Abstract
Utricles are vestibular sense organs that encode linear head movements. They are composed of a sensory epithelium with type I and type II hair cells and supporting cells, sitting atop connective tissue, through which vestibular nerves project. We characterized utricular Cre expression in 11 murine CreER lines using the ROSA26tdTomato reporter line and tamoxifen induction at 6 weeks of age. This characterization included Calbindin2CreERT2, Fgfr3-iCreERT2, GFAP-A-CreER™, GFAP-B-CreER™, GLAST-CreERT2, Id2CreERT2, OtoferlinCreERT2, ParvalbuminCreERT2, Prox1CreERT2, Sox2CreERT2, and Sox9-CreERT2. OtoferlinCreERT2 mice had inducible Cre activity specific to hair cells. GLAST-CreERT2, Id2CreERT2, and Sox9-CreERT2 had inducible Cre activity specific to supporting cells. Sox2CreERT2 had inducible Cre activity in supporting cells and most type II hair cells. ParvalbuminCreERT2 mice had small numbers of labeled vestibular nerve afferents. Calbindin2CreERT2 mice had labeling of most type II hair cells and some type I hair cells and supporting cells. Only rare (or no) tdTomato-positive cells were detected in utricles of Fgfr3-iCreERT2, GFAP-A-CreER™, GFAP-B-CreER™, and Prox1CreERT2 mice. No Cre leakiness (tdTomato expression in the absence of tamoxifen) was observed in OtoferlinCreERT2 mice. A small degree of leakiness was seen in GLAST-CreERT2, Id2CreERT2, Sox2CreERT2, and Sox9-CreERT2 lines. Calbindin2CreERT2 mice had similar tdTomato expression with or without tamoxifen, indicating lack of inducible control under the conditions tested. In conclusion, 5 lines—GLAST-CreERT2, Id2CreERT2, OtoferlinCreERT2, Sox2CreERT2, and Sox9-CreERT2—showed cell-selective, inducible Cre activity with little leakiness, providing new genetic tools for researchers studying the vestibular periphery.
Keywords: CreER/loxP, mouse genetics, utricle, fate-mapping, calbindin, FGFR3, GFAP, GLAST, Id2, otoferlin, parvalbumin, Prox1, Sox2, Sox9
Introduction
The utricle is a vestibular sense organ located in the inner ear. The sensory epithelium, or macula, of the utricle is composed of two resident cell types—sensory hair cells, which encode linear acceleration of the head, and non-sensory supporting cells, which resemble glia. The macula is surrounded by a thin non-sensory, or transitional, epithelium. A loose connective tissue, called the stroma, supports the macula. The peripheral neurites of the vestibular nerve, whose cell bodies are located in the vestibular ganglion, course through the stroma and into the macula. The stroma also contains Schwann cells, endothelial cells, fibroblasts, immune cells, and other cell types.
Although there is a growing understanding of the cellular processes underlying utricular development, maturation, and homeostasis, we have limited tools to define the genes that control these processes. Genetically engineered mice that allow cell type-specific and temporal control of gene expression are very powerful tools to dissect the molecular mechanism of biological pathways, as well as to fate-map cells during development, regeneration, and repair. The Cre-loxP system allows for control of gene expression in select cells by using a cell type-specific promoter or enhancer to drive expression of Cre, a DNA recombinase, and floxed target alleles, which enable deletion of targeted DNA segments and activation or repression of specific genes (Kwan 2002). When a modified estrogen receptor is added to the Cre enzyme, creating a CreER fusion protein (e.g., CreER™, CreERT2, or iCreERT2), temporal control of gene expression is achieved via tamoxifen administration (Feil et al. 1996; Hayashi and McMahon 2002).
More than 30 Cre or CreER alleles have been described for the organ of Corti, the sensory organ for hearing, which like the utricular macula is located in the inner ear and composed of hair cells and supporting cells. In the cochlea, these Cre/CreER lines have been used to manipulate gene expression in different cell types and to fate-map sensory progenitor cells, with studies resulting in significant advances for the auditory field (reviewed in Cox et al. 2012). However, few CreER lines have been characterized for vestibular end organs, including the utricle. Four lines—Atoh1-CreER™, Lgr5eGFP-CreERT2, Plp-CreERT2, and Sox2CreERT2—have been used to study hair cell regeneration or postnatal hair cell addition in neonatal utricles (Gómez-Casati et al. 2010; Burns et al. 2012a, b; Wang et al. 2015; Lin et al. 2015; Wu et al. 2016). However, only two CreER lines have been described in the adult vestibular system. After tamoxifen injection at 6 weeks of age, Plp-CreERT2 mice targeted the majority of supporting cells, as well as a small number of Schwann cells and cells in the transitional epithelium, while Atoh1-CreER™ mice targeted ~ 40 % of type II hair cells. Both CreER lines were recently used to demonstrate hair cell turnover (cell loss and cell addition) in normal adult utricles (Bucks et al. 2017).
Here, we characterized the Cre activity pattern of 11 CreER lines after tamoxifen induction at 6 weeks of age in the mouse utricular macula. We found that five lines—GLAST-CreERT2, Id2CreERT2, OtoferlinCreERT2, Sox2CreERT2, and Sox9-CreERT2—had inducible Cre activity in utricular hair cells and/or supporting cells, which constitute new genetic tools for investigators to manipulate gene expression or to fate-map cells in the vestibular periphery of adult mice.
Materials and Methods
Mouse Models
Calbindin2CreERT2 (stock no. 13730, Taniguchi et al. 2011), GLAST-CreERT2 (stock no. 12586, Wang et al. 2012), Id2CreERT2 (stock no. 16222, Rawlins et al. 2009), ParvalbuminCreERT2 (stock no. 10777, Taniguchi et al. 2011), Sox2CreERT2 (stock no. 17593, Arnold et al. 2011), Sox9-CreERT2 (stock no. 18829, Kopp et al. 2011), and ROSA26tdTomato (also called Ai14, stock no. 7908, Madisen et al. 2010) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Fgfr3-iCreERT2 mice (Rivers et al. 2008; Young et al. 2010) were provided by Dr. William Richardson (University College London, UK) and are now available at The Jackson Laboratory (stock no. 25809). GFAP-A-CreER™ and GFAP-B-CreER™ mice (Chow et al. 2008) were provided by Dr. Suzanne Baker (St. Jude Children’s Research Hospital, Memphis, TN). OtoferlinCreERT2 mice were provided by Dr. Ulrich Müller (The Scripps Research Institute, La Jolla, CA) and are now available from the Mutant Mouse Resource and Research Centers (MMRRC) as stock no. 032782-MU. Prox1CreERT2 mice (Srinivasan et al. 2007) were provided by Dr. Guillermo Oliver (St. Jude Children’s Research Hospital, Memphis, TN) and are now available from The Jackson Laboratory (stock no. 22075). ROSA26tdTomato and all CreER lines were used as heterozygotes for all experiments. Genotyping was performed by Transnetyx, Inc. (Cordova, TN) or as described previously (Cox et al. 2014). Both genders were used in all studies. All procedures were conducted in accordance with approved animal protocols from the Institutional Animal Care and Use Committees at St. Jude Children’s Research Hospital (Memphis, TN) and Southern Illinois University School of Medicine (Springfield, IL).
Mouse Strain Background
Each CreER line was on a different mixed genetic background based on how the line was generated and bred before it was deposited at The Jackson Laboratory or imported to our mouse colony. Specifically, Calbindin2CreERT2 mice were on a C57BL/6J:B6(Cg)-Tyrc-2J/J background, Fgfr3-iCreERT2 mice were on a C57BL/6J:CBA background, GFAP-A-CreER™ and GFAP-B-CreER™ mice were on a FVB/NJ background, GLAST-CreERT2 mice were on a C57BL/6J:C57BL/6N:SJL/J background, Id2CreERT2 mice were on a C57BL/6J:C57BL/6N background, OtoferlinCreERT2 mice were on a C57BL/6J:NZB:SJL/J background, ParvalbuminCreERT2 mice were on a C57BL/6J:C57BL/6N background, Prox1CreERT2 mice were on a NMRI:C57BL/6J:129Sv/Jae background, Sox2CreERT2 mice were on a C57BL/6J:129Sv/Jae background, and Sox9-CreERT2 mice were on a C57BL/6J:BALBc:CD1 background. The ROSA26tdTomato reporter line was purchased from The Jackson Laboratory in 2013 on a C57BL/6J background and has been maintained in our colony since then by breeding with these lines as well as other CreER lines not described in this manuscript. Every 8–12 months, new ROSA26tdTomato breeders were obtained from Cre-negative pups from different breeding strategies and therefore the strain background of individual ROSA26tdTomato breeders varied.
Drug Treatments
Tamoxifen [9 mg/40 g, intraperitoneal injection (IP); Sigma-Aldrich (St. Louis, MO)] was injected once a day on two consecutive days (~ 20–24 h apart) in 6-week-old mice, and samples were collected 1 week after tamoxifen injection, at 7 weeks of age. Controls were age-matched littermates that expressed CreER and ROSA26tdTomato alleles but did not receive tamoxifen injection and were housed separately from the tamoxifen-treated experimental mice.
Immunofluorescent Staining
Temporal bones were removed and post-fixed in electron microscopy grade 4 % paraformaldehyde (Polysciences, Inc., Warrington, PA) overnight at room temperature. After fixation, temporal bones were stored in 10 mM phosphate-buffered saline [PBS; Sigma-Aldrich (St. Louis, MO)] until whole utricles were dissected out of temporal bones and placed in 96-well plates for free-floating immunofluorescent labeling. Utricles and, in some cases, ampullae were immunofluorescently stained as described in Bucks et al. (2017). For cell typing, hair cells were labeled with rabbit anti-myosin VIIa primary antibodies (1:300; Proteus Biosciences, Inc.; cat no. 25–6790). The following additional primary antibodies were used: rabbit anti-calbindin2 (1:500; EMD Millipore cat. no. AB5054), rabbit anti-GLAST (1:1000; Abcam cat. no. ab416), chicken anti-neurofilament 200 kD (1:500; Millipore cat. no. AB5539), mouse anti-otoferlin (1:500; Abcam cat. no. AB181781), mouse anti-parvalbumin (1:500; Sigma-Aldrich cat. no. P3088), goat anti-Sox2 (1:500; Santa Cruz Biotechnology cat. no. sc-17320), rabbit anti-Sox9 (1:500; Millipore cat. no. AB5535), and rabbit anti-IBA1 (1:1000; Wako Pure Chemical Industries cat. no. 019-19741). For samples immunostained with anti-GLAST antibodies, an antigen retrieval step was added prior to the blocking/permeabilization step. Here, samples were incubated with a high pH antigen unmasking solution (1:100; Vector labs cat. no. H-3301) at 95 °C for 45 min in a hybridization oven, followed by three PBS washes before proceeding with the rest of the staining procedure. All primary antibodies were detected with Alexa Flour-conjugated secondary antibodies [1:400, Invitrogen (Carlsbad, CA)]. Nuclei were labeled by 4′,6′-diamidine-2-phenylindole (DAPI; Sigma-Aldrich) at 1 μg/mL in 10 mM PBS. Following staining, whole utricles were mounted in Fluoromount-G (Southern Biotech, Birmingham, AL).
