 3B).
Figure 3.
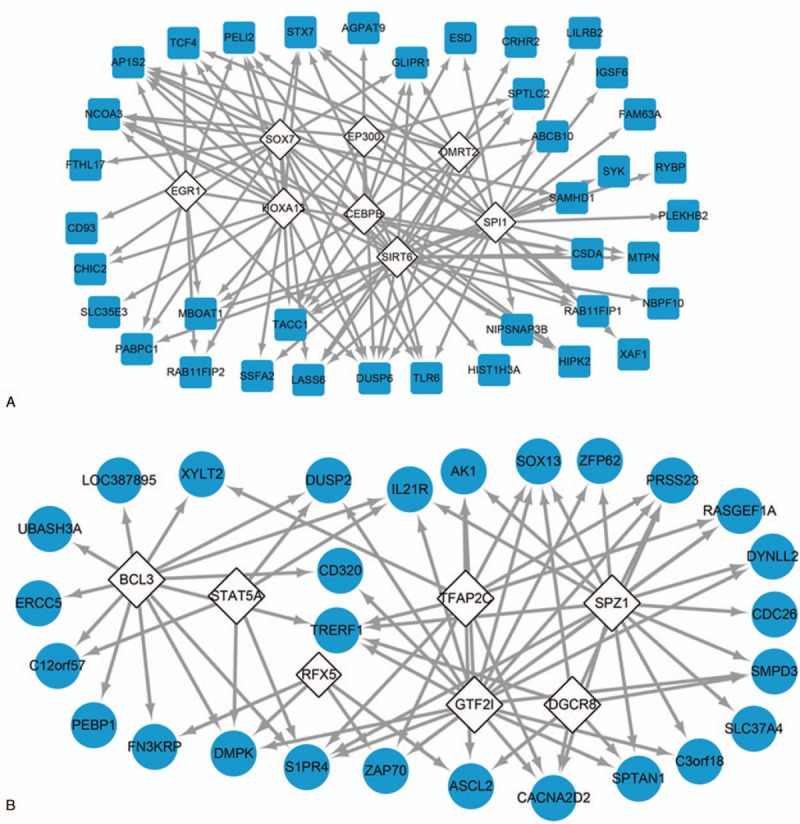
TF analysis of the 86 key DEGs. TF regulatory network of the 52 key DEGs. B, TF regulatory network of the 34 key DEGs. White diamond: TF; blue square: key DEG up-regulated in PMO-KY but down-regulated after LDP treatment; blue circle: key DEG down-regulated in PMO-KY but up-regulated after LDP treatment. DEG(s) = differentially expressed gene(s), LDP = Liuwei Dihuang pill, PMO-KY = postmenopausal osteoporosis with kidney-Yin deficiency, TF = transcription factor.
3.5. Known PMO-related genes
Based on CTD database, 43 of the 86 key DEGs had been previously found to be associated with PMO (Table 2). Among the 43 known PMO-related genes, 37 genes were present in TF-DEG regulatory networks, such as NCOA3, TCF4, and DUSP6. In addition, PELI2 and STX7 were not known PMO-related genes, indicating that they were novel genes identified in this study.
Table 2.
The 43 known PMO-related genes among the 86 key DEGs.
4. Discussion
As a worldwide health problem associated with population ageing, PMO seriously impairs the quality of life and causes disability and mortality in postmenopausal women.[2] LDP, a traditional Chinese medicine, can effectively cure PMO and PMO-KY.[9–12] However, its therapeutic mechanisms underlying the treatment of PMO-KY have not been fully understood. Based on bioinformatics approaches, we re-analyzed the microarray data about PMO-KY and LDP treatment. As a result, 86 potential target genes of LDP in the treatment of PMO-KY were identified. Specially, NCOA3, TCF4, and DUSP6 were known PMO-related genes, whereas PELI2 and STX7 were newly-identified genes that might be related to PMO-KY. Additionally, NCOA3, TCF4, DUSP6, PELI2, and STX7 were predicted to be targeted by HOXA13.
