Abstract
Barrier activity of skin and internal barrier-forming epithelial linings are conferred by a lipid-corneocyte structure (stratum corneum in skin).The integrity of the corneocytes depends on the outer cornified envelope and is essential for maintenance of barrier function. During epidermal development and differentiation, proteins are sequentially incorporated into the envelope via action of epidermal transglutaminases in a well documented process. However, recent knockouts of major cornified envelope constituents have failed to disrupt barrier function significantly, suggesting that additional unidentified components are involved. We report a new gene cluster in the epidermal differentiation complex at human 1q21 encoding a family of 18 proteins that are substrates for epidermal transglutaminases. These proteins incorporate into the cornified envelope late in development and late in the process of envelope maturation during epidermal differentiation. The genes cluster within the epidermal differentiation complex according to expression pattern, i.e., epidermally expressed proteins cluster together while proteins from internal barrier-forming epithelia also cluster. We propose that these proteins modulate barrier activity over the surface of the animal, in a manner analogous to that proposed for the well characterized cornified envelope precursors, the small proline-rich proteins. To emphasize the incorporation of these proteins late in envelope assembly, we call the human proteins late envelope proteins.
Epidermis provides a barrier to the external environment, an essential component of which is the keratinocyte cornified envelope. Epidermal keratinocytes undergo terminal differentiation involving keratin aggregation, nuclear degradation, and replacement of the plasma membrane with a tough, insoluble proteinaceous envelope that is cross-linked to extracellular lipid providing barrier function (reviewed in ref. 1). Cornified envelope assembly is well documented (2); however, recent knockout reports show that protein components previously considered integral are dispensable (3, 4), raising the possibility that additional proteins contribute to the envelope.
Models for cornified envelope structure and assembly have been proposed whereby the structural proteins involucrin, envoplakin, and periplakin are sequentially cross-linked to form an initial scaffold (2, 5, 6). Incorporation is catalyzed by epidermal transglutaminases (TGases) that promote formation of disulphide and (γ-glutamyl)lysine isopeptide bonds (reviewed in ref. 1). Subsequently, proteins such as loricrin, elafin, S100, and small proline-rich region proteins (SPRRs) are added to form a mature envelope.
SPRRs are a family of proteins that are substrates for TGase-mediated cross-linking of structural proteins into the cornified envelope (reviewed in refs. 1 and 7).The SPRR family comprises four groups that are differentially expressed in barrier-providing epithelia. Differential expression of SPRR proteins may modulate barrier activity over the surface of the animal (refs. 8–10 and references within) through modulation of envelope biomechanical properties (11). In addition, expression of some SPRR members is responsive to environmental stimuli [e.g., UV light, phorbol 12-myristate 13-acetate (PMA), cigarette smoke; reviewed in refs. 1 and 7], suggesting a role in modulating barrier response to environmental insult, reinforcing the notion that these proteins affect barrier quality.
There have been persistent reports of additional genes/proteins with structures homologous to cornified envelope proteins. These include XP5, XP31, XP32 (12), the newly identified component of the EDC (NICE-1; ref. 13), protein products of a range of annotated expressed sequence tags (ESTs; ref. 14), and Eig3 protein (15). We show that at least some of these proteins are encoded by a previously undetected gene cluster in the human epidermal differentiation complex (EDC) at 1q21 (16), with homologues detected in mouse (12, 14, 16). We show that these genes encode proteins, which are new cornified envelope constituents distinct from SPRRs. Like SPRRs, the genes are differentially expressed in different types of barrier epithelia. The human genes form subclusters on the chromosome associated with expression in either epidermal or internal epithelia.
These proteins are induced during keratinocyte terminal differentiation and relocate to the envelope very late in terminal differentiation where they are substrates for TGase-mediated envelope incorporation. During epidermal terminal differentiation, the proteins locate to the envelope much later than loricrin, among the last proteins to be incorporated (1). During development, the genes are expressed after SPRR, previously shown to be the last known structural envelope gene induced during fetal barrier formation (17).These proteins are detected only after assembly of the cornified envelope is advanced and after first barrier acquisition. Hence, we call the new family members late envelope proteins (LEPs) and propose that LEPs link to cornified envelope proteins and mediate differences in barrier quality, perhaps through interaction with cytoplasmic components of the cornified cell.