Confocal Microscopy and Image Analysis
An Olympus FV-1000 microscope (Olympus, Center Valley, PA) was used to acquire images of fluorescent labeling. Z-series images were taken of whole-mounted utricles or ampullae with either a × 20 air objective or a × 60 oil objective, starting at the hair cell stereocilia and ending a few microns in the stroma. Z steps ranged from 0.5 to 1 μm increments. Images were analyzed using Fiji (http://fiji.sc/). We examined tdTomato labeling in the macula, the transitional epithelium, and the stroma. Images for the publication were generated using Adobe Photoshop CS4. For quantitative analyses, we generated four sets of z-series images—two from the peripheral macula and two from the central macula. The approximate positions of the sampled regions for every animal are indicated in Fig. 1d. The central regions were enriched for striolar epithelium, and the peripheral regions were enriched for lateral extrastriolar epithelium, although some overlap existed. We did not sample much of the medial extrastriola. All images were × 120 (created using a 60× oil objective with × 2 zoom), and when added together, the four images comprised 24 % of the area of sensory epithelium. Form several CreER lines, tdTomato-positive cells in each image were counted and classified as either type I or type II hair cells or supporting cells (described in the next section) using the Fiji Cell Counter plug-in. For each set of images, we calculated the number of labeled cells per 10,000 μm2, or cell density. To estimate the number of tdTomato-positive hair cells or supporting cells per utricle, we did the following. First, we summed cell numbers from the four images. Then, we multiplied this value by 4.16 (or 100 %/24 %), to reflect the proportion of the macula that all images collectively sampled. To estimate the percentage of each cell type labeled per utricle, we divided the estimated number of tdTomato-positive cells per utricle by either 3800 (for hair cells) or 4000 (for supporting cells), based on the assumption that each utricle had 3800 hair cells and 4000 supporting cells (Golub et al. 2012). Averages, standard deviations, and animal numbers, as well as raw data and gender, for each animal in which quantitative analyses were performed are presented in the tables.
Fig. 1.
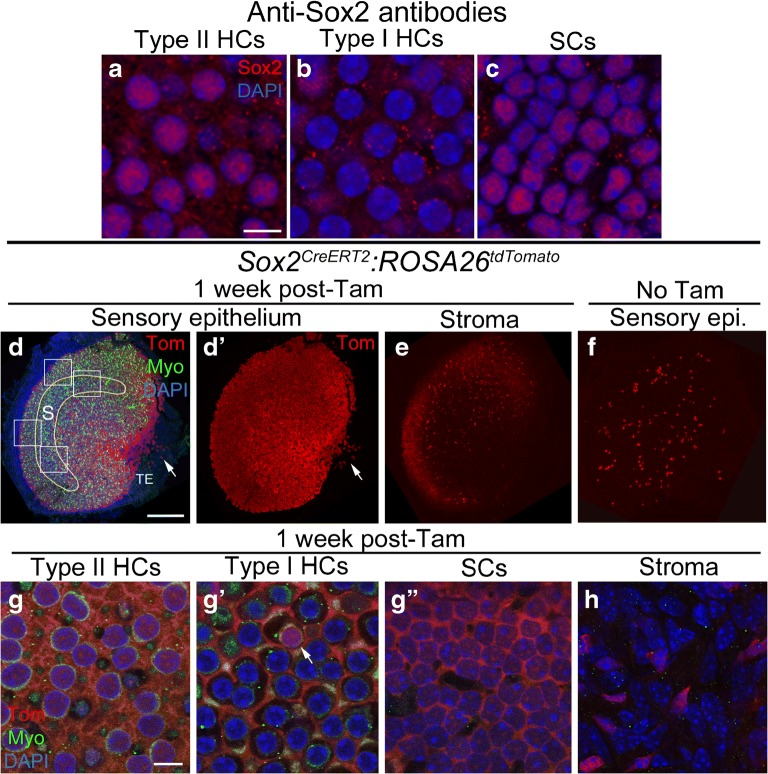
Sox2CreERT2 mice have inducible Cre activity in type II hair cells and supporting cells. a-c Confocal images showing immunolabeling for Sox2 (red) and DAPI nuclear labeling (blue) in slices focused on the nuclei of type II hair cells (HCs) (a), type I HCs (a), and supporting cells (SCs) (c) of the utricular macula. d-e Confocal images of utricles from Sox2CreERT2:ROSA26tdTomato mice injected with tamoxifen, focused on either the macula (d,d) or the stroma (e). d and d show the same view, with labeling for tdTomato (Tom), myosin VIIa (Myo), and DAPI shown in d and Tom only shown in d. The yellow line in d shows the approximate position of the striola (S). The white boxes in d show the approximate regions sampled for cell counts for the Sox2CreERT2 line and for all other lines that were quantitatively analyzed. TE = transitional epithelium. The arrow points to the TE near the notch. e shows Tom labeling in the stroma. f Utricle from a Sox2CreERT2:ROSA26tdTomato mouse that did not receive tamoxifen, focused on the sensory epithelium. g-h Confocal slices through the lateral extrastriolar region, with slices through the type II hair cell (HC) nuclear layer (g), the type I hair cell (HC) nuclear layer (g'), the supporting cell (SC) nuclear layer (g"), or the stroma (h). The arrow in g' points to a Tom-labeled type I HC. Scale bar in a = 5 μm and applies to a-c. Scale bar in d = 150 μm and applies to d-f. Scale bar in g = 5 μm and applies to g-h
Criteria for Cell Typing
Hair cells were classified as type I or II using previously described methods (Rüsch et al. 1998; Eatock and Songer 2011; Pujol et al. 2014; Bucks et al. 2017). Briefly, hair cells were distinguished from supporting cells by myosin VIIa immunoreactivity (Hasson and Mooseker 1997) and the location of their nuclei above the supporting cell nuclear layer. The nuclei of type II hair cells are slightly oblong and larger than type I hair cell nuclei, and they are located in a near-monolayer apical to type I hair cell nuclei. Type II hair cells also have a thick neck and basolateral cytoplasmic processes. Type I hair cells have nuclei that are small and round. They are positioned in a near-monolayer below type II hair cell nuclei and above the supporting cell nuclei, except in the striola, where some type I hair cell nuclei are positioned apical to type II hair cell nuclei. Type I hair cells have a thin neck and lack basolateral cytoplasmic processes. Finally, the cell body of each type I hair cell is encapsulated by a specialized afferent nerve ending called a calyx. By contrast, type II hair cells are contacted by small bouton afferents.
Quantitative Analysis
All statistics were performed using unpaired Student’s t tests on GraphPad Prism version 6.00 (La Jolla, CA. http://www.graphpad.com). Animal numbers, genders for each mouse, and the parental inheritance of the CreER allele for all of the CreER lines that we analyzed quantitatively are presented in the tables. One utricle per animal was assessed, and the “n” value represents the number of mice included in the study.
Results
Structure of the Utricular Macula
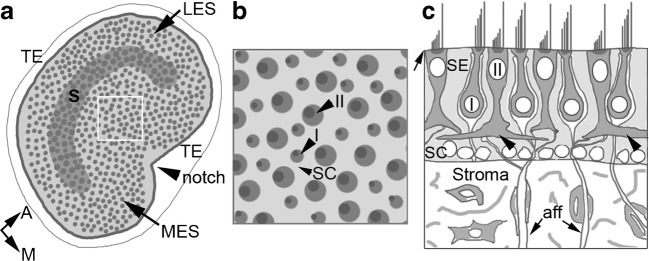
The mouse utricle is a disk-shaped organ with a sensory epithelium (macula) sitting atop a layer of loose connective tissue (stroma). The histology of the mammalian utricle is reviewed in Eatock and Songer (2011). The organ is inundated by processes of the vestibular nerve, each of which is surrounded by glial cells located within the stroma, but not within the macula. The macula houses the cell bodies of supporting cells, type I hair cells, and type II hair cells, the nuclei of which are positioned in discrete layers, as described in the “Materials and Methods” section and illustrated in Fig. 2. The macula has two general zones: the central zone, or striola, and the peripheral zone, or extrastriola (Fig. 2a). The striola is a C-shaped region. It is the only location in the macula in which type I hair cells are innervated by pure calyceal vestibular afferent neurons, which encapsulate two or more type I hair cells. The peripheral zone is further broken into the medial and lateral extrastriolar sub-regions, with the lateral extrastriola having a smooth contour and the medial extrastriola marked by a notch. Hair cells in the two extrastriolar regions—their properties and ratios—appear very similar. Type I and II hair cells are distributed in a mosaic across all zones of the macula (Fig. 2b, c). Hair cells are separated by the cell bodies of supporting cells. Only type II hair cells contact each other, and this occurs only at the level of the basolateral processes (Fig. 2c). The transitional epithelium is a monolayer, several cells in width, which lines the periphery of the macula (Fig. 2a).
Fig. 2.
Histology of the adult mouse utricle. a A top-down view of the macula (light gray) including hair cells (dark gray dots) and the transitional epithelium (TE, white border). The gray C-shaped region is the striola (S), and it is surrounded by the lateral extrastriola (LES) and medial extrastriola (MES). The notch in the medial portion of the macula is indicated by an arrowhead. The anatomical orientations (A = anterior and M = medial) apply to a and b. b A higher magnification view of the apical surface of the utricular macula from a region similar to that boxed in a. Arrowheads point to a supporting cell (SC), the larger-sized apical surface of a type II (II) hair cell and the smaller-sized apical surface of a type I hair cell (I), each with a stererociliary bundle indicated by a dark gray dot on the lateral side of the cell. c A cross-section through the macula (sensory epithelium, or SE) and stroma showing the different cell types in each tissue. Hair cell stereocilia project into the lumenal space from the apical side of the epithelium (arrow). Type II hair cell nuclei lie apical to type I hair cell nuclei. Type I hair cells are surrounded by white nerve calyces of vestibular nerve afferents, whose axons (aff, arrows) travel basally through the stroma to the vestibular ganglion (not shown). Type II hair cells have basolateral processes (arrowheads). The nuclei of supporting cells (SCs) lie along the basal lamina (the thin line at the base of the epithelium, in light gray). In the stroma, several cell types exist, including fibroblasts, macrophages, and myelinating glial cells
CreER Lines Targeting Vestibular Hair Cells and Supporting Cells
Each CreER mouse line was bred to the ROSA26CAG-loxP-stop-loxP-tdTomato (ROSA26tdTomato) reporter line to identify cells with CreER activity. Mice were injected with tamoxifen (9 mg/40 g, IP) at 6 weeks of age (with two injections given ~ 20–24 h apart) and analyzed 1 week later (at 7 weeks of age). To determine if there was any CreER activity in the absence of tamoxifen, a phenomenon called Cre leakiness, we examined control samples that expressed both CreER and ROSA26tdTomato but did not receive tamoxifen and were housed separately from tamoxifen-injected mice.