NCOA3, namely, SRC3 or AIB1, encodes nuclear receptor co-activator 3 that is important in bone and mineral metabolism. NCOA3 interacts with osteoporosis-related nuclear hormone receptors (eg, vitamin D and thyroid hormone receptors) ,[22] and it is associated with lumbar spine BMD.[23] Furthermore, NCOA3 is located within chromosomal genomic regions for femoral neck cross-sectional geometric variables that are risk factors of osteoporosis.[24] Furthermore, NCOA3 has also been reported to be located within quantitative trait locus of BMD in the mouse genome.[25] These studies suggested that NCOA3 was associated with low BMD. In this study, NCOA3 was upregulated in PMO-KY samples, and became downregulated after LDP treatment, indicating that NCOA3 might be a potential target of LDP.
TCF4, also referred to as Tcf7L2, encodes transcription factor 4 (Tcf4), which is a nuclear effector of the Wnt signaling, which integrates with Notch signaling and BMP signaling pathways in osteoblast differentiation.[26]Tcf4 is a final target of Wnt-dependent cellular control in osteoblast survival, growth, proliferation, function, and differentiation.[26,27] In the present study, TCF4 was upregulated in PMO-KY samples, and became downregulated after LDP treatment. A similar previous study has found that TCF4 is up-regulated in human mesenchymal stem cells (hMSC) from osteoporotic donors in responding to 10% uniaxial cyclic tensile strain that is able to promote osteogenesis.[28] Although there is no any other evidence that TCF4 is a responsive gene of LDP so far, we speculate that TCF4 may be a potential target of LDP in the treatment of PMO-KY based on the above results.
DUSP6 encodes dual specificity phosphatase 6 (Dusp6), the expression level of which in adipocytes that share the same cell lineage with osteoblasts is regulated by a local expression quantitative trait locus and correlates with BMD in mice ,[29] indicating the association of DUSP6 with osteoporosis. A recent study has reported that the expression of one homolog of DUSP6, DUSP12, in the neuroendocrine immunomodulation network is changed by Liuwei Dihuang decoction.[30] Therefore, we speculate that DUSP6 may be also a responsive gene of LDP.
In this study, PELI2 and STX7 were newly-identified genes that might be related to PMO-KY. PELI2 encodes pellino-2 protein, a member of Pellino protein family, proteins in which can catalyze the poly-ubiquitylation of interleukin-1 receptor-associated kinases in the Toll-like receptor signaling pathway, which plays key roles in cytokines production and innate immune system.[31] The important role of immunity in PMO has been previously demonstrated by multiple studies.[32,33] Furthermore, SNP rs398652 next to PELI2 is significantly related to leukocyte telomere length, and its variant alleles are associated with longer telomere length.[34] Telomere shortening causes cell apoptosis in osteoblasts and hMSCs and thus contributes to bone aging.[35] Defects in Wrn, a telomere maintenance molecule, inhibit osteoblast differentiation and thus promote osteoporosis.[35] Collectively, PELI2 may be closely related to PMO. STX7 encodes syntaxin 7, a member of soluble-N-ethylmaleimide-sensitive-factor accessory-protein receptors that participate in membrane fusion and cytokine release in almost all aspects of innate and adaptive immune responses.[36,37] As mentioned above, immunity plays key roles in PMO, thus, STX7 might be associated with PMO. Currently, there is no evidence to support the direct linkage between PELI2/STX7 and LDP, whereas, both PELI2 and STX7 were significantly up-regulated in PMO-KY compared with healthy controls, and downregulated after LDP treatment. Therefore, PELI2 and STX7 might be potential targets of LDP.
Strikingly, all of NCOA3, TCF4, DUSP6, PELI2, and STX7 were regulated by the TF HOXA13, a member of homeobox proteins. SNPs in homeobox A cluster are robustly associated with cortical volumetric BMD at the femoral neck trabecular and lumbar spine.[38]HOXA13 is located on 11q14-25, a well-replicated osteoporosis susceptibility loci, and it is a likely candidate osteoporosis susceptibility gene based on bioinformatics tools.[39] Therefore, HOXA13 might play a role in PMO-KY pathogenesis and LDP treatment via regulating the expressions of NCOA3, TCF4, DUSP6, PELI2 and STX7.
Despite the aforementioned results, there were several limitations in this study. The predicted results should be confirmed by laboratory data. Furthermore, the included samples for analysis should be more. In our further studies, more PMO-KY samples will be collected to validate the expression levels of the potential key genes and their relationships with HOXA13.
