Abstract
CRAC channel regulator 2A (CRACR2A) is expressed abundantly in T cells and acts as a signal transmitter between T cell receptor stimulation and activation of the Ca2+-NFAT and JNK-AP1 pathways. CRACR2A has been linked to human diseases in numerous genome-wide association studies and was shown to be one of the most sensitive targets of the widely used statin drugs. However, the physiological role of CRACR2A in T cell functions remains unknown. In this study, using transgenic mice for tissue-specific deletion, we show that CRACR2A promotes Th1 responses and effector function of Th17 cells. CRACR2A was abundantly expressed in Th1 and Th17 cells. In vitro, deficiency of CRACR2A decreased Th1 differentiation under non-polarizing conditions, while presence of polarizing cytokines compensated this defect. Transcript analysis showed that weakened TCR signaling by deficiency of CRACR2A failed to promote Th1 transcriptional program. In vivo, conditional deletion of CRACR2A in T cells ameliorated Th1 responses to acute lymphocytic choriomeningitis virus infection and imparted resistance to experimental autoimmune encephalomyelitis. Analysis of central nervous system from experimental autoimmune encephalomyelitis-induced mice showed impaired effector functions of both Th1 and Th17 cell types, which correlated with decreased pathogenicity. Collectively, our findings demonstrate the requirement of CRACR2A-mediated TCR signaling in Th1 responses as well as pathogenic conversion of Th17 cells, that occurs at the site of inflammation.
INTRODUCTION
Human Ca2+ release-activated Ca2+ channel regulator 2A (CRACR2A, or alternatively EFCAB4B) gene is located on chromosome 12. Links between polymorphism in CRACR2A and human diseases have been identified from numerous genome-wide association studies (GWAS) of Parkinson’s disease, non-alcoholic fatty liver disease (NAFLD), atrial fibrillation (AF), and chronic infection of human immunodeficiency virus type 1 (HIV-1) (1-4). However, the mechanisms underlying this link are largely unknown due to lack of information on the physiological role of CRACR2A. Recent studies have shed some light on the potential role of CRACR2A in T cell-mediated immunity. Engagement of T cell receptors (TCRs) with cognate antigens induces clustering and activation of enzymes and signaling adaptors including phospholipase C-γ1 (PLCγ1) and Vav1 at the immunological synapse, which are responsible for activation of downstream signaling cascades such as the Ca2+-nuclear factor of activated T cells (NFAT) and mitogen-activated protein kinase (MAPK) pathways (5-8). PLCγ1 produces a second messenger inositol 1,4,5-trisphosphate (InsP3) that depletes endoplasmic reticulum (ER) Ca2+ stores and triggers activation of extracellular Ca2+ entry via Ca2+ release-activated Ca2+ (CRAC) channels in a process termed as store-operated Ca2+ entry. Elevated cytoplasmic Ca2+ concentration activates the downstream calcineurin-NFAT pathway. Vav1 is a guanine nucleotide exchange factor that recruits small G proteins to activate the c-Jun N-terminal kinase (JNK) and p38 MAPK pathways that eventually turn on gene transcription by the activator protein 1 (AP1) transcription factors (9). Previously, we reported a function of CRACR2A in regulation of the Ca2+-NFAT and JNK MAPK signaling pathways (10, 11). The short, cytoplasmic isoform of CRACR2A, CRACR2A-c stabilizes CRAC channels by interaction with its key components, Orai1, the plasma membrane (PM) pore subunit and STIM1, the ER Ca2+ sensor necessary for activation of Orai1 channels. Differently from CRACR2A-c, the long isoform, CRACR2A-a is a component of vesicles. It is a member of the unique large Rab GTPase family that also includes Rab44 and Rab45 (11). CRACR2A-a contains multiple functional domains including the N-terminal domain that is identical with CRACR2A-c, a proline-rich protein-interacting domain, and a C-terminal Rab GTPase domain. GTP binding and prenylation are essential for localization of CRACR2A in vesicles while its interaction with Vav1 is necessary for activation of the JNK signaling pathway. Another interesting aspect of CRACR2A-a is its high sensitivity to statin drugs, that inhibit 3-hydroxyl-3-methyl-glutaryl-CoA (HMG-CoA) reductase, a key rate-liming enzyme in cholesterol biosynthesis pathway. Statin treatment-induced de-prenylation causes dissociation of CRACR2A-a from vesicles, leading to its degradation, thereby impairing T cell activation. Although many GWAS have uncovered CRACR2A for susceptibility to various human diseases (1-4), the physiological role of CRACR2A proteins largely remains unknown due to a lack of appropriate animal models.
For a productive immune response, T cells need to be activated by a combination of signals from TCRs, co-stimulatory receptors (e.g., CD28) and receptors for polarizing cytokines. Integration of these signals is essential for lineage determination of effector T cells. Strong TCR signaling blocks the function and expression of GATA3 resulting in inhibition of Th2 programs, and thus induces preferential differentiation of Th1 cells while weakened TCR signaling favors differentiation into Th2 cells by the default expression of GATA3 in naïve T cells (12-14). Consistently, defects in key TCR signaling pathways including the NFAT or JNK signaling pathways favors Th2 differentiation (15-19). In addition to the strength of TCR stimulation, presence of polarizing cytokines including IL-12 and IL-4 skew T cells into Th1 and Th2 cells, respectively. Th17 cells produce high amounts of IL-17A, IL-17F and IL-22, and have an essential role in host defense against pathogens as well as autoimmunity. Differentiation and effector functions of Th17 cells require optimal strength of TCR signaling as demonstrated by decreased Th17 differentiation after deletion of TCR signaling molecules including Itk, PKCθ, and Orai1 (20-22). Differentiation of Th17 cells requires various polarizing cytokines (e.g., IL-1α/β, IL-6, IL-23 or TGFβ). After differentiation at the priming sites (e.g. lymph nodes), Th17 cells become plastic and transit into a highly pathogenic state after migration to the sites of inflammation in distinct tissues. After transition, these ex-Th17 cells produce granulocyte macrophage colony-stimulating factor (GM-CSF) and IFN-γ that play crucial roles in pathogenicity of these cells. GM-CSF plays an important role in recruitment of myeloid lineage innate immune cells (e.g. monocytes). It was shown that deficiency of GM-CSF almost completely abolished the onset of experimental autoimmune encephalomyelitis (EAE) and conversely, GM-CSF-producing cells regardless of effector T cell subsets, were sufficient to induce EAE (23-27). In addition, the transition of Th17 cells into IFN-γ-producing ex-Th17 cells is important for pathogenicity of these cells and this transition requires Th1-polarizing cytokines (e.g., IL-12) and transcription factors (e.g., T-bet and Runx1/3) (28-34). These observations suggest that transition of Th17 cells into highly pathogenic T cells at the inflamed site requires at least, a part of the Th1 program.
Here we show that CRACR2A-mediated signaling plays a key role in Th1 cell differentiation. In vitro, in the absence of any polarizing cytokines and antibodies, deficiency of CRACR2A decreased differentiation of Th1 cells, and correspondingly increased differentiation of Th2 cells. Under in vivo conditions using acute lymphocytic choriomeningitis virus (LCMV) infection model we observed impaired Th1 response in T cell-specific CRACR2A knockout mice. Further, T cell-specific deletion of CRACR2A also imparted resistance to experimental autoimmune encephalomyelitis (EAE). In this model, in addition to Th1 cells, Cracr2afl/fl/Cd4-Cre Th17 cells also exhibited defects in effector functions at the central nervous system (CNS). Furthermore, results from analysis of adoptive transfer of Th17 cells and in vitro conversion model showed that Cracr2afl/fl/Cd4-Cre Th17 cells had weakened pathogenicity as judged by reduced clinical symptoms in recipients of Cracr2afl/fl/Cd4-Cre Th17 cells due to impaired transition into Th1-like Th17 cells that produce IFN-γ and GM-CSF cytokines. Therefore, our findings demonstrate that TCR signaling mediated by CRACR2A is crucial for Th1 differentiation as well as local effector function of Th17 cells.
MATERIALS AND METHODS
Chemicals.
Phorbol 12-myristate 13-acetate (PMA), and ionomycin were purchased from EMD Millipore. Brefeldin A was purchased from ThermoFisher Scientific.
Plasmids and cells.
pLKO.1 plasmids encoding scrambled or human CRACR2A – targeting shRNAs (5 different shRNAs) were purchased from Dharmacon (TRC Lentiviral Human CRACR2A shRNA). The shRNA sequences are described in Table S1. Human HEK293T and Jurkat T cells were purchased from the American Type Culture Collection (ATCC).
Mice.
Generation of Cracr2afl/fl animals has been previously described (11). These mice were bred with Cd4-Cre mice (from The Jackson Laboratory) for 2 generations to obtain T cell-specific deletion of Cracr2a. 8-10-week old age and sex matched control (Cracr2afl/fl) and Cracr2afl/fl/Cd4-Cre littermate animals were used for all in vivo experiments. For passive EAE experiments, age matched donor and Rag2−/− recipients (purchased from The Jackson Laboratory) were used. All animals were maintained in pathogen-free barrier facilities and used in accordance with protocols approved by the Institutional Animal Care and Use Committee at the University of California, Los Angeles.
T cell purification, differentiation, stimulation, and staining.