Sox2 is a high-mobility group (HMG) box domain transcription factor that is required for hair cell development in the mammalian inner ear (Kiernan et al. 2005; Dvorakova et al. 2016). Prior studies demonstrated that Sox2 immunolabeling is detected in all supporting cells and in most or all type II hair cells in utricles from adult CBA/CaJ mice (Oesterle et al. 2008). Using the same antibody in utricles from mice on a mixed background strain (see the “Materials and Methods” section for more detail), we found a similar cell-selective pattern of Sox2 protein expression (Fig. 1a–c). The Sox2CreERT2 line is a knock-in allele where CreERT2 replaced the Sox2 open reading frame and therefore only one copy of Sox2 is expressed in these mice (Arnold et al. 2011). We noted no difference in the density or distribution of hair cells or supporting cells, or any change in motor behavior, that would indicate a phenotypic abnormality due to Sox2 haploinsufficiency. After tamoxifen induction at 6 weeks of age in Sox2CreERT2:ROSA26tdTomato mice, we estimated that tdTomato was expressed in the cytoplasm and/or nucleus of 5137 (± 1 standard deviation) per utricle, of which 3945 ± 238 were supporting cells and 1192 ± 99 were hair cells (Fig. 1d, d’, g-g”; Table 1). These estimates were made by analyzing about 24 % of the macula, sampling from two peripheral and two central regions, which are shown in Fig. 1d for the Sox2CreERT2 line. This approach was applied for all CreER lines when calculating tdTomato-positive cell counts per utricle. We estimated that Cre was active, on average, in 99 % of supporting cells. We detected no difference in the density of labeled supporting cells in peripheral and central regions (unpaired Student’s t test, t(4) = 0.25, p = 0.815). TdTomato was also detected in approximately 32 % of hair cells, 93 % of which were type II hair cells and 7 % of which were type I hair cells (Fig. 1g-g’). Since 93 % of type II hair cells were tdTomato-positive, and type II hair cells comprise a little under 50 % of the utricular hair cell population (Desai et al. 2005), we anticipated we would find that ~ 50 % of hair cells were tdTomato-positive. However, we found that only 32 % of hair cells were tdTomato-positive. This finding is likely due to the fact that we oversampled the striola, which has a smaller proportion of type II hair cells than the extrastriola (Desai et al. 2005). Densities of tdTomato-labeled type II HCs were slightly higher in the peripheral regions than the central regions (unpaired Student’s t test, t(4) = 7.53, p = 0.002). There was also a small number of tdTomato-positive cells observed in the transitional epithelium (Fig. 1d, d’). We were unable to attain accurate counts of labeled cells in this region because we could not reliably image it in every utricle. However, we estimated that each utricle had 10–100 tdTomato-labeled transitional epithelial cells, with the highest density in the medial notch. Numerous tdTomato-positive cells were detected in the connective tissue (stroma) (Fig. 1e, h), which lies underneath the utricular macula. In Sox2CreERT2:ROSA26tdTomato mice that were not treated with tamoxifen, we detected 62.2 ± 38.81 labeled supporting cells and 6.4 ± 5.86 labeled hair cells per utricle (Fig. 1f; Table 1).
Table 1.
Quantification of tdTomato-labeled hair cells and supporting cells in the 5 CreER lines with cell-selective and inducible Cre activity
| Mouse linec | Sox2 CreERT2 | Otoferlin CreERT2 | GLAST-CreER T2 | Id2 CreERT2 | Sox9-CreER T2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| #Cells per utricled | #Cells per 10,000 μm2 | #Cells per utricle | #Cells per utricled | #Cells per 10,000 μm2 | #Cells per utricled | #Cells per 10,000 μm2 | #Cells per utricled | #Cells per 10,000 μm2 | ||
| Tam at 6 weeks | Hair cells (HCs) | 1192 ± 99 | 65.5 ± 5.33 | 403 ± 41 | 6.6 ± 10.9 | 0.41 ± 0.50 | 13.9 ± 24.0 | 0.68 ± 1.17 | 138.3 ± 64.9 | 6.73 ± 3.16 |
| % of HCs labeleda | 32 % | 11 % | 0.2 % | 0.4 % | 4 % | |||||
| Type I HCs/ut (% Type I) | 4.4 ± 1.32 (7 %) | 70 ± 52 (17 %) | 4.7 ± 10.5 (0.1 %) | 0 ± 0 (0 %) | 24.5 ± 31.6 (0.6 %) | |||||
| Type II HCs/ut (% Type II) | 60.08 ± 4.65 (93 %) | 334 ± 38 (83 %) | 1.6 ± 3.5 (0 %) | 13 ± 22.5 (0.3 %) | 109.4 ± 122.9 (0.3 %) | |||||
| Supporting cells (SCs) | 3945 ± 238 | 0 | 2270 ± 1408 | 1159 ± 416 | 3483 ± 556 | |||||
| % of SCs labeleda | 99 % | 0 % | 57 % | 29 % | 87 % | |||||
| Central SCs | 227 ± 13 | 132 ± 82 | 48 ± 26 | 171 ± 48 | ||||||
| Peripheral SCs | 199 ± 17 | 114 ± 71 | 61 ± 27 | 205 ± 15 | ||||||
| %tdTom+ cells that are HCs | 23 % | 100 % | 1 % | 1 % | 4 % | |||||
| %tdTom+ cells that are SCs | 77 % | 0 % | 99 % | 99 % | 96 % | |||||
| Number of mice analyzedb | 3 | 3 | 5 | 3 | 4 | |||||
| No Tam | Hair cells | 6.4 ± 5.86 | 0 | 2.8 ± 5.50 | 0.33 ± 0.58 | 2.3 ± 0.58 | ||||
| Supporting cells | 62.2 ± 38.81 | 0 | 6.3 ± 10.59 | 0.67 ± 0.58 | 1.3 ± 1.53 | |||||
| Number of mice analyzedb | 5 | 4 | 4 | 3 | 3 | |||||
Data represent mean ±1 standard deviation (except for %)
aEstimate of the % of all utricular HCs or SCs labeled with tdTomato, assuming 4000 SCs/utricle and 3800 HCs/utricle
bOne utricle per animal was assessed
cSox2, Otoferlin, and Id2 CreER lines are knock-ins and designated by the following nomenclature: XXXCreERT2. GLAST and Sox9 CreER lines are transgenics and designated by the following nomenclature: XXX-CreERT2
dEstimate of numbers of tdTomato-labeled cells per utricle, based on sampling 24 % of the utricle
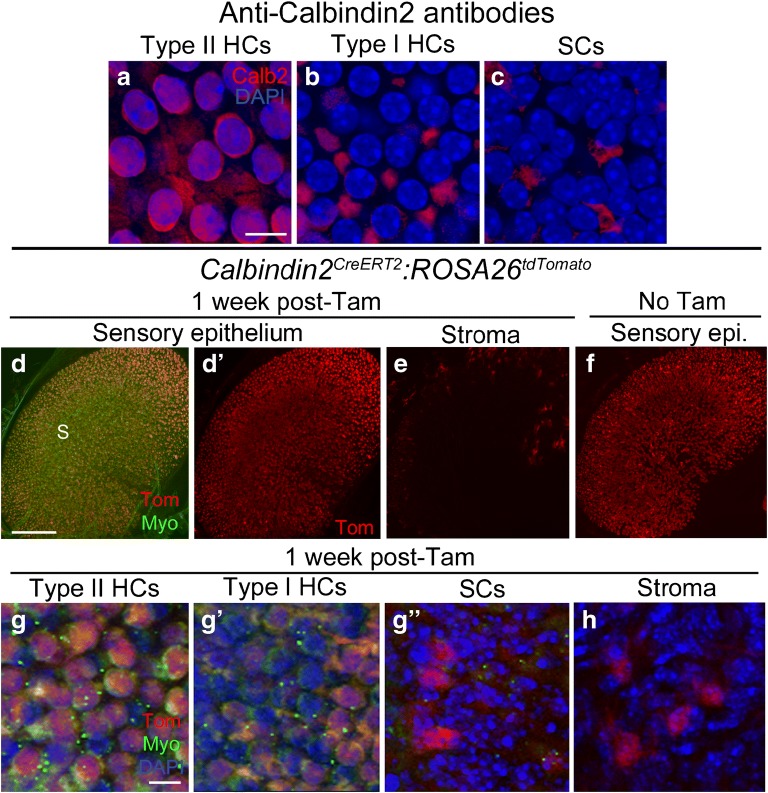
Calbindin2 (also called calretinin) is a calcium-binding protein that is detected by immunostaining primarily in type II hair cells and striolar calyceal afferents in utricles from adult CBA/J mice (Dechesne et al. 1991; Desai et al. 2005; Pujol et al. 2014). Desai et al. (2005) found that anti-calbindin2 antibodies labeled the cytoplasm of ~ 64 % of type II hair cells and ~ 5 % of type I hair cells in adult mouse utricles. Our immunolabeling in mixed background strain mice showed similar results, with nuclear and cytoplasmic labeling of many type II hair cells, including their basolateral processes, and little or no labeling of type I hair cells or supporting cells (Fig. 3a–c). Calbindin2CreERT2 is a knock-in allele, in which CreERT2 was inserted into the initiation codon of the endogenous Calbindin2 locus and therefore only one copy of Calbindin2 is expressed in these mice (Taniguchi et al. 2011). We observed no difference in density or distribution of hair cells or supporting cells, or any change in motor behavior, which would indicate a phenotypic abnormality due to Calbindin2 haploinsufficiency. Analysis of Calbindin2CreERT2:ROSA26tdTomato mice showed that there was no apparent difference in the tdTomato expression pattern (numbers or cell types) between samples that were injected with tamoxifen at 6 weeks of age and controls that did not receive tamoxifen (Fig. 3d, d’, f, g-g”). Since this finding demonstrates that Calbindin2CreERT2 is not behaving in an inducible manner in the adult utricle, we did not quantify tdTomato-positive cells. However, qualitative analysis showed in both tamoxifen-injected and control mice that tdTomato was expressed in a large proportion of type II hair cells and in a very small proportion of type I hair cells and supporting cells. No tdTomato expression was detected in neural elements, which was surprising, since antibody labeling has shown calbindin2 to be a selective marker for calyx-only afferents in the striola (e.g., Desai et al. 2005). No tdTomato was detected in the transitional epithelium (Fig. 3d, d’), and a small number of cells in the stroma were tdTomato-positive (Fig. 3e, h).
Fig. 3.
