Abstract
Fluorescence resonance energy transfer provides information about protein structure and dynamics. Single-molecule analysis can capture the information normally lost through ensemble averaging of heterogeneous and dynamic samples. Immobilization of single molecules, under conditions that retain their biological activity, allows for extended observation of the same molecule for tens of seconds. This can capture slow conformational transitions or protein binding and unbinding cycles. Using an open geometry for immobilization allows for direct observation of the response to changing solution conditions or adding ligands. Here we provide detailed methods for immobilization and observation of fluorescently labeled single proteins using total internal reflection microscopy that are widely applicable to the study of intrinsically disordered proteins.
Keywords: Intrinsically disordered proteins, Single-molecule fluorescence, FRET, Vesicle, Encapsulation, Reconstitution
1. Introduction
Although depleted in hydrophobic amino acids, which cause chain collapse in folded proteins, intrinsically disordered proteins (IDPs) are prone to intramolecular interactions that affect chain dynamics (1). The balance between net charge and hydropathy has been shown to determine the compaction of the polypeptide (2, 3) ranging from extended random coils to disordered globules that are closer in volume to folded proteins (4). Some IDPs can be induced to fold in the presence of ligands or protein-binding partners while others are perpetually disordered. Under native buffer conditions, an ensemble IDP sample contains a dynamic mixture of structures. The distribution of conformations under native conditions and their importance is an open question. Conformational selection of rarely populated states may contribute to the biological function of IDPs (5).
The distance dependence of fluorescence resonance energy transfer (FRET) has been used for decades to characterize polypeptide dynamics (6, 7). The fluorescent donor and acceptor are attached to the polypeptide, through synthesis or mutagenesis, with a defined separation in the primary sequence. In a dynamic polypeptide chain, FRET efficiency can be related to the root mean squared (rms) displacement in any direction, , or absolute distance in stable structures. Such methods have been used to examine polymer models of the denatured state (8) and IDPs under native conditions (9, 10). Ensemble FRET can provide valuable insights into IDPs, but single-molecule FRET (smFRET) can resolve sample heterogeneity and to some extent probe dynamics.
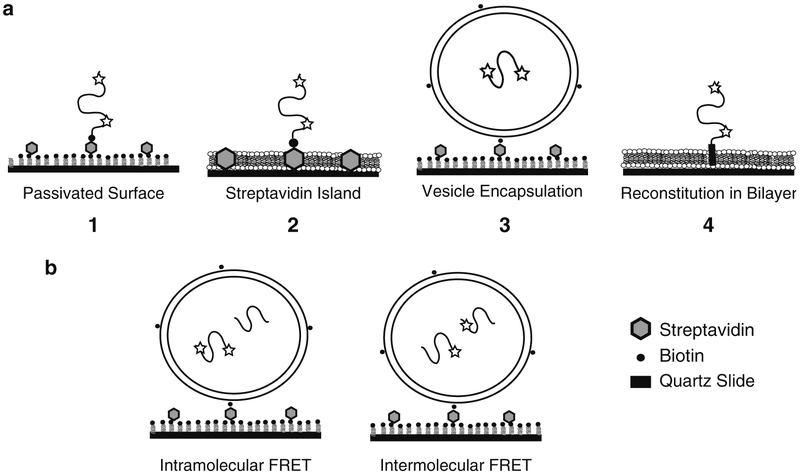
FRET can measure distances and conformational fluctuations on the scale of 2–8 nm (7). For a disordered random coil, this corresponds to fluorophore separations of 50–175 residues in the primary sequence. Conformational dynamics occur on a variety of timescales. Dynamics of extended random coils are faster than the time resolution of current Electron Multiplied Charge Coupled Device (EMCCD) cameras, which leads to a narrow distribution of FRET values. Thus even at the single-molecule level, the details of molecular behavior are time-averaged and are equivalent to ensemble measurement of Rrms. Dynamics approaching the exposure time lead to peak broadening in the FRET distribution (11). EMCCD detection is best at resolving subpopulations, capturing dynamics >10/s, and direct observation of the response to non-equilibrium conditions (e.g., ligand binding). With immobilization, an individual molecule can be recorded for tens of seconds using Total Internal Reflection Fluorescence (TIRF) microscopy with EMCCD camera detection (12). TIRF illumination decays within 100–200 nm of the surface allowing immobilized molecules to be selectively excited. Unfavorable interactions with a surface can destroy a protein sample and must be prevented. In many cases it is possible to retain the biological activity of immobilized proteins. Familiar examples in biochemistry include the interactions between glutathione-S-transferase (GST) and Glutathione Sepharose resin and the biotin-streptavidin system. Retention of activity must be validated by comparison to non-immobilized samples. Here we describe methodology for the immobilization of proteins for TIRF microscopy while retaining biological activity. We have used smFRET to characterize IDPs involved in synaptic transmission, which serve to illustrate our methods for smFRET measurements of immobilized IDPs. We will demonstrate three experimental preparations that we have used for smFRET measurements of IDPs (Fig. 2). First, immobilizing the protein via a biotin-streptavidin interaction on a “passivated” surface provides an open geometry that allows manipulation of buffer conditions or addition of ligands or binding partners. Second, encapsulation confines the protein inside a phospholipid vesicle where it is protected from aggregation and adsorption. Co-encapsulation of multiple proteins within one liposome can be used to study weak protein-protein interactions since the liposome volume is approx. femtoliter. Finally, for membrane proteins, reconstitution into a phospholipid vesicles or deposited phospholipid bilayers can approximate the physiological membrane environment and probe the influence of lipids on IDP conformation.