T cell purification, activation and differentiation were carried out as previously described (22). Briefly, naïve CD4+ T cells were enriched by magnetic sorting from single-cell suspensions generated by mechanical disruption of spleens and lymph nodes of adult mice using MagniSort naïve CD4+ T cell enrichment kit according to manufacturer’s instructions (ThermoFisher Scientific). For effector T cell differentiation, cells were stimulated with 1 µg/ml of anti-CD3 antibody (1452C11, Bio X Cell) and 1 µg/ml of anti-CD28 antibody (Clone 37.51, Bio X Cell) for 48 hours on a plate coated with 0.3 mg/ml of rabbit anti-hamster antibody (MP Biomedicals). CD4+CD25- T cells were cultured with 10 μg/ml anti-IL-4 antibody (Bio X cell), and 10 ng/ml IL-12 for Th1 differentiation; 20 μg/ml anti-IFN-γ antibody (Bio X Cell), 2.5 μg/ml anti-IL-12 antibody and 10 ng/ml IL-4 (Peprotech) for Th2 differentiation; and 10 μg/ml anti-IL-4 antibody, 20 μg/ml anti-IFN-γ antibody, 30 ng/ml IL-6 (Peprotech), 3 ng/ml TGF-β (Peprotech) and 10 ng/ml IL-23 (R&D Systems) for Th17 differentiation. On day 4, differentiated T cells were restimulated with 80 nM PMA and 1 µM ionomycin for 5-6 hours for cytokine analysis. Brefeldin A (1 µg/ml) was added for the last 2 hours. Cells were harvested, washed in phosphate-buffered saline (PBS), permeabilized with 0.5% saponin and stained intracellularly for indicated cytokines. For re-stimulation of Th17 cells, CD4+CD25- T cells cultured under Th17 differentiation conditions (without IL-23) were re-stimulated on day 4 with 0.5 µg/ml of anti-CD3 antibody and 0.5 µg/ml of anti-CD28 antibody for 24 hours on a plate coated with 0.3 mg/ml of goat anti-hamster antibody. Cells were cultured with or without 1 ng/ml of IL-12. On day 7, cells were re-stimulated with 80 nM PMA and 1 µM ionomycin for cytokine analysis. For staining of transcription factors T-bet and GATA-3, differentiated effector T cells were fixed/permeabilized and stained using Transcription Factor Staining Buffer Set (BD Pharmingen). For surface staining, 1 million cells from single cell suspensions of thymus, spleen and lymph nodes were stained with indicated antibodies in PBS plus 1% fetal bovine serum, at 4˚C for 20 mins, washed and used for data acquisition. Data were acquired using FACSCalibur (Becton Dickinson) or BD LSRFortessa cell analyzers and analyzed using FlowJo software (Tree Star).
Immunoblotting.
For immunoblot analyses, naïve CD4+ T cells isolated from C57Bl6/J mice were differentiated into various effector T cells. 4 days after differentiation, cells were stimulated with PMA plus ionomycin and stained for signature cytokines to validate appropriate differentiation. 10 million differentiated effector T cells were then lysed in RIPA buffer (10 mM Tris-Cl pH 8.0, 1% Triton X-100, 0.1% SDS, 140 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate and protease inhibitor cocktail [Roche]) and centrifuged to remove debris. Samples were separated on 8-10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes and subsequently analyzed by immunoblotting with anti-CRACR2A antibody. Polyclonal rabbit antibody for detection of CRACR2A was generated using purified human CRACR2A-c protein (Open Biosystems, Huntsville, AL) and used at 1: 5,000 dilution. Similar pattern of expression was also observed using commercially available anti-CRACR2A antibody (Proteintech, #15206-1-AP and Millipore-Sigma, #SAB5300583). Anti-β-actin antibody was purchased from Santa Cruz Biotechnology (clone I-19). Chemiluminescence images were acquired using an Image reader LAS-3000 LCD camera (FujiFilm).
Cytokine measurements by enzyme-linked immunosorbent assay.
Single cell suspensions of draining lymph nodes from control (Cracr2afl/fl) and Cracr2afl/fl/Cd4-Cre mice immunized with myelin oligodendrocyte glycoprotein (MOG) peptide were stimulated with 20 µg/ml MOG peptide for 72 hours and culture supernatants were used for detection of secreted IFN-γ and IL-17A cytokines using sandwich enzyme-linked immunosorbent assay (ELISA) (Ready-SET-Go! ELISA kit, eBioscience).
Human peripheral blood mononuclear cell culture and analysis.
Peripheral blood mononuclear cells (PBMCs) were obtained under federal and state regulations from the CFAR Virology core Laboratory at UCLA that were prepared from buffy coats from healthy, unidentified adult donors using FicollPAQUE gradients. PBMCs were activated with anti-CD3/CD28 beads (Miltenyi Biotech) and cultured in T cell media (DMEM containing 20% fetal bovine serum and 1% Pen-Strep) supplemented with 20 U/ml IL-2 (Peprotech). Depletion of CRACR2A was carried out by lentiviral infection of cells on days 1 and 2. 48 hours after infection, cells were selected with 1 µg/ml of puromycin for 2 days and then expanded for 3 days with fresh media. On day 7, cells were activated with 80 nM of PMA, 1 µM of ionomycin, and Brefeldin A (3 µg/ml) for 5 hrs, surface stained with anti-CD4 antibody-FITC, and intracellularly stained with anti-IL-4 antibody-APC and anti-IFN-γ antibody-PE. CD4 antibody-FITC-positive cells were gated for analysis. For flow cytometry, the following human specific antibodies were used: anti-CD4 antibody-FITC (OKT4, eBioscience), anti-IL-4 antibody-APC (8D4.8, eBioscience) and anti-IFN-γ antibody-PECy7 (45.B3, eBioscience).
shRNA-mediated depletion.
To generate lentiviruses for transduction, HEK293T cells were transfected with plasmid(s) encoding shRNA and packaging vectors (pMD2.G and psPAX2, Addgene) using calcium phosphate transfection method. Culture supernatants were harvested at 48 and 72 hrs post transfection and used for infection of human PBMCs together with polybrene (8 µg/ml) using the spin-infection method. Cells were selected with puromycin (1 µg/ml) for 48 hrs post infection. Initially stable Jurkat T cells were generated to identify the shRNA that gave maximal depletion of CRACR2A proteins and shRNA#2 was then used for depletion of CRACR2A in human PBMCs.
RNA-seq analysis.
For RNA-seq experiments naïve T cells from control and Cracr2afl/fl/Cd4-Cre mice were purified and stimulated on plate-coated anti-CD3 and anti-CD28 antibodies for 48 hours. After 48 hours, cells were taken off the plate and cultured in medium containing 10 U/ml recombinant human IL-2 (NCI Preclinical repository) for two days. At day 4 post isolation, ~10 million control and CRACR2A KO ThN cells (non-polarized) were stimulated with 20 nM PMA and 1 µM ionomycin for 5 hours and harvested in TriZol reagent (Invitrogen) for RNA preparation. RNA was isolated using RNA purification kit (ThermoFisher Scientific), quantified and used for purification of mRNA, cDNA synthesis and library preparation using TruSeq RNA Sample preparation kit (Illumina) following manufacturer’s instructions. The library of four samples was multiplexed and sequenced using Illumina Hiseq 2000 platform. After demultiplexing, each sample got ~30 million 50 b.p. single-end reads. Tophat (35) was used to align reads to mouse genome GRCm38/mm10 with Ensembl 74 annotation with a cut-off of two maximum mismatches. Only uniquely mapped reads were used for downstream analysis. Htseq-count from HTSeq (version 0.5.3p9) was used to quantify the number of reads aligned per gene with option intersection-nonempty and Ensembl 74 gene sets. Differential expression of genes was performed with DESeq v1.14 (36, 37). Genes with 0 reads across all samples were removed. Replicates were grouped and comparison was done with default DESeq parameters. Genes with adjusted p value < 0.001 and > 2-fold change were considered differentially expressed.
LCMV infection, major histocompatibility complex tetramer and intracellular cytokine staining.
Mice were infected intraperitoneally with 2 × 104 plaque-forming units of LCMV-Armstrong. Viral stocks were prepared and viral titers were quantified as described (38). The absolute number of virus-specific T cells was determined by multiplying the frequency of tetramer+ or cytokine producing CD4 T cells by the total number of cells in the spleen. Splenocytes were stained immediately after isolation with tetramers recognizing the MHC class II-IAb restricted LCMV-gp67 peptide (amino acids 61-80 of LCMV glycoprotein) and for surface expression of CD4. Splenocytes were also stained with tetramers for H-2Db and gp33 (amino acids 33-41 of LCMV glycoprotein) and for surface expression of CD8. Tetramers were obtained from the Tetramer Core Facility of the US National Institutes of Health. For analysis of cytokine expression, splenocytes were stimulated for 5 hours with 5 µg/ml of the MHC class II-restricted LCMV GP61-80 peptide or MHC class I-restricted LCMV GP33-41 peptide, in the presence of 50 U/ml recombinant mouse IL-2 (R&D Systems) and 1 mg/ml brefeldin A (Sigma-Aldrich). Cells were stained for surface expression of CD4 or CD8, then were fixed, permeabilized and stained with anti-TNF, anti-IFN-γ and anti-IL-2 antibodies. Flow cytometry was performed with a FACSVerse (Becton Dickinson) analyzer and data analyzed using FlowJo software (Tree Star).
EAE induction and analyses.
