Abstract
Transgenic mice were used to locate the cis-acting DNA elements that are essential for tissue-specific and inducible expression of the rat proline-rich protein gene, R15. Chimeric genes with up to 10 kb of R15 5′-flanking region fused to chloramphenicol acetyl transferase (CAT) or polyomaviral large T-antigen (PyLT) reporter genes were tested. Our results demonstrate that (1) the isoproterenol/tannin-inducible, parotid-specific transgene expression requires an upstream cis-regulatory domain, namely the parotid control region, which extends from −6 to −1.7 kb of the R15 gene; (2) this parotid control region functions with a heterologous promoter and is indispensable for achieving a reproducible chromosomal position-independent transgene expression; (3) deletion of the R15 5′-flanking region up to −1.7 kb results in a pleiotropic effect on the transgene expression, which includes ectopic (nonsalivary) reporter expression and lack of inducibility by either the β-agonist isoproterenol or dietary tannin stimulation; (4) when the −10 to −6 kb region from the R15 gene is deleted in the construct, the inducible expression in the parotid glands of the transgenic mice decreases by over 30-fold, but position-independent and tissue-specific transgene expression is retained. Moreover, the mechanism of induction by either catecholamine isoproterenol or dietary tannin appears to be through a β1-adrenergic receptor-mediated pathway for both normal (non-transgenic) and transgenic animals.
In multicellular organisms, each tissue exhibits a distinct phenotype corresponding to its specialized function. This is a consequence of differential expression of tissue-specific genes. Most current studies on the molecular mechanism of cell type- or cell stage-specific and inducible gene expression aim to analyze the cis-DNA sequences and their trans-regulatory factors involved in promoter, enhancer, and silencer functions by utilizing transiently transfected cultured cells or in vitro transcription systems. However, the regulatory elements may be located a great distance from the proximal promoter (Nitsch and Schutz, 1993; Grosveld et al., 1987), and/or the higher order chromatin structure may be regulated in a tissue-specific manner (for review see van Holde, 1989), thus limiting the application of in vitro assays. This has prompted us to investigate the expression of the salivary-specific and inducible proline-rich protein (PRP) R15 gene in transgenic mice, a system which will allow us to explore the regulatory hierarchy governing gene expression.
The transgenic mice model provides a unique approach to the elucidation of the genomic configuration needed for recapitulating the tissue-specific, developmentally regulated, or inducible gene expression and mechanisms regulating such expression in vivo. First, the transgene integrates at the one-cell embryo stage, presumably before heterochromatin formation. Unlike stable transfection, there is no selectable marker used in the transgenic approach, so the transgene integration need not be biased in favor of the expression of selectable markers. It is thus more likely that the distribution of transgene integration sites is random. Second, unlike transiently transfected expression constructs, the integrated transgenes are subjected to the same processes (replication, chromatin packaging, etc.) as the host genome. All the nuclear regulatory processes acting on the host genome will also affect the integrated transgene expression. Thus, it provides a more rigorous environment to dissect the regulatory regions that are necessary and sufficient to mediate tissue-specific and inducible transgene expression. However, the potential complication of such an approach is that the integration of transgenes commonly leads to dramatic differences in both the tissue specificity of expression and level of expression (Palmiter and Brinster, 1986; Al-Shawi et al., 1990). These differences are now attributed to the sequences flanking the site of integration, commonly called the position effect (Al-Shawi et al., 1990). The inability to predetermine the sites of integration has proven to be an obstacle for many experiments in studying gene expression and regulation (for review see Palmiter and Brinster, 1986). If one can achieve position-independent chromosomal transgene expression, it would facilitate the identification of the cis-elements that are essential for either modulating or directing tissue-specific, developmental stage-specific, or inducible gene expression.
Proline-rich protein gene expression has been studied as a classical tissue-specific and inducible system for more than 20 years (reviewed by Carlson, 1993). The expression of PRPs is confined to the salivary acinar cells. The salivary glands, like the pancreas, contain discrete groups of terminally-differentiated acinar and duct cells that display exocrine function (Hardman and Spooner, 1992). In the rat and mouse, PRP gene expression is dramatically induced by chronic injection of the β-agonist isoproterenol (Ann et al., 1987). After 6 to 10 days of daily isoproterenol injection, the PRPs comprise up to 70% of the total soluble protein in the rat and mouse parotid extracts. Presumably, isoproterenol induces the expression of PRP through a second messenger, cAMP. Our previous transient transfection analyses have demonstrated that there is a cell type-specific cAMP-inducible element in the proximal 5′ flanking region of the PRP promoter (Lin and Ann, 1992; Lin et al., 1993). This cAMP-inducible enhancer also contains an E-box to which protein factors – salivary cAMP-response element binding proteins (SCBPs) belonging to the basic helix-loop-helix (bHLH) transcription factor/DNA-binding protein superfamily (Lin et al., 1993) – bind.
A related finding is that feeding tannins (pro-anthocyanidins) to rats and mice causes an induction of PRPs in the parotid gland, which mirrors that caused by chronic isoproterenol treatment (Carlson, 1993). It has been hypothesized that induction of PRPs by dietary tannins is also mediated by a rise in the intracellular level of cAMP. The functional significance of tannin-induced PRP expression may relate to the affinity of PRPs for the tannin (Mehansho et al. 1987b). As suggested by Mehansho et al. (1987b), the salivary PRPs may bind to and neutralize multihydroxylated phenols such as tannins that are naturally found in sorghum, tea, and other foods. Interestingly, when hamsters are fed with a 2% tannin diet, the PRPs are not induced, and significant inhibition of growth of these hamsters occurs (Mehansho et al., 1987a). This unusual growth inhibition by a diet with 2% tannin in hamsters deficient in PRP induction supports the hypothesis that the expression of PRPs is beneficial to humans and rodents. Humans, in particular, may benefit from the induction of PRPs, since tannin and related phenolic materials are prevalent in a variety of human diets. For example, sorghum grain probably feeds more people than any other crop with the exception of rice (Mehansho et al., 1987b).
In light of these observations, experiments in this report were designed to analyze the genomic organization of the upstream 5′-flanking regions needed for the complex regulatory features that modulate PRP expression in vivo. We have undertaken a study in transgenic mice using hybrid genes containing a reporter gene and a progressively 5′-truncated rat salivary-specific R15 gene 5′-flanking region. Our results reveal a parotid control region located between −6 and −1.7 kb upstream of the R15 promoter that is capable of directing parotid-specific and isoproterenol-inducible expression of a heterologous promoter construct, and its presence is essential for achieving reproducible chromosomal position-independent transgene expression. Moreover, studies with fusion genes containing this region have confirmed that the parotid control region along with its promoter is capable of recapitulating isoproterenol/tannin-dependent transgene expression in the parotid acinar cells.