Calbindin2CreERT2 mice have Cre activity in type II hair cells and supporting cells. a-c Confocal images showing immunolabeling for calbindin2 (Calb2, red) and DAPI nuclear labeling (blue) in slices focused on the nuclei of type II hair cells (HCs) (a), type I HCs (b), and supporting cells (SCs) (c) of the utricular macula. d-e Confocal images of utricles from Calbindin2CreERT2:ROSA26tdTomato mice injected with tamoxifen, focused on either the macula (d,d') or the stroma (e). d and d' show the same view, with labeling for tdTomato (Tom) and myosin VIIa (Myo) shown in d and Tom only shown in d'. In d, S = approximate position of the striola. e shows Tom labeling in the stroma. f Utricle from a Calbindin2CreERT2:ROSA26tdTomato mouse that did not receive tamoxifen, focused on the sensory epithelium. g-h Confocal slices through the lateral extrastriolar region, with slices through the type II hair cell (HC) nuclear layer (g), the type I hair cell (HC) nuclear layer (g'), the supporting cell (SC) nuclear layer (g"), or the stroma (h). Scale bar in a = 5 μm and applies to a-c. Scale bar in d = 150 μm and applies to d-f. Scale bar in g = 5 μm and applies to g-h
CreER Lines Targeting Vestibular Hair Cells
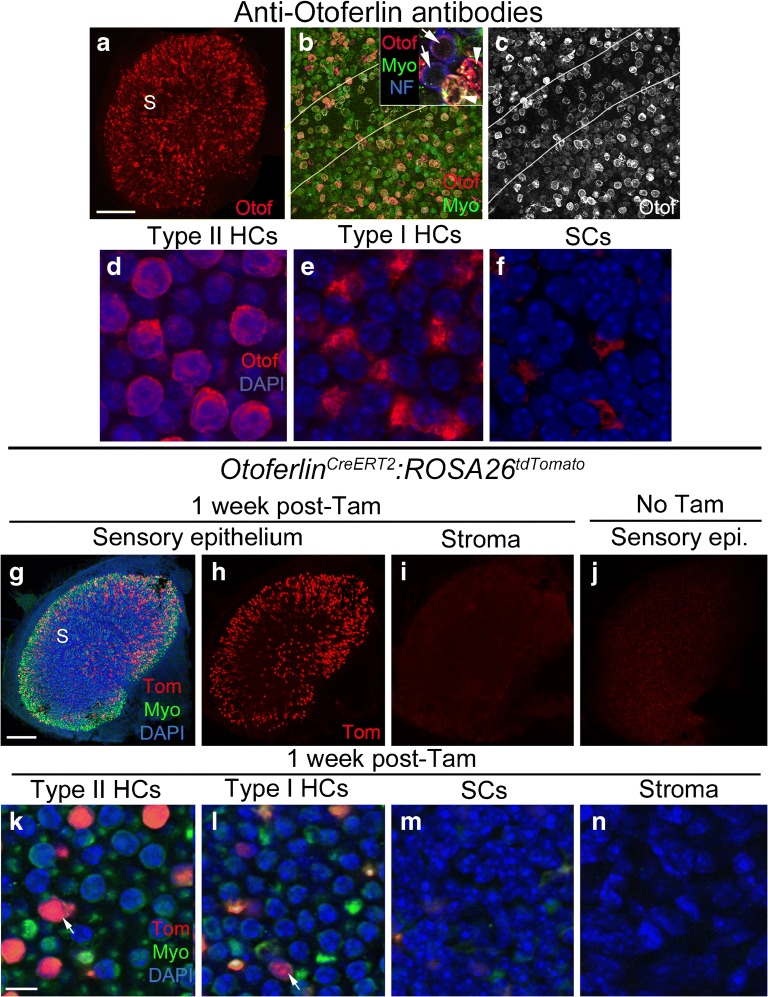
Otoferlin is a calcium-binding protein that regulates SNARE-mediated vesicle fusion in cochlear and vestibular hair cells (Roux et al. 2006; Schug et al. 2006). Immunolabeling in rodents has shown that otoferlin is expressed in type I and II vestibular hair cells (Schug et al. 2006; Dulon et al. 2009). Our immunolabeling in utricles from mixed background strain mice showed a similar result, although labeling was not consistent across hair cells (Fig. 4a–e). The strongest labeling was seen in the perinuclear cytoplasm and basolateral processes of type II hair cells, but not all type II hair cells were labeled. Labeling in type I hair cells was usually much weaker or not detected (Fig. 4b, inset; e). OtoferlinCreERT2 is a knock-in allele where CreERT2 was inserted into the endogenous Otoferlin locus and therefore only one copy of Otoferlin is expressed. It was generated as part of the NIH Neuroscience Blueprint Cre-Driver Network (http://www.credrivermice.org/index). We observed no difference in density or distribution of hair cells or supporting cells, or any change in motor behavior, which would indicate a phenotypic abnormality due to Otoferlin haploinsufficiency. After tamoxifen induction at 6 weeks of age in OtoferlinCreERT2:ROSA26tdTomato mice, tdTomato was expressed in 403 ± 41 hair cells per utricle, which is roughly 11 % of all hair cells (Fig. 4g, h, k, l; Table 1). Of labeled hair cells, 83 % were type II and 17 % were type I (Table 1). Although labeling was seen throughout the utricle, some samples appeared to have a higher density of tdTomato-positive hair cells in peripheral regions compared to central regions, although this was not quantified. No expression of tdTomato was observed in supporting cells (Fig. 4m), transitional epithelial cells (Fig. 4g, h), or cells in the stroma (Fig. 4i, n). In control samples that did not receive tamoxifen, no tdTomato-positive sensory epithelial cells were observed (Fig. 4j; Table 1).
Fig. 4.
OtoferlinCreERT2 mice have inducible Cre activity in many type II hair cells and some type I hair cells. a–f Confocal images showing immunolabeling for otoferlin (Otof, red), myosin VIIa (Myo, green), and/or 200 kD neurofilament (NF, blue) in utricular hair cells. a Whole-utricle view of otoferlin immunolabeling. S = the approximate position of the striola. b, c Magnified view of the region spanning the striola (demarcated by lines), with otoferlin labeling only shown in c. Arrows in the inset in b point to two type I hair cells with a light ring of otoferlin immunoreactivity in the cytoplasm (red) and NF labeling of the nerve calyx (blue). Arrowheads point to two strongly labeled type II hair cells. d–f Confocal images showing otoferlin immunolabeling (red) and DAPI nuclear labeling (blue) in slices focused on the nuclei of type II hair cells (HCs) (d), type I HCs (e), and supporting cells (SCs) (f) of the utricular macula. g–i Confocal images of utricles from OtoferlinCreERT2:ROSA26tdTomato mice injected with tamoxifen, focused on either the macula (g, h) or the stroma (i). g and h show the same view, with labeling for tdTomato (Tom, red), myosin VIIa (Myo, green), and DAPI (blue) shown in g and Tom only shown in h. In g, S = the approximate position of the striola, and TE = transitional epithelium. i Tom labeling in the stroma. j Utricle from an OtoferlinCreERT2:ROSA26tdTomato mouse that did not receive tamoxifen, focused on the sensory epithelium. k–n Confocal images through the lateral extrastriolar macula, with slices through the type II hair cell (HC) nuclear layer (k), the type I hair cell (HC) nuclear layer (l), the supporting cell (SC) nuclear layer (m), or the stroma (n). Arrows in k, l point to tdTomato-labeled cells. Scale bar in a = 150 μm for a, 25 μm for c, d, and 8 μm for d–f. Scale bar in g = 150 μm and applies to g–j. Scale bar in k = 5 μm and applies to k–n
CreER Lines Targeting Vestibular Supporting Cells
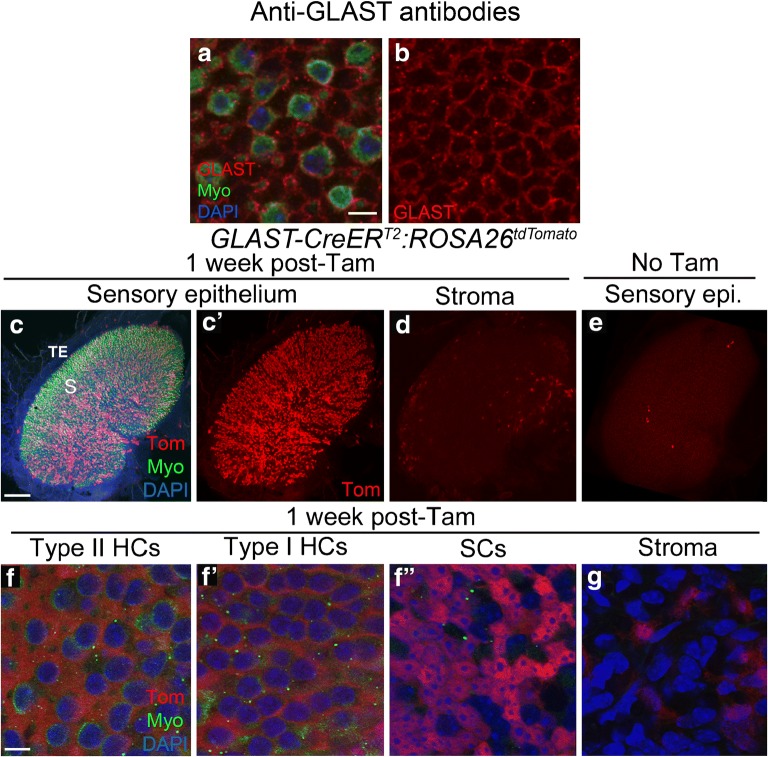
Glutamate-aspartate transporter (GLAST or SLC1A3) is a high-affinity reuptake pump for glutamate and aspartate often expressed in glia (reviewed in Takayasu et al. 2009). In the vestibular system, GLAST is expressed in the membranes of supporting cells (Takumi et al. 1997; Hartman et al. 2010). With antibodies, we localized GLAST to the membranes of supporting cells (Fig. 5a, b), but given the nature of the labeling, we were unable to determine numbers of supporting cells that were labeled or if GLAST was also expressed in the membranes of hair cells. GLAST-CreERT2 is a transgenic line that used a bacterial artificial chromosome (BAC) containing the GLAST promoter (Wang et al. 2012). After tamoxifen induction at 6 weeks of age in GLAST-CreERT2:ROSA26tdTomato mice, tdTomato was expressed in 2276 ± 1418 cells per utricle, of which 2270 ± 1408 were supporting cells and 6.6 ± 10.9 were hair cells, mostly type Is (Fig. 5c, c’, f-f”; Table 1). We estimated that on average, Cre was active in ~ 57 % of supporting cells. The high variability in tdTomato labeling of supporting cells occurred because one of the five samples had a ~ 10-fold lower number of tdTomato-positive cells. Also, tdTomato-positive hair cells were only detected in two of the five samples. We detected no significant difference in the density of tdTomato-labeled cells in peripheral regions versus central regions (unpaired Student’s t test, t(7) = 0.343, p = 0.742). Some cells in the stroma (Fig. 5d, g) and transitional epithelium (Fig. 5c, c’) also expressed tdTomato. We observed 6.3 ± 10.59 labeled supporting cells and 2.8 ± 5.50 labeled hair cells per utricle in controls that were not given tamoxifen (Fig. 5e; Table 1).
Fig. 5.
GLAST-CreERT2 mice have inducible Cre activity in supporting cells. a Confocal images showing immunolabeling for GLAST (red), myosin VIIa (Myo, green), and nuclei (DAPI, blue) in slices focused on the type II hair cell nuclear layer. b GLAST labeling only in same field as shown in a. c-d Confocal images of the utricle from GLAST-CreERT2:ROSA26tdTomato mice injected with tamoxifen, focused on either the macula (c,c') or the stroma (d). c and c' show the same view, with labeling for tdTomato (Tom, red), myosin VIIa (Myo, green), and DAPI (blue) shown in c and Tom only shown in c'.S = approximate position of the striola, TE = transitional epithelium. e Utricle from a GLAST-CreERT2:ROSA26tdTomato mouse that did not receive tamoxifen, focused on the sensory epithelium. f-g Confocal slices through the lateral extrastriolar region, with slices through the type II hair cell (HC) nuclear layer f), the type I hair cell (HC) nuclear layer (f'), the supporting cell (SC) nuclear layer (f"), or the stroma (g). Scale bar in a = 5 μm and applies to a,b. Scale bar in c = 150 μm and applies to c-e. Scale bar in f = 5 μm and applies to f-g
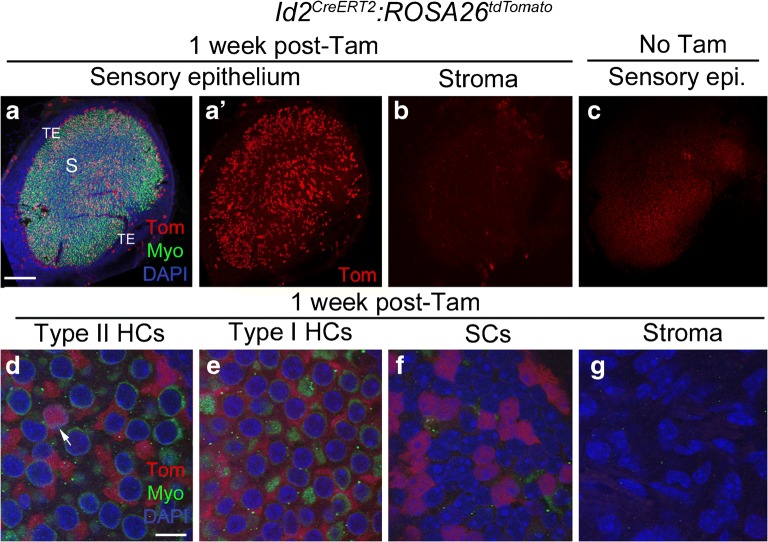
Inhibitor of DNA binding 2 (Id2) is a basic helix loop helix transcription factor that regulates cell differentiation (reviewed in Ling et al. 2014). Its expression pattern has not been described for the mammalian vestibular system. However, Id2 transcripts have been detected in cochlear supporting cells in mice (Jones et al. 2006) and in the chicken utricular macula (Ku et al. 2014). We were unable to find working antibodies to detect Id2 in whole-mount utricles. Id2CreERT2 is a knock-in allele where CreERT2 was inserted into the initiation codon of the endogenous Id2 locus and therefore only one copy of Id2 is expressed (Rawlins et al. 2009). We observed no difference in the density or distribution of hair cells or supporting cells, or any change in motor behavior, which would indicate a phenotypic abnormality due to Id2 haploinsufficiency. The expression pattern of Id2CreERT2:ROSA26tdTomato mice after tamoxifen induction at 6 weeks of age was similar to GLAST-CreERT2:ROSA26tdTomato, but significantly fewer cells were labeled. In Id2CreERT2:ROSA26tdTomato mice, tdTomato was expressed in 1172 ± 440 cells per utricle, of which 1159 ± 416 were supporting cells and 13.9 ± 24.0 were type II hair cells (Fig. 6a, a’, d–f; Table 1). Approximately 29 % of supporting cells per utricle were tdTomato-positive. We detected no significant difference in the density of tdTomato-labeled cells in central regions compared to peripheral regions (unpaired Student’s t test, t(4) = 0.617, p = 0.570). A small number of cells in the stroma (Fig. 6b, g) and the transitional epithelium (Fig. 6a, a’) also expressed tdTomato. In controls that were not given tamoxifen, we observed 0.67 ± 0.58 tdTomato-labeled supporting cells and 0.33 ± 0.58 labeled hair cells per utricle (Fig. 6c; Table 1).