Mice were immunized subcutaneously on day 0 with 100 µg of MOG35-55 peptide (N-MEVGWYRSPFSRVVHLYRNGK-C, Genscript) emulsified in complete Freund’s adjuvant (CFA, Difco) supplemented with 5 mg/mL of Mycobacterium tuberculosis H37Ra (Difco). The mice were also injected i.p. with pertussis toxin (200 ng/mouse, List Biological Laboratories) on days 0 and 2. For passive EAE, donor control and Cracr2afl/fl/Cd4-Cre mice were first immunized with MOG35-55 subcutaneously as described above. At the first sign of EAE symptoms (score 0.5), draining lymph node cells were collected and cultured under Th17-polarizing conditions - with 20 ug/ml of MOG35-55 peptide, IL-23 (10 ng/mL), IL-1β (10 ng/ml), anti-IFN-γ antibody (10 µg/ml), anti-IL4 antibody (10 µg/ml) and anti-IL-12 antibody (10 µg/ml) for 3 days. At day 3, 5 × 106 cells were injected intravenously into Rag2−/− mice. EAE severity was scored according to the following clinical scoring system: 0, no clinical signs; 1, paralyzed tail; 2, partial hind leg paralysis; 3, complete hind leg paralysis or partial hind and front leg paralysis; 4, complete hind and partial front leg paralysis; 5, complete hind and front leg paralysis (moribund). When a mouse was euthanized because of severe paralysis, a score of 4 was entered for that mouse for the rest of the experiment.
Isolation and analysis of cells from the CNS in EAE-induced mice.
To isolate mononuclear cells from spinal cords and brains, tissues were digested with collagenase and DNase I (Roche) for 30 min at 37°C, and cells were separated on a 40-80% Percoll gradient by centrifugation at 500g for 30 min. Cells at the 40-80% interface were collected. Cells were also isolated from draining lymph nodes by passing through nylon mesh, followed by lysis of blood cells and washing with PBS. For intracellular cytokine staining, cells were stimulated with 80 nM PMA and 1 µM ionomycin in the presence of 3 μg/ml brefeldin A for 5 hours and stained for CD4, IFN-γ, IL-17A and GM-CSF.
Real-time quantitative PCR.
For real-time quantitative PCR (qPCR), total RNA from cells isolated from the CNS and draining lymph nodes was extracted using the Direct-zol RNA Prep kit (Zymo Research) as per the manufacturer’s instructions. cDNA was synthesized from total RNA using oligo(dT) primers and Superscript IV First-Strand cDNA synthesis kit (Invitrogen). Quantitative real-time PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) on an iCycler IQ5 system (Bio-Rad) using the primers described in Table S1. Threshold cycles (CT) for all the candidate genes were normalized to the CT values for β-actin to obtain ΔCT and further normalized to the values obtained for control samples to obtain ΔΔCT. The specificity of primers was examined by melt-curve analysis and agarose gel electrophoresis of PCR products.
Statistical analysis.
Statistical analysis was carried out using the Mann-Whitney U test to assess the significance between EAE-induced groups and two-tailed Student’s t-test for other analyses. Differences were considered significant when p values were < 0.05.
RESULTS
CRACR2A signaling regulates differentiation of Th1 cells under non-polarizing conditions
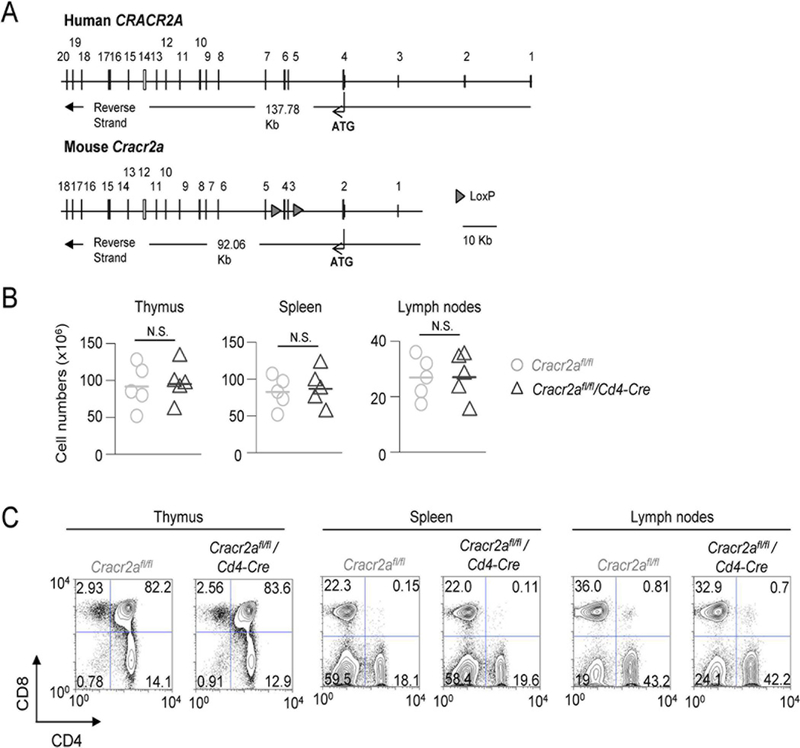
Human and mouse genome encode two transcriptional isoforms of CRACR2A, CRACR2A-a (∼80 kDa) and CRACR2A-c (∼45 kDa). Previously, we showed that CRACR2A-a and CRACR2A-c have overlapping role in the regulation of CRAC channels and CRACR2A-a also uniquely regulates JNK activation after translocation from the trans Golgi network to the immunological synapse via vesicles (10, 11). We also observed predominant expression of CRACR2A-a in the lymphoid organs including spleen and lymph nodes. To determine the physiological role of CRACR2A proteins, we generated Cracr2afl/fl/Cd4-Cre mice by deleting exons 3 and 4, resulting in loss of expression of both the isoforms, CRACR2A-a and CRACR2A-c (Fig. 1A). We showed that Cracr2afl/fl/Cd4-Cre T cells had defects in both the Ca2+-NFAT and JNK-AP1 signaling pathways after TCR stimulation (11). Here we sought to examine the physiological outcomes of reduced Ca2+-NFAT and JNK signaling pathways in Cracr2afl/fl/Cd4-Cre T cells. Development of CD4+ and CD8+ T cells in Cracr2afl/fl/Cd4-Cre mice did not show significant differences from their control counterparts in terms of cell numbers and frequencies (Fig. 1B and C). In addition, steady state T cell activation levels were similar between control and Cracr2afl/fl/Cd4-Cre T cells, when judged by surface expression of CD62L, CD44, CD25 and CD69 in CD4+ and CD8+ cells collected from the spleen and lymph nodes (Suppl. Fig. 1A and B). Finally, FOXP3 expression was also similar between control and Cracr2afl/fl/Cd4-Cre CD4+ T cells isolated from the thymus and lymph nodes (Suppl. Fig. 1C). Collectively, these results suggest that CRACR2A proteins do not play a major role in development or homeostasis of conventional and regulatory T cells.
Figure 1. CRACR2A deletion does not affect development of T cells in vivo.
(A) Schematic showing exon-intron structure of human CRACR2A and mouse Cracr2a gene. For generation of Cracr2afl/fl/Cd4-Cre mice, exons 3 and 4 were flanked with loxP sites (triangles). Alternative splicing from exon 2 to exon 5 causes frameshift mutation, resulting in loss of expression of both the isoforms, CRACR2A-a and CRACR2A-c.
(B) Total cell numbers from thymi, spleen, and lymph nodes of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice. Each symbol represents data from an independent animal. N.S.-not significant.
(C) Representative flow plots showing frequency of CD4+ and CD8+ populations from lymphoid organs of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice. Data are representative of three independent experiments.
To examine its function in different effector T cell populations, we first checked expression of CRACR2A proteins in differentiated T cells. Naïve T cells were cultured in vitro in the presence of appropriate cytokines and antibodies to differentiate into Th1, Th2, Th17 and iTreg cells, in addition to ThN cells, which refer to T cells activated in the absence of any polarizing cytokines or antibodies. Immunoblot analysis showed that both CRACR2A-a (∼80 kDa) and CRACR2A-c (∼45 kDa) were abundantly expressed in ThN, Th1 and Th17 cells and lesser in Th2 and regulatory T cells (Fig. 2A). This expression analysis suggests a more selective role of CRACR2A proteins in functions of inflammatory Th1 and Th17 cells. To check their role in effector T cell functions, we differentiated control and Cracr2afl/fl/Cd4-Cre naïve T cells into various effector T cell populations and examined their cytokine production profiles. Cracr2afl/fl/Cd4-Cre T cells cultured under non-polarizing conditions showed ~50% reduction in IFN-γ expression and almost ~threefold increase in IL-4 expression when re-stimulated with either anti-CD3 and anti-CD28 antibodies or PMA and ionomycin (Fig. 2B). Interestingly, under Th1-polarizing conditions in the presence of IL-12, which activates STAT4-mediated signaling pathway independently from JNK and Ca2+-NFAT pathways triggered by TCR stimulation (39), Cracr2afl/fl/Cd4-Cre T cells did not show any impairment in IFN-γ expression, affirming the previous observation that the defect in Th1 differentiation in Cracr2afl/fl/Cd4-Cre T cells specifically stems from weakened TCR signaling (11). In support of our protein expression analysis, we did not observe any defect in Th2 differentiation in Cracr2afl/fl/Cd4-Cre T cells as judged by IL-4 expression. Surprisingly, even though both CRACR2A-a and CRACR2A-c were abundantly expressed in Th17 cells, their deficiency did not impair Th17 differentiation in the presence of saturating amounts of polarizing cytokines, IL-6, TGF-β and IL-23, as judged by expression of IL-17A (Fig. 2B).
Figure 2. CRACR2A deficiency impairs Th1 differentiation in vitro.
