Abstract
High-temperature stress during the ripening stage leads to quality deterioration due to an increase in chalky grains in brown rice (Oryza sativa L.). In a previous study, we identified a QTL for Appearance quality of brown rice 1 (Apq1) using chromosome segment substitution lines of the indica cultivar ‘Habataki’ in the japonica cultivar ‘Koshihikari’ background and narrowed down the locus to a 48-kb region on chromosome 7. To verify the function and mechanisms of this QTL in grain appearance, in this study, we fine-mapped the gene and conducted high-temperature tolerance tests. As a result of the genetic mapping, we narrowed down the candidate region of Apq1 to a 19.4-kb region including three predicted genes. Among these, the temporal expression pattern of sucrose synthase 3 (Sus3) corresponded well with the high temperature-sensitive period during ripening, and expression of the ‘Habataki’ allele of Sus3 was increased under high-temperature condition. In addition, we transformed the ‘Habataki’ Sus3 gene into ‘Nipponbare’, and the transformants obtained high-temperature tolerance. Therefore, we conclude that the causal gene underlying the QTL Apq1 is the thermo-responsive Sus3 allele, and the increase in Sus3 expression under high-temperature condition during ripening leads to high-temperature tolerance in rice.
Keywords: Apq1, appearance quality, high-temperature tolerance, sucrose synthase 3, rice (Oryza sativa L.)
Introduction
The higher average temperatures caused by global warming affect crop yields. Lyman et al. (2013) estimated that a 1°C increase in average growing season temperature reduces total rice milling revenue by 8.1% to 11.0%. The reduction in brown rice production caused by high temperatures is due to spikelet sterility and decreases in rice weight and quality (Folsom et al. 2014, Hakata et al. 2012, 2017, Morita et al. 2016, Peng et al. 2004). A high temperature during the rice ripening period causes an increase in grain chalkiness (Terashima et al. 2001). Chalky grain phenotypes include white back, basal white, milky white, white core, and white belly (Ebata 1961, Nagato and Ebata 1965, Tashiro and Ebata 1975). In Japan, the market value of brown rice is determined by grain appearance, particularly, the ratio of chalky grains. Therefore, the development of novel cultivars with tolerance to high temperature during ripening is required.
Several studies have identified quantitative trait loci (QTLs) or genes related with chalky grain phenotypes. Chalk5, encoding a vacuolar H+-translocating pyrophosphatase, was detected as a major QTL for grain chalkiness (Li et al. 2014). QTLs for white-back, basal-white, and milky-white grains have also been reported (Ebitani et al. 2008, Kobayashi et al. 2013, Miyahara et al. 2017, Tabata et al. 2007, Wada et al. 2015, Wakamatsu et al. 2010). Yamakawa et al. (2007) and Hakata et al. (2012) observed increased expression of the α-amylase genes Amy1A, Amy1C, Amy3A, and Amy3B during ripening under high temperature and showed that the suppression of these genes decreases the chalky grain rate. Although several chalky grain-related genes have been identified, the molecular mechanism underlying chalky grains is still incompletely understood.
Previously, we identified a QTL for appearance quality of brown rice (Appearance quality of brown rice 1, Apq1) on chromosome 7 by using chromosome segment substitution lines of the indica cultivar ‘Habataki’ in the japonica cultivar ‘Koshihikari’ genetic background (Murata et al. 2014). We also demonstrated that the nearly isogenic line (NIL) of Apq1 (Apq1-NIL) showed a significantly higher percentage of perfect grains (PPG) under field condition, and the candidate region of Apq1 was delineated to a 48-kb region. Further, Apq1-NIL showed higher tolerance to high-temperature stress than ‘Koshihikari’ during the ripening period, with reduced ratios of basal-white and white-back grains. In the current study, we carried out further fine mapping to identify the causal gene of the QTL and we determined the high temperature-sensitive period during ripening to understand the mechanisms of high-temperature tolerance of Apq1.
