Abstract
Background
Bladder cancer is among the common human malignancies that show a heavy mutational load and copy number variations of numerous chromosomes, which makes them a target for diagnostic explorations.
Objective
We aimed to design a multicolor fluorescence in situ hybridization (FISH) test referred to as the quartet test for the detection of bladder cancer in urine.
Design, setting, and participants
We performed genome-wide copy number variation analysis on cohorts from the University of Texas MD Anderson Cancer Center (n = 40) and The Cancer Genome Atlas (n = 129), and identified the most frequently amplified chromosomal regions. These data were used to select four of the amplified regions to design a multicolor FISH test, referred to as the quartet test. Assay validation was performed on urine samples from 98 patients with bladder cancer: 56 with low-grade papillary, 42 with high-grade invasive disease, and 48 benign controls.
Intervention
The quartet test can be used in clinical practice for noninvasive detection of bladder cancer.
Outcome measurements and statistical analysis
We initially analyzed samples using a fraction of abnormal cell scores and then by the quantitative score, which included not only the proportion of cells with abnormal copy numbers, but also the proportion of cells with numbers of altered copies and degree of amplification. We used receiver operator characteristic (ROC) curves to identify cutoff values for the scores at which performances of sensitivity and specificity were maximized.
Results and limitations
The copy number status assessed by probes detected in voided urine reflected the amplification status of the primary tumor. An ROC curve summarizing the proportion of assayed cells with any abnormal copy numbers gave specificity of 93.8% and sensitivity of 78.6% using the proportion of cells with abnormal copy numbers. The quantitative score giving extra weight to cells with multiple simultaneous amplifications provided 95.8% specificity and 76.8% sensitivity. Both percentage of abnormal cells and quantitative scores were highly effective for assessing the grade of the tumor. The full spectrum of potential clinical applications was not explored in the current study, and further validation studies are needed.
Conclusions
The quartet test shows promising specificity and sensitivity results, but it requires validation on a larger multi-institutional cohort of samples.
Patient summary
The quartet test can be used for noninvasive detection of bladder cancer in voided urine. It can also be used to assess the grade of the tumor and tumor recurrence as well as post-treatment effects.
Keywords: Fluorescence in situ hybridization, Copy number variation, Bladder tumor
1. Introduction
Malignant cells frequently acquire chromosomal instability, manifested as increased or decreased copy numbers of entire chromosomes and their segments, referred to as aneuploidy [1]. Aneuploidy has been proposed to drive tumor development by altering cellular phenotypes responsible for such fundamental aspects of malignant transformation as increased cellular proliferation, invasion, and metastasis [2]. Aneuploidy emerges early in the process of cancer development and can be detected in in situ preneoplastic conditions such as dysplasia and carcinoma in situ, or even in adjacent microscopically normal tissue referred to as field effects [3–7]. Therefore, aneuploidy represents an attractive target for explorations of novel early diagnostic and preventive strategies [8–10].
Bladder cancer is among the most common human malignancies that show pronounced features of genomic instability, exhibiting a heavy mutational load and widespread copy number variations (CNVs) affecting numerous chromosomes, which makes it an obvious target for diagnostic explorations [11–13]. Studies from our group and others have shown that anomalies in genes linked to organelles responsible for segregation of chromosomes and their regulatory mechanisms, such as Aurora kinase A (AURKA), contribute to the genomic instability and aggressive behavior of bladder cancer [14]. AURKA copy numbers have also been used as effective biomarkers for bladder cancer detection in voided urine. AURKA copy numbers and other multicolor fluorescence in situ hybridization (FISH) tests assessing chromosomal copy numbers (based predominantly on assays of centromeric probes) in exfoliated cells of voided urine samples have been used in clinical practice as noninvasive detection tests for bladder cancer, as has urine cytology [15–19]. They have also been applied to monitor the recurrence and progression of patients with nonmuscle-invasive bladder tumors. The sensitivity and specificity of the existing tests are not, however, sufficiently high to eliminate or even reduce the frequency of the need for invasive cystoscopy and tissue biopsy to rule out clinically evident bladder cancer [17,20]. The most popular FISH-based test was designed nearly 2 decades ago and was based on pregenomic data [21]. It utilizes predominantly centromeric probes and a probe for 9p deletions. Recent genomic analyses provide comprehensive molecular characterization of bladder cancer and permit the identification of multiple specific chromosomal loci amplified in bladder cancer, which may be used as biomarker targets [11,13]. We hypothesized that a test developed with such biomarkers may offer enhanced diagnostic performance and may be subjected to improvements by selecting multiple alternative combinations of the diagnostic probes.
2. Patients and methods
2.1. Patients and tissue samples
All human tissues used in this study were collected under protocols reviewed and approved by the Institutional Review Board of the University of Texas MD Anderson Cancer Center (MDACC) and collaborating institutions. Informed consent was obtained from all participants who provided tissue samples and urine for this study. All studies were performed in accordance with the relevant guidance and regulations. Genome-wide CNVs were initially assayed in paired fresh frozen bladder tumor and normal peripheral blood samples from 40 patients, including 14 with low-grade papillary (LGPUC) disease (Ta–T1a) and 26 with high-grade invasive (HGUC) disease (T1b–higher) tumor samples. The copy number variants identified in the MDACC cohort were then validated in the initially published The Cancer Genome Atlas (TCGA) cohort containing 129 samples from high-grade muscle-invasive conventional urothelial carcinoma (UC) of the bladder [11]. Performance of the probes and their specificity for respective chromosomal loci as well as standardization of the hybridization procedures were initially tested on normal human peripheral blood lymphocytes and microscopically normal appearing urothelial cells from ureters in nephrectomy specimens, as well as on human metaphase chromosomes (Applied Genetics Laboratories, Inc., Melbourne, FL, USA) as previously described [15,16]. The initial testing of standardized mixtures of four FISH probes were performed on paired samples of touch prints from 53 bladder tumor samples (19 LGPUC and 34 HGUC) obtained by transurethral resection and voided urine typically collected 2–3 d before cystoscopy. Final validation of the quartet test was performed on voided urine samples from 98 cancer patients, including 56 with LGPUC (Ta–T1a) and 42 with HGUC (T1b–higher), and 48 controls, including 18 healthy controls and 30 patients with various non-neoplastic disorders. For patients with bladder cancer, the diagnosis was confirmed by cystoscopy and microscopic examination of the tumor samples. Cancer patients were randomly selected from the pathology data files for the availability of voided urine and pathological samples. For the control group of patients with benign urological disorders, bladder cancer was ruled out by clinical evaluation and standard urological diagnostic work-up. Healthy controls were clinically asymptomatic volunteer donors of urine samples. The clinical and pathological data for 146 human individuals used to assess the performance of the quartet test are summarized in Table 1 and Supplementary Figure 1. All bladder tumor samples and voided urine from patients with bladder cancer were collected at MDACC (Houston, TX, USA). Voided urine samples of healthy controls and patients with non-neoplastic disorders were collected at the University of Texas Southwestern Medical Center (Dallas, TX, USA).