Materials and Methods
Animals and Cells.
Time-mated ICR strain fetuses were generated under U.K. Home Office License 40/1566, as described (17). Normal human epidermal keratinocytes from neonatal foreskin were cultured in KGM media (BioWhittaker). Cells were maintained and transfected (Lipofectamine Plus, Invitrogen) in the presence of low calcium (0.09 mM), and terminal differentiation was induced by raising calcium to 1.5 mM.
DNA Analysis.
I.M.A.G.E. ESTs (18) were from the U.K. Human Genome Mapping Project Resource Centre (HGMP-RC), and sequence was analyzed using biology workbench software (San Diego Supercomputer Centre). An LEP-glutathione S-transferase (GST) fusion protein, comprising most of the coding sequence (amino acids 2–131 of the 134-residue protein) of a murine Group 1 LEP homologue (2310037L11Rik; Table 1), was generated by using standard techniques (19). The primers 5′-GCGGGATCCCAGCAAAGCCAACAGCAGTG-3′ and 5′-GCGGAATTCCGTCCCCAGACTGTTGGCTAC-3′ were used and cloned in-frame in pGEX-4T2 vector (Amersham Pharmacia). SPRR1a fusion protein was generated by using primers 5′-GCG GGA ATT CCG AGT TCC CAC CAG CAG AAG-3′ and 5′-GCG GCT CGA GCC TTC TGC TTT GTC TTC TGC TG-3′. pGEX-4T2-BclX2 and pGEX-4T2-Fak C terminus were donated by A. Gilmore (Univ. of Manchester). Flag-fusion proteins were cloned into p3xFlag-CMV-10/14 (Sigma). DNA was checked for PCR errors by sequencing. Human LEP expression was detected by reverse transcription–PCR, using gene-specific primer pairs (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org) to amplify human RNA from adult heart, skin (external epithelium, Stratagene), and esophagus (internal epithelium; GenPak, New Milton, U.K.). cDNAs were tested for genomic contamination (19).
Table 1.
Relationship between murine LEP gene expression and human genes
| Gene symbol (14) (murine) | GenBank/TIGR accession no. | I.M.A.G.E. ID (18) | SPRR-like name (15) | Murine expression | Human LEP group |
|---|---|---|---|---|---|
| 1110058A15Rik | NM025413/AI604448 | 1228375 | — | Epidermal | Group 1 (LEP 1–6) |
| 1110029C13Rik | TC146632 | 2135373 | SPRR-I 5 | Epidermal | |
| 2130069N01Rik | NM029667 | 318311 | — | Epidermal | |
| 1110031B11Rik | AA792425 | 1151518 | — | Epidermal | |
| — | W18229 | 333214 | SPRR-I 8 | ND | |
| 2310037L11Rik | TC146634 | 482882 | SPRR-I 7 | Epidermal | |
| 1110029C13Rik | AA061750 | 483600 | SPRR-I 4 | ND | |
| 1110008K04Rik | W40830 | 351941 | — | ND | Group 2 (LEP7, -8, and -18) |
| 2310066F03Rik | W82736 | 404039 | — | ND | |
| 1110019L16Rik | TC153554 | 1884947 | SPRR-I 10 | Undetected | |
| — | TC143173 | 330044 | SPRR-I 6 | ND | Group 3 (LEP 9–12) |
| 1110004E04Rik | TC108289 | 1023461 | SPRR-I 2 | ND | |
| 2310003A15Rik | TC108553 | 1150992 | SPRR-I 3 | ND | |
| 2310020L01Rik | TC139332 | 481703 | SPRR-I 9 | ND | |
| 2300007B01Rik | W37034/AF176515 | 334939 | SPRR-I 1/Eig3 | Internal | Group 4 (LEP 13–17) |
| 2310002A05Rik | AK009081 | 480989 | — | Internal |
ND, not determined; I.M.A.G.E., integrated molecular analysis of genomes and their expression; TIGR, the Institute for Genomic Research.