Materials and Methods
Plant materials and growth conditions
Apq1-NIL was developed in our previous study (Murata et al. 2014) and it possesses less than a 2.8-Mb ‘Habataki’ chromosome segment (spanning 25.8–28.6-Mb on chromosome 7) of Apq1 locus in the ‘Koshihikari’ genetic background. Two recombination lines were screened using two PCR markers, Tak6166-3 and RM21971, from approximately 10,000 F2 plants derived from a cross between ‘Koshihikari’ and Apq1-NIL and the genotypes of these plants were determined by using markers ERF2-1, ERF2-2, and AP2-indel. The sequences of the primers used for genetic mapping are shown in Supplemental Table 1. The seedlings used for genetic mapping were transplanted to a paddy field at the Toyama Agricultural Research Institute in Toyama, Japan (36.4°N, 137.1°E) at the late April and were grown under natural climate condition until yielding. The average temperature of the ripening period was 27.1°C. Seedlings of ‘Koshihikari’ and Apq1-NIL were transplanted to a paddy field at the Research Center for Bioresources Development in Fukui, Japan (36.2°N, 136.2°E) at the beginning of May and were grown under natural climate condition. These plants were transferred from the paddy field into a low-temperature climate chamber (25°C from 07:00 to 19:00 and 23°C from 19:00 to 07:00) at 7 days before heading. Transgenic plants were grown in a closed climate chamber at 28°C from 09:00 to 18:00 and 25°C from 18:00 to 09:00 until one week before heading. To test high-temperature tolerance during the ripening stage for Koshihikari, Apq1-NIL and transgenic plants, plants were grown at low (25°C from 07:00 to 19:00 and 23°C from 19:00 to 07:00) and high temperatures (34°C from 07:00 to 19:00 and 26°C from 19:00 to 07:00) until yielding.
To delineate the high temperature-sensitive period, we subjected plants to three days of high-temperature treatment. At least 100 spikelets/plant were marked at the flowering day under the low-temperature condition. The start of the three-day treatment varied from 1 DAF to 30 DAF, and after treatment, the plants were returned to the low-temperature condition and grown until yielding. Three plants were used for each treatment.
Phenotype evaluation
Perfect grains were defined as non-chalky, with a normal shape. Grains with unusual shapes or colors were left out of the evaluation. Chalky grains were classified as basal-white kernels, white-back kernels, white-berry kernels, milky-white kernels, or white-core kernels as described by Ebitani et al. (2008). At least 100 grains per plant and three biological replicates were used for evaluation.
Production of transgenic plants
The genomic clone of Sus3Haba was amplified using the primers Tak6166-3-Asc1-U (5′-ATGGCGCGCCAC ATCCGATAATGGAGTCGA-3′) and Asc1-Sus3-L 5′-GTAAGCGCCAAGCTCGCTGAGGCGCGCCAT-3′) with PrimeSTAR Max DNA Polymerase (TAKARA Bio, Tokyo, Japan) and was cloned into the AscI site of the binary vector pYL-TAC7 (Liu et al. 1999), provided by the RIKEN BioResource Center. This clone contains a 6195-bp fragment upstream from the transcriptional start site of the Sus3 gene and a 757-bp fragment downstream from the end of the Sus3 3′UTR. The binary vector was transformed into Agrobacterium tumefaciens strain EHA105 (Hood 1993) by electroporation, and ‘Nipponbare’ plants were transformed as reported previously (Ashikari et al. 2005). ‘Nipponbare’ plants transformed with empty vector were used as controls. Transgenic plants carrying the transgene were selected by PCR using hygromycin resistance gene (HPT)-specific primers, HPT-L: 5′-CGTATATGCTCCGCATTGGT-3′ and HPT-R: 5′-ATTTGTGTACGCCCGACAGT-3′.
RNA isolation and quantitative reverse-transcription (qRT)-PCR
Total RNAs were extracted from the ovary at 0 day after flowering (DAF), endosperm (immature grain includes endosperm and embryo) at 1, 4, 7, 10, 13, 16, 19 DAF, and flag leaf at 4 DAF. using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). cDNA was synthesized from total RNA using SuperScript III (Invitrogen, Carlsbad, CA, USA). For quantification of mRNA, qRT-PCR was carried out using SYBR Premix Ex TaqTM II (TAKARA Bio). Primer sequences used for qRT-PCR are shown in Supplemental Table 2. OsUbi1 was used as an internal control. The Thermal Cycler Dice Real Time System (TAKARA Bio) was used for qRT-PCR.