Fig. 2.
Experimental approaches to immobilized single protein molecules. (a) From left to right, (1) shows a schematic of a biotinylated single molecule directly immobilized on a passivated surface via streptavidin. Dyes are indicated by stars. The passivation can use bBSA or PEG (Subheadings 3.1.1 and 3.1.3). (2) Shows a protein immobilized on a streptavidin island surrounded by a deposited lipid bilayer (Subheading 3.1.2). (3) Shows a protein encapsulated within a lipid vesicle (Subheading 2.3.2), which is immobilized to a passivated surface (Subheading 3.2). (4) Shows a membrane protein reconstituted via the transmembrane domain into lipid bilayer (Subheading 2.3.3). A deposited bilayer is formed from vesicles containing reconstituted membrane proteins (Subheading 3.3). (b) Schematic showing co-encapsulation experiments to study protein binding. Intramolecular FRET (left) can monitor changes in a protein induced by binding to an unlabeled ligand protein. Intermolecular FRET (right) between two proteins, each singly labeled with donor or acceptor, can directly report on protein binding events.
2. Materials
2.1. Reagents and Supplies
1 in. × 3 in. Quartz microscope slide (G. Finkenbeiner, Inc., Waltham, MA).
Diamond grinding bit, 0.029 in. tip diameter, #115005 (Star-lite Industries Inc., Rosemont, PA).
400 XPR Rotary Tool with Model 220 stand (Dremel, Racine, WI).
24 × 30 mm #1.5 micro cover glass (VWR, Radnor, PA).
Double and single-sided tape (Scotch/3M, St. Paul, MN).
5 min epoxy (ITW Devcon, Danvers, MA).
Optical adhesive 63 (Norland Products, Cranbury, NJ).
Electro-lite CS-410 UV Light Curing System (Thorlabs Inc., Newton, NJ).
Benchtop UV Transilluminator (UVP, Upland, CA).
Egg PC and 18:1 Biotinyl Cap PE in chloroform (Avanti Polar Lipids, Alabaster, AL).
Hand-held Mini-Extruder Kit (Avanti Polar Lipids, Alabaster, AL).
TBS buffer (20 mM Tris, 150 mM NaCl, pH 7.5).
Sepharose CL-4B packed in a NAP-5 column (GE Healthcare, Piscataway, NJ).
Biotinylated bovine serum albumin (bBSA) (Sigma-Aldrich, St. Louis, MO).
Escherichia coli strain AVB101 (Avidity LLC, Aurora, CO).
Streptavidin (Invitrogen, Carlsbad, CA).
Beveled pipette tips (VWR #53503-566).
MATLAB Software (The Mathworks, Nattick, MA).
2.2. Observation Chambers for TIRF Microscopy
Here we provide a procedure to prepare flow channels for single-molecule observations from quartz microscope slides, which assumes the use of prism-based TIRF excitation. The use of quartz or fused silica is paramount to avoid fluorescent impurities found in glass (see Note 10).
2.2.1. Preparation and Cleaning
Submerge the quartz slide in water. Drill holes using the diamond bit with the Rotary tool set to 35,000 rpm. To avoid breaking the slides, replace worn out bits often. Up to five flow chambers can be made on a slide as long as their total size does not exceed the width of the cover glass (see Note 1).
Before use, slides are carefully cleaned by sequential sonication in acetone, ethanol, and 1 M KOH. Place slides in a standard staining jar and cover with solvent. Bath sonicate for 30 min. Use a separate container for each solvent and dedicated tweezers to move slides. Finally, slides are extensively rinsed with deionized water.
2.2.2. Construction of Flow Channels
Make sure work area is clean. Briefly pass the slide through the flame of a propane torch until dry. Allow the slide to cool by placing it on a metal ring stand with the slide interior surface facing down (see Note 1). We reuse the ~2 in. metal lids from chemical and restriction enzyme containers for this purpose.
Cut tape to form flow channel walls. Use double-sided tape unless lipid bilayers are being used. Attach a ~2 in. piece of tape lengthwise on a clean glass microscope slide. Cut with a razor blade into 1–2 mm wide strips. Trim to ~1.5 in. in length.
Immediately before assembling the chamber, turn the slide face up to expose the interior surface (see Note 1). Place strips of tape on the slide as spacers between the drilled holes to form the flow chambers (Fig. 1). Do not allow the tape to touch the channel interior. Avoid air bubbles beneath the tape as this will result in variations in the channel thickness. Work out any bubbles using pressure from the tweezers or a similar tool.
To remove antistatic coating and dust, quickly pass both sides of the cover glass through a propane flame (< 1 s). Cover glasses can easily warp or crack if held in the flame too long and should be discarded if this happens. Allow to air cool for 30 s while holding with tweezers.
Align the long edge of the cover glass over the slide to cover all the chambers. Lower the cover glass onto the slide in a continuous motion. Once stuck to the tape, the cover glasses are difficult to remove. Use pressure from the tweezers or a similar tool to work out any bubbles trapped between the glass and the tape. Cut off any overhanging tape with a new razor blade.
Combine the 5 min epoxy and allow 1–2 min for partial curing. If the epoxy is not sufficiently viscous, capillary action will draw epoxy into the channel, which can possibly cover the drilled holes. Using a pipette tip, apply a thin bead of epoxy to both ends of the flow chamber. Avoid covering holes as this will render the chamber unusable. To avoid leakage, ensure that each chamber has a continuous seal. Allow to cure completely.