Materials and methods
Construction of R15/reporter fusion genes
Three R15/CAT fusion genes containing various lengths of the 5′-flanking region of rat parotid-specific PRP gene, R15, were constructed in this study. The 5′-flanking regions of R15 were derived from either cosmid R15 or plasmid R15BH (Lin and Ann, 1991). Plasmid R15BH was constructed from the 6 kb BamH I/Hind III fragment of cosmid R15, which includes 1.7 kb of 5′-flanking region, three exons, and the polyadenylation site. An R15 oligo, which corresponds to the complementary sequence of the −21 to +2 region of R15 (Lin and Ann, 1991) was synthesized. This R15 oligo primer together with the M13 reverse primer was used in a polymerase chain reaction to amplify the −1.7 kb to +2 DNA fragment from R15BH. Subsequently, the reaction product was cloned into pUC19, and the resulting construct was digested with Sal I and blunt-ended by Klenow. After BamH I digestion, the released insert was ligated into the BamH I/Bgl II (blunted by Klenow) sites of pBLCAT3; the Bgl II site was regenerated in the process. This construct was designated −1.7R15/CAT. The second construct, −10R15/CAT, containing approximately 10 kb of the 5′-upstream region of R15, was engineered by ligating an 8.3 kb Hind III/BamH I fragment of cosmid R15 (Lin and Ann, 1991) into the Hind III and BamH I sites of −1.7R15/CAT. The third construct, −6R15/CAT, was engineered by ligating a 4.3 kb Pst I/BamH I DNA fragment of approximately −6 to −1.7 kb from cosmid R15 (Lin and Ann, 1991) into the Pst I and BamH I sites of −1.7R15/CAT. Another construct, R15PCR/tk/CAT, driven by a heterologous promoter, was created by inserting a Pst I/BamH I fragment of R15 parotid control region (PCR) upstream of a thymidine kinase promoter (tk, from nt −105 to +51) that drives the CAT reporter gene.
To construct the R15/PyLT fusion genes, we used plasmid pPXMT-PyLT (kindly provided by Dr. Lorraine Chalifour, McGill University) containing a Cla I/Xho I to Bgl II fragment of the mouse metallothionein I (MT-1) promoter and a Bgl II to BamH I fragment of the cDNA for polyoma viral large T-antigen (PyLT; Chalifour et al., 1990). First, plasmid pBLCAT3 was cleaved with Ava I, filled in with Klenow, and then cut to completion with BstY I. A 0.7 kb fragment containing the SV40 small t splicing and poly(A) signal was purified and ligated into the BamH I/Sal I (filled in with Klenow) sites of pPXMT-PyLT, regenerating the Sal I site. This plasmid was designated pPyLT17. Subsequently, the plasmids −1.7R15/CAT and −6R15/CAT were cut to completion with Sal I and Bgl II, respectively, and the fragments containing the R15 regulatory regions −1.7 kb to +2 and −6 kb to nt +2 were purified and ligated into the Xho I/Bgl II sites of pPyLT17 separately to replace its original MT-1 promoter. These two constructs were designated −1.7R15/PyLT and −6R15/PyLT, respectively.
Transgenic mice
The R15/CAT DNA fragments for microinjection were prepared by digestion with Kpn I to release the 3′ ends and an appropriate enzyme to release the 5′ ends. The R15/PyLT DNA fragments were separated from vector sequences after digestion with Cla I and Sal I. After agarose gel electrophoresis, the desired DNA fragments were purified through a Gene-Clean Kit (Bio 101) according to the manufacturer’s instructions. The DNA concentration was measured and diluted to approximately 2 ng/μl for microinjection. FVB/N mouse pronuclei were each injected with about 1–2 pl DNA solution and were subsequently transferred to pseudo-pregnant females, as described by Hogan et al. (1986). Founder mice were identified by polymerase chain reaction analyses of tail DNA using primers designed according to the sequences of CAT or PyLT. Positive founders were bred to establish the transgenic lines, and two to six transgenic lines were established for each construct. The F1 and F2 offspring were used for this study. Transgene copy numbers were determined in Southern analyses with 32P-labeled CAT or PyLT probes by comparing the intensities of labeled bands from tail DNA with those of known amounts of CAT or PyLT internal standards (Hogan et al., 1986). Southern analyses indicated that the transgenes were inserted in a head-to-tail array at a single integration site (data not shown).
Animal treatment and tissue collection
Rats, transgenic mice, and their non-transgenic litter mates were normally fed with Purina Lab Chow ad libitum. The regulation of the R15 transgenes was studied under three metabolic conditions: control (fed with Purina Lab Chow only), Ipr-injected (repeatedly injected with isoproterenol for defined periods), and tannin-fed (fed with a 6% tannin-containing diet for 6 days). Isoproterenol was administered as described previously (Ann et al., 1987); each mouse or rat received a daily intraperitoneal injection of 2 or 5 mg, respectively, of dl-isoproterenol HC1 in 0.2 ml of 140 mM NaCl for 10 days, unless otherwise indicated.
For tannin feeding, mice were kept on Purina Lab Chow before starting the feeding experiments. Sorghum grain with high tannin content, IS-8260, was ground and incorporated into the diet to a final tannin content of 6%, as described previously (Mehansho et al., 1987a). Food and water were provided ad libitum. Total RNA and soluble extracts were prepared from salivary and nonsalivary tissues of at least three transgenic animals in each of the three treatment groups (i.e., at least nine animals from each R15/reporter pedigree) and assayed for reporter expression.
In the experiments on the role of the β-adrenergic receptor in the regulation of R15 gene expression, a β-adrenergic receptor non-selective blocker [(±)-propranolol and its optical isomers (+)- or (−)-propranolol], a β1-selective blocker (metoprolol), and a β2-selective blocker (butoxamine) were used. Each selected blocker was individually mixed with the tannin-containing sorghum at the ratio of 1 mg per gram of sorghum mixture. The mice were fed with this mixture for 6 days, and the CAT activity in the parotid extracts was compared to that from the above-mentioned tannin-fed group.
For the treatment combining isoproterenol and a β-blocker, mice were injected daily for six days with 0.1 ml of 140 mM NaCl solution containing 1 mg each of dl-isoproterenol and one of the selected β-blockers mentioned above.
For all the experiments, animals were anesthetized with CO2 and sacrificed by exsanguination. The dissected organs were snap-frozen in liquid nitrogen and stored at −80°C until use.
CAT assay
To determine CAT activity, tissues were thawed and homogenized in 0.1 to 0.2 ml of 250 mM Tris (pH 7.5) and 0.5 mM PMSF. After three cycles of freezing and thawing, the homogenates were spun in a microcentrifuge for 15 minutes, and protein concentrations of the supernatants were determined with a Bio-Rad protein assay kit. Subsequently, the samples were heated at 65°C for 10 minutes to inactivate endogenous deacetylases, and 1 to 100 μg of the protein extracts were assayed for CAT activities, as described previously (Lin et al., 1991). The inclusion of 0.5 mM of the protease inhibitor PMSF in the tissue preparation and reaction (Pothiec et al., 1992) is critical for a quantitative CAT assay using extracts prepared from the salivary glands of transgenic mice. The CAT enzyme reaction products were separated by thin-layer chromatography. Acetylated forms of [14C]chloramphenicol were excised individually and counted in a scintillation counter. For quantification, these assays were demonstrated to be within linear range by varying the incubation time and amount of extracts used.
Reverse transcriptase-coupled polymerase chain reaction and Northern blot analyses
Total RNA was extracted from frozen mouse tissues as described by Chomczynski and Sac-chi (1987). Approximately 1 μg of total RNA was incubated with RNase-free DNase (U. S. Biochemical) according to the manufacturer’s instructions. Subsequently, the DNA-free RNA was reverse-transcribed in a total volume of 10 μl with random hexamers and M-MLV reverse transcriptase (BRL). Following the cDNA synthesis, 4 pmol each of a pair of specific oligomers corresponding to mouse glyceraldehyde-3-phosphate dehydrogenase (GAD) or PyLT were added to 4 μl of the cDNA reaction mixture, respectively, for the polymerase chain reaction. Each 20 μl reaction mixture included 10 mM Tris (pH 8.3), 50 mM KC1, 2.5 mM MgCl2, 0.01% gelatin, 200 μM dNTPs, and 0.5 unit of Taq DNA polymerase (Promega). Sixteen cycles of amplification reactions were performed using the following profile: 92°C, 30 seconds; 55°C, 30 seconds; and 72°C, 30 seconds. Ten μl from each reaction were analyzed for the presence of either transgene or GAD cDNA by Southern analyses with 32P-labeled GAD or PyLT probes. These conditions were carefully selected so that the message levels were in proportion to the levels of the amplified product. Also, negative controls were included in each assay. For Northern analyses, approximately 10 μg total RNA for each sample were electrophoresed, blotted, and hybridized with 32P-labeled specific probes as previously described (Ann et al., 1987).