Table 1.
Summary of clinical and pathological data of human individuals used to assess the performance of the quartet test (n = 146)
| Number | Gender | |||
|---|---|---|---|---|
| Urine samples | of samples | F | M | Mean age ± STDEV |
| Control samples | 48 | 22 | 26 | 60.70 ± 11.12 |
| Healthy individuals | 18 | 9 | 9 | 59.17 ± 10.14 |
| Benign disorders | 30 | 13 | 17 | 61.62 ± 11.74 |
| Hematuria | 2 | 2 | 0 | 44.34 ± 3.51 |
| Elevated PSA | 2 | 0 | 2 | 74.08 ± 8.66 |
| Hyperlipidemia/hypercholesterolemia | 7 | 1 | 6 | 66.52 ± 9.35 |
| Kidney stone | 1 | 0 | 1 | 55.31 |
| Renal failure | 1 | 0 | 1 | 67.96 |
| Stricture of ureter | 1 | 0 | 1 | 46.39 |
| Neurogenic bladder | 3 | 3 | 0 | 50.35 ± 7.95 |
| Others | 13 | 7 | 6 | 65.96 ± 9.24 |
| Tumor samples | 98 | 23 | 75 | 66.00 ± 12.56 |
| LGPUC (Ta–T1a) | 56 | 16 | 40 | 64.48 ± 12.57 |
| HGUC (T1b–higher) | 42 | 7 | 35 | 68.01 ± 12.41 |
| Total | 146 | 45 | 101 | 64.26 ± 12.32 |
F = female; HGUC = high-grade urothelial carcinoma; LGPUC = low-grade papillary urothelial carcinoma; M = male; PSA = prostate-specific antigen; STDEV = standard deviation.
UCs were classified according to the histological tumor grading system of the World Health Organization and were dichotomized as low- or high-grade tumors [22,23]. The growth pattern of papillary versus nonpapillary or solid tumors, and the depth of invasion were also recorded. Levels of invasion were defined according to the tumor–node–metastasis staging system. T1 tumors were substaged as T1a or T1b to divide them into superficial (Ta–T1a) or invasive (T1b and higher), as previously described [15,16]. Tumors from both MDACC and TCGA datasets included only pure conventional UCs. The bladder cancer variants were not included in this study.
2.2. CNV analysis and design of quartet test
CNV analysis was performed using two-sample Illumina Human 1M-Duo V1 DNA Analysis BeadChips, which interrogate >1.1 million loci per sample. For CNV analysis, DNA was extracted from paired fresh frozen bladder tumor and peripheral blood samples from 40 cancer patients as previously described. Arrays were prepared according to the Infinium II Assay protocol and scanned the same day using an Illumina BeadArray Reader 500G. The microarray data from the MDACC and TCGA cohorts were imported into the single nucleotide polymorphism genotyping module in the Illumina Genome Studio software to perform CNV analysis. These analyses identified the most frequently amplified chromosomal loci; we combined these with their respective gene content to design the quartet test. In selecting the chromosomal loci to interrogate with FISH probes, we used both the frequency and the degree of their amplification in bladder cancer as well as their genomic content in terms of the specific genes and repetitive sequences. In addition, other published data concerning the CNV analysis were included in the selection of the probes [24]. Using these factors, probes for the following regions (listed with their respective fluorescent tags) were selected and provided by Kreatech/Leica (Buffalo Grove, IL, USA): 6p22, E2F3-CDKAL1, 525KB, green, PlatinumBright495; 8q22. PABPC1-ZNF706, 480 KB, gold, PlatinumBright530; 11q13, FGF19-FGF3, dark red, PlatinumBright590; and 20q11.2, MAPRE1, 610KB, blue, PlatinumBright415. The probes were produced from their respective BAC clones using REPEAT-FREE FISH technology and were labeled with their respective fluorochromes by the universal linkage system method (http://www.leicabiosystems.com/ihc-ish-fish/ish-probes-molecular-pathology/kreatech-fish-probes/repeat-freetm-technology/).
2.3. Tumor samples and urine analyzed by FISH
Voided urine specimens (approximately 200 ml) were collected and prepared for FISH analysis as previously described [15,16]. In brief, the urine was centrifuged for 15 min at 200g, and the resulting pelleted material containing exfoliated tumor cells was resuspended in 2 ml of Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA, USA) containing 10% dimethyl sulfoxide and stored at –70°C. For FISH analysis, frozen sediment samples containing exfoliated cells were defrosted, washed three times in phosphate buffered saline, and cytospun onto slides. The cytospin preparations were fixed in methanol:acetic acid (3:1), pretreated in 2× saline sodium citrate (SSC) buffer at 37°C for 30 min, and then dehydrated in increasing concentrations of ethanol. The slides were heated at 90°C for 5 min to denature the DNA and then incubated overnight at 37°C with a mixture of the four FISH probes. After hybridization, the slides were washed with 0.5× SSC with 0.1% sodium dodecyl sulfate at 65°C, counterstained with 4′,6-diamidino-2-phenylindole (Invitrogen), mounted with an antifade solution (Roche Diagnostics, Mannheim, Germany), and coverslipped. Fluorescent signals were counted and photographed using a Leica Fluorescent Microscope and Image Analysis System (CytoVision DM5500; Leica).
2.4. Data analysis
We scored samples for abnormality in two ways. The first, “abnormal proportion” (AP) score, is simply the proportion of the cells examined that showed any visible abnormality. The second, “quantitative score” (QS), includes not only the proportion of cells with abnormal copy numbers, but also the proportions of cells with numbers of altered copies and their degree of amplification calculated as follows:
We used receiver operator characteristic (ROC) curves to identify cutoff values for the scores at which the joint performances (sensitivity + specificity) were maximized. We also used the area under the ROC curve (AUC) to assess the performance of the quartet test to detect bladder cancer. Interval estimates for the AUC values were computed using 1000 bootstrap simulations. In each simulation, one bootstrap sample was drawn from the group of controls, another bootstrap sample was drawn from the group of cancer cases, and the AUC for these two groups was computed. The 1000 values obtained were sorted, and the 25th and 975th values comprise the interval reported. We used a Wilcoxon-Mann-Whitney rank sum test to compare the case and control groups. Interval estimates for proportions (sensitivity and specificity) used the 2.5th and 97.5th percentiles of a beta distribution proportional to the likelihood function. In follow-up analyses comparing abnormality scores between cohorts, statistical significance of the differences between mean values was tested by unpaired two-sample t tests or Wilcoxon rank sum tests. Comparisons involving three or more groups or multiple factors were performed using analysis of variance. Linear discrimination was used to determine the performance of the proportion of the abnormal cells and QS in the assessment of tumor grades, and leave-one-out cross validation was employed to evaluate the performance of the classifiers. All statistical tests were two sided. A p value ≤0.05 was considered statistically significant. Copy number gains were reported as categorical integer values (roughly log2 scale) by the Illumina Genome Studio software: 0 = diploid; 1 = amplification; and 2 = marked amplification. Data analysis and calculations were performed using R Package Software (version 3.3.2).