Fig. 6.
Id2CreERT2 mice have inducible Cre activity in supporting cells. a-cConfocal images of utricles from Id2CreERT2:ROSA26tdTomato mice injected with tamoxifen, focused on either the macula (a,a') or stroma (b). a and a' show the same view, with labeling for tdTomato (Tom, red), myosin VIIa (Myo, green), and DAPI (blue) shown in a and Tom only shown in a'. S = approximate position of the striola, TE = transitional epithelium. b shows Tom labeling in the stroma. c Utricle from an Id2CreERT2:ROSA26tdTomato mouse that did not receive tamoxifen, focused on the sensory epithelium. d-g Confocal slices showing higher magnification views of the lateral extrastriolar region. d shows a slice through the type II hair cell (HC) layer, e shows a slice through the type I hair cell (HC) layer, f shows a slice through the supporting cell (SC) layer, and g shows a slice through the stroma. Arrow in d indicates a Tomato-positive type II HC. Scale bar in a = 150 μm and applies to a-c. Scale bar in d = 5 μm and applies to d-g
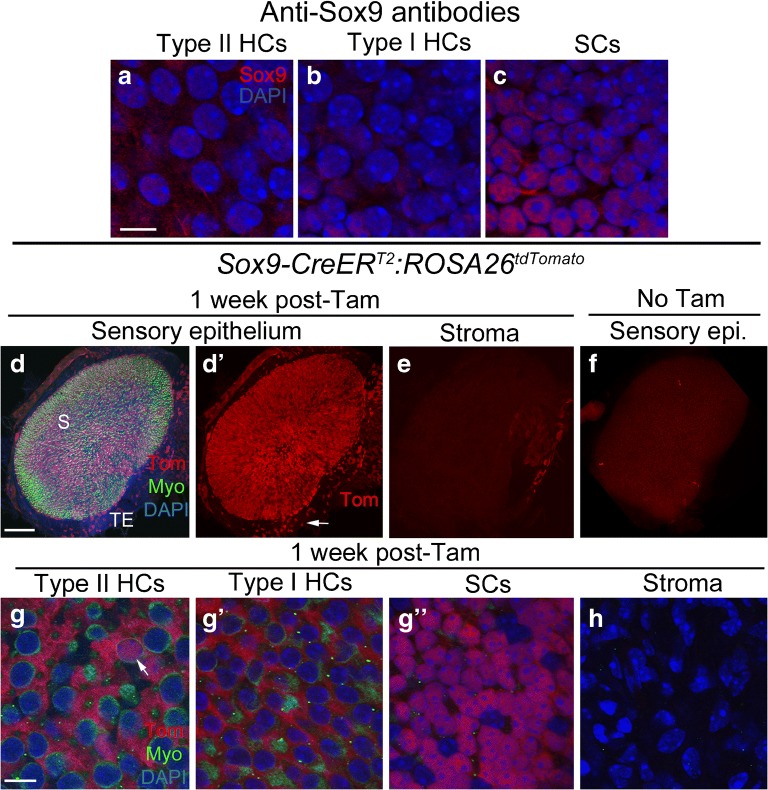
Sox9 is a HMG box domain transcription factor that has been immunolocalized to supporting cells in utricles of mature NMRI outbred mice (Loponen et al. 2011). Immunolabeling of utricles from mixed background mice using the same antibody revealed labeling in all supporting cell nuclei, with no significant labeling in hair cells (Fig. 7a–c). Sox9-CreERT2 is a transgenic line that used a BAC containing the entire mouse Sox9 sequence with CreERT2 inserted after the start codon (Kopp et al. 2011). Similar to GLAST-CreERT2:ROSA26tdTomato and Id2CreERT2:ROSA26tdTomato mice, the vast majority of tdTomato-positive cells were supporting cells in Sox9-CreERT2:ROSA26tdTomato mice following tamoxifen injection at 6 weeks of age. In Sox9-CreERT2:ROSA26tdTomato mice, tdTomato was expressed in 3621 ± 620 cells per utricle, of which 3483 ± 556 were supporting cells and 138.3 ± 64.9 were hair cells, mostly type IIs (Fig. 7d, d’, g–g”; Table 1). We estimated that ~ 87 % of supporting cells per utricle were tdTomato-labeled. Analysis of densities revealed no difference between central versus peripheral regions (unpaired Student’s t test, t(6) = 1.485, p = 0.188). A significant number of cells in the transitional epithelium expressed tdTomato (Fig. 7d, d’), but there was no tdTomato expression in stromal cells (Fig. 7e, h). In controls that were not given tamoxifen, we observed 2.3 ± 0.58 labeled hair cells and 1.3 ± 1.53 labeled supporting cells per utricle (Fig. 7f; Table 1).
Fig. 7.
Sox9-CreERT2 mice have inducible Cre activity in supporting cells. a-c Confocal images showing immunolabeling for Sox9 (red) and nuclei (DAPI, blue) in slices focused on the nuclei of type II hair cells (HCs) (a), type I hair cells (HCs) (b), and supporting cells (SCs) (c) of the utricular macula. d-eConfocal images of the surface of utricle from Sox9-CreERT2:ROSA26tdTomato mice injected with tamoxifen, focused on either the sensory epithelium (d,d') or stroma (e). d and d' show the same view, with labeling for tdTomato (Tom, red), myosin VIIa (Myo, green), and DAPI (blue) shown in d and Tom only shown in d'. S = approximate position of the striola, TE = transitional epithelium. Arrow in d' points to labeled cells in the TE. eshows Tom labeling in the stroma. f Utricle from a Sox9-CreERT2:ROSA26tdTomato mouse that did not receive tamoxifen, focused on the macula. g-h Confocal slices through the lateral extrastriolar region, with slices through the type II hair cell (HC) nuclear layer (g), the type I hair cell (HC) nuclear layer (g'), the supporting cell (SC) nuclear layer (g"), or the stroma (h). Arrow in g indicates a Tomato-positive type II HC. Scale bar in a = 5 μm and applies to a-c. Scale bar in d = 150 μm and applies to d-f. Scale bar in g = 5 μm and applies to g-h
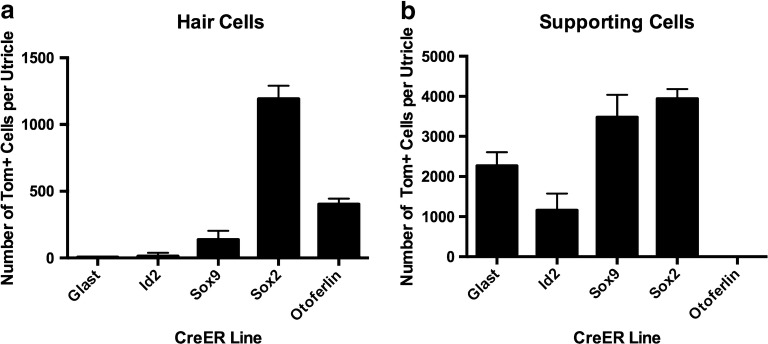
Comparisons of CreER Efficiencies in Utricular Hair Cells and Supporting Cells in GLAST-CreERT2, Id2CreERT2, Sox9-CreERT2, Sox2CreERT2, and OtoferlinCreERT2 Mouse Lines
Figure 8 and Table 1 show quantitative data for tdTomato-positive hair cells and supporting cells in the five best-performing CreER lines we examined, which we define as having the highest cell specificity, the fewest non-macular cells with Cre activity, and the smallest level of Cre leakiness. Of the two lines with substantial Cre activity in hair cells (Sox2CreERT2 and OtoferlinCreERT2), Sox2CreERT2 had the highest efficiency (Fig. 8a), but it also labeled nearly 100 % of supporting cells (Fig. 8b) and had some Cre leakiness (Fig. 2f; Table 1). By contrast, OtoferlinCreERT2 had no Cre activity in supporting cells (Fig. 8b) and no Cre leakiness (Fig. 4j; Table 1).
Fig. 8.
Graphs comparing Cre efficiencies in hair cells and supporting cells for several lines. Each column represents average number of tdTomato-labeled cells per utricle (+ 1 standard deviation) for hair cells (a) and supporting cells (b) from each CreER line. TdTomato-positive cells were quantified from two central regions and two peripheral regions, which together comprised 24 % of the total macula’s area. Conversion to number of labeled cells per utricle is described in the “Materials and Methods” section
All five lines, with the exception of OtoferlinCreERT2, showed Cre activity in supporting cells. Cre activity in GLAST-CreERT2, Id2CreERT2, and Sox9-CreERT2 was restricted largely to supporting cells (Fig. 8b), with very few Cre-expressing hair cells (Fig. 8a), while in Sox2CreERT2 mice, ~ 32 % of hair cells (mostly type IIs) and ~ 99 % of supporting cells had Cre activity (Fig. 8). Furthermore, GLAST-CreERT2, Id2CreERT2, and Sox2CreERT2 had some Cre-expressing cells in the stroma (Figs. 2e, h; 5d, g; 6b, g); while Sox9-CreERT2 had few, if any Cre-positive cells in the stroma (Fig. 7e, h).
CreER efficiencies may vary according to the gender of the parent from which the CreER allele was inherited (Lomeli et al. 2000; Hayashi et al. 2003; Cochrane et al. 2007; Gallardo et al. 2007; Heffner et al. 2012). We tracked parental inheritance for all mice upon which we performed quantitative data analysis (Table 2). However, we could not effectively test correlations between inheritance and CreER activity in any of the lines, with or without tamoxifen treatment, because animal numbers were too small. Similarly, although we tracked the gender of each mouse, we could not assess effects of gender upon Cre efficiency, given the small animal numbers.
Table 2.