Promoter-swap analysis
Promoter sequences of Sus3 were amplified with the primers BamHI-pSus3-U (5′-ggatccCAGTTTAAACCGCTAATAGTCA-3′) and pSus3-L (5′-AGAAGACAGCCACAAGCTCA-3′) using PrimeSTAR Max DNA Polymerase and genomic DNA of ‘Koshihikari’ and ‘Habataki’ as templates. Coding sequences of Sus3 were amplified with the primers Sus3-mRNA-5UTR-U (5′-GCATCCATCGGTTCTCTGCT-3′) and Sus3-mRNA-3UTR-NosT-L (5′-ttgaacgatcCGGGTAAGGCCAGATCATTT-3′) using PrimeSTAR Max DNA Polymerase and cDNA of ‘Koshihikari’ and ‘Habataki’ as templates. The Nos terminator sequence was amplified with the primers Sus3-Nos-T-U (5′-GCCTTACCCGgatcgttcaaacatttggca-3′) and NosT-Not1-L (5′-GCGGCCGCcccgatctagtaacatagat-3′) using PrimeSTAR Max DNA Polymerase and pYL-TAC7 DNA as a template. The primers Sus3-mRNA-3UTR-NosT-L and Sus3-Nos-T-U were designed to overlap each other by 20 bp. The promoter sequences were cloned into the pCR4 vector using Zero Blunt TOPO PCR Cloning Kit for Sequencing (Life Technologies, CA, USA). The coding sequences and Nos terminator sequence were fused by PCR using PrimeSTAR max DNA Polymerase and the concatenated product was cloned into the pCR4 vector using Zero Blunt TOPO PCR Cloning Kit for Sequencing. Promoter and coding + Nos terminator sequences were digested using the restriction enzymes BamHI and NotI, respectively. The fragments obtained were cloned into BamHI- and NotI-digested pYL-TAC7 vector. Transgenic plants carrying the transgene were selected by PCR using the HPT-specific primers described above.
Results
Apq1 reduces the basal-white and white-back grain ratios
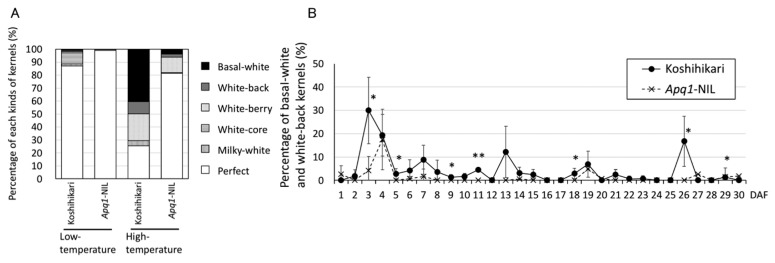
To confirm the function of Apq1 in grain appearance, we compared the appearance ratios of perfect grains and each type of chalky grains between ‘Koshihikari’ and Apq1-NIL plants grown under low and high temperatures during ripening. Both ‘Koshihikari’ and Apq1-NIL showed high PPG under low-temperature condition (89% and 98%, respectively) (Fig. 1A). In line with our previous report, the PPG of ‘Koshihikari’ decreased to 25%, while that of Apq1-NIL remained high (81%), under high temperature (Fig. 1A). Consistently, the basal-white and white-back grain ratio was restored in Apq1-NIL (Fig. 1A). To delineate the high temperature-sensitive period, we subjected plants to three days of high-temperature treatment throughout ripening. As a result, we observed the highest percentage of basal-white and white-back (PBW) kernels in ‘Koshihikari’ exposed to high temperature at 3 DAF (Fig. 1B). Apq1-NIL showed significantly lower PBW at 3 DAF. This result indicates that PBW in ‘Koshihikari’ is the most sensitive to high temperature at 3–5 DAF, and the QTL Apq1 might provide high-temperature tolerance in this period.
Fig 1.
Confirmation of the role of Apq1 in high-temperature tolerance. (A) Percentages of indicated kernel phenotypes of ‘Koshihikari’ and Apq1-NIL grown under low- and high-temperature conditions during the ripening stage. (B) Percentages of basal-white and white-back kernels, measured to compare high-temperature tolerance. Each spikelet was marked at opening and fertilization day under low-temperature condition and incubated for three days under high temperature. The numbers in the X-axis indicate initial day of three days high-temperature treatment. Error bars indicate standard deviation. ** p < 0.01, * p < 0.05 (Student’s t-test).