If using lipid bilayers, single-sided tape is used in conjunction with UV curable optical adhesive to form the channel. Lipid bilayers extract fluorescent impurities from double-sided tape.
Prepare the optical adhesive ahead of time. Remove the plunger and fill a 1-ml syringe with adhesive. Attach at 30G1/2 needle and insert plunger driving glue to tip.
All procedures up to step 4 of Subheading 2.2.2 are the same only using single-sided tape. Apply a thin bead of adhesive along the center of each strip of single-sided tape (Fig. 1). There should be just enough adhesive to cover the tape after compressing the cover glass to the slide. Too much adhesive will fill the chamber and may cover the holes while too little will expose the sample chamber to the edge of the tape.
Apply the cover glass as described in Subheading 2.2.2, step 4. Take care as the cover glass can only be applied once. Mild pressure can be used to direct adhesive flow and completely cover the tape edges. Use judicial application of the UV light curing system to harden the adhesive in each chamber for 30 s. The goal is not to fully cure the adhesive but only to fix the cover glass in place and insure optimal coverage of the tape.
Finally seal the ends of the flow chamber with adhesive. Avoid covering the holes with excess adhesive by capillary action. Place the slide cover glass up on a UV light box. Use ~2 mm wooden dowels to elevate the slide off the surface. To insure a uniform chamber thickness, a glass microscope slide is placed atop the cover glass with ~70 g of weight applied to the center (e.g., a 3 in. optical post). Expose to UV for 30 min to fully cure the adhesive.
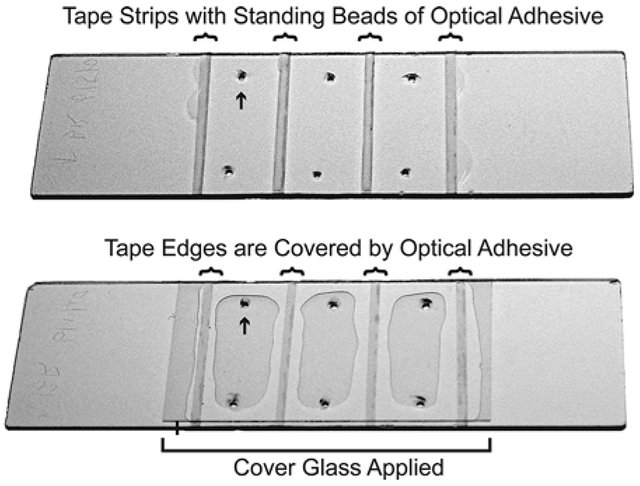
Fig. 1.
Assembly of multichannel flow chambers using UV adhesive. (Top) A quartz slide is shown with six holes drilled (e.g., arrow) to create three flow chambers. Strips of single-sided tape (curved brackets) demark the walls of the flow chambers. A thin bead of UV adhesive sits on the center of each strip of tape but does not exceed the width of the tape. (Bottom) The same slide is shown after the cover glass has been applied (straight bracket denotes edges of the cover glass). The adhesive has flowed to cover the tape edges and now constitutes the walls of the flow chamber. The flow chambers are then cured with UV light.
2.3. Preparation of Phospholipid Vesicles
Phospholipids can be used to encapsulate soluble proteins in vesicles, reconstitute membrane proteins or to form a deposited bilayer on the microscope slide surface. A wide variety of phospholipids are available to mimic biological membranes, prevent nonspecific interactions or add functional groups or affinity tags including biotin and Ni-NTA. We typically use phosphatidylcholine extracted from chicken egg (egg PC), which shows low levels of fluorescent impurities and minimal nonspecific binding of proteins.
2.3.1. Formation of Unilamellar Vesicles by Extrusion
Place 30 mg of lipid in a chloroform solution in a 13 × 100-mm glass tube. Inside a chemical fume hood, dry the lipid under a gentle stream of argon or dry nitrogen gas while rotating. Form a thin but compact film as this is easier to recover during resuspension. Once lipids are dried, place the tube under vacuum for 30–60 min to remove residual chloroform. A pump is recommended as house vacuum is often insufficient.
Add 1 ml of TBS to make a 30 mg/ml lipid solution. The high concentration of lipids facilitates encapsulation and reconstitution. Vortex for 30 s or until all dry lipid is suspended.
Use 4–5 freeze-thaw cycles to aid in the solubilization of the lipids. The goal is to take the sample above and below the lipid phase transition temperature, which depends on the lipids in use. For each cycle, flash freeze the lipid suspension in liquid nitrogen and thaw in a 37°C water bath followed by 30 s vortex.
Assemble the extruder according to the manufacturer’s instructions. The membrane filter pore size depends on the experiment. A 50 nm diameter is used for deposited bilayers while 100–200 nm is used for encapsulation. Test everything for leakage with TBS buffer before extruding.
Draw lipids into the first syringe. Go slowly for the first few passes to avoid rupturing the membrane. Extrude with 20–30 passes back and forth through the extruder. The opacity will decrease noticeably depending on the vesicle size.
Collect liposomes from the second syringe (i.e., NOT the first syringe used to initially draw up the lipids). This insures that the collected material has passed through the extruder. Spin the sample at 14,000 × g in a microcentrifuge and collect the supernatant containing vesicles.
2.3.2. Encapsulation of Soluble Proteins in Vesicles
For immobilized vesicles, prepare a stock of egg PC with 0.1 mol% of 18:1 Biotinyl Cap PE in chloroform. Otherwise, egg PC in chloroform is sufficient. Dry lipids as described in Subheading 2.3.1, step 1.