3. Results
The overall plan of probe identification, design, and testing is outlined in Figure 1.
Fig. 1.
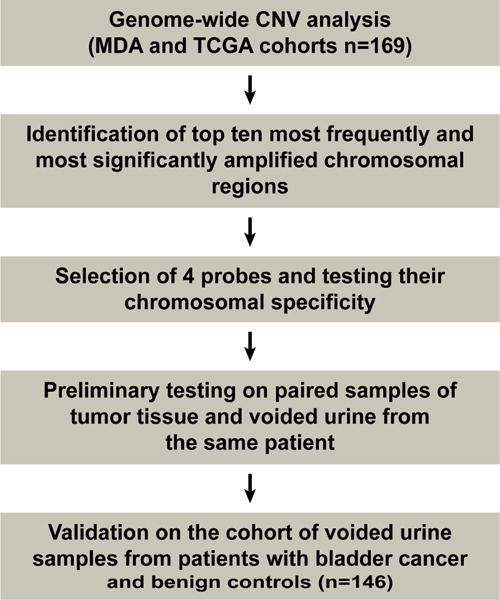
Overall plan for the development and testing of the quartet test.
CNV = copy number variation; MDA = MD Anderson; TCGA = The Cancer Genome Atlas.
3.1. CNV analysis and probe design
The purpose of the copy number analysis was to identify the most frequently amplified (regions scored 1 or 2 in the Genome Studio output) and markedly amplified (regions scored 2) chromosomal regions in order to design a multicolor FISH test. The goal of these analyses was to select the four best possible probes. The number of probes was restricted to four because there were four commercially available fluorochromes that permitted their clear distinction at distinct wavelengths using a specific set of filters under a fluorescent microscope. Initial analysis of paired bladder tumor and peripheral blood samples from 40 cancer patients, 14 with superficial LGPUC (Ta–T1a) and 26 with invasive HGUC (T1b–higher) disease, identified the 10 most frequently and the 10 most markedly amplified chromosomal regions (Fig. 2A, and Supplementary Fig. 2A and 2B). These most frequently and markedly amplified regions were validated on 129 tumor samples from the original published TCGA cohort (Fig. 2B and 2C). These two sets of analyses were somewhat overlapping but not identical. In general, the positions of amplified peaks were virtually identical in both cohorts, but the relative frequencies of amplification were different. Using these data, we selected the four chromosomal regions to design the probes for the quartet test. These regions included amplified chromosomal segments showing distinct frequency peaks on chromosomes 6, 8, 11, and 20 (Fig. 2D). The specific chromosomal positions, their target genes, and the labeling fluorochromes included the following: 6p22, E2F3-CDKAL1, 525KB, green, PlatinumBright495; 8q22, PABPC1-ZNF706, 480 KB, gold, PlatinumBright530; 11q13, FGF19-FGF3, dark red, PlatinumBright590; and 20q11.2, MAPRE1, 610KB, blue, PlatinumBright415 (Supplementary Fig. 3). We initially tested the specificity of the probes both individually and in mixture, on normal human metaphase cells, which showed that all FISH probes specifically hybridized to their respective chromosomal loci (Fig. 3A and 3B). A mixture of the four probes generated the expected diploid eight signals for their respective fluorescent tags in normal peripheral blood lymphocytes and urothelial cells (Fig. 3C). Preliminary testing on touch prints from high-grade bladder carcinoma samples revealed gross aneuploidy with multiple (>2) copy numbers for all probes in practically all tumor cells (Fig. 3D).
Fig. 2.
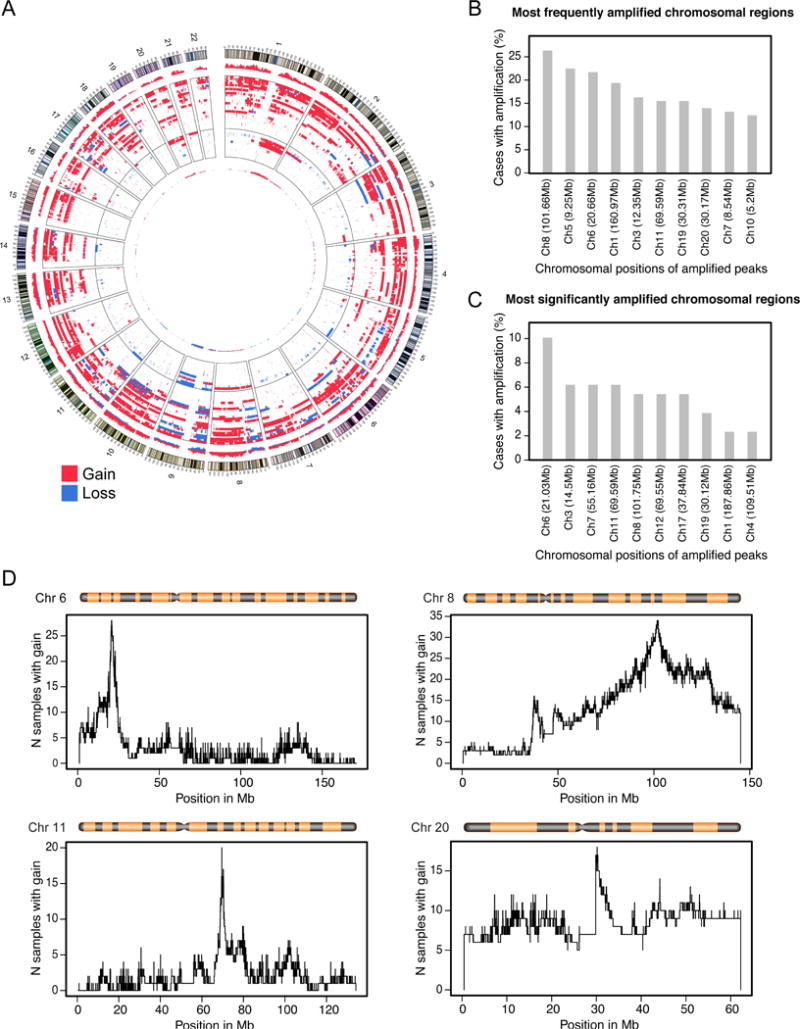
Copy number analysis of bladder cancer cohorts. (A) Circos diagram summarizing CNV analysis of bladder cancer tumor samples from the MDACC cohort (n = 40). (B) Top 10 most frequently amplified chromosomal regions validated in the TCGA cohort (n = 126). (C) Top 10 most significantly amplified chromosomal regions validated in the TCGA cohort. (D) Frequency histograms of amplifications of four chromosomes selected to design the FISH probes for the quartet test.