Raw data, gender, and CreER inheritance for each animal in which quantitative analysis was performed
| CreER Lineb | No Tam | Tam at 6 weeks | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample no. | Gender | Labeled SCsa | Labeled HCsa | CreER inheritance | Sample no. | Gender | Labeled SCs | Labeled HCs | CreER inheritance | |
| GLAST-CreER T2 | 3324 | F | 0 | 11 | Maternal | 3480 | M | 1635 | 25 | Paternal |
| 3325 | F | 0 | 0 | Maternal | 3483 | F | 3399 | 0 | Paternal | |
| 3327 | F | 22 | 0 | Maternal | 3305 | F | 3910 | 0 | Paternal | |
| 3943 | M | 3 | 0 | Maternal | 3942 | F | 404 | 8 | Maternal | |
| 3945 | F | 2001 | 0 | Maternal | ||||||
| Id2 CreERT2 | 4345 | M | 0 | 1 | Paternal | 3860 | M | 1614 | 0 | Maternal |
| 4347 | M | 1 | 0 | Paternal | 3861 | M | 799 | 0 | Maternal | |
| 4348 | F | 1 | 0 | Paternal | 3895 | M | 1065 | 42 | Maternal | |
| Sox9-CreER T2 | 3435 | M | 0 | 3 | Maternal | 3434 | M | 3790 | 220 | Maternal |
| 3441 | M | 3 | 2 | Maternal | 3283 | F | 3960 | 158 | Maternal | |
| 4113 | M | 1 | 2 | Maternal | 4109 | F | 2704 | 100 | Maternal | |
| 4112 | F | 3478 | 75 | Maternal | ||||||
| Sox2 CreERT2 | 4680 | F | 68 | 16 | Paternal | 5438 | M | 3698 | 1140 | Paternal |
| 4684 | M | 120 | 6 | Paternal | 5436 | F | 4172 | 1131 | Paternal | |
| 5439 | F | 67 | 5 | Paternal | 5434 | M | 3964 | 1306 | Paternal | |
| 5440 | F | 40 | 5 | Paternal | ||||||
| 5442 | F | 16 | 0 | Paternal | ||||||
| Otoferlin CreERT2 | B46 | F | 0 | 0 | Maternal | B38 | M | 0 | 416 | Maternal |
| B315 | F | 0 | 0 | Maternal | B346 | F | 0 | 436 | Maternal | |
| B516 | M | 0 | 0 | Maternal | B273 | M | 0 | 358 | Maternal | |
| B528 | F | 0 | 0 | Maternal | Maternal | |||||
aEstimated total number of labeled cells per utricle
bSox2, Otoferlin, and Id2 CreER lines are knock-ins and designated by the following nomenclature: XXXCreERT2. GLAST and Sox9 CreER lines are transgenics and designated by the following nomenclature: XXX-CreERT2
CreER Lines with Sparse Expression Patterns in the Vestibular Sensory Epithelium
Prospero Homeobox 1 (Prox1) is a homeodomain transcription factor that plays a role in the development of many organs and in tumorigenesis (reviewed in Elsir et al. 2012). Prox1 protein is detected in utricular supporting cells at embryonic but not postnatal ages (Bermingham-McDonogh et al. 2006; Kirjavainen et al. 2008). Prox1CreERT2 is a knock-in allele where CreERT2 was inserted after an internal ribosome entry site (IRES) in intron 2 of endogenous Prox1 (Srinivasan et al. 2007). Therefore, both copies of Prox1 should be expressed; however, the level of Prox1 expression may be reduced. We observed no difference in the density or distribution of hair cells or supporting cells, or any change in motor behavior, which would indicate a phenotypic abnormality due to Prox1 haploinsufficiency. After tamoxifen induction at 6 weeks of age in Prox1CreERT2:ROSA26tdTomato mice, we observed 1 ± 1 tdTomato-positive hair cell (Fig. 9a). However, we did a quick analysis of the lateral ampulla and found that several tdTomato-positive hair cells and supporting cells were detected in the crista (Fig. 9b). These data were not quantified. In controls that were not given tamoxifen, no tdTomato-positive cells were observed (data not shown).
Fig. 9.
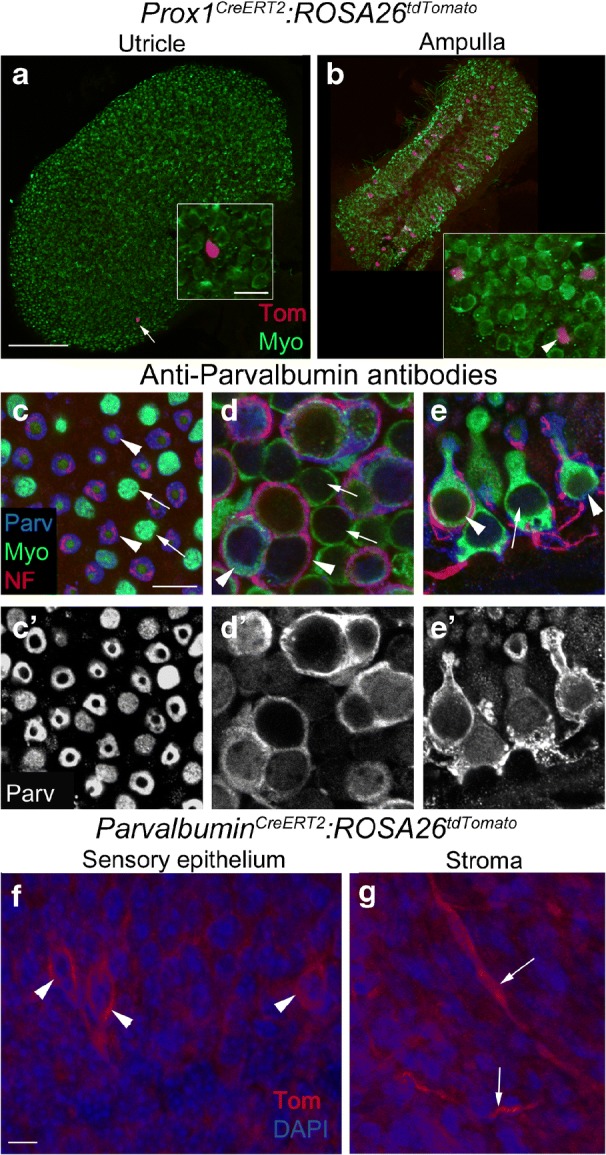
Cre activity is minimal in the utricular macula in Prox1CreERT2 and ParvalbuminCreERT2 lines. a,b Confocal images of the utricle (a) and the horizontal ampullae (b) from Prox1CreERT2:ROSA26tdTomato mice injected with tamoxifen and labeled with tdTomato (Tom, red) and antibodies to myosin VIIA (Myo, green). Arrow in a points to one Tom-labeled cell. Inset shows the same cell, which is located in the type II hair cell layer and is Myo-positive, although the strong Tom labeling obscures the Myo labeling. Inset in b shows an area from the ampulla at higher magnification, with three labeled cells (2 hair cells and 1 supporting cell—arrowhead). c-e' Confocal slices showing immunolabeling for parvalbumin (Parv, blue), myosin VIIa (Myo, green), and 200 kD neurofilament (NF, red), focused on the tops of the hair cells (c), type I and II hair cell nuclei in the striola (d), and the edge of the lateral extrastriola, which affords side views of type I and II hair cells (e). Arrowheads in c-e point to type I hair cells, while arrows point to type II hair cells. c'-e' show parvalbumin (Parv, white) staining alone. f,g Confocal slices from utricles of ParvalbuminCreERT2:ROSA26tdTomato mice injected with tamoxifen, showing tdTomato (Tom, red) labeling in calyces (arrowheads) in the sensory epithelium (f) and nerve fibers (arrows) in the stroma (g). Scale bar in a = 150 μm and applies to a,b. Scale bar in a inset = 10 μm and applies to insets in a,b. Scale bar in c= 5 μm and applies to c-e'. Scale bar in f = 5 μm and applies to f,g
Parvalbumin is a calcium-binding protein that has been localized by immunostaining to centrally located type I hair cells of the vestibular organs of neonatal rodents, as well as in vestibular ganglion neurons and neurites in developing and mature rodents (Demêmes et al. 1993; Raymond et al. 1993; Pujol et al. 2014). In adult mouse utricles from a mixed background strain, antibodies to parvalbumin labeled afferent fibers and endings, as well as some hair cell nuclei (Fig. 9c–e’; Bucks et al. 2017). ParvalbuminCreERT2 is a knock-in line where CreERT2 was inserted into the endogenous Parvalbumin locus and therefore only one copy of Parvalbumin is expressed (Taniguchi et al. 2011). We observed no difference in the density or distribution of hair cells or supporting cells, or any change in motor behavior, which would indicate a phenotypic abnormality due to Parvalbumin haploinsufficiency. After tamoxifen induction at 6 weeks of age in ParvalbuminCreERT2:ROSA26tdTomato mice, we observed tdTomato expression in 2 to 19 afferent nerve calyces that surrounded type I hair cells in the striola, including some likely multi-calyx afferents (Fig. 9f), and their associated nerve fibers. No tdTomato-positive hair cells or supporting cells were observed and there were a small number of tdTomato-positive cells in the stroma (Fig. 9g). In controls that were not given tamoxifen, no tdTomato-positive cells were observed (data not shown).
CreER Lines with No Expression in the Vestibular Sensory Epithelium
Fibroblast growth factor receptor 3 (Fgfr3) is one of five receptors in the FGF family, which plays a key role in morphogenesis and sensorineural differentiation in the developing ear (reviewed in Fritzsch et al. 2006). Fgfr3 is expressed in supporting cells in the mouse cochlea (e.g., Hayashi et al. 2007). Its expression pattern has not been described for the mammalian vestibular system. However, in the chicken utricle, Fgfr3 was detected by RNA-Seq (Ku et al. 2014), but not by in situ hybridization (Bermingham-McDonogh et al. 2001). Fgfr3i-CreERT2 is a transgenic line where an improved version of CreER (iCreER) was inserted into the first exon of Fgfr3 in a phage artificial chromosome (PAC) (Rivers et al. 2008; Young et al. 2010). After tamoxifen induction at 6 weeks of age in Fgfr3-iCreERT2:ROSA26tdTomato mice, no tdTomato-positive cells were observed in the sensory epithelium of the utricle, but numerous star-shaped tdTomato-positive cells were observed in the stroma (data not shown). Considering the possibility that the labeled cells could be macrophages, we labeled utricles with an antibody to ionized calcium-binding adaptor molecule 1 (Iba1), a macrophage marker, but there was no co-labeling with tdTomato. Thus, the identity of these cells is not currently known.
Glial fibrillary acidic protein (GFAP) is an intermediate filament protein that is expressed in astrocytes (reviewed in Pekny and Pekna 2004). It is also expressed in utricular supporting cells in the peripheral, extrastriolar region at postnatal ages (Rio et al. 2002). We investigated the expression pattern of two transgenic CreER lines that use the human GFAP promoter to drive CreER expression but have different insertion sites in the genome (GFAP-A-CreER™ and GFAP-B-CreER™) (Chow et al. 2008). There were no tdTomato-positive cells observed in the utricle of either GFAP-A-CreER™:ROSA26tdTomato mice or GFAP-B-CreER™:ROSA26tdTomato mice after tamoxifen induction at 6 weeks of age (data not shown).
Discussion
CreER/loxP is a powerful genetic system that allows dissection of gene functions that may differ across development and in maturity. Deletion or overexpression of specific genes with CreER/loxP occurs in a cell type-specific and time-controlled manner, which allows for further understanding of tissue development and gene function. Fate-mapping of different cells types can also be achieved using CreER/loxP and is important for understanding development, repair, and regeneration processes. The present study characterized the Cre expression pattern of 11 CreER alleles in the utricle of adult mice. We found that five lines—GLAST-CreERT2, Id2CreERT2, OtoferlinCreERT2, Sox2CreERT2, and Sox9-CreERT2—had cell-selective, inducible Cre activity with little leakiness, comprising effective new tools for researchers who wish to employ conditional and inducible genetic manipulation to study the vestibular periphery in adult mammals. We also report that the other 6 CreER lines we studied have little to no CreER activity in the adult utricle or, in the case of Calbindin2CreERT2 mice, are not inducible in adult mice under the conditions used in the study.