In situ hybridization
The in situ hybridization was carried out as previously described (Lazowski et al., 1992). Briefly, a 400 bp CAT fragment was subcloned into the riboprobe vector pGEM-4 (Promega) in an orientation permitting transcription of CAT antisense RNA from the SP6 promoter. The transcripts were labeled with [35S]UTP. Frozen sections of submandibular glands from control and trans-genic mice were thaw-mounted onto slides coated with 3-aminopropyltriethoxysilane (TESPA). The slides were stored at −70°C until processed. Tissue sections were fixed in 4% formaldehyde in 1× PBS for 5 minutes, washed, and acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine-HCl, pH 8.0, for 10 minutes at room temperature. After washing in 2× SSC, sections were dehydrated in a graded series of ethanol, delipidated in 100% chloroform, and air-dried. Subsequently, sections were hybridized for 18 hours at 52°C in a solution containing 50% formamide, 0.6 M NaCl, 10 mM Tris-HCl, 1× Denhardt’s solution, 1 mM EDTA, 0.01% salmon sperm DNA, 0.05% total yeast RNA, 10% dextran sulfate, 100 mM DTT, 0.1% SDS, 0.1% sodium thiosulfate, and 2 × 104 cpm/μl of 35S-labeled probe. Following hybridization, slides were washed for 1 hour in several changes of 2× SSC/10 mM DTT to remove the unhybridized probe, treated for 30 minutes at room temperature with 20 μg/ml RNase, and washed another 30 minutes with RNase buffer, twice in 2× SSC at room temperature, 1 hour in 2× SSC at 50°C, and finally in 0.2× SSC for 1 hour each at 55°C and 60°C. After dehydration in an ascending series of ethanol (50%, 70%, 90%, and 95%) containing 0.3 M ammonium acetate and in 100% ethanol, slides were air-dried and exposed to Hyperfilm-βmax (Amersham) to estimate the approximate time of exposure for the emulsion autoradiography. Emulsion autoradiography was performed at 4°C using Kodak NTB2 emulsion. Tissue sections were exposed for the times estimated as described above, developed with Kodak D-19 developer, fixed, and counterstained with hematoxylin-eosin.
Results
Inducible and tissue-specific expression of rat PRP genes, R15 and R4
The dramatic changes in rodent salivary gland gene expression induced by the catecholamine isoproterenol (Ipr) and tannins in the diet suggest that this is a useful model system to study overall regulation of tissue-specific gene expression. As demonstrated in Figure 1A, R15 mRNA from nontransgenic rats is too low to be detected in salivary glands of control (untreated) animals and nonsalivary tissues such as liver of both control and Ipr-injected animals. However, upon isoproterenol injection of the nontransgenic rats, the R15 messages are tremendously increased in the salivary glands. This induction of R15 expression is more pronounced in the parotid glands than in the submandibular glands (Fig. 1A, lanes 4 and 6), in contrast to the PRP R4 messages (Lin and Ann, 1991) that are dramatically induced in both the parotid and submandibular glands (Fig. 1A, lanes 8 and 10). In order to dissect regulatory element(s) directing the tissue-specific and inducible PRP R15 gene expression in vivo, we generated various constructs consisting of 5′-flanking regions of R15 and one of the reporters, CAT (chloramphenicol acetyltransferase) or PyLT (polyomaviral large T-antigen), as shown in Figure 1B.
Figure 1.

Northern analysis of rat parotid and submandibular glands and liver PRP expression and schematic diagram of R15/reporter transgene constructs. A. RNA (10 μg) from parotid (Pa), submandibular (Su) glands, and liver (Li) of control (C) and isoproterenol-treated (I) rats were electrophoresed on a 1.5% denaturing agarose gel, blotted onto nitrocellulose filters, and probed with two PRP gene-specific probes (R15 and R4). The blot was first probed with 32P-labeled R15-specific probes. After removing the hybridized R15 probes, the same filter was reprobed with R4-specific probes. The indicated molecular weight (left) was estimated from the 0.16–1.77 kb RNA ladder (BRL) analyzed in the same gel. Autoradiography was performed at −80°C for 4 hours with two intensifying screens, for both R15 and R4 probes. B. Top heavy line is a schematic diagram of the rat proline-rich protein gene, R15. Numbers indicate the distance from the transcription initiation site (↱) in kilobases, except that 2* represent nt +2. Genomic fragments (dotted box) derived from cosmid R15 were inserted into the promoterless expression vector (open box) containing either the CAT or PyLT coding region with splicing/poly(A) signals (t.A.) to create constructs used in the transgenic animal studies. The R15/reporter transgenes with progressively truncated 5′-flanking DNA are terminated with a fixed 3′ endpoint at nt +2 (2*).
Distal sequences are required for salivary-specific and isoproterenol/tannin-dependent expression of R15/reporter genes in transgenic animals
To test whether the proximal regulatory region of the R15 gene is sufficient to target PRP expression in the salivary glands, constructs containing the rat R15 proximal regulatory region (−1.7 kb to nt +2) and either the CAT (−1.7R15/CAT) or PyLT (−1.7R15/PyLT) were used to generate transgenic animals. Total RNA and tissue extracts prepared from either Ipr-injected or tannin-fed transgenic mice were analyzed and compared to that prepared from control transgenic mice. As shown in Figure 2, among the 11 tissues surveyed from −1.7R15/CAT transgenic mice, significant levels of CAT activity were detected in brain, thymus, trachea, and submandibular glands. Except for the submandibular glands, PRP is not normally found in brain, thymus, and trachea. Despite the report that PRPs or related proteins have been identified immunologically in the human trachea (Warner and Azen, 1984), there is no evidence that the endogenous PRP genes are expressed or respond to the β-agonist isoproterenol in the rodent trachea (Carlson, 1993). Also of note is a decrease in CAT activity upon either chronic isoproterenol injection (Fig. 2A) or tannin feeding (not shown) in the submandibular glands. However, no significant change in CAT activities was detected in the brain, thymus, and trachea of these treated −1.7R15/CAT transgenic animals. The decrease of CAT activity in the submandibular glands may be explained as an indirect side effect of the isoproterenol or tannin treatment. For example, the great increase in the submandibular endogenous PRP content upon treatment with the β-agonist isoproterenol may, in effect, dilute the measured CAT-specific activity. Further examination of tissues from four independent −1.7R15/PyLT transgenic lines by RT-polymerase chain reaction analyses revealed ectopic (nonsalivary) transgene expression and lack of isoproterenol-inducibility in either parotid or submandibular glands (Fig. 2B). No apparent change of −1.7R15/PyLT transgene messages was observed in these tested tissues, which also supports our explanation of the above-mentioned decrease of CAT activity in the submandibular glands. Clearly, with two different transgene reporters, CAT and PyLT, the 1.7 kb of R15 5′-flanking information is insufficient to confer either isoproterenol or tannin inducibility in the tissues studied.
Figure 2.