(A) Schematic showing the domain structure of human CRACR2A-a and CRACR2A-c (top). CRACR2A-a and CRACR2A-c share EF-hand motifs and a coiled-coil domain that interacts with CRAC channels to regulate Ca2+ entry. CRACR2A-a contains additional proline-rich domain (PRD) and a Rab GTPase domain with a prenylation site at the C terminus. Immunoblot analysis showing expression of CRACR2A-a (∼80 kDa) and CRACR2A-c (∼45 kDa) in various effector T cell subsets (bottom). Naïve CD4+CD25- T were differentiated into effector T cells under non-polarizing (ThN) conditions or in the presence of cytokines/antibodies to induce polarization into indicated effector T cell populations and cellular extracts were analyzed for expression of CRACR2A-a and CRACR2A-c using β-actin as a loading control. Bar graph (right) shows densitometric analysis of CRACR2A-a in various effector T cell populations after normalization to that in ThN cells.
(B) Naive T cells from Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice differentiated under ThN conditions were stimulated with anti-CD3 and anti-CD28 antibodies or PMA plus ionomycin for 5 hours. Representative flow plots showing expression of IFN-γ and IL-4 are shown (left). Graph on the right shows normalized frequencies (± s.e.m.) of IFN-γ + or IL-4+ population from T cells cultured under non-polarizing conditions, IFN-γ+ population from Th1 cells, IL-4+ population from Th2 cells, and IL-17A+ population from Th17 cells. Cytokine frequencies are normalized to those of littermate controls. Each symbol represents data obtained from an independent animal.
(C) Levels of secreted IFN-γ and IL-4 cytokines from supernatants of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre naïve T cells stimulated with anti-CD3 and anti-CD28 antibodies under ThN conditions for 48 hours.
(D) Representative flow plots (left) and quantification graphs (right) showing expression of T-bet and GATA-3 in Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre T cells stimulated under ThN conditions. Each symbol represents data obtained from an independent animal.
(E) Representative flow plots (left) and quantification graphs (right) showing IFN-γ and IL-4 expression among CD4+ T cells in human PBMCs transduced with lentiviruses encoding control (Scr) or CRACR2A-targeting shRNAs after re-stimulation with PMA plus ionomycin. Each symbol represents data obtained from an independent donor. * p < 0.05, ** p < 0.005, *** p < 0.0005.
Since many small Rab GTPases are known to be involved in protein secretion, we examined whether CRACR2A proteins play any role in cytokine secretion. We expected to observe greater reduction in extracellularly secreted cytokines compared to those intracellularly, if CRACR2A proteins are also required for secretion. However, we observed similar reduction in intracellular and secreted cytokine levels from control and Cracr2afl/fl/Cd4-Cre T cells (Figs. 2B and C), suggesting that CRACR2A proteins play a primary role in signaling pathways involved in transcription of cytokine genes, rather than cytokine secretion. Furthermore, consistent with the cytokine profile, we observed reduced T-bet and increased GATA-3 expression in Cracr2afl/fl/Cd4-Cre T cells cultured under non-polarizing conditions (Fig. 2D). These results suggested that CRACR2A proteins play an active role in transcriptional programs of Th1 differentiation by promoting T-bet expression and suppressing GATA-3 function.
To examine whether the role of CRACR2A is conserved in human T cells, we depleted its expression using shRNA in human PBMCs cultured under non-polarizing conditions. Depletion of CRACR2A proteins led to a profound reduction in IFN-γ and increased production of IL-4, similar to our observations with murine Cracr2afl/fl/Cd4-Cre T cells (Fig. 2E). In summary, our results suggest an important role of CRACR2A proteins in Th1 differentiation in humans and mice, especially in the absence of saturating concentration of polarizing cytokines, potentially via their known functions in regulation of the Ca2+-NFAT and JNK-AP1 pathways.
CRACR2A-mediated signaling contributes to the transcriptional program to drive Th1 differentiation
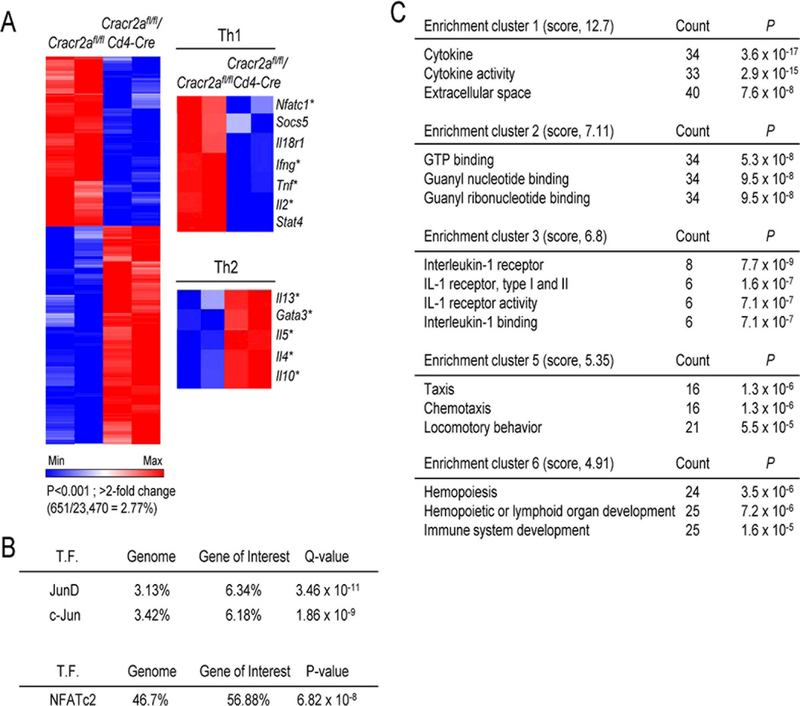
To understand how CRACR2A-mediated Ca2+-NFAT and JNK-AP1 pathways influence T cell differentiation, we performed transcriptome analyses from control and Cracr2afl/fl/Cd4-Cre T cells cultured under non-polarizing conditions. Transcription of < 3% of genes was significantly influenced in Cracr2afl/fl/Cd4-Cre T cells (P < 0.001 and > 2-fold change, 651/23,470 genes = 2.77% of genome; Fig. 3A). This gene list included cytokines, cytokine receptors, signaling molecules and transcription factors involved in Th1 and Th2 differentiation. Consistent with the role of CRACR2A proteins in the Ca2+-NFAT and JNK pathways, a majority of these genes including Il2, Tnf, Il4, Il5, Il13, Ifng, Gata3 and Nfatc1 are known to be regulated by the NFAT and/or AP1 transcription factors (40-42). Furthermore, among the differentially expressed genes in Cracr2afl/fl/Cd4-Cre T cells, using the ENCODE ChIP-Seq Significance Tool (http://encodeqt.stanford.edu/hyper/) or by calculation of static potential, we also observed enrichment of AP1 and NFAT-binding sites (Fig. 3B). Pathway analysis using the DAVID (Database for Annotation, Visualization and Integrated Discovery) bioinformatics database showed that signaling pathways mediated by CRACR2A proteins influenced expression of cytokines, chemokines, and factors for T cell differentiation (Fig. 3C), enrichment clusters 1, 5, and 6). Other gene clusters included pathways associated with GTP binding proteins and IL-1 receptor signaling. It is possible that there is compensatory expression of other GTP binding proteins in Cracr2afl/fl/Cd4-Cre T cells. One candidate from IL-1 receptor cluster was IL-18 receptor that is important for Th1 differentiation (43). Together, these data indicated that CRACR2A deficiency preferentially altered expression of genes regulated by the Ca2+-NFAT and JNK-AP1 signaling pathways affecting transcriptional programs for T cell differentiation.
Figure 3. CRACR2A is involved in the transcriptional program for Th1 differentiation by modulation of the Ca2+-NFAT and JNK-AP1 pathways.
(A) Heat map of gene expression in control and Cracr2afl/fl/Cd4-Cre CD4+ T cells cultured under ThN conditions and stimulated for 5 hours with PMA plus ionomycin. Heat map shows data from 651 genes (2.77%) differentially expressed with a P value of < 0.001 and a fold change of > 2.0. Right panels show heat map of genes involved in Th1 (top) or Th2 (bottom) differentiation. Data are representative of one experiment performed in duplicate from two independent Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice. * indicates genes known to be regulated by NFAT and/or AP1 transcription factors (see text).
(B) Predicted transcription factor binding sites at the promoters of genes from the entire genome or those differentially expressed in Cracr2afl/fl/Cd4-Cre T cells. Genes with binding sites for transcription factors (T.F.) at transcription start sites (TSS) ± 500 bps are indicated as a percentage of total expressed genes (Genome) or differentially expressed genes in Cracr2afl/fl/Cd4-Cre CD4+ T cells (Gene of Interest). The percentages and the false discovery rates (Q values) were generated using the ENCODE ChIP-Seq Significance Tool (top). The static potential of NFATc2 binding motif was calculated by identifying the sequence matches of the binding motif in the transcription start sites (TSS) ± 500 bps. Murine NFATc2 binding motif (the position frequency matrix, or PFM) was retrieved from bioconductor package MotifDb. The hypergeometric P values were calculated by comparing background (whole genome) to genes of interest to see enrichment of NFATc2 binding site (below).
(C) Pathway enrichment analysis of genes differentially expressed in Cracr2afl/fl/Cd4-Cre ThN cells than their control counterparts, assessed with the DAVID database and presented as total genes meeting that criterion in each pathway (Count), along with P values. Scores in parenthesis indicate the overall enrichment score calculated with the EASE score (modified Fisher’s exact P value) for each term member according to the DAVID database.