Ab Production and Immunohistochemistry.
Polyclonal Ab was raised in New Zealand white rabbits. Serum was affinity-purified against fusion protein by using the Sulfolink Affinity Purification kit (Pierce), tested by Western analysis on epidermal protein extracts, and immunohistochemistry was performed on whole fetal tissue slices. Immunohistochemistry was performed on 5 μm paraffin sections, 5–10 μm frozen sections, or acetone-fixed cells on chamber slides. Ab complexes were detected by silver-staining (AuroProbe/Intense kits, Amersham Pharmacia) or with fluorescein-conjugated secondary Abs, then counterstained with hematoxylin or Hoechst 33342 (Sigma). Loricrin and K14 Abs were from Babco (Richmond, CA). Cornified envelopes from late gestation epidermis [embryonic day (E) 17–18] were isolated by repeated sonication and boiling in extraction buffer (50 mM Tris⋅HCl, pH 8.8/1 mM EDTA/10 mM DTT/2% SDS), collected by centrifugation (14,000 × g), then washed 3–4 times in extraction buffer. Immunolabeling was performed by washing and blocking envelopes in 1% BSA in Tris-buffered saline/1% Tween 20 for 4 h at 4°C. Primary and secondary Ab incubations were performed overnight at 4°C in blocking buffer. Envelopes were collected by centrifugation (14,000 × g) between each step, embedded in Agar100 resin (Agar Scientific, Essex, U.K.), and processed for standard transmission electron microscopy.
TGase assay was as described (20) using GST fusion proteins as substrates. LEP-GST and SPRR1a-GST were tested as TGase substrates, and Fak-GST, Bcl2-GST, and GST alone were negative controls. Monodansylcadaverine, a known TGase substrate, was a positive control (20). Frozen sections (5 μm) of fresh mouse footpad epidermis were used. After the TGase reaction, sections were washed extensively, and bound GST fusion protein was detected with anti-GST Ab (Amersham Pharmacia) and fluorescein-conjugated secondary Ab (Jackson ImmunoResearch). For some experiments, washing steps included a 15-min wash at 65°C with 1% SDS before immunological detection.
Riboprobes and in Situ Hybridization (ISH).
ESTs corresponding to murine SPRR1a cDNA (GenBank accession no. AA1726615) and murine LEP cDNAs (see Table 1 for accession nos. used in ISH) were obtained from the U.K. Human Genome Mapping Project Resource Centre and sequenced to confirm identity and orientation. Riboprobe generation and whole-mount ISH (21) were carried out at high stringency by using both full-length and 3′-specific probes. Stringency of the technique is demonstrated by the fact that full-length (coding sequence-containing) murine LEP (1110019L16Rik, Group 2 homologue) with 82–89% homology in the coding domain to murine Group 1 skin members cannot be detected in skin. Presence of hybridization in skin was confirmed with 3′ untranslated region-specific probes for skin LEPs.
Results
Identification of Barrier Markers.