In conclusion, the above discussed genes (eg, NCOA3, TCF4, DUSP6, PELI2, and STX7) might be the therapeutic targets of LDP in the treatment of PMO-KY, and their expressions might be regulated by HOXA13. The potential identified target genes might provide a novel direction for future drug design. However, beyond the scope of this study, further investigation should be performed to fully evaluate the roles of these genes in PMO-KY and LDP treatment.
Author contributions
Conceptualization: Feng Xu, Feng Gao.
Data curation: Feng Xu.
Formal analysis: Feng Xu.
Methodology: Feng Xu, Feng Gao.
Supervision: Feng Gao.
Writing – original draft: Feng Xu.
Writing – review & editing: Feng Gao.
Supplementary Material
Footnotes
Abbreviations: AU = approximately unbiased, BMD = bone mineral density, BP = bootstrap probability, CTD = Comparative Toxicogenomics Database, DEGs = differentially expressed genes, Dusp6 = dual specificity phosphatase 6, hMSC = human mesenchymal stem cells, LDP = Liuwei Dihuang pill, PMO = postmenopausal osteoporosis, PMO-KY = PMO with kidney-Yin deficiency, TFs = transcription factors.
Supplemental Digital Content is available for this article.
The authors declare no conflicts of interest.
References
- [1].Raisz LG. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J Clin Invest 2005;115:3318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Burge R, Dawson-Hughes B, Solomon DH, et al. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J Bone Miner Res 2007;22:465–75. [DOI] [PubMed] [Google Scholar]
- [3].An S, Li E, Tong X. Study on relationship between estrogen receptor gene polymorphism and syndrome differentiation typing of female postmenopausal osteoporosis in traditional Chinese medicine. Zhongguo Zhong Xi Yi Jie He Za Zhi 2000;20:907–10. [PubMed] [Google Scholar]
- [4].Shen Ziyin WW. Dialectical reference standards of traditional Chinese medicine deficiency syndrome. Clin Focus 1987;4:189. [Google Scholar]
- [5].Cianferotti L, D’Asta F, Brandi ML. A review on strontium ranelate long-term antifracture efficacy in the treatment of postmenopausal osteoporosis. Ther Adv Musculoskelet Dis 2013;5:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tsai JN, Uihlein AV, Lee H, et al. Teriparatide and denosumab, alone or combined, in women with postmenopausal osteoporosis: the DATA study randomised trial. Lancet 2013;382:50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jiang Y, Zhang ZL, Zhu HM, et al. Menatetrenone versus alfacalcidol in the treatment of Chinese postmenopausal women with osteoporosis: a multicenter, randomized, double-blinded, double-dummy, positive drug-controlled clinical trial. Clin Interv Aging 2014;9:121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maeda SS, Lazaretti-Castro M. An overview on the treatment of postmenopausal osteoporosis. Arq Bras Endocrinol Metabol 2014;58:162–71. [DOI] [PubMed] [Google Scholar]
- [9].Peng S. Study of added decoction of six drugs containing Rehmannia Root on osteoporosis after menopause. Mod J Integr Tradit Chin West Med 2007;16:4592–3. [Google Scholar]
- [10].Xia B, Xu B, Sun Y, et al. The effects of Liuwei Dihuang on canonical Wnt/beta-catenin signaling pathway in osteoporosis. J Ethnopharmacol 2014;153:133–41. [DOI] [PubMed] [Google Scholar]
- [11].Lijing L, Ai-Hua Z, Ning Z, et al. A traditional Chinese medicinal formula, protects against osteoporosis. Pharmaceutica Analytica Acta 2013;4:2. [Google Scholar]
- [12].Xie YM, Yuwen Y, Dong FH, et al. Clinical practice guideline of traditional medicine for primary osteoporosis. Chin J Integr Med 2011;17:52–63. [DOI] [PubMed] [Google Scholar]
- [13].Lihua X, Juan C, Shengqiang L, et al. Effect of Liuweidihuang pill on JAK /STAT signaling pathway gene expression in postmenopausal osteoporosis with the kidney yin deficiency. Chin J Osteoporos 2014;20:741–6. [Google Scholar]
- [14].Lihua X, Chaoling F, Shengqiang L, et al. Influence of Liuwei Dihuang Pills on differentially expressed genes in kidney Yin deficiency patients with postmenopausal osteoporosis. Chin J Osteoporos 2015;21:971–85. [Google Scholar]