CRACR2A regulates Th1 responses to acute infection with lymphocytic choriomeningitis virus
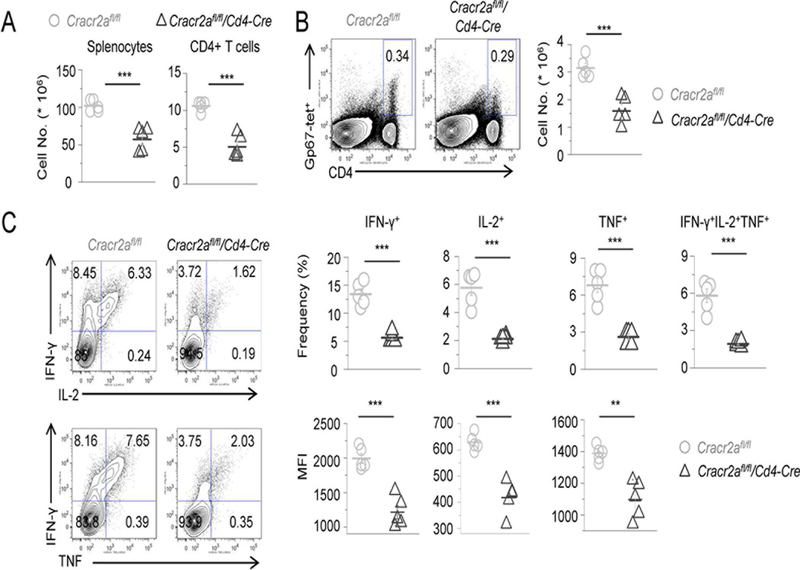
Results from in vitro T cell culture showed that CRACR2A deficiency influenced Th1 differentiation, but not in the presence of polarizing cytokines (Fig. 2B). To determine if CRACR2A signaling is required for efficient Th1 responses to virus infection in vivo, we infected control and Cracr2afl/fl/Cd4-Cre mice with the Armstrong strain of LCMV to induce acute infection. In response to acute LCMV infection, CD4+ T cells predominantly differentiate into Th1 or T follicular helper cells (44). In this infection model, CD4+ Th1 immunity is characterized by the expression of IFN-γ, tumor necrosis factor (TNF) and IL-2 (38). Eight days after infection (the peak of the antiviral effector response), we observed reduced numbers of total splenocytes and CD4+ T cells in Cracr2afl/fl/Cd4-Cre mice compared to littermate controls (Cracr2afl/fl; Fig. 4A). Furthermore, the numbers of LCMV-specific CD4+ T cells were substantially decreased in Cracr2afl/fl/Cd4-Cre mice (Fig. 4B). Consistent with decreased Th1 response, Cracr2afl/fl/Cd4-Cre mice had reduced frequencies and numbers of IFN-γ, IL-2 and TNF-producing LCMV-specific CD4+ T cells, as well as a large reduction in poly-functional LCMV-specific CD4+ T cells capable of simultaneously producing multiple cytokines (Fig. 4C). In addition, the mean fluorescence intensity (MFI) of IFN-γ, IL-2 and TNF produced by Cracr2afl/fl/Cd4-Cre CD4+ T cells was decreased, indicating decreased function and Th1 responses at a cellular level. Similar with CD4+ cells, we also observed reduced numbers of total and LCMV-specific CD8+ cells (Suppl. Figs. 2A and B). In addition, frequencies and numbers of IFN-γ, IL-2 and TNF-producing LCMV-specific CD8+ T cells were also reduced (Suppl. Fig. 2C). Overall, these data demonstrate an important role of CRACR2A proteins in Th1 responses and cytokine production to acute virus infection in vivo.
Figure 4. CRACR2A deficiency impairs Th1 response to acute infection with LCMV.
(A) Number of total splenocytes (left) and splenic CD4+ T cells (right) on day 8 after LCMV-Armstrong infection of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice. Each symbol represents data obtained from an independent animal.
(B) Representative flow plots showing the frequency of LCMV-specific IAb-gp67 tetramer-positive CD4+ T cells (gp67-tet). Graph shows absolute numbers of gp67-tetramer-positive LCMV-specific CD4+ T cells from independent animals.
(C) Representative flow plots showing the frequency of IFN-γ and IL-2 (top), or IFN-γ and TNF (bottom)-producing control and Cracr2afl/fl/Cd4-Cre CD4+ T cells on day 8 after LCMV infection following ex vivo stimulation for 5 hours with LCMV-GP61–80 peptide. Graphs (right panels) show quantification of absolute numbers of IFN-γ+, IL-2+, TNF+ or polyfunctional IFN-γ+IL-2+TNF+ CD4+ T cells (top) and mean fluorescence intensity (MFI, bottom) of IFN-γ+, IL-2+, TNF+ CD4+ T cells (bottom) from independent animals. These data are representative of three independent LCMV infection experiments with 4–6 animals in each experiment.
** p < 0.005, *** p < 0.0005.
Deletion of CRACR2A ameliorates Th1 and Th17 cell-mediated experimental autoimmune encephalitis
Expression analysis showed abundant CRACR2A expression in T cells cultured under Th17-polarizing conditions (Fig. 2A). However, we could not detect any defect in IL-17A expression in Cracr2afl/fl/Cd4-Cre T cells, in vitro in the presence of polarizing cytokines (Fig. 2B). To determine if this was consistent in vivo, we utilized an in vivo model of autoimmunity, EAE in which Th17 cells are important for the onset and pathogenicity of the disease (45).
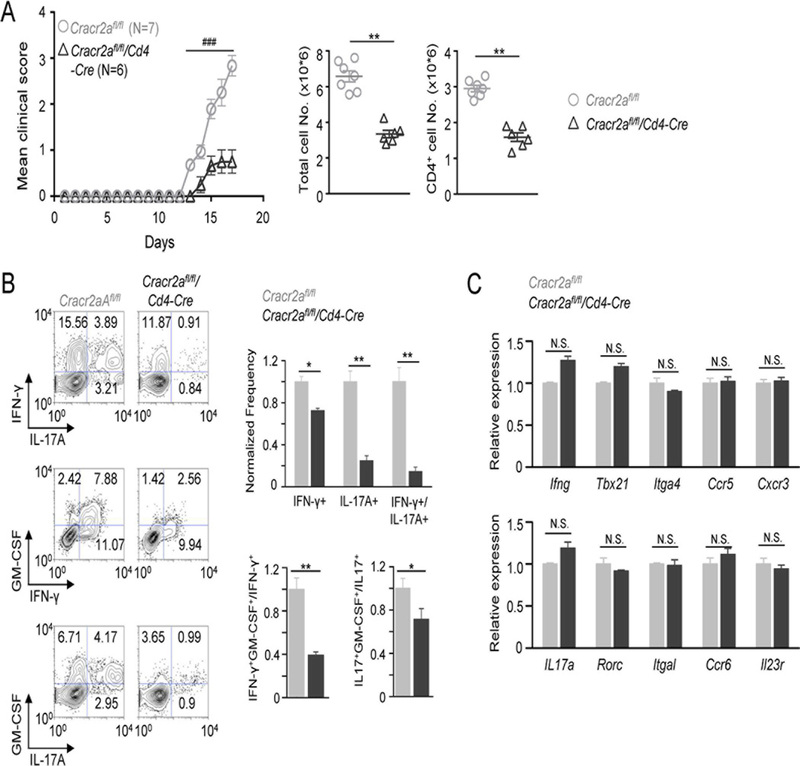
In vivo, EAE-induced Cracr2afl/fl/Cd4-Cre mice showed a marked amelioration of clinical symptoms and accordingly, reduced numbers of total mononuclear cells and CD4+ T cells infiltrated into the CNS when compared to littermate controls (Fig. 5A). We also observed significantly reduced frequency of IFN-γ- and IL-17A-producing CD4+ cells in the CNS of Cracr2afl/fl/Cd4-Cre mice compared to those of control mice (Fig. 5B). Interestingly, the frequency of IFN-γ+IL-17A+ T cells (IFN-γ-producing Th17 cells or Th1-like Th17 cells) was dramatically decreased in the CNS from EAE-induced Cracr2afl/fl/Cd4-Cre mice. This IFN-γ+IL-17A+ subset is derived from Th17 cells and is known to contribute to the pathogenicity of Th17 cells in many autoimmune diseases (29, 31, 34). GM-SCF has also been identified as an important cytokine that contributes to the pathogenicity of inflammatory T cells in EAE by inducing activation and recruitment of innate immune cells (25, 26). In support of our clinical observations, we observed a significant decrease in the frequency of GM-CSF-producing Th1 and Th17 cells isolated from the CNS of EAE-induced Cracr2afl/fl/Cd4-Cre mice compared to that of control mice.
Figure 5. CRACR2A deficiency ameliorates EAE by decreasing the effector function of Th1 and Th17 cells at the central nervous system.
(A) Time course of mean clinical score of EAE in indicated genotypes of mice. For detailed scoring please refer the method section. The graph shows averages ± s.e.m. from one of three independent repeats of experiments with 4–8 mice per trial. Scatter plots (right) show the numbers of total mononuclear (left) or CD4+ (right) cells isolated from the CNS of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice at the peak of the disease. Each symbol represents data obtained from an independent animal. Statistical significance was calculated using the Mann-Whitney U test (### p < 0.0001).