CNV = copy number variation; FISH = fluorescence in situ hybridization; MDACC = MD Anderson Cancer Center; TCGA = The Cancer Genome Atlas.
Fig. 3.
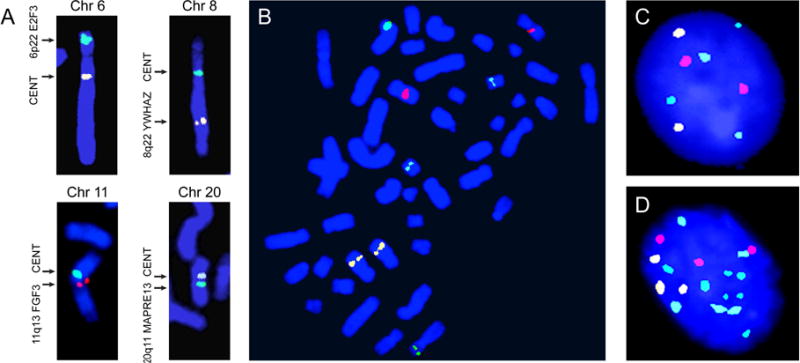
Testing of specificity for chromosomal FISH probes selected to design the quartet test. (A) Dual color FISH with centromeric and respective chromosomal probes 6p22 E2F3, 8q22 YWHAZ, 11q13 FGF3, and 20q11 MAPRE13. (B) Hybridization signals for a mixture of four probes (6p22, E2F3, green, PlatinumBright495; 8q22, YWHAZ, gold, PlatinumBright530; 11q13, FGF3, dark red, PlatinumBright590; and 20q11, MAPRE13, blue, PlatinumBright415) showing the hybridization signals on their respective human metaphase chromosomes. (C) Hybridization signals obtained with a mixture of the four probes listed above (the quartet test) in normal human urothelial cells. (D) Hybridization signals obtained with a mixture of four probes comprising the quartet test in a cell from a touch print prepared from a tumor tissue of high-grade bladder cancer.
FISH = fluorescence in situ hybridization.
3.2. Quartet test study in voided urine
Performance of the quartet test was initially analyzed on paired samples of voided urine typically collected 2–3 d before cystoscopy and tumor tissue from a cohort of 53 patients: 19 patients with LGPUC and 34 patients with HGUC (Fig. 4A–D). In every instance, abnormal copy number levels were detected in touch print preparations of the tumor tissue and the corresponding paired voided urine samples from the same patient. Although there were some discrepancies, in the majority of cases the percentage of cells with abnormal copy numbers for one or more FISH probes were similar in tumor and urine samples from the same patient. It was also evident that the proportions of cells with abnormal copy numbers in both tumor and voided urine samples were significantly higher in HGUC than in LGPUC.
Fig. 4.
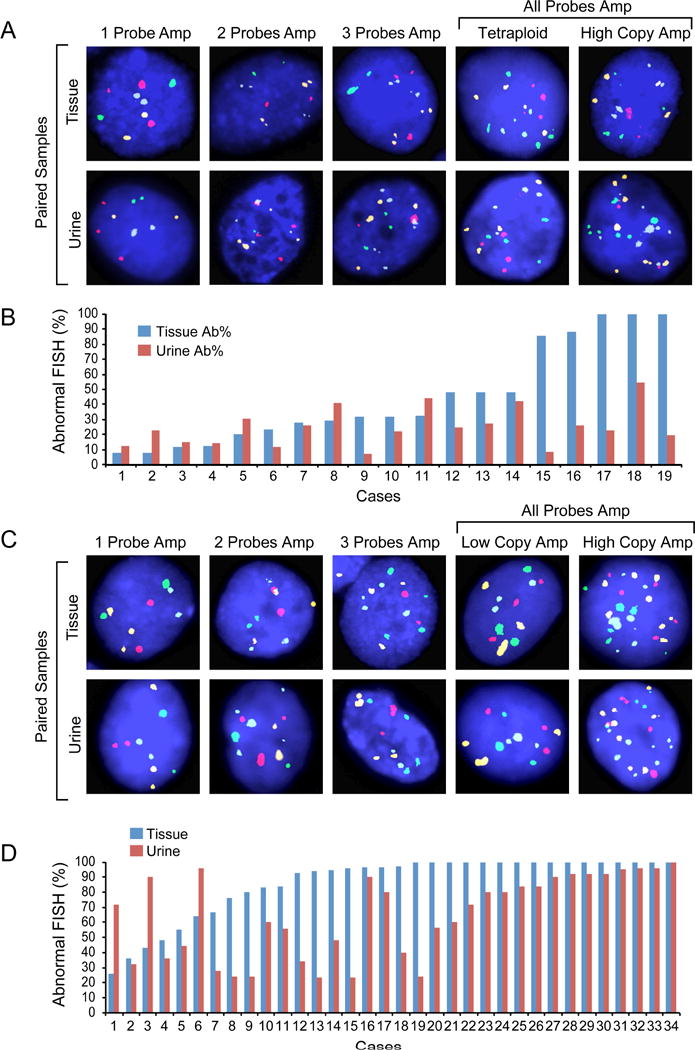
Quantitative assessment of abnormal cells in pairs of tumor tissue and voided urine of the same patients in low- and high-grade urothelial carcinoma by the quartet test. (A) Representative FISH images of low-grade papillary urothelial carcinoma. Upper row shows representative images of tissue samples. Lower row shows representative images of paired voided urine samples of the same patient. (B) Quantitative assessment of the percentage of cells with abnormal copy numbers in paired samples of low-grade papillary urothelial carcinoma and voided urine of the same patient. (C) Representative FISH images of high-grade urothelial carcinoma. Upper row shows representative images of tissue samples. Lower row shows representative images of paired voided urine samples of the same patient. (D) Quantitative assessment of the percentage of cells with abnormal copy numbers in paired samples of high-grade urothelial carcinoma and voided urine of the same patient.
Ab% = percentage of cells with abnormal copy number; FISH = fluorescence in situ hybridization.
Performance of the quartet test was then evaluated in a blinded fashion in voided urine samples from 98 patients with bladder tumors and 48 controls (Fig. 5A–D). We only used samples that contained at least 10 intact cells with measurable, clearly defined fluorescent signals, but in the majority of cases the number of analyzed cells was ≥20. A small fraction of samples could not be analyzed because of an insufficient number of intact cells with fluorescent signals available for microscopic inspection in cytological preparations of urine sediments (14.5% of samples in control group and 7.1% of samples in the cancer group). The initial analysis of the control samples disclosed that the majority of them contained a small fraction of cells with an increased copy number of individual probes. They typically involved extra numeral signals in a range of three to four copies restricted to one probe. The extra numeral copy signals involving two and four probes were present only in two cases of the control group. The AP value in the control group ranged from 2% to 20% (8.69 ± 5.33%) and strongly correlated with age (r = 0.971; Supplementary Fig. 4). It was significantly higher (p = 0.002) in males (10.8 ± 5.6%) when compared with females (6.1 ± 3.7%).