Supporting cells in the adult utricle were targeted by GLAST-CreERT2, Id2CreERT2, Sox2CreERT2, and Sox9-CreERT2. However, each CreER line varied in the amount of supporting cells with CreER activity. Specifically, we estimate that Sox2CreERT2, Sox9-CreERT2, GLAST-CreERT2, and Id2CreERT2 had inducible CreER activity in ~ 99, ~ 87, ~ 57, and ~ 29 % of supporting cells, respectively. This variability across lines may reflect slightly different expression patterns of these 4 genes and could suggest that not all supporting cells in the utricle are the same. This is most plausible with the two knock-in lines—Id2CreERT2 and Sox2CreERT2—since Cre expression is controlled by endogenous regulatory elements in these lines. However, it is also possible that the tdTomato labeling pattern is not an accurate reflection of native gene expression in Sox9-CreERT2 and GLAST-CreERT2 lines, which are transgenics where the promoter used to drive CreER may not contain all regulatory elements used by the endogenous genes. Also, positional effects may have occurred where the transgene insertion site within the genome is near repressor or enhancer elements that affect CreER expression (Jaenisch et al. 1981; Wilson et al. 1990). For GLAST-CreERT2, Sox2CreERT2, and Sox9-CreERT2 lines, tdTomato expression was generally consistent with antibody labeling, which supports the notion that Cre expression reflects native gene expression. However, we were unable to make this assessment with Id2CreERT2, since we were not able to find antibodies for immunolabeling of Id2.
Very little is known about vestibular supporting cell properties and whether there are subpopulations of supporting cells with discrete functional, molecular, or physiological properties. Indeed, supporting cells across the adult utricle share many common features, including cytoplasmic ultrastructure, nuclear shape and position, chromatin and nucleolar properties. A few markers show non-uniform distribution across the utricle. For instance, a Hes5 reporter mouse showed higher levels of reporter expression in supporting cells located in the central region of the utricle in maturity (Hartman et al. 2009). It is interesting that hair cells are regenerated mainly in the lateral extrastriolar region in adult mouse utricles after near-complete hair cell destruction (Golub et al. 2012), which suggests that supporting cells in different regions may have variable capacity to serve as hair cell progenitors. Unique regional capacities for regeneration have been reported among supporting cells in other hair cell epithelia, including lateral line neuromasts (Romero-Carvajal et al. 2015) and avian auditory organs (Cafaro et al. 2007).
In mice that received tamoxifen, we detected small numbers of tdTomato-labeled hair cells in Sox9-CreERT2, GLAST-CreERT2, and Id2CreERT2 lines. Numbers were greater than those seen in no-tamoxifen controls. These labeled hair cells may reflect supporting cell conversion into hair cells, which occurs normally in adult mouse utricles (Bucks et al. 2017). Because there is some Cre leakiness in each line, in both hair cells and supporting cells, it is possible that labeled hair cells were derived from tdTomato-positive supporting cells over long periods of time, rather than simply during the week after tamoxifen injection. It is also possible that some supporting cells with induced Cre converted into hair cells in the 7-day survival time post-tamoxifen. Finally, small number of hair cells may actually express each gene.
Our observation that only ~ 11 % of hair cells expressed tdTomato after tamoxifen in OtoferlinCreERT2 mice was surprising. Otoferlin regulates fusion of synaptic vesicles at the ribbon synapse (Moser and Starr 2016). In rodents, mature utricular hair cells—type I and II—have ribbon synapses (e.g., Rüsch and Eatock 1996; Nouvian et al. 2006; Pujol et al. 2014), and prior studies showed that otoferlin is immunolabeled in both hair cell types (Schug et al. 2006; Dulon et al. 2009). Since OtoferlinCreERT2 is a knock-in allele, we expected many more hair cells to be tdTomato-positive. We also detected otoferlin by immunostaining in hair cells throughout the utricle, but not all hair cells were labeled strongly and some seemed to lack otoferlin labeling. It is possible that the otoferlin protein has a slow turnover rate and the activity of the Otoferlin promoter may be low. Since we only injected tamoxifen twice (on two consecutive days) and the half-life of tamoxifen is ~ 16 h (Robinson et al. 1991), only cells with active Otoferlin promoters at the time of injection would show CreER activity. It is also possible that some vestibular hair cells use an alternative protein to otoferlin to mediate vesicle fusion. However, this is unlikely, because studies have shown that vesicle exocytosis in hair cells in Otoferlin−/− mice have slower kinetics, reduced calcium sensitivity, and nonlinear calcium dependence (Dulon et al. 2009), suggesting otoferlin function is critical in many vestibular hair cells. Similar explanations could be made for the small number of Cre-positive neurons in ParvalbuminCreERT2 mice, in which we expected to see many afferent neurons labeled, based on immunolabeling here and in prior papers (Demêmes et al. 1993; Pujol et al. 2014; Raymond et al. 1993).
The absence of CreER activity prior to tamoxifen induction is crucial for conditional gene studies because the effects of leakiness can accumulate over time and complicate the interpretation of results. Therefore, our characterization studies also included samples that expressed each CreER allele and the ROSA26tdTomato reporter line but did not receive tamoxifen to assess the level of leakiness. While the ligand-binding region of ER fused to Cre has been mutated to decrease its affinity for endogenous estrogen, it still retains a low affinity for estrogen which may cause leakage in some cells. CreER™ contains a single point mutation (G525R), while CreERT2 has three point mutations (G400V/M543A/L544A) in this region (Danielian et al. 1998; Indra et al. 1999). Therefore, one might predict that CreER™ alleles are more likely to be leaky. However, our results suggest that leakiness varies among individual CreER lines, because of the nine CreERT2 alleles we investigated, there were three different results. No Cre leakiness was observed in Fgfr3-iCreERT2, OtoferlinCreERT2, ParvalbuminCreERT2, and Prox1CreERT2 mice; a small degree of leakiness was seen in GLAST-CreERT2, Id2CreERT2, Sox2CreERT2, and Sox9-CreERT2 lines; and the same pattern of tdTomato expression with or without tamoxifen was observed in CalbindinCreERT2. These differences may have been caused by the expression level of CreER. Previous studies showed that CreER lines with higher levels of promotor activity and thus high levels of CreER protein are more likely to be leaky (Buelow and Scharenberg 2008). Others have reported evidence of proteolysis, where ER is cleaved from Cre in some cells, as the source of leakiness (Nakamura et al. 2006). There are also reports of differential CreER leakiness based on parental inheritance where Cre recombination occurs spontaneously in the germline of one parent. Several studies have shown this to occur more frequently when the female parent transmits Cre to the offspring (Lomeli et al. 2000; Hayashi et al. 2003; Cochrane et al. 2007; Gallardo et al. 2007; Heffner et al. 2012). However we were not able to assess this in our study due to small sample size (Table 2). Regardless of the cause, our results suggest that CalbindinCreERT2 is not inducible in the adult utricle and additional controls not injected with tamoxifen are needed for experiments using GLAST-CreERT2, Id2CreERT2, Sox2CreERT2, and Sox9-CreERT2 lines due to a low level of leakiness.
Four of the CreER lines we investigated—Id2CreERT2, OtoferlinCreERT2, Prox1CreERT2, and Sox2CreERT2—are knock-in alleles, where CreER replaced the coding region of the endogenous gene. Since we used heterozygotes for this study, only one copy of the endogenous gene is present and if these lines were bred as homozygotes, they would in essence be a germline knockout. While we did not detect any obvious differences in utricle histology or motor behavior from these CreER lines, we did not test vestibular function and others have reported haploinsufficient phenotypes in some knock-in Cre lines (Siegenthaler et al. 2008; Atkinson et al. 2018). Therefore, if any of these lines are used to delete or overexpress a gene, Cre-positive controls (either without tamoxifen injection or without the floxed allele) would be important to make proper conclusions of the data.
In summary, our study has identified five CreER lines with cell type-specific and inducible expression of CreER in the adult utricle in supporting cells and/or hair cells. These lines constitute new genetic tools for investigators to manipulate gene expression or to fate-map cells in the vestibular periphery of adult mice.
Acknowledgements
This work was supported by the National Institutes of Health (F32 DC013695 to SAB, R01 DC013771 to JSS, R01 DC03696 to JSS, R01 DC014441 to BCC, and P30 DC04661 to the UW Research Core Center), the Office of Naval Research (N00014-13-1-0569 to BCC), the Office of the Assistant Secretary of Defense for Health Affairs (W81XWH-15-1-0475 to BCC), and the Wellcome Trust (089015 to MML). We thank Tot Nguyen, Irina Omelchenko, and Jialin Shang from the University of Washington, and Michelle Randle and Kaley Graves from Southern Illinois University School of Medicine, for technical assistance. We are grateful to Dr. Suzanne Baker (St. Jude Children’s Research Hospital) for sharing GFAP-A-CreER™ and GFAP-B-CreER™ mice; Dr. Ulrich Müller (The Scripps Research Institute) for sharing OtoferlinCreERT2 mice; Dr. Guillermo Oliver (St. Jude Children’s Research Hospital) for sharing Prox1CreERT2 mice; and Dr. William Richardson (University College London) for sharing Fgfr3-iCreERT2 mice. We thank Dr. Jian Zuo (St. Jude Children’s Research Hospital) for his support of the project which began when Marcia Mellado Lagarde and Brandon Cox were postdoctoral fellows in his lab.
Compliance with Ethical Standards
Conflict of Interest
Brandon C. Cox is a consultant with Turner Scientific, LLC. No other authors have any professional or financial affiliations that may be perceived as a conflict of interest.