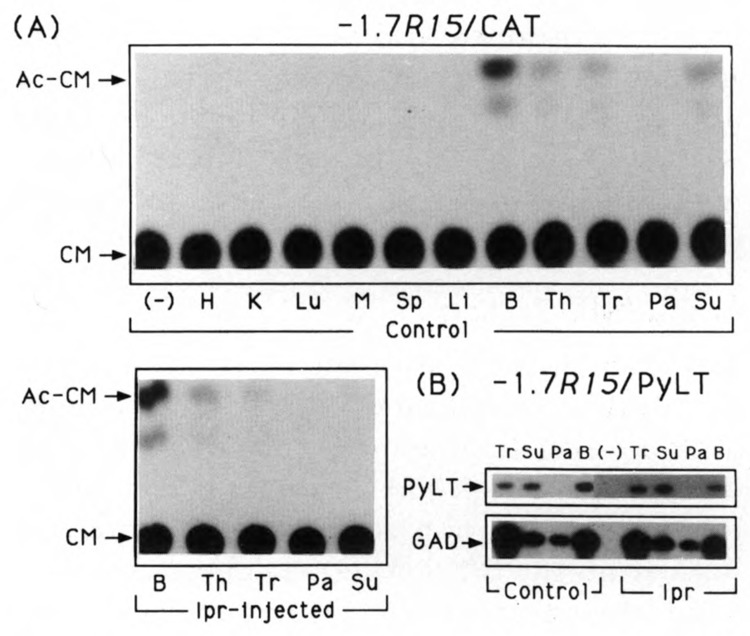
Representative tissue profiles of −1.7/R15/CAT and −1.7R15/PyLT transgene expression in transgenic mice. A. Protein extracts were prepared from the indicated tissues of −1.7R15/CAT transgenic mice fed Purina Lab Chow (control) or injected with isoproterenol for 10 days (Ipr-injected; see Materials and Methods for details). The CAT assays were performed using 100 μg of soluble protein for each sample, and the reactions were at 37°C for 14 hours. H, heart; K, kidney; Lu, lung; M, muscle; Sp, spleen; Th, thymus; Tr, trachea; Li, liver; B, brain; Pa, parotid; Su, submandibular; (−), protein extract from parotid glands of nontransgenic mice as a negative control; CM, chloramphenicol; Ac-CM, acetylated chloramphenicol. The percent conversions were: B, 11%; Th, 3%; Tr, 2%; Su, 4%; Ipr-B, 10%; IprTh, 4%; Ipr-Tr, 1%; <1% for Ipr-Su and the remaining tissue extracts. B. One microgram total RNA from the indicated tissues of control and Ipr-treated −1.7R15/PyLT transgenic mice was reverse transcribed using random hexamers as primers. Subsequently, equal amounts of cDNA were subjected to amplification with PyLT or glyceraldehyde-3-phosphate dehydrogenase (GAD)-specific primer pairs in polymerase chain reactions, electrophoresed through a 1.5% agarose gel, blotted to Nytran membrane, and hybridized with 32P-labeled probes as indicated. The minus RNA as a negative control is denoted as (−). The autoradiography was performed with double intensifying screens at −80°C for 16 hours.
We next examined four independent transgenic lines generated from a −10R15/CAT construct that carries 10 kb of contiguous rat R15 upstream sequences. Shown in Figure 3 is a representative tissue CAT activity profile from one of these lines. In the control (untreated) transgenic mice generated with the −10R15/CAT construct, no significant CAT activity could be detected in any tissue. This is similar to the expression pattern of the endogenous R15 gene, for which basal expression could not be detected (Fig. 1A). Furthermore, like the endogenous R15 gene activity, the CAT transgene activity was dramatically induced in the parotid glands from both the Ipr-injected and tannin-fed transgenic mice (Fig. 3). The induction of transgene expression in the submandibular glands by isoproterenol was much less than that in the parotid glands, which also resembles the pattern of endogenous R15 rather than R4 gene induction (Fig. 1A, lanes 4 and 6, 8 and 10). However, a high-tannin diet induced transgene expression only in the parotid, and not the submandibular glands. A moderate CAT induction by isoproterenol treatment in the trachea, as shown in Figure 3, was not always reproducible, even in the same line of transgenic mice. The overall expression pattern among the four independent lines of −10R15/CAT was quantitatively similar to the one shown. Together, these results indicate that regulatory elements upstream of the R15 proximal regulatory region are critically important in facilitating a high level of isoproterenol/tannin-dependent transgene expression in the parotid glands and in preventing the ectopic transgene expression observed in the transgenic animals harboring the −1.7R15/reporter constructs.
Figure 3.

Tissue-specific induction of −10R15/CAT transgene expression by catecholamine isoproterenol and dietary tannin. F2 progeny of transgenic line −10R15/CAT-01 were fed Purina Lab Chow (Control), fed a diet containing IS-8260 high-tannin content sorghum for 6 days (Tannin-fed), or injected with isoproterenol for 10 days (Ipr-injected; see Materials and Methods for details). Soluble protein (50 μg) was assayed for CAT activity at 37°C for 2 hours. The CAT assays using protein extracts from the parotid (Pa) glands of Ipr-injected and tannin-fed −10R15/CAT-01 transgenic mice were over-reacted in order to demonstrate a weak induction in the submandibular (Su) glands by isoproterenol, and not by tannin. See Figure 2 for details.
1.7 kb of 5′-flanking region is able to direct acinar cell type-specific expression
To determine the cell-type specificity of R15 transgene expression within the salivary glands, cross sections of salivary tissue from selected transgenic mice were analyzed by in situ hybridization with anti-CAT cRNA. Because the endogenous PRP expression occurs at high levels in the parotid glands of Ipr-injected rats and mice (Ann et al., 1987), and the highest levels of CAT enzyme activity were also observed in the induced parotid glands of −10R15/CAT transgenic mice (Fig. 3), we sought to identify the cell types expressing the transgenes within the induced parotid glands. As shown in Figure 4E-H, the signals for the −10R15/CAT transgene induced by either the catecholamine isoproterenol or dietary tannin were primarily seen in the acinar cells of parotid glands (open arrows), which is consistent with the previous report on the localization of the induced endogenous PRP messages (Lazowski et al., 1992).
Figure 4.
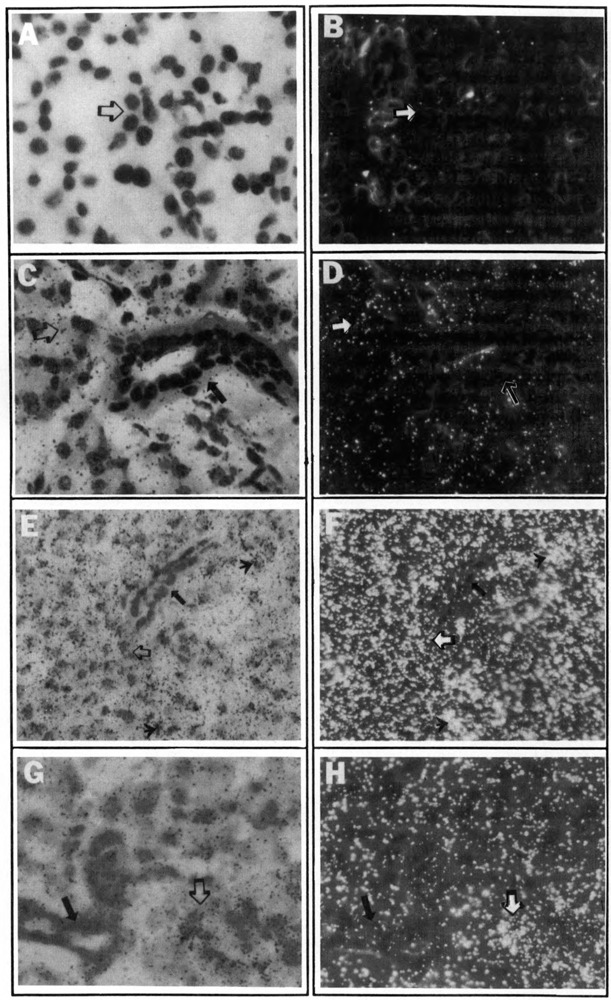
Localization of CAT messages in the salivary acinar cells of transgenic mice using in situ hybridization. Tissue sections of submandibular glands from non-transgenic litter-mate (A and B) and −1.7R15/CAT transgenic (C and D) mice, as well as parotid glands of Ipr-injected (E and F) and tannin-fed −10R15/CAT transgenic mice (G and H) were prepared as described in Materials and Methods. In situ hybridization was performed using 35S-labeled CAT cRNA probes. Both bright field (A, C, E, G) and dark field (B, D, F, H) are shown. All the sections were displayed at 400× magnification. Open arrows indicate the acinar cells, and dark arrows indicate the duct cells. The arrowheads shown in E and F indicate the patchy appearance of transgene expression.