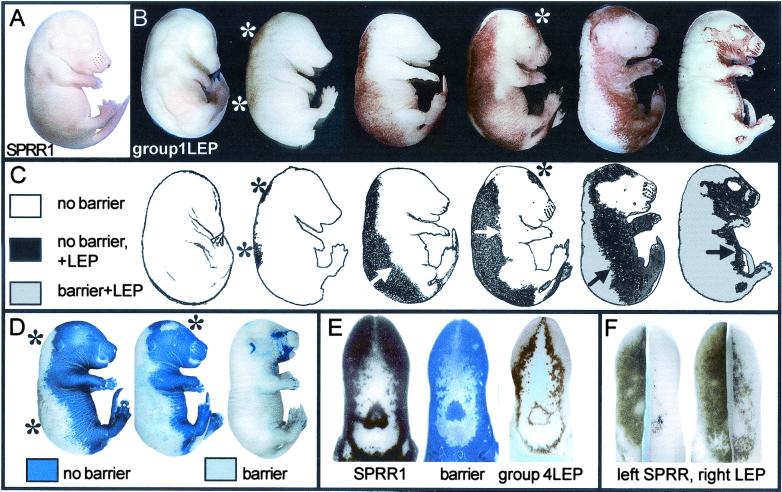
We recently showed that SPRR1a/b are molecular markers for barrier induction in internal keratinizing epithelia, expressing just before barrier formation in the same pattern (17). However, SPRR1 gene expression is undetectable in epidermis, apart from follicles (Fig. 1A), consistent with previous reports (22). While searching the sequence databases for an analogous epidermal barrier marker, we found a group of related murine ESTs (Table 1, Group 1), many deriving from a skin cDNA library (Stratagene). Whole-mount ISH with these ESTs showed that they express in fetal epidermis in a pattern analogous to SPRR1 genes in oral epithelia (Fig. 1 B and C), i.e., are expressed just before barrier formation in the epidermal barrier formation pattern (Fig. 1D; ref. 17). Hence, we had found markers for epidermal barrier formation with apparent expression similarities to SPRR genes. Expression of the new epidermal genes was undetectable in oral epithelia by using ISH, i.e., they show an opposite expression pattern to SPRR1a/b, which are abundant in oral epithelia and excluded from epidermis. The genes predict proteins of similar size to SPRR proteins, with sequence similarity to SPRRs and loricrin, thus our initial hypothesis was that they were epidermal equivalents of SPRR1a/b.
Figure 1.
(A) SPRR1 is not detected in epidermis by ISH, except for whisker follicles. Later, it is expressed in the pelage hair follicles. (B and C) In contrast, LEP Group 1genes (Table 1) are expressed abundantly in fetal epidermis (left to right, increasing gestational age) before barrier formation and in the fetal barrier formation pattern (D). Arrows show the direction of “fronts” of LEP gene expression, then barrier formation, that appear to cross fetal skin. Asterisks show “initiation sites” where change is first detected. Emerging barrier excludes hybridization probes and prevents detection of gene expression (C). LEP expression pattern is reminiscent of SPRR1a/b in internal epithelia (12). (E) ISH of age-matched tongues showing abundant SPRR1a/b expression, barrier assay, and expression of an oral (Group 4) LEP by ISH (2300007B01Rik, Table 1). Emerging barrier is apparent in the central area where dyes and ISH probes are excluded. LEP expression precedes barrier formation and is in the barrier pattern but appears later than SPRR. (F) Direct demonstration that LEP expression follows SPRR expression in single tongues cut sagittally before ISH. SPRR is abundantly expressed (left), whereas LEP expression lags (right).
However, further searches revealed closely related homology groups deriving largely from rodent tongue and forestomach libraries (ref. 14; Table 1), which include homologues to human XP5 (12) and the recently reported murine Eig3 (15). Whole-mount expression analysis confirms that the ESTs from internal libraries are expressed prominently in internal stratified keratinizing epithelia and are at very low levels, or undetectable, in epidermis (Fig. 1E).
These internal ESTs coexpress with SPRR1a/b in oral epithelia. We show that the new ESTs are expressed later than SPRR genes during development and just before fetal barrier induction (Fig. 1F). Hence, the new group of genes must encode proteins distinct from SPRR proteins, as suggested for Eig3 (15). Based on homology with known envelope proteins (loricrin, involucrin, and SPRR), it is probable that the new proteins are components of the cornified envelope (12, 14, 15). These new genes are very abundant in skin libraries, indicating that the encoded proteins are major products of differentiating keratinocytes and their presence in skin was confirmed with 3′ untranslated region probes. Furthermore, their expression indicates that they are not needed until very late in envelope assembly.
Human LEP Genes.