For encapsulation, labeled protein(s) can be added to the lipid samples at Subheading 2.3.1, step 1 or after step 3 (see Note 2). Freeze-thaw cycles can facilitate encapsulation efficiency but are not tolerated by all proteins. Aggregation of proteins during repeated freezing should be tested.
Remove unencapsulated proteins by desalting on Sepharose CL-4B. Vesicles elute at the void volume while free proteins are retained. Columns are created by replacing the Sephadex G-25 in commercial NAP-5 columns with Sepharose CL-4B to a bed height of 25–30 mm and replacing the frit. Measure the void volume with extruded vesicles. The presence of lipids can be confirmed with a spectrophotometer by measuring scattering at 400 nm.
Equilibrate a CL-4B desalting column in TBS. Load 200 μl of sample. Allow all liquid to drain before adding additional buffer. Use two applications of 100 μl TBS to rinse the frit. Add additional TBS to bring the total added volume (including sample) within 100 μl of the measured void volume of the column. Collect 200-μl fractions. Sample is typically recovered in 400–600 μl and spun at 14,000 × g in a microcentrifuge. Light scattering by vesicles makes measurement of protein concentration difficult.
2.3.3. Detergent-Assisted Insertion to Reconstitute Membrane Proteins in Vesicles
Membrane proteins are exchanged into TBS containing 100 mM β-D-octyl glucoside (βOG) by affinity chromatography. Mix the protein sample at a 1:4 ratio with 30 mg/ml extruded 50 nm liposomes in TBS. Incubate on ice for 30 min.
Dilute sample 1:1 with detergent-free TBS. Incubate at room temperature for 5 min.
Remove detergent and unincorporated protein by desalting with Sepharose CL-4B as described in Subheading 2.3.2, step 4. This method can lead to uniform orientation of membrane proteins with their cytoplasmic domain facing outwards. Orientation can be tested by proteolysis in the presence and absence of 1% Triton X-100.
3. Methods
To monitor protein conformational changes or protein interactions over time, molecules must be immobilized so they will not diffuse out of the observation volume. Here we describe three different approaches to immobilize single molecules. First, soluble proteins are biotinylated and immobilized directly to the surface using a biotin-streptavidin linkage (Fig. 2a (1, 2)). Before immobilization, the surface must be “passivated” through the use of blocking agents to prevent adsorption and loss of activity (13). Second, vesicles containing encapsulated soluble proteins are immobilized to the surface using a biotin-streptavidin linkage (Fig. 2a (3)). Most proteins are an order of magnitude smaller than the vesicle diameter, so they undergo free diffusion within the vesicle interior. The vesicle protects the encapsulated protein from interactions with the surface and other proteins. Third, proteins containing a transmembrane domain are reconstituted into vesicles, which are then used to form a planar lipid bilayer on the slide surface (Fig. 2a (4)). Although imperfect, deposited bilayers can be used to examine the effect of lipids on conformation or binding studies. We describe methods where rinsing and sample application is implemented using hand-activated pipettes although automated buffer exchange schemes are also possible (see Note 10).
3.1. Direct Immobilization of Proteins to a Passivated Surface
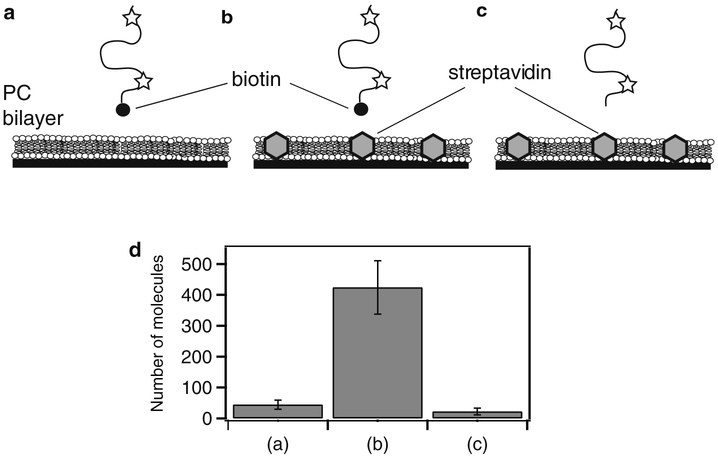
Proteins can be biotinylated enzymatically or during recombinant expression using commercial reagents and the AVB101 bacterial strain. The effectiveness of a surface passivation strategy must be tested for each protein. First, the level of nonspecific binding is tested by omitting streptavidin or forgoing biotinylation (Fig. 3). Ultimately the results of studies using the immobilized protein should be compared to encapsulated or freely diffusing controls.
Fig. 3.
Determining the extent of nonspecific binding to a passivated surface. Data is shown for streptavidin islands (Subheading 3.1.2) but the same method can be used to test any surface preparation. (a) As a control, biotinylated, fluorescently labeled protein was applied to a deposited lipid bilayer containing no streptavidin. After 5 min incubation, the proteins were rinsed out and the extent of binding was assessed. (b) The same protein solution in (a) applied to a streptavidin island surface and rinsed. (c) As an additional control, the same protein was expressed, purified, and labeled without biotinylation and applied to the streptavidin island surface. (d) To assess the degree of binding, the number of molecules retained on the surface was counted. Shown are the average number of molecules per field of view for the experiments shown in (a)–(c), respectively. The degree of nonspecific binding can be assessed by comparing retention of the protein to the streptavidin islands to the surface without streptavidin and/or protein lacking biotin (see Note 9).