Fig. 5.
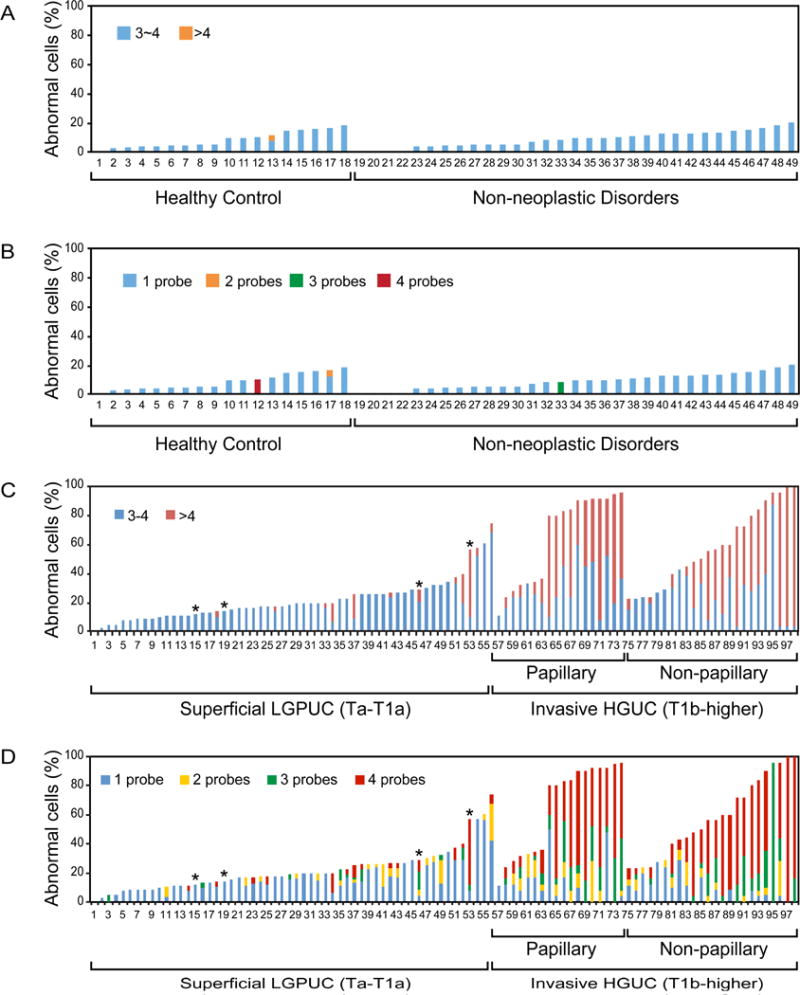
Quantitative assessment of cells with abnormal copy numbers by the quartet test in voided urine samples (n = 146). (A) Percentage of cells with abnormal copy numbers in individual samples of 48 benign controls dichotomized into groups with low levels of amplification (three to four copies for at least one probe) and high levels of amplification (more than four copies for at least one probe). (B) Percentage of cells with abnormal copy numbers in individual samples of 48 controls separated into four groups according to the numbers of probes with abnormal copy numbers. (C) Percentage of cells with abnormal copy numbers in individual samples of 98 voided urines from patients with bladder cancer dichotomized into groups with low levels of amplification (three to four copies for at least one probe) and high levels of amplification (more than four copies for at least one probe). (D) Percentage of cells with abnormal copy numbers in individual samples of 98 voided urines from patients with bladder cancer separated into four groups according to the number of probes with abnormal copy numbers.
HGUC = high-grade urothelial carcinoma; LGPUC = low-grade papillary urothelial carcinoma.a T1a tumors.
By analyzing the ROC curve for the AP score (Fig. 6A), we identified the optimal cutoff score as 0.16. Samples in which >16.4% of the cells examined contained at least one probe with three or more copies were flagged as likely having bladder cancer. With these criteria, the AP quartet test was positive for 77/98 samples of patients with bladder cancer, corresponding to a sensitivity of 78.6% (approximately 95% coverage interval 0.694, 0.855). The AP quartet test was positive for three of 48 control samples, corresponding to a specificity of 93.8% (0.831, 0.977). The AUC is 0.902 (95% confidence interval [CI] = 0.855–0.949; p < 0.001; Fig. 6A). Overall, the proportion of cells with abnormal copy numbers was significantly higher (p < 0.001) in patients with high-grade tumors (61.20 ± 28.46) when compared with patients with low-grade tumors (21.72 ± 14.48) and benign controls (8.69 ± 5.33; Fig. 6B).
Fig. 6.
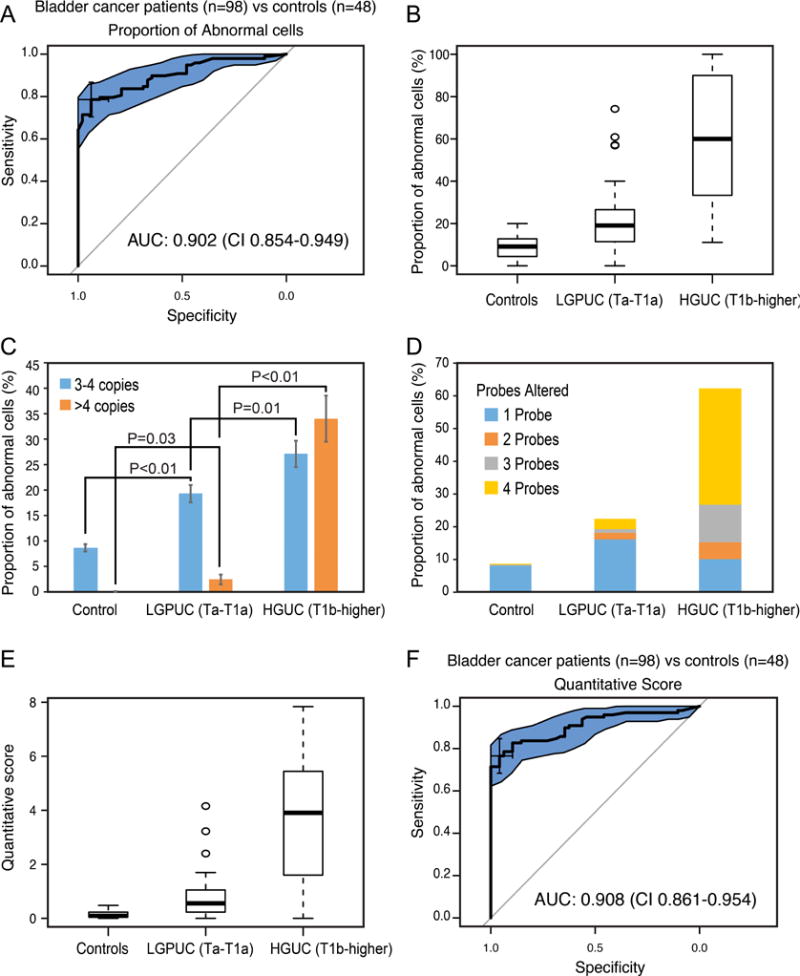
Detection of bladder cancer in voided urine by the quartet test (n = 146). (A) Receiver operating characteristic curve (ROC) based on the proportion of cells with abnormal copy numbers for the set consisting of 98 urine samples from patients with bladder cancer and 48 urine samples from control individuals (18 healthy controls and 30 with benign non-neoplastic disorders of the urinary tract). The quartet test for the detection of bladder cancer showed an area under the ROC curve (AUC) of 0.902 (95% confidence interval [CI] = 0.854–0.949). (B) Box plot analysis of mean percentage of abnormal cells in benign control samples and two groups of bladder cancer dichotomized into a superficial low-grade papillary carcinoma (LGPUC, Ta–T1a) and high-grade invasive carcinoma (HGUC, T1b–higher). (C) Average proportion of cells in voided urine showing three to four or more than four copies for at least one probe in benign controls (LGPUC [Ta–T1a] and HGUC [T1b–higher] groups of samples). (D) Average proportion of cells with increased copy numbers of one to four probes in benign controls (LGPUC [Ta–T1a] and HGUC [T1b–higher] groups of samples). (E) Box plot analysis of the QS values in benign controls (LGPUC [Ta–T1a] and HGUC [T1b–higher] groups of samples). (F) ROC based on the QS values for the set consisting of 98 urine samples from patients with bladder cancer and 48 urine samples from control individuals (18 healthy controls and 30 with benign non-neoplastic disorders of the urinary tract). The QS values of the quartet test for the detection of bladder cancer showed an AUC of 0.908 and CI = 0.861–0.954.