Genetic mapping of Apq1
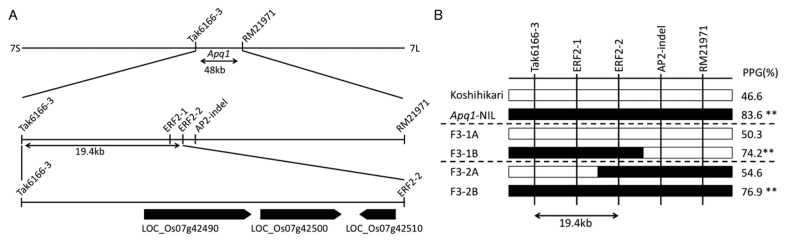
To determine the causal gene of Apq1, we selected two plants possessing chromosome recombination in the previously delineated candidate 48-kb region between PCR markers Tak6166-3 and RM21971 from F2 plants derived from a cross between ‘Koshihikari’ and Apq1-NIL (Fig. 2A). For finer gene mapping, we selected the homozygous genotypes from each F3 line. By comparing the PPG of the two genotypes, the candidate region of Apq1 was narrowed down to a 19.4-kb region between Tak6166-3 and ERF2-2, and this region harbors three predicted genes in the ‘Nipponbare’ genome (Fig. 2A, 2B, Rice Genome Annotation Project, http://rice.plantbiology.msu.edu/). According to gene prediction, LOC_Os07g42490, LOC_Os07g42500, and LOC_Os07g42510 encode sucrose synthase 3, FYVE zinc finger domain-containing protein, and AP2 domain-containing protein, respectively. As a result of DNA sequencing of this 19.4-kb candidate region of ‘Koshihikari’ and ‘Habataki’ and gene prediction using FGENISH (http://www.softberry.com/berry.phtml, Solovyev et al. 2006), we revealed that there are no variety specific genes, and detected amino-acid substitutions in the coding regions of LOC_Os07g42490 and LOC_Os07g42510 between the varieties. In LOC_Os07g42500, only a synonymous substitution was detected.
Fig. 2.
Genetic mapping of Apq1. (A) Location of Apq1 on the genetic map. Box arrows indicate the predicted gene locus. (B) Genotypes and phenotypes of the recombinants. White and black bars indicate homozygous chromosomes of ‘Koshihikari’ and ‘Habataki’, respectively. ** p < 0.01, * p < 0.05 (Student’s t-test).
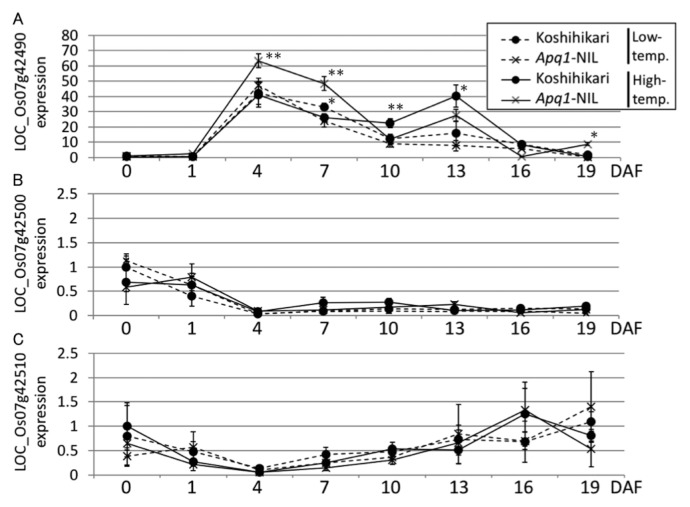
To determine when these three genes are expressed, qRT-PCR was performed using RNA extracted from ovaries and endosperms at various developmental stages. LOC_Os07g42490 was highly expressed from the early endosperm developmental stages and its expression reached a peak at 4 DAF (Fig. 3A). Although the expression level of LOC_Os07g42490 was not different between ‘Koshihikari’ and Apq1-NIL grown under low-temperature condition, significantly higher expression was detected in Apq1-NIL than in ‘Koshihikari’ under high temperature at 4 and 7 DAF (Fig. 3A). LOC_Os07g42500 and LOC_Os07g42510 showed no significant differences at any developmental stage (Fig. 3B, 3C). Because LOC_Os07g42490 expression peaked at 4 DAF, which corresponds with the high-temperature sensitive period, and was promoted by high temperature, we speculated that LOC_Os07g42490, sucrose synthase 3 (Sus3), is the causal gene of Apq1.
Fig. 3.