3.1.1. Biotinylated Bovine Serum Albumin Surface (14)
Use the flow cell shown in Subheading 2.2.2. Using a beveled pipette tip to form a tight seal, wash the channel with 4 × 200 αl of deionized water followed by TBS (see Note 5).
Add 100 μl of 0.2-μm filtered 1 mg/ml bBSA to the channel (see Note 4). After the incubation, rinse the channel with 4 × 200 μl TBS to remove unbound bBSA.
Nonspecific binding to bBSA can be further reduced by adding 100 μl egg PC vesicles (50 nm at 15 mg/ml). BSA is a fatty acid-binding protein so phospholipids can block exposed hydrophobic sites. Incubate and rinse as described above.
Add 100 μl 0.2 μm filtered 0.1 mg/ml streptavidin (SA). Incubate and rinse as described above.
The fluorescently labeled, biotinylated protein sample should be diluted in TBS to ~pM (see Note 4). Incubate 5 min and rinse with TBS (see Note 5). The sample is now immobilized and can now be imaged or subjected to manipulations of the external solution (Fig. 2a (1)).
3.1.2. Streptavidin Islands in a Deposited Lipid Bilayer (15)
Use the optical adhesive flow chamber described in Subheading 2.2.2, step 7. Equilibrate the flow channel with TBS as described in Subheading 3.1.1, step 1. Inject 100 μl of 1 nM SA. Incubate 5 min to allow the sparse deposition of individual SA molecules, which retain specific binding activity when adsorbed. Rinse with TBS.
Inject 100 μl 50 nm egg PC vesicles at 3 mg/ml in TBS. Incubate 1 h in a humid environment to allow the spontaneous condensation of a lipid bilayer around the adsorbed SA molecules (see Note 3). Rinse slowly with TBS (see Note 5).
Nonspecific binding can be reduced by further incubation with 1 mg/ml BSA and/or 30 mg/ml egg PC liposomes.
The fluorescently labeled, biotinylated protein sample should be diluted in TBS to ~pM (see Note 4). Incubate 5 min and rinse with TBS. The sample is now immobilized and can now be imaged or subjected to manipulations of the external solution (Fig. 2a (2)).
3.1.3. Biotinylated Poly (ethylene glycol) Surface
Poly(ethylene glycol) (PEG) coated surface is an effective and popular way of preventing nonspecific binding. A detailed protocol is described elsewhere (16).
A major difference from the protocols above is that PEG is applied to both the quartz slide and cover glass before the observation chamber is constructed. The interior surfaces ofthe quartz slide and the cover glass are amine functionalized by incubation in 1% Vectabond in acetone for 5 min. Rinse exhaustively with deionized water. Dry with dry nitrogen or under vacuum.
Assemble the optical adhesive flow chamber as described in Subheading 2.2.2, step 7. Take care to protect the interior surfaces (see Note 1).
Dissolve 100 mg of 99 mol% m-PEG-SPA: 1 mol% biotin-PEG-NHS in 1 ml 100 mM sodium bicarbonate. Quickly inject 100 μl directly into each of the flow channels. Incubate for 1–2 h in a humidity-controlled environment (see Note 3). Wash exhaustively with deionized water to remove unbound PEG. PEG coated slides can be stored after drying in air for a few days.
When ready to use, exchange channel into TBS. Add 100 μl 0.1 mg/ml SA and incubate for 5 min. Wash with TBS. Now the surface is ready to immobilize biotinylated samples. Non-specific binding can be further reduced by incubation with 1 mg/ml BSA prior to the streptavidin (17).
The fluorescently labeled, biotinylated protein sample should be diluted in TBS to ~pM (see Note 4). The sample is now immobilized and can now be imaged or subjected to manipulations of the external solution (Fig. 2a (1)).
3.2. Immobilization of Vesicles to a bBSA Surface (12)
Use the epoxy flow cell shown in Subheading 2.2.2. Prepare the bBSA and SA surface as described in Subheading 3.1.1.
Fluorescently labeled protein(s) should be vesicle encapsulated as described in Subheading 2.3.2. To study the effects of intramolecular interactions on IDP conformation, an unlabeled ligand protein can be encapsulated with the labeled sample using the procedure described in Subheading 2.3.2, step 2. (Fig. 2b, left). The concentrations of the unlabeled ligand can vary depending on the binding affinities to monitor a partial or a full effect on the conformational changes of the labeled samples. To monitor intermolecular interactions, two singly labeled proteins (one labeled with donor and the other with acceptor) are co-encapsulated at a 1:1 ratio using the procedure described in Subheading 2.3.2, step 2 (Fig. 2b, right).
Dilute the encapsulated sample ~1:104 in TBS (see Note 2). Inject 100 μl into the flow channel and incubate for 5 min (see Note 3). Remove free vesicles by gently rinsing with TBS (see Note 5). The sample is now immobilized and ready for measurement (Fig. 2a (3)).
3.3. Formation of a Deposited Bilayer Containing Membrane Proteins (see Note 8)
Use the optical adhesive flow chamber described in Subheading 2.2.2, step 7. Equilibrate the flow channel with TBS.
Membrane proteins are reconstituted as described in Subheading 2.3.3. Empirical dilution with unlabeled 50 nm egg PC liposomes may be required to achieve optical resolution (see Note 4). Inject 100 μl of protein containing vesicles. Incubate 15 min to allow the spontaneous condensation of a lipid bilayer at the surface of the slide.
Rinse the channel gently with TBS (see Note 5). An additional incubation with 15 mg/ml protein-free 50 nm egg PC liposomes for 1 h can block defects remaining after step 2.