HGUC = high-grade urothelial carcinoma; LGPUC = low-grade papillary urothelial carcinoma; QS = quantitative score.
There were striking differences in the degree of amplification among high- and low-grade tumor patients as well as benign controls. The proportion of cells with a low degree of amplification (three to four copies for at least one probe) was significantly higher in patients with high-grade tumors (27.11 ± 16.74) when compared with patients with low-grade tumors (19.32 ± 12.91) and benign controls (8.61 ± 5.31). There were striking differences among the proportions of cells with a high degree of amplification (greater than four copies for at least one probe) in high-grade tumors (34.02 ± 29.48) when compared with low-grade tumors (2.4 ± 7; Fig. 6C). Urothelial cells in voided urine samples from benign controls had virtually no cells with more than four copies for any of the probes. A similar pattern of changes was observed when the proportions of cells with large numbers of altered probes were analyzed (Fig. 6D). High-grade tumors were characterized by large populations of cells that showed increased copy numbers for more than two probes. In fact, on average, >80% of the cells in these tumors showed alterations of all four probes. These data suggest that a pattern of amplifications detected by the four probes in exfoliated cells from voided urine reflects the degree of increased copy numbers in the tested chromosomal loci of bladder cancer cells. In order to provide a quantitative assessment reflecting the degree of this change, we designed a weighted QS incorporating the numbers of altered probes seen per cell as well as binary altered/unaltered calls. With the formula for QS described in the Patients and methods section, the maximum score is 8. The QS values for patients with high-grade tumors (3.73 ± 2.32) were strikingly higher than those for patients with low-grade tumors (0.77 ± 0.76) and benign controls (0.14 ± 0.13; Fig. 6E). There was a minimal overlap of the QS values between high- and low-grade tumors. Examination of the ROC curve for the QS quartet test shows an optimal cutoff score of 0.41. Using this rule, the QS quartet test was positive for 75/98 samples of patients with bladder cancer, corresponding to a sensitivity of 76.5% (approximate 95% coverage interval 0.672, 0.838). The QS quartet test was positive for two of the 48 control samples, corresponding to a specificity of 95.8% (0.860, 0.987). The AUC is 0.908 (95% CI = 0.861–0.954; p < 0.001; Fig. 6F). We also assessed the negative and positive predictive values of the quartet test. The negative predictive value calculated using AP was 68.2% and that calculated using QS was 66.7%. The positive predictive value for AP was 96.3% and for QS was 97.4%.
Since both AP and QS values appeared effective in grade assessment, we compared their performance by AUC and logistic linear regression. The AUC analyses identified 27% and 1.7 cutoff points for the AP and QS values, respectively, as the most optimal for the assessment of low- and high-grade tumors (Supplementary Fig. 5A and 5B). Both parameters were equally effective and the overall accuracy of grade assessment was 83.7%. Both AP and QS values predicted high grade of the tumors with 95% specificity. Correlation analysis showed that both parameters were closely related to each other and performed in a similar fashion, thus combining them did not improve the accuracy of the classifier (Supplementary Fig. 5C).
Finally, we assessed the interobserver variability in which 19 cases from the cancer group were independently analyzed by four observers who received brief training concerning the identification of cells in cytological preparations of voided urine and quantitative inspection of fluorescent signals. The data disclosed some variability among individual observers, which was typically within a 10% range of the mean value, and the differences among the individual observers were statistically insignificant (Supplementary Fig. 6A–F).
4. Discussion
The ubiquitous presence of genomic copy number alterations in many human cancers, including bladder cancer, makes measures of aneuploidy tempting as effective detection biomarkers [1]. For studies of biomarkers, bladder carcinoma is an ideal disease model because its development and progression can be monitored using noninvasive and minimally invasive techniques. Mucosa of the bladder can be examined, and biopsies can be obtained via endoscopic procedures. In addition, the morphology and molecular aberrations of exfoliated urothelial cells as well as secreted products can be scrutinized in urine at no risk to the patient [25].
FISH has already been shown to be an effective technique for the detection of urinary tract malignancies, as it is capable of identifying individual abnormal cells against a background of heterogeneous normal cell populations [17,19,26]. The multicolor FISH test introduced here, the quartet test, comprises four distinct chromosomal probes aimed at the specific amplified chromosomal regions of 6p22, 8q22, 11q13, and 20q11.2. These regions were selected by genome-wide CNV analysis of the MDA and TCGA cohorts. The data from these two cohorts were overlapping but not identical. As the TCGA cohort is restricted to high-grade muscle-invasive bladder cancer, it is not ideal to validate the chromosomal amplicons involved in the full spectrum of bladder cancers. In order to address this issue, we included in the selection process the data from Chekaluk et al [24], who analyzed genome-wide CNVs of both invasive and superficial papillary subsets of bladder cancers, which contain three of the four amplified regions identified in the MDA and TCGA cohorts. Most importantly, it included an amplicon on chromosome 11q13. Test results were analyzed using either AP or QS. The AP quartet test detected bladder cancer with 93.8% specificity and 78.6% sensitivity. The QS quartet test provides additional information concerning tumor grade, and was helpful in identifying patients with high- and low-grade bladder cancer. The QS quartet test detected bladder cancer with 95.8% specificity and 76.85% sensitivity.