Expression analysis of Apq1 candidate genes. Relative expression of three candidate genes of Apq1 in endosperms grown under low- and high-temperature conditions was calculated using OsUbi1 as an internal control and was normalized to the expression level in samples of ‘Koshihikari’ at 0 DAF. LOC_Os07g42490, LOC_Os07g42500, and LOC_Os07g42510 are shown in (A), (B), and (C), respectively. Error bars indicate standard deviations. Student’s t-test was performed between ‘Koshihikari’ and Apq1-NIL under the same temperature condition. ** p < 0.01, * p < 0.05.
Complementation test of Apq1
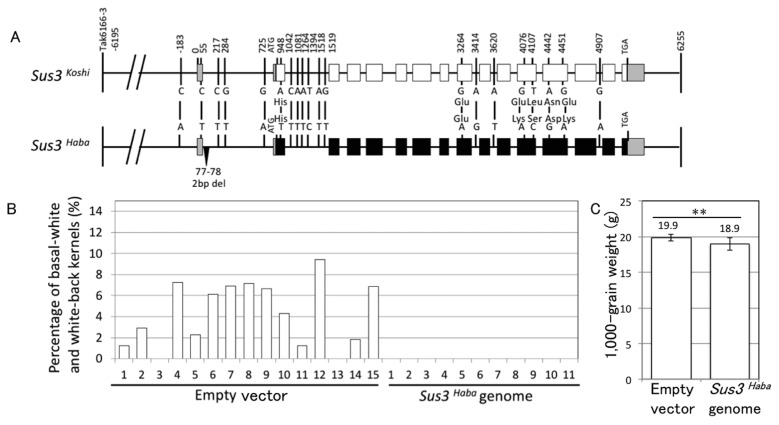
Based on genome sequence comparison, we detected 21 polymorphisms around the Sus3 gene between ‘Koshihikari’ and ‘Habataki’ (Fig. 4A). Six single nucleotide polymorphisms were detected in the coding sequence of Sus3, four of which result in amino-acid substitutions. These four amino-acid changes are not located in any conserved domain of sucrose synthase (Huang et al. 2016). Six polymorphisms were detected in the region upstream from the first ATG, including the UTR, and in an intron of Sus3. To confirm that Sus3 is the causal gene for Apq1, we cloned a 12.5-kb fragment of the ‘Habataki’ allele of Sus3 (Sus3Haba), containing all polymorphisms, into the binary vector pYL-TAC7 and transformed the construct into plants of the japonica cultivar ‘Nipponbare’ by the Agrobacterium-mediated method. The Sus3 gene sequence of ‘Nipponbare’ is identical to that of ‘Koshihikari’. All Sus3Haba-transgenic plants showed 0% PBW after high-temperature treatment during ripening, while basal-white and white-back kernels appeared in most empty-vector control plants (Fig. 4B). Therefore, we concluded that Sus3 is the causal gene of Apq1, and polymorphisms detected in the ‘Habataki’ allele of Sus3 offer tolerance to high temperature during the ripening stage. Furthermore, we measured 1,000-grain weight of these transgenic plants because we observed that it was slightly decreased in Apq1-NIL (Murata et al. 2014). As a result, Sus3Haba-transgenic plants showed significantly lower 1,000-grain weight than that of Empty vector plants (Fig. 4C).
Fig. 4.
Complementation test of Apq1. (A) DNA sequence polymorphisms of Sus3 in ‘Koshihikari’, Sus3Koshi, and ‘Habataki’, Sus3Haba. White and black boxes indicate protein-coding regions of ‘Koshihikari’ and ‘Habataki’, respectively. Gray boxes indicate UTRs. Numbers indicate positions of polymorphism from the transcription start site of Sus3. (B) High-temperature treatment test of Sus3Haba-transformed plants. Empty vector- and Sus3Haba-transformed ‘Nipponbare’ were grown under high-temperature condition the during ripening stage. The percentages of basal-white and white-back kernels were measured to compare high-temperature tolerance. Independently obtained empty vector-transformed (n = 15) and Sus3Haba-transformed (n = 11) plants were tested. (C) Average of 1,000-grain weight (dry-weight) of transgenic plants. ** p < 0.01 (Student’s t-test).