For membrane proteins, the integrity of the bilayer is critical. Diffusion of fluorescently labeled lipids can be tested by Fluorescence Recovery After Photobleaching (FRAP) or single particle tracking. Similarly, defects can be identified by incubation of the bilayer with fluorescently labeled proteins or liposomes and assessing the degree of retention. Protein orientation, which may differ from vesicle samples, can be checked with proteolysis (Fig. 2a (4)).
3.4. Data Collection and Analysis
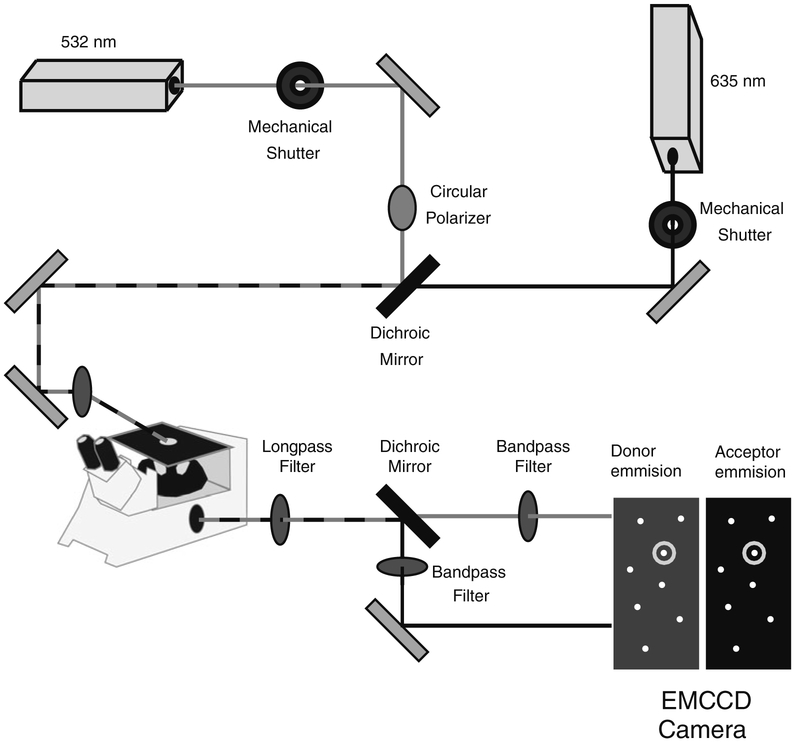
The assembly of prism TIRF microscopy instrumentation is beyond the scope of this chapter and has been thoroughly described elsewhere (16). A prism-based TIR instrument can be constructed from a standard inverted microscope and commercial optomechanical components with a small amount of custom milling (Fig. 4). Briefly, excitation of the Alexa555 (or Cy3) donor and measurement of FRET efficiency uses a circularly polarized 532-nm laser, while direct excitation of the Alexa647 (or Cy5) acceptor uses a 635-nm laser. The lasers are brought to the microscope stage by means of standard geometrical optics and introduced to the sample through a quartz prism (e.g., PLBC 5.0-79.5-SS CVI Laser) coupled to the quartz slide with an index matching oil (e.g., Cargille Type FF). Immediately before imaging, samples are exchanged into imaging buffer (see Note 6) to stabilize fluorescence emission. The microscope image is passed through a series of optical elements to separate the donor and acceptor emission (Fig. 4). This is achievable using commercial image splitters (e.g., Optosplit II; Cairn Research Ltd., Kent, UK), which results in separate donor and acceptor images of the sample. Fluorescent beads (e.g., Fluo-spheres; Invitrogen, Carlsbad, CA) are used to determine the pixel registry between the donor and acceptor images, which allows identification of corresponding donor and acceptor fluorophores attached to a single molecule. Since the protein is randomly labeled with acceptor (A) and donor (D) dyes, all combinations are possible [AA, DA, AD, DD]. Also, more than one molecule can be encapsulated inside a single vesicle. To distinguish these behaviors, we used alternating laser illumination (18) to determine the number of dyes in diffraction-limited spot (see Note 7).
Fig. 4.
Schematic of a prism-based total internal reflection microscope for single-molecule spectroscopy. A circularly polarized 532-nm laser and a linear polarized 635-nm laser are combined using a dichroic mirror and routed to a quartz prism on the microscope stage. Mechanical shutters control the excitation wavelength. The fluorescence emission is split by color into two images using a dichroic mirror, which are passed through additional optical elements to isolate donor and acceptor signals. The two replicate images are collected by an EMCCD camera and are processed using MATLAB to identify single molecules containing a donor and acceptor fluorophore.
3.4.1. Extraction of smFRET Efficiency from the Microscope Image
Each EMCCD camera exposure can be treated as a numerical array and is readily processed using MATLAB. Single molecules appear as diffraction-limited spots, which are easily detected as pixels of maximum intensity separated by five or more pixels from any neighboring maxima. A ten-frame average taken under 635 nm illumination is used to locate active acceptor fluorophores. Once identified, these pixels are reexamined at each frame of the movie.
Using a 60 ×, 1.2 NA water immersion objective with a 512 × 512 pixel EMCCD camera, a single molecule is contained within a 3 × 3 pixel array about the most intense pixel. Fluorescence intensity for a single molecule can be determined simply by taking the sum of the 3 × 3 matrix. In practice, most of the single-molecule intensity falls on the top four of the pixels so tracking this smaller set of pixels is sufficient. Such undersampling decreases the width of the single-molecule intensity distribution (unpublished data) reflecting lower noise.