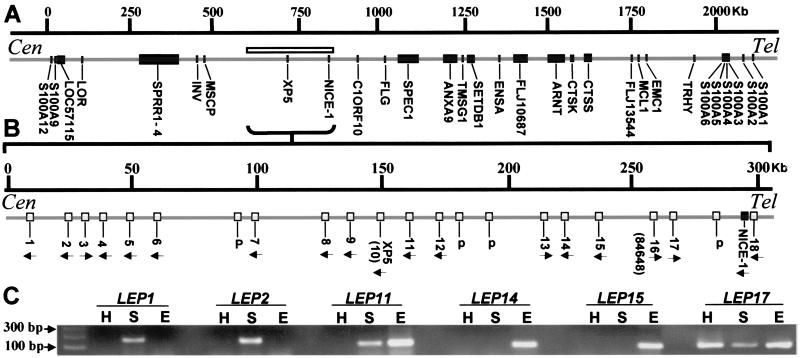
Analysis of the human genome revealed human homologues of these genes in the EDC clustered around XP5 on the telomeric side of the IVL/SPRR/LOR group near NICE-1 (Fig. 2A). We detect distinct cDNAs for 18 predicted genes and there are, additionally, 4 pseudogenes (Figs. 2B and 7, which is published as supporting information on the PNAS web site, www.pnas.org). Because the genes encode probable cornified envelope constituents and are not expressed until very late during fetal barrier development, we propose naming the human predicted proteins as LEPs, and have named the human genes sequentially as LEP 1-18 (excluding the 4 pseudogenes). Other authors have recently given a subset of the murine homologues multiple alternative names (Table 1; refs. 14 and 15).
Figure 2.
(A) Distribution of human LEP genes (white bar) in the EDC (1q21). (B) Fine location of LEP genes (white boxes) and NICE-1. Arrows show direction of transcription and “p” indicates pseudogenes. XP5 is LEP10 and loc84648 is LEP 16. IVL, involucrin; LOR, loricrin; other abbreviations are as described (National Center for Biotechnology Information). Human LEP coding sequence and protein are compared in Table 2 and Figs. 7 and 8. The proteins fall into four structural groups, and group-specific regions are shaded. There is little homology with NICE-1. (C) Reverse transcription–PCR analysis of human LEP shows gene location/expression pattern relationship. LEP1 and -2 (Group 1), LEP11 (Group 3), LEP14, -15, and -17 (Group 4) expression is shown, and remaining LEP expression is in Table 2. H, heart; S, skin (external epithelium); and E, esophagus (internal epithelium).
Human LEPs fall into distinct structural groups (Fig. 8, which is published as supporting information on the PNAS web site), and genes encoding structurally similar proteins cluster on the genome. Interestingly, each group has distinct amino acid sequences (Fig. 8) that may confer distinct properties on the protein.
Comparison with mouse LEPs shows that Group 1, comprising LEP 1-6, corresponds structurally with mouse genes expressed predominantly in epidermis (Table 1). Group 2 comprises LEP7, -8, and -18 and is structurally the least homogenous group, as well as being scattered on the chromosome. Group 3 comprises LEP 9-12 and includes XP5 (corresponding to LEP10). Group 4, comprising LEP 13-17, corresponds with the mouse genes expressed most prominently in internal epithelia, including Eig3 (15). Hence, based on homology with mouse genes and murine expression patterns, it is predicted that epidermal expression will be most prominent in Group 1 and internal gene expression most common in Group 4.
The predicted relationship between human LEP gene structural grouping and expression was tested by reverse transcription (RT)-PCR in skin (as an example of external epithelium) and esophagus (internal epithelium) (Fig. 2C; Table 2). Group 1 members LEP 1-5 are strongly skin-specific, as predicted (Fig. 2C). LEP 6 (on the most telomeric side of the Group 1 cluster) also has prominent expression in skin, but low level expression is detected in internal epithelia by RT-PCR. Expression of Group 2 members (LEP7, -8, -18) is variable. Group 3 members (including XP5-LEP10) express strongly in skin, as has been reported for XP5 (12). As predicted, members of Group 4, in particular LEP14 and -15, are expressed strongly in internal epithelia, with LEP14 and -15 expressing exclusively in internal epithelia. LEP17 seems to have lost tissue specificity and is detected in all tissues tested (Fig. 2C, data not shown).
LEP Genes Are Induced After Calcium-Mediated Differentiation and Late in Differentiation, LEP Associates with the Cell Periphery.