(B) Representative flow plots showing cytokine profile of CD4+ cells isolated from the CNS of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice at the peak of the disease. Bar graphs (right) show averages of normalized frequency (± s.e.m.) of IFN-γ+, IL-17+, and IFN-γ+IL-17+ cells (top) and ratio of IFN-γ+GM-CSF+/IFN-γ+ or IL-17+GM-CSF+/IL-17+ cells (bottom). The frequencies and ratio values of CNS cells from Cracr2afl/fl/Cd4-Cre mice were normalized to those of control Cracr2afl/fl mice.
(C) Quantitative mRNA expression analysis (± s.d.m.) of indicated cytokines, transcription factors and homing receptors from the draining lymph node cells of EAE-induced Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice at early stages of disease severity (clinical score of ~0.5 in control Cracr2afl/fl mice).
* p < 0.05, ** p < 0.005, N.S. – not significant.
To determine whether the reduced infiltration of T cells in Cracr2afl/fl/Cd4-Cre mice was derived from impairment in differentiation of Th1 and Th17 cells, we analyzed T cells isolated from the draining lymph nodes of EAE-induced animals at the early stages. Surprisingly, our transcript analysis did not show any marked difference between control and Cracr2afl/fl/Cd4-Cre CD4+ T cells in terms of expressions of Th1- (Ifng, Tbx21, Integrina4, Ccr5 and Cxcr3), or Th17-specific genes (Il17a, Rorc, Integrin al, Ccr6 and Il23r) (Fig. 5C). These results suggest that the decreased production of IFN-γ and IL-17 by CNS-infiltrated CD4+ cells in EAE-induced Cracr2afl/fl/Cd4-Cre mice is unlikely due to impairment in differentiation or expression of homing receptors necessary for infiltration into the CNS. Taken together, our results suggest that the reduced clinical symptoms in EAE-induced Cracr2afl/fl/Cd4-Cre mice are caused by impairment in re-stimulation and effector functions (e.g., cytokine production and conversion) of Th1 and Th17 cells at the inflamed tissue.
CRACR2A signaling is important for the transition of Th17 cells into pathogenic Th1-like cells
Even though, Cracr2afl/fl/Cd4-Cre T cells did not show impairment in Th17 differentiation in vitro (Fig. 2B), they showed profound reduction in IL-17A production and transition to pathogenic Th1-like Th17 cells in the CNS of EAE-induced Cracr2afl/fl/Cd4-Cre mice. To further dissect the role of CRACR2A-mediated signaling specifically in Th17 cell functions, especially at the site of inflammation, we carried out adoptive Th17 cell transfer-mediated EAE. Because cells cultured under Th17-expansion conditions are transferred, this model is more focused on re-stimulation and effector function of Th17 cells at the CNS by bypassing the differentiation steps. We isolated cells from the draining lymph nodes of EAE-induced control and Cracr2afl/fl/Cd4-Cre mice, cultured them under Th17-expansion conditions for 3 days, and confirmed similar levels of IL-17A expression in CD4+ cells (Fig. 6A). Rag2−/− recipient mice reconstituted with draining lymph node cells from Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice developed EAE with similar onset time (Fig. 6B). However, the clinical symptoms were significantly milder in Rag2−/− recipients that received Cracr2afl/fl/Cd4-Cre cells cultured under Th17-expansion conditions when compared with those receiving control cells cultured under the same condition. In support of these data, total mononuclear and CD4+ T cell numbers were much lesser in the CNS of recipients of Cracr2afl/fl/Cd4-Cre T cells. It is known that Th17 cells have high plasticity and they transit into Th1-like cells, especially after being transferred into immune-compromised hosts (46). In consistence with this observation, we found that a majority of the transferred T cells transited to IFN-γ-producing cells (Fig. 6C and D). In recipients of Cracr2afl/fl/Cd4-Cre T cells, the population of IFN-γ-producing cells decreased. In addition, the populations of IL-17A+ cells as well as IFN-γ+IL-17A+ cells were also reduced in recipients of Cracr2afl/fl/Cd4-Cre T cells. Importantly, expression of the pathogenic cytokine, GM-CSF was significantly reduced in all the CD4+ T cells in the CNS of recipients of Cracr2afl/fl/Cd4-Cre T cells (Fig. 6C and D). Transcription factors T-bet, Runx1 and Runx3 play an important role in transition of Th17 cells into pathogenic Th1-like Th17 cells (34, 47). In consistence with our CNS cytokine expression analysis, we observed a significant reduction in expression of all the three transcription factors in the mononuclear cells isolated from the CNS of recipients of Cracr2afl/fl/Cd4-Cre T cells (Fig. 6E).
Figure 6. CRACR2A deficiency suppresses effector functions of Th17 cells in passive EAE-induced mice.
(A) For passive EAE, donor control and Cracr2afl/fl/Cd4-Cre mice were immunized with MOG35–55 peptide. At the first sign of EAE symptoms, draining lymph node cells were collected and cultured under Th17-expansion conditions for 3 days. Representative FACS plots show cytokine levels in CD4+ cells before transfer.
(B) Time course of mean clinical score of EAE in Rag2−/− mice after adoptive transfer of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre lymph node cells cultured under Th17-expansion conditions. The graph shows averages ± s.e.m. from one of three independent experiments with 4–8 mice per trial. Scatter plots (right) show the numbers of total mononuclear cells (left) or CD4+ (right) cells isolated from the CNS of the Rag2−/− recipients at the peak of the disease. Statistical significance was calculated using the Mann-Whitney U test (### p < 0.0001).
(C) Representative FACS plots showing indicated cytokine profile of CD4+ T cells isolated from the CNS of recipients of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre cells at the peak of the disease.
(D) Bar graphs show normalized cytokine levels (average ± s.e.m.) of IFN-γ+ and IL-17A+ cells and normalized ratio of IFN-γ+-GM-CSF+/IFN-γ+ or IL-17+-GM-CSF+/IL-17+ cells.
(E) Quantitative mRNA expression analysis (± s.d.m.) of indicated transcription factors in the mononuclear cells isolated from the CNS of Rag2−/− recipients of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre cells at the peak of the disease.
* p < 0.05, ** p < 0.005, *** p < 0.0005.
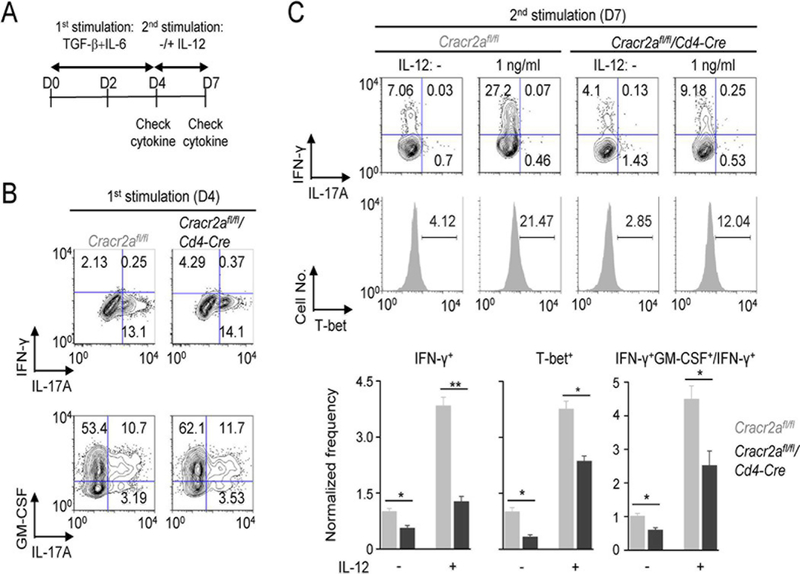
To validate the role of CRACR2A in transition of Th17 cells into Th1-like Th17 cells, we took advantage of in vitro transition model of Th17 cells (34). In this model, naïve T cells cultured under Th17-polairzing conditions for four days are re-differentiated into IFN-γ-producing cells with re-stimulation in the absence of any cytokines or in the presence of IL-12 (Fig. 7A). First, we cultured naïve T cells isolated from Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre mice under Th17-polarizing conditions. In consistence with the previous results, CRACR2A deficiency did not influence IL-17A or GM-CSF production (Fig. 7B). Re-stimulation of T cells cultured under Th17-polarizing conditions induced transition into IFN-γ-producing cells, that was further enhanced by addition of low concentration (1 ng/ml) of polarizing cytokine, IL-12 (Fig. 7C). In support of our in vivo observations, Cracr2afl/fl/Cd4-Cre T cells cultured under Th17-polarizing conditions showed impairment in transition into IFN-γ-producing cells as well as in production of GM-CSF. Re-stimulation under non-polarizing conditions induced T-bet expression in T cells pre-cultured under Th17-polarizing conditions, that was further enhanced by inclusion of IL-12. However, T-bet expression in the absence and presence of IL-12 treatment was significantly decreased in Cracr2afl/fl/Cd4-Cre T cells. Together, we concluded that CRACR2A proteins play an important role in transition of Th17 cells into IFN-γ and GM-CSF-producing cells, without significantly impairing differentiation of Th17 cells.
Figure 7. CRACR2A-mediated TCR signaling is important for transition of Th17 cells.
(A) Schematic of the protocol for conversion of Th17 cells. CD4+CD25- Naïve T cells cultured under Th17-polarizing conditions for four days were re-stimulated with anti-CD3 and anti-CD28 antibodies in the absence or presence of low concentration of IL-12 (1 ng/ml). After three days, cytokine levels were determined by intracellular staining after 5-hour stimulation with PMA plus ionomycin.
(B) Representative flow plots showing cytokine profile of Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre CD4+ cells cultured under Th17-polarizing conditions for four days as indicated in panel A. At day 4, cells were stimulated with PMA plus ionomycin for 5 hours. Data are representative of three independent experiments.