Sus3 promoter-swap analysis
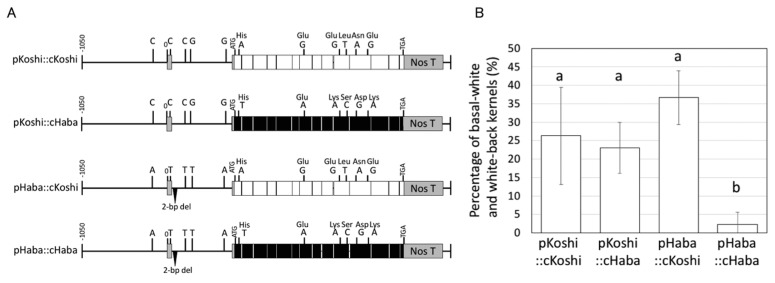
To determine whether polymorphisms detected in the region upstream of the first ATG of Sus3 are involved in tolerance to high temperature, we performed a promoter-swap analysis using four constructs. First ATG-upstream regions were cloned from genomic DNA of ‘Koshihikari’ (pKoshi) and ‘Habataki’ (pHaba) and fused to coding regions cloned from cDNA of ‘Koshihikari’ (cKoshi) and ‘Habataki’ (cHaba) (Fig. 5A). Interestingly, only pHaba::cHaba plants showed low PBW among the four types of transgenic plants (Fig. 5B). This result indicated that both the promoter and the coding region of the ‘Habataki’ allele of Sus3 are required for high-temperature tolerance during ripening. From these results, we concluded that Apq1 provides high-temperature tolerance via higher expression of Sus3 at the early stage of endosperm development.
Fig. 5.
Promoter-swap analysis of Sus3. (A)Vector constructs for promoter-swap analysis. Promoter sequences of Sus3Koshi (pKoshi) and Sus3Haba (pHaba) were fused to the coding cDNA sequences of Sus3Koshi (cKoshi) and Sus3Haba (cHaba). All four constructs were fused with the Nos terminator at the end of the coding sequence. (B) High-temperature treatment test of promoter-swap plants. The four constructs indicated in (A) were transformed into ‘Nipponbare’. Transformants were grown under high-temperature condition during the ripening stage. The percentages of basal-white and white-back kernels were measured to compare high-temperature tolerance. Error bars indicate standard deviations. Different letters above the bars indicate a significant difference at 5% (Tukey’s HSD test), n > 3.
Expression analysis of other Sus genes
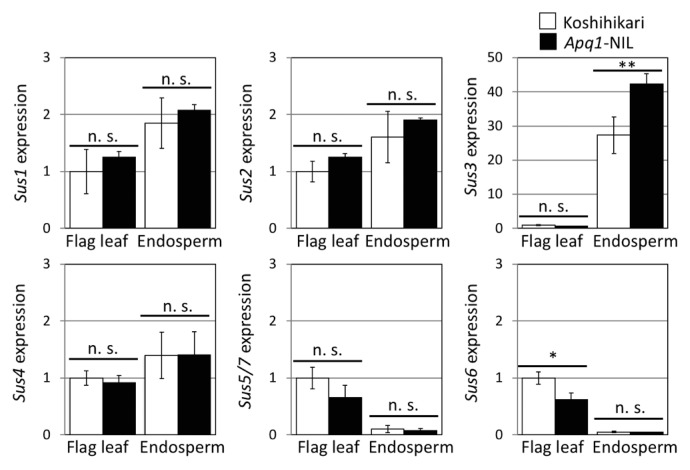
The rice genome contains seven conserved Sus genes, which are suspected to have compensatory functions (Cho et al. 2011, Hirose et al. 2008). We analyzed the expression of all Sus genes under high-temperature condition at 4 DAF. While Sus6 was significantly lower expressed in the flag leaf of Apq1-NIL than in that of ‘Koshihikari’, the Sus genes did not show significant differences in expression in the endosperm between ‘Koshihikari’ and Apq1-NIL at 4 DAF stage, except for Sus3 (Fig. 6). This result indicated that only Sus3Haba is responsive to high temperature and substantiated that this gene is responsible for the high-temperature tolerance during ripening conferred by Apq1.
Fig. 6.
Expression analysis of Sus genes in rice. Expression levels of all seven Sus genes in flag leaf and endosperm under the high-temperature condition were compared at 4 DAF. White and black bars indicate ‘Koshihikari’ and Apq1-NIL, respectively. Because of the highly similar mRNA sequences of Sus5 and Sus7, the expression was measured using a common sequence primer set. Gene expression was calculated using OsUbi1 as an internal control and was normalized to the expression level in the flag leaf of ‘Koshihikari’. Error bars indicate standard deviations. ** p < 0.01, * p < 0.05 (Student’s t-test), n > 3.