For each molecule, the donor and acceptor intensity in each frame of the movie is plotted as a function of the frame number to examine the time dependence of fluorescence emission (Fig. 5). These time traces are first analyzed to confirm that the diffraction-limited spot contains a single molecule labeled with one donor and one acceptor. Single fluorophores are confirmed by the observation of a single step in the photobleaching decay to baseline (Fig. 5). If labeling efficiency is high and molecules are optically well resolved, two single fluorophores in a diffraction-limited spot can be attributed to a doubly labeled protein. To characterize a sample, observations of hundreds to thousands of single molecules are collected.
The hallmark of smFRET is anticorrelated donor intensity during acceptor photobleaching (Fig. 5). A critical normalization factor, γ, is taken from the magnitude of this change and used to adjust for differences in quantum yield and detection efficiency, which can vary between samples and even between molecules (19). To calculate smFRET efficiency, E, requires correction for the leakage of donor emission into the acceptor channel, β. This value can be obtained from control experiments and are constant as long as the dyes and instrument are unchanged. Depending on the experimental setup, correction for direct excitation of the acceptor may also be required but is not significant at the laser powers used in our system.
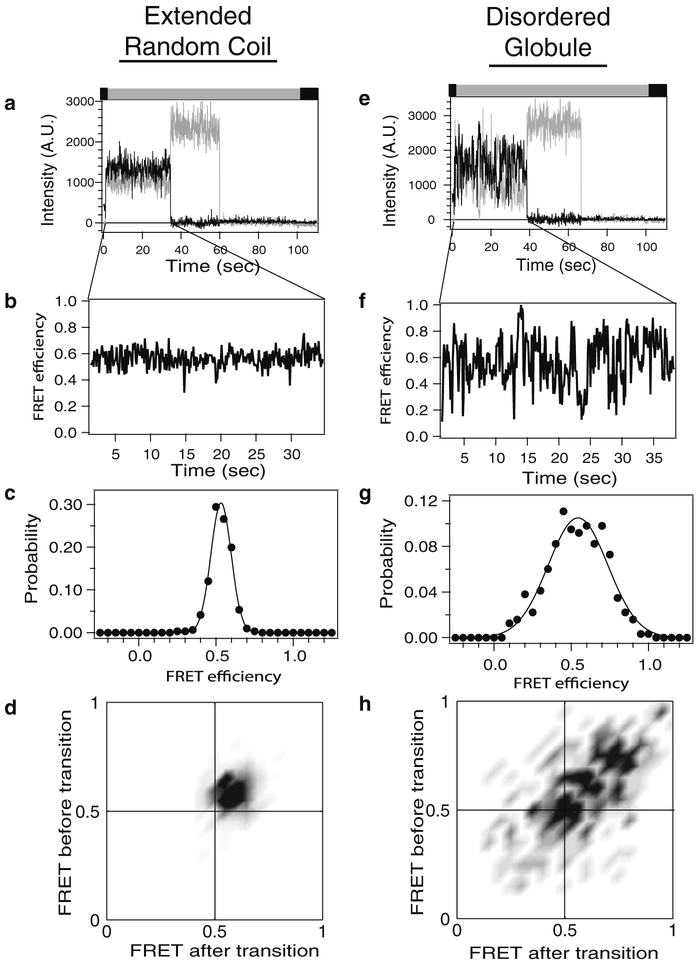
Fig. 5.
Analysis of single-molecule fluorescence intensity time traces. Representative data analysis is shown for two classes of IDP that have been labeled with donor and acceptor. Left: Extended random coil IDP. Right: Disordered globular IDP. (a and e) Background corrected fluorescence intensity of a single molecule. Donor emission is colored gray while acceptor is colored black. Note the single-step photobleaching of each dye to baseline, which confirms a single molecule. Colored bar above the panel describes the alternating laser excitation (see Note 7). (b and f) Intensities above have been converted to FRET efficiencies for the period before the acceptor dye molecule photobleached. Note the steady FRET of the random coil compared to the stochastic switching in a disordered globule. (c and g) FRET efficiency at each time point is compiled into histograms to examine the probability distribution. These histograms contain data from only one molecule, while a typical experiment would compile hundreds to thousands of molecules. Random coils give a single peak of shot noise-limited width. (d and h) The time series of changes in FRET are examined using transition density plots, which plot the FRET state before a transition (y-axis) against the FRET state after that transition (x-axis) for molecules (a) and (e), respectively. The darker areas indicate more favored transitions between the indicated FRET states. The random coils show only one peak while the globular IDP makes transition to many different FRET states.
3.4.2. Analysis of smFRET Efficiency Time Traces
The most straightforward analysis is to compile all the observed smFRET efficiency values into a histogram that represents the probability distribution. For a random coil, rapid conformational dynamics lead to the appearance of a single peak that is well described by a single Gaussian function using nonlinear least squares fitting. The mean FRET efficiency and the distribution width both provide information about IDP structure and dynamics. For compact globular IDPs, more complicated smFRET distributions are observed that require multiple functions to describe (Fig. 5).
The averaging of rapid intramolecular dynamics in random coil-like IDPs gives rise to steady FRET efficiency levels over time (Fig. 5a–d). The resultant FRET distributions typically show a single Gaussian peak of shot noise-limited width. Because of the time-averaging, this single-molecule data can be related only to ensemble properties using polymer models.