(C) Representative flow plots showing cytokine profile of re-stimulated Cracr2afl/fl or Cracr2afl/fl/Cd4-Cre CD4+ cells in the absence or presence of IL-12 at day 7. Cells were stimulated with PMA plus ionomycin for 5 hours, and cytokine levels (top panels) or transcription factor expression (histograms, middle panels) were checked by intracellular staining. Flow plots are representative of three independent experiments. Bar graphs (bottom panels) show averages ± s.e.m. of IFN-γ+, T-bet+ or ratio of IFN-γ+GM-CSF+/IFN-γ+ positive cells, normalized to those of controls cells without IL-12 treatment.
* p < 0.05, ** p < 0.005
DISCUSSION
CRACR2A gene is located on Chromosome 12 in humans, and GWAS has identified its link to diseases including Parkinson’s disease, NAFLD, atrial fibrillation, and chronic HIV infection, that are potentially related with immune functions (1-4). However, the role of CRACR2A proteins in these diseases is largely unknown due to minimal information about their physiological functions. Our previous studies uncovered a role for these proteins in T cells, providing a potential link between anomalies in CRACR2A function and T cell-mediated human diseases. The short isoform CRACR2A-c was originally identified to act as a cytoplasmic factor to facilitate CRAC channel function (10). Later, a long isoform CRACR2A-a, that localizes to the trans Golgi network, was found to be involved in regulation of CRAC channels and the JNK-MAPK pathway triggered by TCR stimulation (11). The C-terminal GTPase domain of CRACR2A-a closely resembles that of the Rab GTPases, known to play a crucial role in vesicular transport between organelles. CRACR2A-a translocates into the immunological synapse via vesicles upon TCR stimulation, induces activation of the Ca2+-NFAT and JNK MAPK pathways, and finally be inactivated by GTP hydrolysis and degradation. Our physiological studies using T cell-specific Cracr2afl/fl/Cd4-Cre mice here further define its important role in highly inflammatory Th1 and Th17 cells, and for the first time, uncover their potential functions in human inflammatory diseases.
TCR stimulation plays a pivotal role in fate determination of T cells including their differentiation into Th1 or Th2 cells (48, 49). Strong TCR signaling blocks intrinsic Th2 programs of naïve T cells by inhibiting the function and expression of GATA3 (12, 13), and hence plays an important role in Th1 differentiation (49). In our previous work, we demonstrated an important role of CRACR2A proteins in activation of the NFAT and JNK pathways downstream of TCR signals (10, 11). Our current data based on intracellular staining and the genome-scale transcript analysis by RNA-seq suggest that weakened TCR signaling by the absence of CRACR2A proteins fails to adequately attenuate the Th2 transcriptional program. Consistent with this view, CRACR2A deficiency only influenced T cells cultured in the absence of any polarizing cytokines, where the role of TCR stimulation is emphasized. Accordingly, we also observed an impairment in Cracr2afl/fl/Cd4-Cre T cells in Th1 differentiation in response to LCMV infection. However, Cracr2afl/fl/Cd4-Cre T cells did not show any defect in Th1 differentiation at the draining lymph nodes, while the Th1 responses were substantially decreased in the CNS of EAE-induced mice. Currently, the reason underlying this discrepancy between animal models is not clear, but it is possible that the contribution of TCR signaling with peptide injection to induce EAE can be relatively small compared to that of TCR signaling triggered by LCMV infection. It is also possible that the concentration of Th1-polarizing cytokines (e.g., IL-12 or IL-18) at the draining lymph nodes of EAE-induced mice is higher than the spleen of LCMV-infected mice and these cytokines may compensate for the weakened TCR signaling observed due to CRACR2A deficiency. Nonetheless, commonly in both LCMV infection and EAE models, we observed that CRACR2A deficiency substantially decreased effector T cell function at the primary sites of infection and inflammation, the spleen and CNS, respectively.
An important role of CRACR2A in Th1 differentiation was expected based on its abundant expression in ThN and Th1 cells, and its known role in regulation of the strength of TCR signaling (10, 11). However, although expression of CRACR2A proteins in T cells cultured under Th17-polarizing conditions was similar to that in Th1 cells, we could not detect any defect in Th17 differentiation in Cracr2afl/fl/Cd4-Cre CD4+ cells in vitro. In support of these data, in vivo Th17 differentiation was unaffected as judged by expression of signature transcription factors, cytokines and homing receptors in cells isolated from draining lymph nodes of EAE-induced Cracr2afl/fl/Cd4-Cre mice (Fig. 5C). However, we found that Cracr2afl/fl/Cd4-Cre Th17 cells that migrated into the CNS of EAE-induced Cracr2afl/fl/Cd4-Cre mice had defects in effector T cell functions at the inflamed tissue, CNS, as judged by reduced frequency of IL-17A+ and GM-CSF+ cell populations. To completely exclude any minor contribution from differentiation defect, we confirmed these results with adoptive transfer of T cells cultured under Th17-expansion conditions into Rag2−/− recipients using the animal model of passive EAE. These results showed that Cracr2afl/fl/Cd4-Cre T cells had less pathogenic potential and exhibited a defect in transition to cells producing IFN-γ and GM-CSF (Fig. 6). Mechanistically, CRACR2A deficiency decreased expression of transcription factors that are involved in the transition to pathogenic Th17 cells and GM-CSF production (34). These results re-emphasize the role of CRACR2A-mediated TCR signaling triggered by local antigen interaction in induction of the Th1 transcriptional program during the conversion of Th17 cells.
Our previous studies showed that CRACR2A-a is a prominent target of the widely used statin drugs for treatment of high blood cholesterol levels. In the presence of very low amounts of atorvastatin that had minimal effects on localization of small Rab GTPases, CRACR2A-a underwent de-prenylation that caused its detachment from vesicles, and induced its degradation, thereby decreasing the strength of TCR signaling (11). Our in vivo data showing reduced Th1/Th17 responses in Cracr2afl/fl/Cd4-Cre mice match well with those obtained from human studies where consumption of atorvastatin and simvastatin ameliorates autoimmune diseases by decreasing Th1 and Th17 responses (50-52). Our observations of a Th2-biased differentiation and impairment in Th17 cell effector functions due to CRACR2A deficiency suggest that CRACR2A may be one of the key targets of statin to reduce Th1 and Th17 responses in autoimmune disease patients who consume statin drugs for therapeutic purposes. Interestingly, T cell-specific loss of CRACR2A did not influence development or homeostasis of CD4+, CD8+ and regulatory T cells. When combined with the results from GWAS, a selective function of CRACR2A in peripheral T cells, especially the ones involved in proinflammatory Th1 and Th17 responses, makes it an ideal drug target to suppress autoimmunity. Future drug designs to specifically inactivate CRACR2A protein(s) may minimize the undesired side effects of statin drugs that are administered to autoimmune disease patients.
In summary, our study has identified CRACR2A proteins as an intracellular signaling module activating two important proximal TCR signaling pathways, Ca2+-NFAT and JNK-AP1, to selectively affect Th1 differentiation. Further, in this study we found that the Th1 transcriptional program driven by CRACR2A is also important for the transition of Th17 cells into pathogenic Th1-like Th17 cells. Considering the frequent identification of CRACR2A in multiple GWAS, our research uncovers the underlying molecular mechanisms and further identifies CRACR2A proteins as a lucrative therapeutic target to suppress local effector T cell functions to ameliorate autoimmune symptoms.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Benjamin Segal and Mr. Patrick Duncker (University of Michigan) for kind advice and sharing protocols for establishing passive EAE animal model experiments. We also thank Dr. Guillaume Calmettes for advice on statistical analysis.
This work was supported by the National Institute of Health grants AI083432 (Y.G.) and AI130653 (S.S). Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by the JCCC grant P30 CA016042 and 5P30 AI028697. Peripheral blood mononuclear cells (PBMCs) were obtained from the CFAR Virology core Laboratory at UCLA that is supported by 5P30 AI028697.
Footnotes
AUTHOR CONTRIBUTIONS
S.S. and Y.G. designed research. J.S.W performed in vitro T cell analysis and active/passive EAE experiments. S.S. generated and maintained Cracr2afl/fl animals with help from J.L. and performed in vitro T cell experiments and RNA-seq analysis (in collaboration with Jing.L. and M.P.). K.D.K. analyzed development, homeostasis and in vitro T cell phenotypes from Cracr2afl/fl animals. LCMV infection experiments were done by H.E., S.S, and D.G.B. Y.G., S.S, and J.S.W. wrote the manuscript with input from other authors. Y.G. supervised the project.
DISCLOSURES
The authors have no financial conflicts of interests.