Discussion
Sus3 confers high-temperature tolerance during the ripening stage in rice
In this study, Sus3Haba was demonstrated to provide high-temperature tolerance during the ripening stage in the japonica genetic background. Sus is a key enzyme that catalyzes the first step of starch synthesis, namely, the conversion of sucrose and uridine diphosphate (UDP) into fructose and UDP-glucose, in the rice endosperm (Huang et al. 2016). Sus enzyme activity was reported to lower under high-temperature conditions in rice and maize (Chaturvedi et al. 2017, Wilhelm et al. 1999). The higher expression of Sus3Haba might compensate for the reduced activity of Sus under high temperature. In a previous study, we observed slightly decreased 1,000-grain weight and culm length (Murata et al. 2014). We detected smaller 1,000-grain weight of Sus3Haba-transgenic plants and this is consistent with the previous study (Fig. 4C). Hence, Apq1 seems to slightly decrease grain weight also under high-temperature stress conditions. Although we did not focus to culm length or other agronomic traits of Apq1 in this study, the information on the pleiotropic effect is also important. We will clarify this point in future studies.
Sus3 polymorphisms underlying high-temperature tolerance
In this study, we detected 21 polymorphisms around the Sus3 gene and we found that both the promoter and the coding region are required for high-temperature tolerance (Fig. 5B). Although four amino-acid substitutions were detected, we could not conclude which substitution is important for high-temperature tolerance because none of the four substitutions are in the conserved domain of Sus. However, since pHaba::cHaba plants acquired high-temperature tolerance, at least polymorphisms in the intron regions might not be important. We identified five single nucleotide polymorphisms and a 2-bp deletion upstream of the first ATG of Sus3 between ‘Koshihikari’ and ‘Habataki’ (Fig. 4A). One of these polymorphisms might be involved in the high expression of Sus3 in response to high temperature.
Application of Apq1 in breeding programs
Apq1 is predicted to be an economically important QTL to increase the PPG of the elite rice variety ‘Koshihikari’. Because its effect was confirmed in ‘Nipponbare’ background, Apq1 might be applicable in a wide range of japonica rice varieties. Although Apq1 was originally identified from ‘Habataki’, an indica variety, ‘Habataki’ shows low PPG (Murata et al. 2014). Thus, the effect of Apq1 seems to depend on the genetic background. In this study, although we succeeded in identifying the thermo-responsive Sus3 as the causal gene of Apq1, the underlying mechanism is still unclear. Clarifying this mechanism should further contribute to the application of high-temperature tolerance in future breeding programs.
Supplementary Information
Acknowledgments
This study was financially supported by the Ministry of Agriculture, Forestry and Fisheries, Japan through a research project entitled “Development of technologies for mitigation and adaptation to climate change in Agriculture, Forestry and Fisheries”.
We thank Dr. Tatsuro Hirose for helpful comments on this study.
Literature Cited
- Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., Angeles, E.R., Qian, Q., Kitano, H. and Matsuoka, M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Chaturvedi, A.K., Bahuguna, R.N., Shah, D., Pal, M. and Jagadish, S.V.K. (2017) High temperature stress during flowering and grain filling offsets beneficial impact of elevated CO2 on assimilate partitioning and sink-strength in rice. Sci. Rep. 7: 8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J., Kim, H., Kim, C., Hahn, T. and Jeon, J. (2011) Identification and characterization of the duplicate rice sucrose synthase genes OsSUS5 and OsSUS7 which are associated with the plasma membrane. Mol. Cells 31: 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebata, M. (1961) Studies on white-core rice kernel. Jpn. J. Crop Sci. 29: 409–411. [Google Scholar]
- Ebitani, T., Yamamoto, Y., Yano, M. and Funane, M. (2008) Identification of quantitative trait loci for grain appearance using chromosome segment substitution lines in rice. Breed. Res. 10: 91–99. [Google Scholar]
- Folsom, J.J., Begcy, K., Hao, X., Wang, D. and Walia, H. (2014) Rice Fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakata, M., Kuroda, M., Miyashita, T., Yamaguchi, T., Kojima, M., Sakakibara, H., Mitsui, T. and Yamakawa, H. (2012) Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotech. J. 10: 1110–1117. [DOI] [PubMed] [Google Scholar]