More complex, stochastic conformational transitions have been observed in globular IDPs or protein complexes involving IDPs. The transitions are slow; occurring within a single frame (Fig. 5e–h). Data of this type can be analyzed with Hidden Markov models (20) or Edge detection strategies (21). From such analyses one obtains the distribution of FRET states (Fig. 5c, g), the dwell times for each state (i.e., state lifetime) and the sequence of state transitions. Each of these parameters can be analyzed to extract information about the sequence and nature of conformational fluctuations as well as the stability of intermediate states.
For both types of IDP, the use of immobilized molecules with EMCCD detection allows one to capture individual binding events and the instantaneous response to changes in solution conditions.
4. Notes
A three channel slide will need three holes at the top and bottom (Fig. 1). When drilling slides, the exit hole can be slightly irregular, which prevents the formation of a good seal with the pipette during rinsing. This side of the slide is used as the channel interior. As such, it should be protected from contacting anything during assembly.
Encapsulation efficiency depends on the absolute concentration of protein and lipids. Efficiency is highly sample dependent and typically <10%. Micromolar protein concentrations at the lipid concentration described results in most liposomes being empty or containing a single molecule. Membrane protein reconstitution is similarly variable.
Longer incubations are carried out in a humidified, vapor-saturated chamber, such as an empty pipette tip box with deionized water in the bottom. This prevents samples from drying in the chamber. A drop of TBS is placed over each hole of the chamber to further reduce evaporation. Should this dry, apply a drop of TBS to one hole to force air out the other before rinsing.
The goal is to empirically achieve spacing between immobilized molecules larger than the optical resolution by adjusting the sample concentration (typically <250 molecule/field). Because reconstitution and encapsulation efficiency can vary, the dilutions need to achieve optical resolution of samples may vary.
It is critical to remove any trapped air bubbles, which present an air-water interface, and prevent the introduction of bubbles at all future steps. Similarly, rinsing should be carried out slowly to avoid excessive shear forces at the surface. Once active enzymes or deposited bilayers are present, either bubbles or shear can destroy the sample.
Imaging buffer is TBS with 1% glucose and includes oxygen scavengers (20 U/ml glucose oxidase and 1,000 U/ml catalase), which extend fluorophore lifetime and 100 μM cyclooctate-traene, which prevents fluorophore blinking.
First we illuminate with 635 nm (0–1 s) to excite acceptor-labeled molecules; next 532 nm illumination is used (1.5–100 s) to observe FRET events. Finally, 635 nm (100.5–110 s) is used to verify whether the acceptor dye molecules have photobleached.
This technique is best for type 1 or 2 (single spanning) membrane proteins that only protrude from one side of the membrane. In these cases, free diffusion within the plane of the bilayer is observed. There is only ~2 nm between the deposited bilayer and the slide surface. Polypeptide protruding beneath the bilayer adsorbs to the quartz and becomes immobilized or denatured.
With multichannel slides, the channels are used sequentially to prevent cross contamination. The slide is tilted vertically along its long axis for rinsing and the bottom channel is used first so outflow does not cross the other channels.
The presence of fluorescent contamination can be checked at any point using the microscope. Contamination can be introduced at any step and is checked by testing all reagents and processes involved.
References
- 1.Uversky VN, Dunker AK (2010) Understanding protein non-folding. Biochim Biophys Acta 1804(6):1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao AH et al. (2010) Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA 107(18):8183–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller-Spath S et al. (2010) Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci U S A 107(33):14609–14614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uversky VN (2002) What does it mean to be natively unfolded? Eur J Biochem 269(1):2–12 [DOI] [PubMed] [Google Scholar]
- 5.Boehr DD, Nussinov R, Wright PE (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5 (11):789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flory JP (1969) Statistical mechanics of chain molecules Interscience, New York, NY [Google Scholar]
- 7.Stryer L, Haugland RP (1967) Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci USA 58(2):719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Rhoades E (2008) Fluorescence characterization of denatured proteins. Curr Opin Struct Biol 18(4):516–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreon AC et al. (2009) Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci U S A 106(14):5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weninger K et al. (2008) Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure 16(2):308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalinin S et al. (2010) On the origin of broadening of single-molecule FRET efficiency distributions beyond shot noise limits. J Phys ChemB 114(18):6197–6206 [DOI] [PubMed] [Google Scholar]
- 12.Boukobza E, Sonnenfeld A, Haran G (2001) Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J Phys Chem B 105(48):12165–12170 [Google Scholar]
- 13.Rasnik I, McKinney SA, Ha T (2005) Surfaces and orientations: much to FRET about? Acc Chem Res 38(7):542–548 [DOI] [PubMed] [Google Scholar]
- 14.Ha T et al. (1999) Ligand-induced conformational changes observed in single RNA molecules. Proc Natl Acad Sci U S A 96(16):9077–9082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graneli A et al. (2006) Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir 22(1):292–299 [DOI] [PubMed] [Google Scholar]
- 16.Roy R, Hohng S, Ha T (2008) A practical guide to single-molecule FRET. Nat Methods 5(6):507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pertsinidis A, Zhang Y, Chu S (2010) Subnanometre single-molecule localization, registration and distance measurements. Nature 466 (7306):647–651 [DOI] [PubMed] [Google Scholar]
- 18.Lee NK et al. (2005) Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys J 88(4):2939–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCann JJ et al. (2010) Optimizing methods to recover absolute FRET efficiency from immobilized single molecules. Biophys J 99 (3):961–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinney SA, Joo C, Ha T (2006) Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J 91 (5):1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sass LE et al. (2010) Single-molecule FRET TACKLE reveals highly dynamic mismatched DNA-MutS complexes. Biochemistry 49 (14):3174–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]