- [15].Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Warnes GR, Bolker B, Bonebakker L, et al. gplots: Various R programming tools for plotting data. R package version 2009;2 [Google Scholar]
- [17].Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006;22:1540–2. [DOI] [PubMed] [Google Scholar]
- [18].Wang JH, Zhao LF, Lin P, et al. GenCLiP 2.0: a web server for functional clustering of genes and construction of molecular networks based on free terms. Bioinformatics 2014;30:2534–6. [DOI] [PubMed] [Google Scholar]
- [19].Rekin's J, Annelien V, Hana I, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 2014;10:e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Davis AP, Grondin CJ, Lennon-Hopkins K, et al. The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res 2015;43(D1):D914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen H, Lin RJ, Schiltz RL, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 1997;90:569–80. [DOI] [PubMed] [Google Scholar]
- [23].Zmuda J, Cauley J, Ferrel R. Nuclear receptor coativator-3 (NCO A3/AIB1/SRC3) alleles are a strong correlate of bioavailable testosterone and vertebral bone mass in older men. J Bone Miner Res 2004;19:S387. [Google Scholar]
- [24].Xiong DH, Shen H, Xiao P, et al. Genome-wide scan identified QTLs underlying femoral neck cross-sectional geometry that are novel studied risk factors of osteoporosis. J Bone Miner Res 2006;21:424–37. [DOI] [PubMed] [Google Scholar]
- [25].Xiong Q, Han C, Beamer WG, et al. A close examination of genes within quantitative trait loci of bone mineral density in whole mouse genome. Crit Rev Eukaryot Gene Expr 2008;18:323–43. [DOI] [PubMed] [Google Scholar]
- [26].Lin GL, Hankenson KD. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J Cell Biochem 2011;112:3491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene 2004;341:19–39. [DOI] [PubMed] [Google Scholar]
- [28].Charoenpanich A, Wall ME, Tucker CJ, et al. Cyclic tensile strain enhances osteogenesis and angiogenesis in mesenchymal stem cells from osteoporotic donors. Tissue Eng Part A 2014;20:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mesner LD, Ray B, Hsu YH, et al. Bicc1 is a genetic determinant of osteoblastogenesis and bone mineral density. J Clin Invest 2014;124:2736–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou W, Cheng X, Zhang Y. Effect of Liuwei Dihuang decoction, a traditional Chinese medicinal prescription, on the neuroendocrine immunomodulation network. Pharmacol Ther 2016;162:170–8. [DOI] [PubMed] [Google Scholar]
- [31].Moynagh PN. The Pellino family: IRAK E3 ligases with emerging roles in innate immune signalling. Trends Immunol 2009;30:33–42. [DOI] [PubMed] [Google Scholar]
- [32].Faienza MF, Ventura A, Marzano F, et al. Postmenopausal osteoporosis: the role of immune system cells. Clin Dev Immunol 2013;2013:575936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhao R. Immune regulation of osteoclast function in postmenopausal osteoporosis: a critical interdisciplinary perspective. Int J Med Sci 2012;9:825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gu J, Chen M, Shete S, et al. A genome-wide association study identifies a locus on chromosome 14q21 as a predictor of leukocyte telomere length and as a marker of susceptibility for bladder cancer. Cancer Prev Res (Phila) 2011;4:514–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pignolo RJ, Suda RK, McMillan EA, et al. Defects in telomere maintenance molecules impair osteoblast differentiation and promote osteoporosis. Aging Cell 2008;7:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012;151:1256–69. [DOI] [PubMed] [Google Scholar]
- [37].Stow JL, Manderson AP, Murray RZ. SNAREing immunity: the role of SNAREs in the immune system. Nat Rev Immunol 2006;6:919–29. [DOI] [PubMed] [Google Scholar]
- [38].Yerges LM, Klei L, Cauley JA, et al. Candidate gene analysis of femoral neck trabecular and cortical volumetric bone mineral density in older men. J Bone Miner Res 2010;25:330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang QY, Li GH, Cheung WM, et al. Prediction of osteoporosis candidate genes by computational disease-gene identification strategy. J Hum Genet 2008;53:644–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