Analysis of submandibular cross sections from a transgenic mouse harboring the −1.7 R15/CAT construct revealed that the transgene was detected unambiguously in most submandibular cells (Fig. 4C and D), whereas there were practically no detectable signals in those from non-transgenic litter mates (Fig. 4A and B). Furthermore, these CAT messages, like that from parotid glands of the −10R15/CAT transgenic mice, were localized in the acinar cells (open arrows, Fig. 4C and D), but not in the duct cells (dark arrows, Fig. 4C and D) of the submandibular glands. These results suggest that there are cis-elements located in the proximal 1.7 kb of the 5′-flanking region of R15 that are able to direct the basal transgene expression unambiguously to most of the salivary acinar cells (Fig. 4C and D). However, this 1.7 kb region alone does not contain the necessary elements to confer β-adrenergic or tannin inducibility on the transgene expression in either the parotid or submandibular glands of transgenic mice (Fig. 2).
The parotid control region is located between −6 and −1.7 kb of R15
To localize the sequences within the 5′-distal region that are required for parotid-specific and isoproterenol/tannin-dependent R15/reporter transgene expression, we generated six and five lines of transgenic mice harboring 6 kb of the R15 5′-flanking region with either the CAT or PyLT reporter gene, respectively. No detectable CAT activity was seen in 11 tissues from the control −6R15/CAT transgenic mice, whereas significant CAT induction was observed solely in the parotid glands of both the Ipr-injected and tannin-fed transgenic mice (Fig. 5A). The induction level was, however, significantly less than that seen in the −10R15/CAT transgenic mice, taking into account the amounts of soluble protein and incubation times used in the CAT assays (see Figures 3 and 5A). To partially account for the effect of the transgene integration sites, expression of the −6R15/PyLT transgenes was analyzed by Northern blots. As shown in Figure 5B, a single band, which corresponds to the predicted size of PyLT transcript, was detected in the parotid glands of Ipr-injected −6R15/PyLT transgenic mice. No similar band or band of different size was detected in the RNA isolated from tissue of control transgenic mice or from the brain, liver, and submandibular glands of treated −6R15/PyLT mice (Fig. 5B). These results suggest that the distal −10 to −6 kb region of R15 is not required for the observed tissue-specific transgene expression; however, it may enhance the fold of induction by isoproterenol and tannin.
Figure 5.

Induction of −6R15/CAT and −6R15/PyLT transgene expression by β-receptor agonist, isoproterenol, and high-tannin sorghum. Results from representative independent lines containing either −6R15/CAT or −6R15/PyLT transgene are shown. A. CAT assays were performed using 100 μg of soluble protein at 37°C for 14 hours. The percent conversions were Ipr-Pa, 23%; Tannin-Pa, 62%; <1% for the remaining tissue extracts. B. Northern blot was first probed with PyLT probe for transgene expression (upper panel), and then the same blot was stripped and rehybridized with mouse β-actin cDNA probes (lower panel). The autoradiography was performed with double intensifying screens at −80°C for 4 days (PyLT) and 2 days (β-actin), respectively. The positions of 18S ribosomal RNA, as visualized by EtBr staining, are shown at the right. See Figures 2 and 3 for further details.
The overall pattern of expression among the 21 transgenic lines used in this study is summarized in Table 1. There is no obvious difference in the penetrance of the pedigrees derived from the same series of R15/CAT and R15/PyLT constructs. This suggests that the observed patterns in terms of tissue specificity and inducibility among different transgenic lines with various lengths of R15 5′-flanking regions are not caused by the reporter genes we used, CAT and PyLT, and that the sequences of the −10 to −6 kb region are dispensable for the reproducible parotid-specific transgene expression. Also, since no two transgenic lines are likely to have the same integration sites, the observed phenotypes seem to be independent of the site of integration. Therefore, we have designated the −6 to −1.7 kb R15 region the parotid control region (PCR), due to its ability to direct isoproterenol/tannin-dependent transgene expression exclusively in the parotid glands of all 15 lines harboring constructs containing this region. The levels of isoproterenol- or tannin-induced CAT-specific activity from the −10 and −6R15/CAT constructs, after faking copy-number into consideration, can vary by as much as 10-fold among independent lines derived from the same construct (not shown). In some experiments using the founder transgenic mice (Wilkie et al., 1986), copy number-independent transgene expression may be explained by the supposition that the transgenes in transgenic mice are mosaic; this is not the case in our experiments, since the mice analyzed were either the F1 or F2 offsprings. The “patchier” appearance of transgene expression than the endogenous PRP expression pattern (Lazowski et al., 1992) detected by in situ hybridization, as shown in Figure 4E and F, suggests that this variation in CAT expression may be due to the fact that not all acinar cells responded to isoproterenol stimulation or expressed the transgene equally. A similar observation has been made in other transgenic systems (Simonet et al., 1993).
Table 1.
Summary of R15/reporter transgene studies.
| Construct | Number of transgenic lines | Copy number of transgene | Parotid specificity | Non-salivary ectopic expression | Parotid IPR inducibility | Parotid tannin inducibility |
|---|---|---|---|---|---|---|
| −10R15/CAT | 4 | 1–15 | 4/4 | 0/4 | 4/4 | 4/4 |
| −6R15/CAT | 6 | 1–26 | 6/6 | 0/6 | 6/6 | 6/6 |
| −6R15/PyLT | 5 | 1–19 | 5/5 , | 0/5 | 5/5 | 5/5 |
| −1.7R15/CAT | 2 | 21–45 | 0/2 | 2/2 b | 0/2 | 0/2 |
| −1.7R15/PyLT | 4 | N.D. a | 0/4 | 4/4 b | 0/4 | N.D. a |
Not determined.
Ectopic expression was observed in the brain, trachea, and thymus.
To verify that the parotid control region directs the observed parotid-specific and isoproterenol-dependent transgene expression, we analyzed the expression of transgenes containing CAT reporter gene driven by the heterologous herpes simplex virus thymidine kinase (HSV-tk) promoter. When a 2.9 kb Pst I–BamH I fragment from the PCR, which contains two salivary-specific DNase I-hypersensitive sites (D. K. Ann et al., unpublished data), was ligated upstream of the heterologous tk promoter, a high level of expression of the resulting R15PCR/tk/CAT fusion transgene in the parotid glands of transgenic mice receiving isoproterenol injection was observed (Fig. 6). This isoproterenol-dependent expression is not observed in other tissues, such as liver, or in the parotid glands of transgenic mice bearing tk/CAT transgenes. Thus, the parotid control region is able to direct parotid isoproterenol-dependent expression via a heterologous promoter, confirming that this region contains most of the necessary sequence information to confer isoproterenol-inducible gene expression to the parotid gland.
Figure 6.