- Hakata, M., Wada, H., Masumoto-Kubo, C., Tanaka, R., Sato, H. and Morita, S. (2017) Development of new heat tolerance assay system for rice spikelet sterility. Plant Methods 13: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T., Scofield, G.N. and Terao, T. (2008) An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 174: 534–547. [Google Scholar]
- Hood, E.E., Gelvin, S.B., Melchers, L.S. and Hoekema, A. (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2: 208–218. [Google Scholar]
- Huang, Y., Hsiang, E., Yang, C. and Wang, A. (2016) New insight into the catalytic properties of rice sucrose synthase. Plant Mol. Biol. 90: 127–135. [DOI] [PubMed] [Google Scholar]
- Kobayashi, A., Sonoda, J., Sugimoto, K., Kondo, M., Iwasawa, N., Hayashi, T., Tomita, K., Yano, M. and Shimizu, T. (2013) Detection and verification of QTLs associated with heat-induced quality decline of rice (Oryza sativa L.) using recombinant inbred lines and near-isogenic lines. Breed. Sci. 63: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Fan, C., Xing, Y., Yun, P., Luo, L., Yan, B., Peng, B., Xie, W., Wang, G., Li, X.et al. (2014) Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46: 398–404. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S. and Shibata, D. (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96: 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, N.B., Jagadish, K.S.V., Nalley, L.L., Dixon, B.L. and Siebenmorgen, T. (2013) Neglecting rice milling yield and quality underestimates economic losses from high temperature stress. PLoS ONE 8: e72157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara, K., Wada, T., Sonoda, J., Tsukaguchi, T., Miyazaki, M., Tsubone, M., Yamaguchi, O., Ishibashi, M., Iwasawa, N., Umemoto, T.et al. (2017) Detection and validation of QTLs for milky-white grains caused by high temperature during the ripening period in Japonica rice. Breed. Sci. 67: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, S., Wada, H. and Matsue, Y. (2016) Countermeasures for heat damage in rice grain quality under climate change. Plant Prod. Sci. 19: 1–11. [Google Scholar]
- Murata, K., Iyama, Y., Yamaguchi, T., Ozaki, H., Kidani, Y. and Ebitani, T. (2014) Identification of a novel gene (Apq1) from the indica rice cultivar ‘Habataki’ that improves the quality of grains produced under high temperature stress. Breed. Sci. 64: 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagato, K. and Ebata, M. (1965) Effects of high temperature during ripening period on the development and the quality of rice kernels. Jpn. J. Crop Sci. 34: 59–66. [Google Scholar]
- Peng, S., Huang, J., Sheehy, J.E., Laza, R.C., Visperas, R.M., Zhong, X., Centeno, G.S., Khush, G.S. and Cassman, K.G. (2004) Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 101: 9971–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, V., Kosarev, P., Seledsov, I. and Vorobyev, D. (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7 (Suppl 1): S10.1–S10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata, M., Hirabayashi, H., Takeuchi, Y., Ando, I., Iida, Y. and Ohsawa, R. (2007) Mapping of quantitative trait loci for the occurrence of white-back kernels associated with high temperatures during the ripening period of rice (Oryza sativa L.) Breed. Sci. 57: 47–52. [Google Scholar]
- Tashiro, T. and Ebata, M. (1975) Studies on white-belly rice kernel: IV. Opaque rice endosperm viewed with a scanning electron microscope. Jpn. J. Crop Sci. 44: 205–214. [Google Scholar]
- Terashima, K., Saito, Y., Sakai, N., Watanabe, T., Ogata, T. and Akita, S. (2001) Effects of high air temperature in summer of 1999 on ripening and grain quality of rice. Jpn. J. Crop Sci. 70: 449–458. [Google Scholar]
- Wada, T., Miyahara, K., Sonoda, J., Tsukaguchi, T., Miyazaki, M., Tsubone, M., Ando, T., Ebana, K., Yamamoto, T., Iwasawa, N.et al. (2015) Detection of QTLs for white-back and basal-white grains caused by high temperature during ripening period in japonica rice. Breed. Sci. 65: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu, K., Tanaka, A. and Sasaki, O. (2010) Effects of the high air temperature treatments during the ripening period on the grain quality of brown rice. Rep. Kyushu Br. Crop Sci. Soc. Jpn. 76: 12–14. [Google Scholar]
- Wilhelm, R.P., Mullen, R.E., Keeling, P.L. and Singletary, G.W. (1999) Heat stress during grain filling in maize: effects on kernel growth and metabolism. Crop Sci. 39: 1733–1741. [Google Scholar]
- Yamakawa, H., Hirose, T., Kuroda, M. and Yamaguchi, T. (2007) Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 144: 258–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.