References
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R. Sox2+ adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9(4):317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson PJ, Dong Y, Gu S, Liu W, Udagawa T, Cheng AG (2018) Sox2 haploinsufficiency primes regeneration and Wnt-responsiveness in the mouse cochlea. J Clin Invest in press [DOI] [PMC free article] [PubMed]
- Bermingham-McDonogh O, Stone JS, Reh TA, Rubel EW. FGFR3 expression during development and regeneration of the chick inner ear sensory epithelia. Dev Biol. 2001;238(2):247–259. doi: 10.1006/dbio.2001.0412. [DOI] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, Hayashi T. Expression of Prox1 during mouse cochlear development. J Comp Neurol. 2006;496(2):172–186. doi: 10.1002/cne.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucks SA, Cox BC, Vlosich BA, Manning JP, Nguyen TB, Stone JS. Supporting cells remove and replace sensory receptor hair cells in a balance organ of adult mice. elife. 2017;6:e18128. doi: 10.7554/eLife.18128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelow B, Scharenberg AM. Characterization of parameters required for effective use of tamoxifen-regulated recombination. Edited by Sebastian D. Fugmann. PLoS One. 2008;3(9):e3264. doi: 10.1371/journal.pone.0003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Cox BC, Thiede BR, Zuo J, Corwin JT. In vivo proliferative regeneration of balance hair cells in newborn mice. J Neurosci. 2012;32(19):6570–6577. doi: 10.1523/JNEUROSCI.6274-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, On D, Baker W, Collado MS, Corwin JT. Over half the hair cells in the mouse utricle first appear after birth, with significant numbers originating from early postnatal mitotic production in peripheral and Striolar growth zones. J Ass Res Otolaryngol. 2012;13(5):609–627. doi: 10.1007/s10162-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro J, Lee GS, Stone JS. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev Dyn. 2007;236(1):156–170. doi: 10.1002/dvdy.21023. [DOI] [PubMed] [Google Scholar]
- Chow LM, Zhang J, Baker SJ. Inducible Cre recombinase activity in mouse mature astrocytes and adult neural precursor cells. Transgenic Res. 2008;17(5):919–928. doi: 10.1007/s11248-008-9185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane RL, Clark SH, Harris A, Kream BE. Rearrangement of a conditional allele regardless of inheritance of a Cre recombinase transgene. Genesis. 2007;45(1):17–20. doi: 10.1002/dvg.20259. [DOI] [PubMed] [Google Scholar]
- Cox BC, Liu Z, Mellado Lagarde MM, Zuo J. Conditional gene expression in the mouse inner ear using Cre-loxP. J Ass Res Otolaryngol. 2012;13(3):295–322. doi: 10.1007/s10162-012-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen D-H, Chalasani K, Steigelman KA, Fang J, Rubel EW, Cheng AG, Zuo J. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141(4):816–829. doi: 10.1242/dev.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8(24):1323–13S2. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dechesne CJ, Winsky L, Kim HN, Goping G, Vu TD, Wenthold RJ, Jacobowitz DM. Identification and ultrastructural localization of a calretinin-like calcium-binding protein (protein 10) in the guinea pig and rat inner ear. Brain Res. 1991;560(1–2):139–148. doi: 10.1016/0006-8993(91)91224-o. [DOI] [PubMed] [Google Scholar]
- Demêmes D, Eybalin M, Renard N. Cellular distribution of parvalbumin immunoreactivity in the peripheral vestibular system of three rodents. Cell Tissue Res. 1993;274(3):487–492. doi: 10.1007/BF00314545. [DOI] [PubMed] [Google Scholar]
- Desai SS, Zeh C, Lysakowski A. Comparative morphology of rodent vestibular periphery. I. Saccular and utricular maculae. J Neurophysiol. 2005;93(1):251–266. doi: 10.1152/jn.00746.2003. [DOI] [PubMed] [Google Scholar]
- Dulon D, Safieddine S, Jones SM, Petit C. Otoferlin is critical for a highly sensitive and linear calcium-dependent exocytosis at vestibular hair cell ribbon synapses. J Neurosci. 2009;29(34):10474–10487. doi: 10.1523/JNEUROSCI.1009-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorakova M, Jahan I, Macova I, Chumak T, Bohuslavova R, Syka J, Fritzsch B, Pavlinkova G. Incomplete and delayed Sox2 deletion defines residual ear neurosensory development and maintenance. Sci Rep. 2016;6(1):38253. doi: 10.1038/srep38253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eatock RA, Songer JE. Vestibular hair cells and afferents: two channels for head motion signals. Annu Rev Neurosci. 2011;34:501–534. doi: 10.1146/annurev-neuro-061010-113710. [DOI] [PubMed] [Google Scholar]
- Elsir T, Smits A, Lindström MS, Nistér M. Transcription factor PROX1: its role in development and cancer. Cancer Metastasis Rev. 2012;31(3–4):793–805. doi: 10.1007/s10555-012-9390-8. [DOI] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93(20):10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pauley S, Beisel KW. Cells, molecules and morphogenesis: the making of the vertebrate ear. Brain Res. 2006;1091(1):151–171. doi: 10.1016/j.brainres.2006.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, vasa-Cre. Genesis. 2007;45(6):413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub JS, Tong L, Ngyuen TB, Hume CR, Palmiter RD, Rubel EW, Stone JS. Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J Neurosci. 2012;32(43):15093–15105. doi: 10.1523/JNEUROSCI.1709-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Casati ME, Murtie J, Taylor B, Corfas G. Cell-specific inducible gene recombination in postnatal inner ear supporting cells and glia. J Ass Res Otolaryngol. 2010;11(1):19–26. doi: 10.1007/s10162-009-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman BH, Basak O, Nelson BR, Taylor V, Bermingham-Mcdonogh O, Reh TA. Hes5 expression in the postnatal and adult mouse inner ear and the drug-damaged cochlea. J Ass Res Otolaryngol. 2009;10(3):321–340. doi: 10.1007/s10162-009-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman BH, Reh TA, Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107(36):15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T, Mooseker MS. The growing family of myosin motors and their role in neurons and sensory cells. Curr Opin Neurobiol. 1997;7(5):615–623. doi: 10.1016/s0959-4388(97)80080-3. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Tensen T, McMahon AP. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis. 2003;37(2):51–53. doi: 10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Cunningham D, Bermingham-McDonogh O. Loss of FGFR3 leads to excess hair cell development in the mouse organ of corti. Devl Dyn. 2007;236(2):525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]
- Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, Donahue LR, Eppig JT, Murray SA. Supporting conditional mouse mutagenesis with a comprehensive Cre characterization resource. Nat Commun. 2012;3:1218. doi: 10.1038/ncomms2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the Tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27(22):4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Jähner D, Nobis P, Simon I, Löhler J, Harbers K, Grotkopp D. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell. 1981;24(2):519–529. doi: 10.1016/0092-8674(81)90343-3. [DOI] [PubMed] [Google Scholar]
- Jones JM, Montcouquiol M, Dabdoub A, Woods C, Kelley MW. Inhibitors of differentiation and DNA binding (ids) regulate Math1 and hair cell formation during the development of the organ of Corti. J Neurosci. 2006;26(2):550–558. doi: 10.1523/JNEUROSCI.3859-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KKH, Tang ASP, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KSE. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434(7036):1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kirjavainen A, Sulg M, Heyd F, Alitalo K, Ylä-Herttuala S, Möröy T, Petrova TV, Pirvola U. Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Devl Biol. 2008;322(1):33–45. doi: 10.1016/j.ydbio.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138(4):653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku Y-C, Renaud NA, Veile RA, Helms C, Voelker CCJ, Warchol ME, Lovett M. The transcriptome of utricle hair cell regeneration in the avian inner ear. J Neurosci. 2014;34(10):3523–3535. doi: 10.1523/JNEUROSCI.2606-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis. 2002;32(2):49–62. doi: 10.1002/gene.10068. [DOI] [PubMed] [Google Scholar]
- Lin J, Zhang X, Wu F, Lin W. Hair cell damage recruited Lgr5-expressing cells are hair cell progenitors in neonatal mouse utricle. Front Cell Neurosci. 2015;9:113. doi: 10.3389/fncel.2015.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F, Kang B, Sun X-H. Id proteins: small molecules, mighty regulators. Curr Top Devl Biol. 2014;110:189–216. doi: 10.1016/B978-0-12-405943-6.00005-1. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Ramos-Mejia V, Gertsenstein M, Lobe CG, Nagy A. Targeted insertion of Cre recombinase into the TRAP gene:excision in primordial germ cells. Genesis. 2000;26(2):116–117. [PubMed] [Google Scholar]
- Loponen H, Ylikoski J, Albrecht JH, Pirvola U. Restrictions in cell cycle progression of adult vestibular supporting cells in response to ectopic cyclin D1 expression. PLoS One. 2011;6(11):e27360. doi: 10.1371/journal.pone.0027360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser T, Starr A. Auditory neuropathy—neural and synaptic mechanisms. Nat Rev Neurol. 2016;12(3):135–149. doi: 10.1038/nrneurol.2016.10. [DOI] [PubMed] [Google Scholar]
- Nakamura E, Nguyen M-T, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235(9):2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Nouvian R, Beutner D, Parsons TD, Moser T. Structure and function of the hair cell ribbon synapse. J Memb Biol. 2006;209(2–3):153–165. doi: 10.1007/s00232-005-0854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR. Sox2 and Jagged1 expression in normal and drug-damaged adult mouse inner ear. J Ass Res Otolaryngol. 2008;9(1):65–89. doi: 10.1007/s10162-007-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Pekna M. Astrocyte intermediate filaments in CNS pathologies and regeneration. J Pathol. 2004;204(4):428–437. doi: 10.1002/path.1645. [DOI] [PubMed] [Google Scholar]
- Pujol R, Pickett SB, Nguyen TB, Stone JS. Large basolateral processes on type II hair cells are novel processing units in mammalian vestibular organs. J Comp Neurol. 2014;522(14):3141–3159. doi: 10.1002/cne.23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BLM. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136(22):3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J, Dechesne CJ, Desmadryl G, Dememes D. Different calcium-binding proteins identify subpopulations of vestibular ganglion neurons in the rat. Acta Oto-Laryngol Supp. 1993;503:114–118. doi: 10.3109/00016489309128090. [DOI] [PubMed] [Google Scholar]
- Rio C, Dikkes P, Liberman MC, Corfas G. Glial fibrillary acidic protein expression and promoter activity in the inner ear of developing and adult mice. J Comp Neurol. 2002;442(2):156–162. doi: 10.1002/cne.10085. [DOI] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11(12):1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SP, Langan-Fahey SM, Johnson DA, Jordan VC. Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab Dispos. 1991;19(1):36–43. [PubMed] [Google Scholar]
- Romero-Carvajal A, Acedo JN, Jiang L, Kozlovskaja-Gumbriene A, Alexander R, Li H, Piotrowski T. Regeneration of sensory hair cells requires localized interactions between the notch and Wnt pathways. Devl Cell. 2015;34(3):267–282. doi: 10.1016/j.devcel.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux I, Safieddine S, Nouvian R, Grati M, Simmler M-C, Bahloul A, Perfettini I, et al. Otoferlin, defective in a human deafness form, is essential for exocytosis at the auditory ribbon synapse. Cell. 2006;127(2):277–289. doi: 10.1016/j.cell.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Rüsch A, Eatock RA. A delayed rectifier conductance in type I hair cells of the mouse utricle. J Neurophysiol. 1996;76(2):995–1004. doi: 10.1152/jn.1996.76.2.995. [DOI] [PubMed] [Google Scholar]
- Rüsch A, Lysakowski A, Eatock RA. Postnatal development of type I and type II hair cells in the mouse utricle: acquisition of voltage-gated conductances and differentiated morphology. J Neurosci. 1998;18(18):7487–7501. doi: 10.1523/JNEUROSCI.18-18-07487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug N, Braig C, Zimmermann U, Engel J, Winter H, Ruth P, Blin N, Pfister M, Kalbacher H, Knipper M. Differential expression of otoferlin in brain, vestibular system, immature and mature cochlea of the rat. Eur J Neurosci. 2006;24(12):3372–3380. doi: 10.1111/j.1460-9568.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex. 2008;18(8):1865–1875. doi: 10.1093/cercor/bhm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan RS, Dillard ME, Lagutin OV, Lin FJ, Tsai S, Tsai MJ, Samokhvalov IM, Oliver G. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev. 2007;21(19):2422–2432. doi: 10.1101/gad.1588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayasu Y, Iino M, Takatsuru Y, Tanaka K, Ozawa S. Functions of glutamate transporters in cerebellar Purkinje cell synapses. Acta Physiol. 2009;197(1):1–12. doi: 10.1111/j.1748-1716.2009.02019.x. [DOI] [PubMed] [Google Scholar]
- Takumi Y, Matsubara A, Danbolt NC, Laake JH, Storm-Mathisen J, Usami S, Shinkawa H, Ottersen OP. Discrete cellular and subcellular localization of glutamine synthetase and the glutamate transporter GLAST in the rat vestibular end organ. Neuroscience. 1997;79(4):1137–1144. doi: 10.1016/s0306-4522(97)00025-0. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71(6):995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rattner A, Zhou Y, Williams J, Smallwood PM, Nathans J. Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell. 2012;151(6):1332–1344. doi: 10.1016/j.cell.2012.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chai R, Kim GS, Pham N, Jansson L, Nguyen D-H, Kuo B, May LA, Zuo J, Cunningham LL, Cheng AG. Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in damaged neonatal mouse utricle. Nat Commun. 2015;6:6613. doi: 10.1038/ncomms7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6(1):679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- Wu J, Li W, Lin C, Chen Y, Cheng C, Sun S, Tang M, Chai R, Li H. Co-regulation of the notch and Wnt signaling pathways promotes supporting cell proliferation and hair cell regeneration in mouse utricles. Sci Rep. 2016;6(1):29418. doi: 10.1038/srep29418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KM, Mitsumori T, Pringle N, Grist M, Kessaris N, Richardson WD. An Fgfr3-iCreERT2 transgenic mouse line for studies of neural stem cells and astrocytes. Glia. 2010;58(8):943–953. doi: 10.1002/glia.20976. [DOI] [PMC free article] [PubMed] [Google Scholar]