After calcium-induced terminal differentiation of keratinocytes, there is sequential induction of differentiation markers (23). First keratins 1 and 10, then precursors of the cornified envelope, envoplakin and involucrin, are synthesized, followed by SPRRS, loricrin, and filaggrin (2, 23, 24).
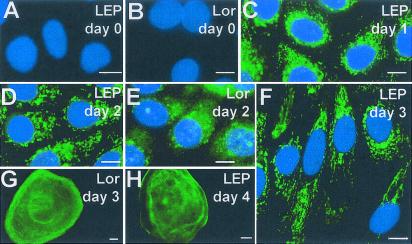
We tested the behavior of LEP protein during calcium-induced differentiation by preparing an affinity-purified Ab to a murine representative of the Group 1 LEPs (2310037L11Rik, Table 1). The Ab is specific to epidermally expressed LEP members as it does not detect protein in internal epithelia (see below). LEP protein production in culture is absolutely calcium-dependent and after calcium-induction of keratinocytes, LEPs are detected and form intracellular aggregates (Fig. 3 A, C, and D). LEP protein, although abundant in keratinocytes, does not colabel with the cornified cells until 4–5 days after calcium induction (Fig. 3H), well after loricrin labels these large cornified cells at days 3–4 (Fig. 3G). The calcium-dependent appearance of endogenous LEPs in culture is the result of gene induction, as transfected FLAG-tagged proteins expressed under the cytomegalovirus promoter are readily detected before calcium induction, where they form similar perinuclear aggregates until after calcium induction (data not shown).
Figure 3.
(A and B) Before calcium-induction (day 0) neither LEP nor loricrin (Lor) is detected. (C and D) LEP gene is induced by calcium, and perinuclear LEP aggregates are abundant 1 and 2 days after calcium induction. (E) One and two days after calcium induction, Lor appears in aggregates. (F) By 3 days LEP is still cellular, although appears associated with filaments. In contrast, Lor (G) has started to associate with the differentiated squames (adherent cornified cells) by day 3. However, LEP does not associate with squames until day 4 (H). [Bar = 5 μm.]
We showed previously that loricrin relocation to the envelope correlates closely with the initial stage of barrier formation during development (25). During development, transcription of LEP genes is delayed until just before initial formation of barrier (Fig. 1), i.e., LEP genes are probably one of the last genes transcribed before nuclear degradation in the granular cell. We use the new Ab to show that LEP protein cannot be detected in fetal epidermis until well after initiation of barrier formation and associated relocation of loricrin to the membrane (E17, data not shown). This finding is consistent with the culture results and suggests that the protein is important for maturation of the cornified envelope or the squame.
LEP Proteins Are TGase Substrates.
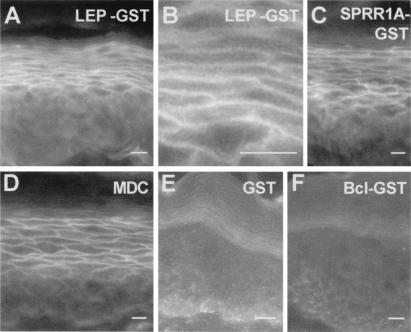
LEP members contain structural similarities to known cornified envelope precursors such as SPRR1 and loricrin, including characteristic N-terminal glutamine residues which are major sites for TGase-mediated cross-linking in SPRRs (10, 15, 26). This similarity strongly suggests that LEP members are TGase substrates. To test this idea, we used an in situ assay for epidermal TGase activity (20) to find whether a typical Group 1 epidermal LEP member (2310037L11Rik, Table 1) could provide a substrate for TGase incorporation into epidermis. We show (Fig. 4 A–C) that both LEP and SPRR sequences stimulate incorporation of a GST tag into the murine epidermal envelope, as does a known TGase substrate monodansylcadaverine (ref. 20; Fig. 4D), whereas control fusion proteins (GST alone, BclX-GST; Fig. 4 E and F) are ineffective. Incorporation occurs at the cell periphery, spatially consistent with incorporation into the envelope. Incorporated protein is also resistant to heat and detergent treatment, consistent with covalent cross-linking into the envelope. Because epidermal TGases are strictly calcium-dependent (1), EDTA was substituted for calcium, resulting in failure to incorporate (data not shown).