Interestingly, a large proportion of individuals in the control group had a small proportion of exfoliated cells in urine, which exhibited an abnormal copy number. These abnormalities were typically restricted to one probe in the range of three to four copies. The AP value showed a strong correlation with patient age, and males had a statistically significantly higher proportion of cells in their voided urine with abnormal copy numbers when compared with females. The smoking status of the control group was unknown and could not be analyzed. These data open an interesting opportunity for future studies to explore the options of screening tests in selected target groups, such as smokers or workers with industry exposure, to evaluate future FISH tests as screening tools.
Many technologies, including measures of various genetic, epigenetic, and protein aberrations, have been applied to detect bladder cancer [25]. They range from quantifications of single molecular targets such as genes and their respective encoded proteins through summaries derived from genomic approaches analyzing the profiles of alterations seen with various platforms. Urine cytology using microscopic inspection of exfoliated urothelial cells is a conventional noninvasive technique for the detection of bladder cancer. It has both high specificity and high sensitivity for high-grade UC, but it is suboptimal for the detection of low-grade papillary bladder cancer [17–19]. Among the FISH-based technologies, UroVysion, which utilizes centromeric probes for three chromosomes and a 9p probe for its deletion, is the most popular and widely used test. In general, it performs in the 70–80% range for both specificity and sensitivity [18,19,26]. It can also be used to predict recurrence of bladder cancer and is therefore used in surveillance [26]. The single gene AURKA FISH test that we developed performs in the same 80% range of specificity and sensitivity [15,16].
Methylation markers have emerged as a new set of diagnostic tools and are typically used in panels that perform in similar ranges to those from UroVysion [27,28]. Proteomic profiling has evolved from initial surface-enhanced laser desorption/ionization–based technologies to high-resolution mass spectrophotometry, and reports increasing specificities and sensitivities over the last decade from initially 60% to recently in upper 80% range [27–29]. Our own proteomic biomarker studies yielded sensitivity and specificity of 80% and 100%, respectively [30]. The most recent reports utilizing methylation markers and mutational analysis combining multiplex polymerase chain reaction and next-generation sequencing report high specificity and sensitivity of >90% [31,32].
The quartet test introduced here shows very promising specificity and sensitivity results, but it requires validation in a larger multi-institutional cohort of samples. The high specificity (>90%) is very promising, but the sensitivity (slightly below 80%) requires improvement. Similarly, the test shows satisfactory positive predictive value of >95%, but the negative predictive value of <70% limits its current application as a cancer exclusion test. The fact that the quartet test was designed based on a genome-wide copy number analysis suggests an opportunity for improvement by replacing the weakest performing probes with alternative probes targeting other chromosomal regions of amplification. Manual reading of FISH preparation is time consuming, requires trained personnel, and is subjected to interobserver variability. Our analysis of interobserver variability identified some differences in scoring of individual samples, which were statistically not significant. Both the efficiency of the test and its objective scoring can be improved further by automated high-throughput, computer-based image analysis [33–35]. In addition, similar to other diagnostic tests utilizing cytology, FISH, and DNA methylation markers, diagnostic efficiency of the quartet test may be improved in future studies by combining it with antibody-based sorting and cell filtration [36–38].
The available data should be considered as preliminary, and the full spectrum of potential clinical applications is not explored in the current report, representing a major limitation of this study. Further validation studies may include assessment of recurrence, response to treatment, as well as diagnostic efficiency for specific pathogenetic and clinical subsets of bladder cancer.
5. Conclusions
In conclusion, we described the development and preliminary performance of a novel multicolor FISH test, referred to as the quartet test, which includes four specific chromosomal probes. It can improve the detection of bladder cancer as well as the management of patients with already diagnosed disease by decreasing the number of invasive cytoscopic surveillance procedures required.
Supplementary Material
Take Home Message.
We have developed a multicolored fluorescence in situ hybridization probe test comprising four probes aimed at the loci 6p22, 8q22, 11q13, and 20q11.2, which detects bladder cancer in voided urine with approximately 95% specificity and nearly 80% sensitivity.
Acknowledgments
Funding/Support and role of the sponsor: This study was supported in part by grants from National Institute of Health (R01 CA151489 and P50 CA91846/Project 1 and Core C) to B. Czerniak.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Bogdan Czerniak had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Czerniak.
Acquisition of data: Lotan, Dinney.
Analysis and interpretation of data: Yao, Baggerly, Bondaruk, Majewski, Czerniak.
Drafting of the manuscript: Czerniak.
Critical revision of the manuscript for important intellectual content: Baggerly.
Statistical analysis: Yao, Wei, Baggerly.
Obtaining funding: Czerniak.
Administrative, technical, or material support: Lotan, Dinney.
Supervision: Czerniak.
Other: S. Zhang and Y. Wang performed FISH test and their validation; Y. Lotan and C. Dinney supervised the collection of urine samples and their clinical data; J. Bondaruk and T. Majewski performed the genomic analysis; and S. Lee, J.G. Lee, and D. Cogdell performed the validation studies and prepared samples for genomic analysis. B. Czerniak wrote the final version of the manuscript and supervised the study.