The parotid control region of the R15 gene is able to direct isoproterenol-inducible and parotid-specific expression from a heterologous promoter. A 2.9 kb Pst I–BamH I fragment (PCR), which is within the −6 to −1.7 kb of the R15 parotid control region, was ligated upstream of a thymidine kinase (tk) promoter that drives a CAT reporter gene. The control tk/CAT fragment and the fusion construct, R15PCR/tk/CAT, were used to generate transgenic mice separately. The parameters used for CAT assays were the same as those described in Figure 5A. The percent conversions were: Ipr-Pa of the R15PCR/tk/CAT transgenic mice, 7% ;<1 % for the other tissue extracts. Representative results from liver (Li), parotid (Pa), and submandibular (Su) glands are shown for each construct. CM, chloramphenicol; Ac-CM, acetylated chloramphenicol.
β1-adrenergic receptor is likely involved in the regulation of R15/CAT transgene expression in the parotid glands
To demonstrate that the parotid control region can recapitulate the regulation of endogenous R15 expression, we examined the time course of CAT induction by either isoproterenol (Fig. 7A) or dietary tannin (Fig. 7B) in two independent lines derived from either the −10R15/CAT(solid curves) or the −6R15/CAT (dashed curves) construct, respectively. As shown in Figure 7A and B, the effect of these treatments on the CAT activities in the parotid glands of two independent −10R15/CAT lines was similar to that in two individual −6R15/CAT lines; they shared a comparable, but not identical, time-response curve and displayed their maximum activities after approximately 3 to 4 days of treatment. Generally, the pattern of induction in the parotid glands of these transgenic mice (Fig. 7A and B) is consistent with the kinetics of endogenous R15 induction by isoproterenol in the rat parotid glands (Fig. 7C). However, with the −10R15/CAT transgenic mice, induced CAT activity was more than 30-fold greater than that induced in the −6R15/CAT transgenic mice by both catecholamine and dietary tannin stimulation (Fig. 7A and B, curves 3/4 versus 1/2). Taken together, these results confirm that most of the isoproterenol/tannin-responsive elements are located within the parotid control region (−6 to −1.7 kb of R15) and suggest that one of the putative function of the DNA sequences between −10 and −6 kb of R15 is to modulate the magnitude of induction mediated by the parotid control region.
Figure 7.

Kinetics of CAT induction by isoproterenol treatment or tannin feeding in transgenic mice, compared to the induction of endogenous rat R15 expression by isoproterenol. CAT activity was measured from a pool of parotid glands (from 3 mice) of two independent lines of −6R15/CAT (−6R15/CAT-01 and −03; curves 1 and 2) and −10R15/CAT (−10R15/CAT-01 and −04; curves 3 and 4) constructs, respectively, with defined periods (days as indicated) of isoproterenol treatment (A) and tannin-feeding (B). The results are expressed as the relative CAT activity. One relative CAT activity is defined as 1% chloramphenicol conversion per copy of transgene when 50 μg of soluble parotid protein extracted from −6R15/CAT transgenic mice were incubated at 37°C for 17 hours. The parotid extracts from −10R15/CAT mice were serially diluted in order to obtain a comparable CAT activity that is in a linear range with that from the −6R15/CAT mice. The calculated relative CAT activities are normalized for the fold-dilution. C. RNA (10 μg) isolated from the parotid glands of rats treated with isoproterenol (days indicated) was separated on a denaturing agarose gel, blotted onto nitrocellulose paper, and probed with 32P-labeled R15-specific probe (upper panel). The autoradiography was performed with double intensifying screens at −80°C for 4 hours. Three micrograms of the same RNA preparation were also analyzed by EtBr staining to demonstrate the quality of the RNA preparation and equal loading (lower panel).
Further investigations were conducted to determine whether a β-adrenergic receptor pathway is involved in regulating R15/CAT transgene expression by a tannin diet in the transgenic mice derived from either −10R15/CAT or the −6R15/CAT construct. As shown in Figure 8, (±) propranolol (a nonspecific β-antagonist), when included in the diet, almost completely blocked the induction of CAT activity by dietary tannin (compare lanes 1 and 2), as it did in the isoproterenol-induced CAT expression (compare lanes 7 and 8) in the parotid glands of −10R15/CAT transgenic mice. Treatment with either metoprolol (β1-selective antagonist, lanes 6 and 10) or (−) propranolol (active optical isomer of propranolol, lane 3) resulted in inhibition of the induced CAT expression comparable to that from treatment with (±) propranolol. In contrast to this marked effect by the β1-adrenergic antagonist on CAT induction, treatment with butoxamine (β2-selective antagonist) or (+) propranolol (inactive optical isomer of propranolol) either enhanced (lane 5) or had little effect (lanes 4 and 9) on the tannin- or isoproterenol-induced CAT activities. Although this enhancement of tannin-induced CAT activity by butoxamine is reproducible, the exact mechanism remains obscure. Similar observations were made in a representative line harboring −6R15/CAT transgenes (not shown). Taken together, comparable inhibition of the isoproterenol- and tannin-induced −10 and −6R15/CAT activities occurred when the β1-selective antagonist was used; therefore, the β-adrenergic receptor involved in the induction pathways by both isoproterenol and tannin is likely to be of the β1 subtype. These findings not only are consistent with the previous hypothesis (Mehansho et al., 1987b) that isoproterenol and dietary tannin regulate PRP expression via a rise in the intracellular level of cAMP, but also suggest that the induction of PRP R15 expression by catecholamine isoproterenol and dietary tannin in the parotid glands may share some common mechanisms, and that R15 expression is regulated, in part, by the parotid control region we have identified: −6 to −1.7 kb of R15.
Figure 8.

β1-adrenergic pathway is involved in modulating R15 expression by either isoproterenol or a high level of dietary tannin. CAT activities from the parotid extracts of −10R15/CAT-01 transgenic mice fed Purina Lab Chow only (lane 11), fed a diet containing IS-8260 high-tannin sorghum for 6 days (lane 1), or injected with isoproterenol for 10 days (lane 7) were assayed. These CAT activities were compared to that from the parotid extracts of the same line of transgenic mice receiving the combined treatment of tannin feeding or isoproterenol injection together with metoprolol (β1-antagonist; lanes 6 and 10), butoxamine (β2-antagonist; lanes 5 and 9), (±) propranolol (nonspecific β-antagonist) or its optical isomers (+) or (−) propranolol (lanes 2–4 and 8), as indicated. See Materials and Methods for further details. The CAT assays were performed at 37°C for 2 hours. The percent conversions were: lane 1, 37%; lane 4, 42%; lane 5, 98%; lane 7, 13%; lane 9, 20%; <1% for lanes 2, 3, 6, 8, 10, and 11. CM, chloramphenicol; Ac-CM, acetylated chloramphenicol.
Discussion
We have studied the expression of various R15/CAT and R15/PyLT transgenes with respect to their tissue specificity and regulation by the (β-agonist isoproterenol and dietary tannin in vivo. Our data suggest that the 6 kb R15 5′-flanking region contains most of the control elements necessary for the position-independent, tissue-specific, inducible transgene expression, but lacks the element(s) required for its high level of expression. Since no transgenic mice were produced with a construct containing the 10 kb R15 5′-flanking region with an internal deletion of the −6 to −1.7 kb sequences linked to the reporter gene, it is difficult to determine whether the observed high expression level in the −10R15/CAT transgenic mice resulted from interaction(s) between elements in distal −10 to −6 kb region and the −6 to −1.7 kb fragment or the basic 1.7 kb promoter region. However, using transient transfection assays, the distal −10 to −6 kb region was shown to function as an enhancer for the heterologous promoter in various non-salivary cells, and several AP1 or API-like sequences are located in this region (D. K. Ann et al., unpublished observation). This suggests that the respective tissue-specific and inducible regulatory elements are located within the R15 −6 to −1.7 kb region, and their activity can be enhanced by sequences outside this region. Parotid-specific and isoproterenol-inducible transgene expression was found in all mouse lines in which the −6 to −1.7 kb region was included in the transgene construct but was not observed in any line without it. Since the parotid-specific and inducible regulation of homologous and heterologous promoters was restored by the presence of the −6 to −1.7 kb R15 region, it has been designated the parotid control region.