References
- 1.Edelman D, Kalia H, Delio M, Alani M, Krishnamurthy K, Abd M, Auton A, Wang T, Wolkoff AW, and Morrow BE. 2015. Genetic analysis of nonalcoholic fatty liver disease within a Caribbean-Hispanic population. Mol Genet Genomic Med 3: 558-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shata MT, Abdel-Hameed EA, Hetta HF, and Sherman KE. 2013. Immune activation in HIV/HCV-infected patients is associated with low-level expression of liver expressed antimicrobial peptide-2 (LEAP-2). J Clin Pathol 66: 967-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, Debette S, Lumley T, Folsom AR, van den Herik EG, Bos MJ, Beiser A, Cushman M, Launer LJ, Shahar E, Struchalin M, Du Y, Glazer NL, Rosamond WD, Rivadeneira F, Kelly-Hayes M, Lopez OL, Coresh J, Hofman A, DeCarli C, Heckbert SR, Koudstaal PJ, Yang Q, Smith NL, Kase CS, Rice K, Haritunians T, Roks G, de Kort PL, Taylor KD, de Lau LM, Oostra BA, Uitterlinden AG, Rotter JI, Boerwinkle E, Psaty BM, Mosley TH, van Duijn CM, Breteler MM, Longstreth WT Jr., and Wolf PA. 2009. Genomewide association studies of stroke. N Engl J Med 360: 1718-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, Cui J, Taylor KD, Wilson L, Cummings OW, Chen YD, Rotter JI, and Nonalcoholic N Steatohepatitis Clinical Research. 2010. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology 139: 1567-1576, 1576 e1561–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Kadlecek TA, Au-Yeung BB, Goodfellow HE, Hsu LY, Freedman TS, and Weiss A. 2010. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb Perspect Biol 2: a002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownlie RJ, and Zamoyska R. 2013. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol 13: 257-269. [DOI] [PubMed] [Google Scholar]
- 7.Srikanth S, Woo JS, Sun Z, and Gwack Y. 2017. Immunological Disorders: Regulation of Ca(2+) Signaling in T Lymphocytes. Adv Exp Med Biol 993: 397-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tybulewicz VL 2005. Vav-family proteins in T-cell signalling. Curr Opin Immunol 17: 267-274. [DOI] [PubMed] [Google Scholar]
- 9.Kyriakis JM, and Avruch J. 2012. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev 92: 689-737. [DOI] [PubMed] [Google Scholar]
- 10.Srikanth S, Jung HJ, Kim KD, Souda P, Whitelegge J, and Gwack Y. 2010. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat Cell Biol 12: 436-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srikanth S, Kim KD, Gao Y, Woo JS, Ghosh S, Calmettes G, Paz A, Abramson J, Jiang M, and Gwack Y. 2016. A large Rab GTPase encoded by CRACR2A is a component of subsynaptic vesicles that transmit T cell activation signals. Sci Signal 9: ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang ES, Szabo SJ, Schwartzberg PL, and Glimcher LH. 2005. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307: 430-433. [DOI] [PubMed] [Google Scholar]
- 13.Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, and Strober W. 2006. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med 203: 755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie D, and Zhu J. 2014. Transcriptional regulatory networks for CD4 T cell differentiation. Curr Top Microbiol Immunol 381: 125-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang CF, D’Souza WN, Ch’en IL, Pages G, Pouyssegur J, and Hedrick SM. 2012. Polar opposites: Erk direction of CD4 T cell subsets. J Immunol 189: 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang G, Shi LZ, and Chi H. 2009. Regulation of JNK and p38 MAPK in the immune system: signal integration, propagation and termination. Cytokine 48: 161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rincon M, and Davis RJ. 2009. Regulation of the immune response by stress-activated protein kinases. Immunol Rev 228: 212-224. [DOI] [PubMed] [Google Scholar]
- 18.Hodge MR, Ranger AM, Charles de la Brousse F, Hoey T, Grusby MJ, and Glimcher LH. 1996. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 4: 397-405. [DOI] [PubMed] [Google Scholar]
- 19.Ranger AM, Oukka M, Rengarajan J, and Glimcher LH. 1998. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development. Immunity 9: 627-635. [DOI] [PubMed] [Google Scholar]
- 20.Kwon MJ, Ma J, Ding Y, Wang R, and Sun Z. 2012. Protein kinase C-theta promotes Th17 differentiation via upregulation of Stat3. J Immunol 188: 5887-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon MJ, Wang R, Ma J, and Sun Z. 2010. PKC-theta is a drug target for prevention of T cell-mediated autoimmunity and allograft rejection. Endocr Metab Immune Disord Drug Targets 10: 367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KD, Srikanth S, Tan YV, Yee MK, Jew M, Damoiseaux R, Jung ME, Shimizu S, An DS, Ribalet B, Waschek JA, and Gwack Y. 2014. Calcium Signaling via Orai1 Is Essential for Induction of the Nuclear Orphan Receptor Pathway To Drive Th17 Differentiation . J Immunol 192: 110-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McQualter JL, Darwiche R, Ewing C, Onuki M, Kay TW, Hamilton JA, Reid HH, and Bernard CC. 2001. Granulocyte macrophage colony-stimulating factor: a new putative therapeutic target in multiple sclerosis. J Exp Med 194: 873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponomarev ED, Shriver LP, Maresz K, Pedras-Vasconcelos J, Verthelyi D, and Dittel BN. 2007. GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J Immunol 178: 39-48. [DOI] [PubMed] [Google Scholar]
- 25.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, and Rostami A. 2011. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol 12: 568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, and Becher B. 2011. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol 12: 560-567. [DOI] [PubMed] [Google Scholar]
- 27.Spath S, Komuczki J, Hermann M, Pelczar P, Mair F, Schreiner B, and Becher B. 2017. Dysregulation of the Cytokine GM-CSF Induces Spontaneous Phagocyte Invasion and Immunopathology in the Central Nervous System. Immunity 46: 245-260. [DOI] [PubMed] [Google Scholar]
- 28.Bending D, De la Pena H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, and Cooke A. 2009. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest 119: 565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, Garefalaki A, Potocnik AJ, and Stockinger B. 2011. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol 12: 255-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nurieva R, Yang XO, Chung Y, and Dong C. 2009. Cutting edge: in vitro generated Th17 cells maintain their cytokine expression program in normal but not lymphopenic hosts. J Immunol 182: 2565-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, and Weaver CT. 2009. Late developmental plasticity in the T helper 17 lineage. Immunity 30: 92-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, and Weaver CT. 2010. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 32: 616-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, Roychoudhuri R, Ferreyra GA, Shen W, Durum SK, Feigenbaum L, Palmer DC, Antony PA, Chan CC, Laurence A, Danner RL, Gattinoni L, and Restifo NP. 2011. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity 35: 972-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, Lee YK, Weaver CT, Yagi R, and Lazarevic V. 2014. The Transcription Factors T-bet and Runx Are Required for the Ontogeny of Pathogenic Interferon-gamma-Producing T Helper 17 Cells. Immunity 40: 355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Pachter L, and Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fioretta ES, Fledderus JO, Baaijens FP, and Bouten CV. 2012. Influence of substrate stiffness on circulating progenitor cell fate. J Biomech 45: 736-744. [DOI] [PubMed] [Google Scholar]
- 37.Anders S, and Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks DG, Teyton L, Oldstone MB, and McGavern DB. 2005. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J Virol 79: 10514-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balasubramani A, Mukasa R, Hatton RD, and Weaver CT. 2010. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol Rev 238: 216-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuvpilo S, Jankevics E, Tyrsin D, Akimzhanov A, Moroz D, Jha MK, Schulze-Luehrmann J, Santner-Nanan B, Feoktistova E, Konig T, Avots A, Schmitt E, Berberich-Siebelt F, Schimpl A, and Serfling E. 2002. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity 16: 881-895. [DOI] [PubMed] [Google Scholar]
- 41.Macian F, Lopez-Rodriguez C, and Rao A. 2001. Partners in transcription: NFAT and AP-1. Oncogene 20: 2476-2489. [DOI] [PubMed] [Google Scholar]
- 42.Scheinman EJ, and Avni O. 2009. Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin-4 receptor and T cell receptor. J Biol Chem 284: 3037-3048. [DOI] [PubMed] [Google Scholar]
- 43.Hoshino K, Tsutsui H, Kawai T, Takeda K, Nakanishi K, Takeda Y, and Akira S. 1999. Cutting edge: generation of IL-18 receptor-deficient mice: evidence for IL-1 receptor-related protein as an essential IL-18 binding receptor. J Immunol 162: 5041-5044. [PubMed] [Google Scholar]
- 44.Crotty S 2011. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29: 621-663. [DOI] [PubMed] [Google Scholar]
- 45.Pierson E, Simmons SB, Castelli L, and Goverman JM. 2012. Mechanisms regulating regional localization of inflammation during CNS autoimmunity. Immunol Rev 248: 205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Zhang Y, Yang XO, Nurieva RI, Chang SH, Ojeda SS, Kang HS, Schluns KS, Gui J, Jetten AM, and Dong C. 2012. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity 36: 23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, and Glimcher LH. 2011. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol 12: 96-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Constant SL, and Bottomly K. 1997. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu Rev Immunol 15: 297-322. [DOI] [PubMed] [Google Scholar]
- 49.Zhu J, Yamane H, and Paul WE. 2010. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 28: 445-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Youssef S, Stuve O, Patarroyo JC, Ruiz PJ, Radosevich JL, Hur EM, Bravo M, Mitchell DJ, Sobel RA, Steinman L, and Zamvil SS. 2002. The HMG-CoA reductase inhibitor, atorvastatin, promotes a Th2 bias and reverses paralysis in central nervous system autoimmune disease. Nature 420: 78-84. [DOI] [PubMed] [Google Scholar]
- 51.Weber MS, Youssef S, Dunn SE, Prod’homme T, Neuhaus O, Stuve O, Greenwood J, Steinman L, and Zamvil SS. 2006. Statins in the treatment of central nervous system autoimmune disease. J Neuroimmunol 178: 140-148. [DOI] [PubMed] [Google Scholar]
- 52.Dunn SE, Youssef S, Goldstein MJ, Prod’homme T, Weber MS, Zamvil SS, and Steinman L. 2006. Isoprenoids determine Th1/Th2 fate in pathogenic T cells, providing a mechanism of modulation of autoimmunity by atorvastatin. J Exp Med 203: 401-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.