Financial disclosures: Bogdan Czerniak certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
References
- 1.Sansregret L, Swanton C. The role of aneuploidy in cancer evolution. Cold Spring Harb Perspect Med. doi: 10.1101/cshperspect.a028373. In press. [DOI] [PMC free article] [PubMed]
- 2.Naylor RM, van Deursen JM. Aneuploidy in cancer and aging. Annu Rev Genet. 2016;50:45–66. doi: 10.1146/annurev-genet-120215-035303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazzucchelli R, Barbisan F, Stramazzotti D, Montironi R, Lopez-Beltran A, Scarpelli M. Chromosomal abnormalities in macroscopically normal urothelium in patients with bladder pT1 and pT2a urothelial carcinoma: a fluorescence in situ hybridization study and correlation with histologic features. Anal Quant Cytol Histol. 2005;27:143–51. [PubMed] [Google Scholar]
- 4.Obermann EC, Junker K, Stoehr R, et al. Frequent genetic alterations in flat urothelial hyperplasias and concomitant papillary bladder cancer as detected by CGH, LOH, and FISH analyses. J Pathol. 2003;199:50–7. doi: 10.1002/path.1259. [DOI] [PubMed] [Google Scholar]
- 5.Czerniak B, Li L, Chaturvedi V, et al. Genetic modeling of human urinary bladder carcinogenesis. Genes Chromosomes Cancer. 2000;27:392–402. [PubMed] [Google Scholar]
- 6.Lee S, Jeong J, Majewski T, et al. Forerunner genes contiguous to RB1 contribute to the development of in situ neoplasia. Proc Natl Acad Sci U S A. 2007;104:13732–7. doi: 10.1073/pnas.0701771104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majewski T, Lee S, Jeong J, et al. Understanding the development of human bladder cancer by using a whole-organ genomic mapping strategy. Lab Invest. 2008;88:694–721. doi: 10.1038/labinvest.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips JL, Richardson IC. Aneuploidy in bladder cancers: the utility of fluorescent in situ hybridization in clinical practice. BJU Int. 2006;98:33–7. doi: 10.1111/j.1464-410X.2006.06189.x. [DOI] [PubMed] [Google Scholar]
- 9.Dinney CP, McConkey DJ, Millikan RE, et al. Focus on bladder cancer. Cancer Cell. 2004;6:111–6. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Czerniak B, Dinney C, McConkey D. Origins of bladder cancer. Annu Rev Pathol. 2016;11:149–74. doi: 10.1146/annurev-pathol-012513-104703. [DOI] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dadhania V, Zhang M, Zhang L, et al. Meta-analysis of the luminal and basal subtypes of bladder cancer and the identification of signature immunohistochemical markers for clinical use. EBioMedicine. 2016;12:105–17. doi: 10.1016/j.ebiom.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540–56.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Jr, Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci. 2013;70:661–87. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park HS, Park WS, Bondaruk J, et al. Quantitation of Aurora kinase A gene copy number in urine sediments and bladder cancer detection. J Natl Cancer Inst. 2008;100:1401–11. doi: 10.1093/jnci/djn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mobley A, Zhang S, Bondaruk J, et al. Aurora kinase A is a biomarker for bladder cancer detection and contributes to its aggressive behavior. Sci Rep. 2017;7:40714. doi: 10.1038/srep40714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dal Moro F, Valotto C, Guttilla A, Zattoni F. Urinary markers in the everyday diagnosis of bladder cancer. Urologia. 2013;80:265–75. doi: 10.5301/urologia.5000041. [DOI] [PubMed] [Google Scholar]
- 18.Glass RE, Coutsouvelis C, Sheikh-Fayyaz S, et al. Two-tiered subdivision of atypia on urine cytology can improve patient follow-up and optimize the utility of UroVysion. Cancer Cytopathol. 2016;124:188–95. doi: 10.1002/cncy.21630. [DOI] [PubMed] [Google Scholar]
- 19.Lavery HJ, Zaharieva B, McFaddin A, Heerema N, Pohar KS. A prospective comparison of UroVysion FISH and urine cytology in bladder cancer detection. BMC Cancer. 2017;17:247. doi: 10.1186/s12885-017-3227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima T, Kawai K, Miyazaki J, Nishiyama H. Biomarkers for precision medicine in bladder cancer. Int J Clin Oncol. 2017;22:207–13. doi: 10.1007/s10147-016-1068-8. [DOI] [PubMed] [Google Scholar]
- 21.Sokolova IA, Halling KC, Jenkins RB, et al. The development of a multitarget, multicolor fluorescence in situ hybridization assay for the detection of urothelial carcinoma in urine. J Mol Diagn. 2000;2:116–23. doi: 10.1016/S1525-1578(10)60625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eble JN, Guido S, Epstein JI, Sesterhenn IA. Pathology and genetics of tumours of the urinary system and male genital organs WHO/OMS. IARC Press; 2004. [Google Scholar]
- 23.Edge S, Byrd DR, Compton CC, Fritz AG, Greene F, Trotti A. AJCC cancer staging handbook: from the AJCC cancer staging manual. 7. New York, NY: Springer-Verlag; 2009. [Google Scholar]
- 24.Chekaluk Y, Wu CL, Rosenberg J, et al. Identification of nine genomic regions of amplification in urothelial carcinoma, correlation with stage, and potential prognostic and therapeutic value. PLoS One. 2013;8:e60927. doi: 10.1371/journal.pone.0060927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czerniak B. Molecular pathology and biomarkers of bladder cancer. Cancer Biomark. 2010;9:159–76. doi: 10.3233/CBM-2011-0175. [DOI] [PubMed] [Google Scholar]
- 26.Seideman C, Canter D, Kim P, et al. Multicenter evaluation of the role of UroVysion FISH assay in surveillance of patients with bladder cancer: does FISH positivity anticipate recurrence? World J Urol. 2015;33:1309–13. doi: 10.1007/s00345-014-1452-9. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61:8659–63. [PubMed] [Google Scholar]
- 28.Kumar P, Nandi S, Tan TZ, et al. Highly sensitive and specific novel biomarkers for the diagnosis of transitional bladder carcinoma. Oncotarget. 2015;6:13539–49. doi: 10.18632/oncotarget.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilz SW, Liu D, Liu C, Yang J. Development of a test to identify bladder cancer in the urine of patients using mass spectroscopy and subcellular localization of the detected proteins. Am J Transl Res. 2015;7:1458–66. [PMC free article] [PubMed] [Google Scholar]
- 30.Majewski T, Spiess PE, Bondaruk J, et al. Detection of bladder cancer using proteomic profiling of urine sediments. PLoS One. 2012;7:e42452. doi: 10.1371/journal.pone.0042452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward DG, Baxter L, Gordon NS, et al. Multiplex PCR and next generation sequencing for the non-invasive detection of bladder cancer. PLoS One. 2016;11:e0149756. doi: 10.1371/journal.pone.0149756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feber A, Dhami P, Dong L, et al. UroMark—a urinary biomarker assay for the detection of bladder cancer. Clin Epigenetics. 2017;9:8. doi: 10.1186/s13148-016-0303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson K, Thompson A, Browne G, Shasserre C, Seelig SA, King W. Automation of fluorescence in situ hybridization pretreatment: a comparative study of different sample types. Mol Diagn. 2000;5:209–20. doi: 10.1054/modi.2000.9731. [DOI] [PubMed] [Google Scholar]
- 34.Smith GD, Bentz JS. “FISHing” to detect urinary and other cancers: validation of an imaging system to aid in interpretation. Cancer Cytopathol. 2010;118:56–64. doi: 10.1002/cncy.20066. [DOI] [PubMed] [Google Scholar]
- 35.Kohler CU, Martin L, Bonberg N, et al. Automated quantification of FISH signals in urinary cells enables the assessment of chromosomal aberration patterns characteristic for bladder cancer. Biochem Biophys Res Commun. 2014;448:467–72. doi: 10.1016/j.bbrc.2014.04.137. [DOI] [PubMed] [Google Scholar]
- 36.Costa VL, Henrique R, Danielsen SA, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16:5842–51. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 37.Macgregor-Ramiasa M, McNicholas K, Ostrikov K, et al. A platform for selective immunocapture of cancer cells from urine. Biosens Bioelectron. 2017;96:373–80. doi: 10.1016/j.bios.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 38.Tzouanas CSYLJ, Wen Y, Thiery JP, Khoo BL. Microdevices for non-invasive detection of bladder cancer. Chemosensors. 2017;5:1–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