Studies of several other mammalian genes in transgenic mice have revealed that regulatory elements in the proximal promoter are sufficient to confer tissue-specific expression, although maximal expression requires a distal upstream or downstream enhancer (Pinkert et al., 1987; Neznanov et al., 1993; Rindt et al., 1993). For instance, 300 bp of the proximal 5′-flanking sequence of the albumin gene are sufficient to direct relatively low levels of transgene expression to the liver; however, high levels of expression require the presence of an enhancer −8.5 to −10.4 kb upstream from the albumin promoter (Pinkert et al., 1987). For the human keratin 18 gene, a major enhancer element located at approximately 4 kb downstream from the transcription start site is required for a high level of expression in the liver and intestine (Neznanov et al., 1993). Further studies have shown that reporter genes driven by the pancreatic-specific elastase (Kruse et al., 1993) and cardiac-specific myosin light chain (Lee et al., 1992) promoters are properly expressed when only 205 or 250 nucleotides of the proximal 5′-flanking region, respectively, are included in the constructs used to generate transgenic mice. In contrast, the proximal 1.7 kb of the 5′-flanking region of R15 appear to be incapable of directing reporter expression to the parotid glands of Ipr-injected or tannin-fed transgenic mice. Although a high level of induced expression was achieved with the −10R15/CAT construct, we cannot rule out the possibility that there may be other regulatory elements, either further upstream from −10 kb or downstream from +2 nt of the R15 gene, involved in governing its expression. This uncertainty stems from the possibility that CAT, whose activity was measured in this study, has a half-life different from that of the PRP, at either the protein or message level. Therefore, it is unclear whether the induced level of −10R15/CAT quantitatively reflects expression of endogenous R15.
Based on our previous results, the lack of isoproterenol inducibility of transgene expression in the salivary glands of mice harboring −1.7R15/reporter constructs is puzzling. In our previous studies, in which several proximal regulatory elements affecting gene transcription were identified (Lin and Ann, 1992), the activity of the PRP promoter had been analyzed by the transient transfection experiments. In those assays, the most critical element directing PRP expression was found to be the SCRE enhancer, located between nt −136 and nt −109 relative to the transcription start site. This SCRE binds to basic helix-loop-helix SCBP proteins; co-expressed SCBP isoforms mimic the cAMP effect in that they enhance expression of the transiently transfected SCRE-reporter constructs (Lin et al., 1993). However, the −1.7R15/CAT and −1.7R15/PyLT constructs containing this SCRE motif failed to display isoproterenol inducibility in any of the tissues that expressed these transgenes (Fig. 2). It has been demonstrated in many cases that when elements identified by transient transfection were tested in assay systems that involve gene integration into chromatin, such as a transgenic animal model, the ability of these elements to potentiate transcription varies considerably. Furthermore, the level of transcription of the transgenes is almost always considerably lower than the level obtained in the gene’s native environment (Palmiter and Brinster, 1986). There are two possible explanations for such a discrepancy between the transient transfection assays and transgenic studies.
The first explanation involves the function of putative boundary elements of eukaryotic expression domains. Hypothetically, these boundary elements flank eukaryotic expression domains and act to insulate the regulatory domains from the influence of neighboring chromatin and possibly to exert topological constraints on the regulatory sequences. The matrix-attachment sites of the chicken lysozyme gene have been proposed as candidates for such boundary elements (Bonifer et al., 1990). They are located both 5′ upstream and 3′ downstream of the chicken lysozyme gene in order to create an environment favorable to the functioning of regulatory domains of the gene. Presumably, the expression of a transgene without boundary elements in transgenic mice is easily affected by the sequence near the transgene integration site, especially for a relatively short transgene. In contrast, such chromosomal boundary elements are not required for transient expression, for which the transgene is not integrated into the genome. Thus, in our −1.7R15/CAT or PyLT transgenic mice, the lack of putative chromosomal boundary elements may have dramatically influenced the functioning of regulatory domains of these relatively short constructs and thus may account for the discrepancy between the transient transfection assays and transgenic mice studies, and the observed ectopic transgene expression.
A second possibility is that the expression of certain genes is mediated by a collection of individual elements surrounding the transcriptional unit, with each individual element having limited activity. However, highly sensitive reporter genes, such as CAT or luciferase, may allow DNA sequences with a very weak effect to be classified as generic major enhancers in transient transfection assays. The enhancer elements of the β-globin gene are good illustrations of this possibility (Grosveld et al., 1987). A series of four DNase I-hypersensitive sites are located within 15 kb at the far 5′ region of the β-globin locus (Grosveld et al., 1987). By itself, each site is capable of enhancing the expression of a linked β-globin gene, albeit at a lower level than with the cumulative effect of the four hypersensitive sites. This synergistic effect of multiple regulatory elements in controlling cell-specific expression of eukaryotic genes has also been demonstrated in many other transgenic studies, including those with the albumin gene (Kruse et al., 1993) and tyrosine aminotransferase gene (Nitsch and Schultz, 1993) in the liver and the myosin heavy chain gene (Rindt et al., 1993) and myosin light chain gene (Lee et al., 1992) in the muscle. Our observation that a high level of induced transgene expression in the salivary glands depends on the inclusion of the DNA sequences from −10 to −6 kb of R15 in the transgene also supports this possibility.
Among independent lines of transgenic mice prepared from different constructs, only transgenic mice made with constructs containing the parotid control region (−6 to −1.7 kb of R15) were capable of displaying predictable transgene expression in the parotid glands. In contrast, all the transgenic mice that harbored constructs without the parotid control region not only showed ectopic expression in nonsalivary tissues but also lacked inducible expression in the parotid glands (Table 1). Taken together, these findings suggest that the R15 parotid control region, along with the R15 promoter, can reproducibly direct isoproterenol/tannin-dependent reporter expression in the parotid glands of 15 out of 15 independent transgenic mice derived from 3 different constructs: −10R15/CAT, −6R15/CAT, and −6R15/PyLT (Table 1). It is unlikely that such high-frequency, reproducible transgene expression is a coincidence of transgene integration. Allen et al. (1988) have found, using an enhancer trap experiment, that the frequency of gene expression/activation through a weak promoter that resulted from random transgene integration was 16 out of 72, or 22%. This is significantly lower than the penetrance achieved by the constructs containing the parotid control region in this report (15 out of 15 transgene-positive lines).
Although the co-integration of parotid control region into the R15 transgene loci resulted in establishing persistently position-independent, tissue-specific, and inducible transgene activity, there is no direct evidence that these parotid control regions are actually the boundaries of its chromatin structure. However, the parotid control region exerts its effect only in the stably transfected cell lines and in transgenic mice described herein, and not in transient transfectants (D. K. Ann et al., unpublished data). Therefore, the identified R15 parotid control region probably functions at the chromatin level.
Clearly, these effects are similar to some of those mediated by the locus control region (LCR) in regulating RNA polymerase II transcription (Dillon and Grosveld, 1993). The LCR is a potent DNA element that confers copy number-dependent and chromosomal position-independent expression to genes that are stably integrated into the genome (Dillon and Grosveld, 1993). The lack of copy number-dependent transgene expression in our experiments (∼10-fold variation per copy of transgene derived from the same construct) may simply reflect a less-than-perfect role assumed by R15 parotid control region. This compromised copy number-dependent transgene expression is also observed in the transgenic studies using the LCR or its homologue of human β-globin gene (Dillon and Grosveld, 1991) and murine immunoglobulin gene (Jenuwein et al., 1993) in their artificial constructs.
One hallmark of LCRs is that they contain DNase I-hypersensitive sites in the chromatin structure and sequences displaying regulatory function (Dillon and Grosveld, 1993). We have observed the parotid-specific DNase I-hypersensitive sites within the parotid control region of the R15 (data not shown), and the presence of regulatory elements in the identified parotid control region is further evidenced by the fact that part of this region is capable of directing isoproterenol-stimulated expression of a heterologous promoter construct in the parotid gland of transgenic animals (Fig. 6).
Another possible role of the parotid control region may be targeting the transgene to active chromatin domains. However, this hypothesis implies that the transgenes are able to determine integration sites, which seems unlikely since the foreign DNA is integrated into the genome of a one-cell (sometimes two-cell) embryo in which the patterns of heterochromatin have not yet been established.
In conclusion, although the exact mechanism of its action calls for further investigation, our data establish that the novel parotid control region is required for the tissue-specific R15 gene induction by isoproterenol and tannin in vivo, and thus serve as a starting point for defining the cis-elements that function in vivo. Identification of these elements should allow for a more accurate definition of the trans-factors responsible for salivary transcriptional regulation of PRP expression. Upon establishing how these elements work in concert and unraveling their DNA sequences, we should be able to design vectors in which transgene expression is predictable and reproducible. Further experiments are also needed to elucidate the putative mechanism by which isoproterenol/tannin treatment leads to the formation of a parotid-specific transcriptional complex that mediates PRP expression via the parotid control region. We favor the bimodal process proposed by Archer et al. (1992), in which the hormone-dependent activation of mouse mammary tumor virus expression goes through two steps: receptor-dependent chromatin remodeling and then transcription factor recruitment. Our transgenic mice system clearly is a valuable tool to investigate the molecular basis of tissue-specific and inducible transgene expression in a chromosomal position-independent manner.
Acknowledgments
We are grateful to Dr. Larry Butler for providing us with the high-tannin sorghum, IS-8260. Many thanks are owed to Dr. Harry Orr for invaluable discussion and helpful comments on this manuscript. We are also in debt to Dr. Marilee Wick for her critical reading of this manuscript, and Sandy Horn for excellent technical assistance.
Research funds from Minnesota Medical Foundation and Graduate School, University of Minnesota, for setting up the transgenic facility are greatly appreciated. This work is supported in part by Public Health Service Grants R29-DE09175, R03-DE10432, and R01-DEI0742, and Research Career Development Award K04-DE00292 to D. K. Ann.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
H. Helen Lin is currently in the Department of Medicine, University of Minnesota Medical School, Minneapolis, MN 55455.
References
- Allen N. D., Cvan D. G., Barton S. C., Hettle S., Reik W., and Surani M. A. (1988), Nature 333, 852–855. [DOI] [PubMed] [Google Scholar]
- Al-Shawi R., Kinnaird J., Burke J., and Bishop J. O. (1990), Mol Cell Biol 10, 1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ann D. K., Clements S., Johnstone E. M., and Carlson D. M. (1987), J Biol Chem 262, 899–904. [PubMed] [Google Scholar]
- Archer T. K., Lefebvre P., Wolford R. G., and Hager G. L. (1992), Science 255, 1573–1576. [DOI] [PubMed] [Google Scholar]
- Bonifer C., Vidal M., Grosveld F., and Sippel A. (1990), EMBO J 9, 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson D. M. (1993), Critical Rev Oral Biol Med 4, 495–502. [DOI] [PubMed] [Google Scholar]
- Chalifour L. E., Gomes M. L., Wang N.-S., and Mes-Masson A. M. (1990), Oncogene 5, 1718–1726. [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi N. (1987), Anal Biochem 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Dillon N. and Grosveld F. (1991), Nature 350, 252–254. [DOI] [PubMed] [Google Scholar]
- Dillon N. and Grosveld F. (1993), Trends Genet 9, 154–157. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., and Kollias G. (1987), Cell 51, 975–985. [DOI] [PubMed] [Google Scholar]
- Hardman P. and Spooner B. S. (1992), in Epithelial Organization and Development (Fleming T. P., ed.), Chapman and Hall, New York, pp. 353–375. [Google Scholar]
- Hogan B., Costantini F., and Lacy E. (1986), Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Jenuwein T., Forrester W. C., Qiu R.-G., and Grosschedl R. (1993), Mol Cell Biol 7, 2016–2032. [DOI] [PubMed] [Google Scholar]
- Kruse F., Rose S. D., Swift G. H., Hammer R. E., and MacDonald R. J. (1993), Genes Dev 7, 774–786. [DOI] [PubMed] [Google Scholar]
- Lazowski K. W., Mertz P. M., Redman R. S., Ann D. K., and Kousvelari E. (1992), Differentiation 51, 225–232. [DOI] [PubMed] [Google Scholar]
- Lee K. J., Ross R. S., Rockmann H. A., Harris A. N., O’Brien T. X., van Bilsen M., Shubeita H. E., Kandolf R., Brem G., Price J., Evans S. M., Zhu H., Franz W.-M., and Chien K. R. (1992), J Biol Chem 267, 15875–15885. [PubMed] [Google Scholar]
- Lin H. H., Kousvelari E., and Ann D. K. (1991), Gene 104, 219–226 [DOI] [PubMed] [Google Scholar]
- Lin H. H. and Ann D. K. (1991), Genomics 10, 102–103. [DOI] [PubMed] [Google Scholar]
- Lin H. H. and Ann D. K. (1992), Gene Expr 2, 365–377. [PMC free article] [PubMed] [Google Scholar]
- Lin H. H., Li W.-Y., and Ann D. K. (1993), J Biol Chem 268, 10214–10220. [PubMed] [Google Scholar]
- Mehansho H., Ann D. K., Butler L. G., Rogler J., and Carlson D. M. (1987a), J Biol Chem 262, 12344–12350. [PubMed] [Google Scholar]
- Mehansho H., Butler L. G., and Carlson D. M. (1987b), Annu Rev Nutr 7, 423–440. [DOI] [PubMed] [Google Scholar]
- Neznanov N., Thorey I. S., Cecéna G., and Oshima R. G. (1993), Mol Cell Biol 13, 2214–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch D. and Schutz G. (1993), Mol Cell Biol 13, 4494–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D. and Brinster R. L. (1986), Annu Rev Genet 20, 465–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkert C. A., Ornitz D. M., Brinster R. L., and Palmiter R. D. (1987), Genes Dev 1, 268–276. [DOI] [PubMed] [Google Scholar]
- Pothiec R., Ouellet M., Julien J., and Guerin S. L. (1992), DNA Cell Biol 11, 83–90. [DOI] [PubMed] [Google Scholar]
- Rindt H., Gulick J., Knotts S., Neumann J., and Robbins J. (1993), J Biol Chem 268, 5332–5338. [PubMed] [Google Scholar]
- Simonet W. S., Bucay N., Lauer S. J., and Taylor J. M. (1993), J Biol Chem 268, 8221–8229. [PubMed] [Google Scholar]
- van Holde K. E. (1989), in Chromatin (van Holde K. E., ed.), Springer Verlag, New York, pp. 355–408. [Google Scholar]
- Warner T. F. and Azen E. A. (1984), Am Rev Respir Dis 130, 115–118. [DOI] [PubMed] [Google Scholar]
- Wilkie T. M., Brinster R. L., and Palmiter R. D. (1986), Dev Biol 118, 9–18. [DOI] [PubMed] [Google Scholar]