Figure 4.
(A) LEP-GST protein is cross-linked into the upper spinous/granular layer of epidermis at the cell periphery (B). SPRR1a-GST (C) and monodansylcadaverine (MDC) (D) are also cross-linked; however, GST alone (E) or additional fusion proteins including BclX-GST (F) or Fak-GST (not shown) are ineffective substrates. [Bar = 10 μm.]
LEP Proteins Are Structural Components of Mature Cornified Envelopes.
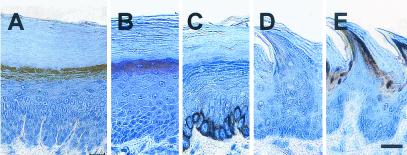
To substantiate epithelial tissue specificity of LEP members, whole-body sections from late gestation fetuses (E18, just before birth) were scanned by immunohistochemistry, showing that anti-LEP activity was confined to the epidermis and nasal epithelium (data not shown). Within the epidermis, the anti-LEP activity was associated with the upper granular layer of skin (Fig. 5A), consistent with a role in formation of the cornified envelope late in differentiation. The anti-LEP Ab does not recognize protein in oral epithelia (Fig. 5D), demonstrating lack of crossreaction to the Group 4 LEPs or SPRR1 proteins (Fig. 1).
Figure 5.
(A) Expression of an LEP protein in mouse epidermis in the upper granular layer. (B and C) Control sections showing loricrin (B) and K14 (C). (D) Dorsal tongue section showing that LEP Ab does not crossreact with the orally expressed Group 4 LEPs or SPRRs; however, (E) loricrin is readily detected. [Bar = 20 μm.]
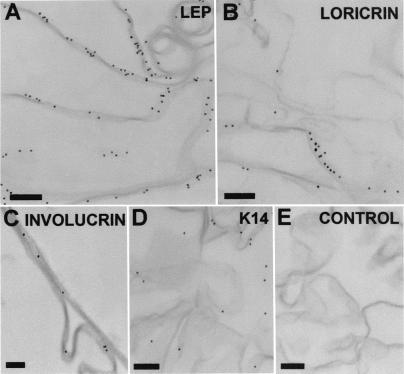
Immunoelectron microscopy of envelopes isolated from E17–E18 skin shows that LEP associates strongly with the envelope (Fig. 6A) as do known envelope proteins loricrin and involucrin (Fig. 6 B and C). In contrast, Abs to abundant nonenvelope epidermal proteins K14 (Fig. 6D) and desmocollin 3 (data not shown) do not label the envelope, demonstrating specificity of LEP association.
Figure 6.
(A) Immunolabeling of isolated cornified envelopes demonstrates that LEP is a structural envelope component labeling isolated envelopes. (B) Loricrin, labels with similar abundance to LEPs. (C) Involucrin, one of the first proteins to be incorporated, localizes further toward the inner envelope face. (D) K14, a very abundant epidermal protein, shows no association with the cornified envelope, nor is there background labeling when primary Ab is omitted (E). [Bar = 150 nm.]
Discussion
In this work, we show that XP5 is one member of a family of human proteins we name LEPs that are major cornified envelope components in barrier-forming epithelia. The proteins are produced very late during envelope assembly. Of the LEP genes, 18 cluster within the EDC at 1q21 in human, and one of these genes, XP5, has been identified (12). Murine Eig3 (15) is most homologous to human LEP13 and -14, which are expressed predominantly in internal epithelia. Human NICE-1 (13), located at the edge of the cluster, is least homologous and, therefore, probably not a member of the group.
Within the EDC, structurally similar and coexpressed LEP genes form subclusters. This clustering could derive from evolution through gene duplication or could reflect coregulation, i.e., there could be an epidermal enhancer at the centromeric end of the LEP cluster and/or an internal epithelial enhancer at the telomeric end. Locus control regions in the EDC have been suggested (12, 16).
It is possible that differential expression of multiple LEP genes modulates barrier quality over the animal surface, as has been proposed for SPRR members. Cabral et al. (9) proposed that SPRR genes encode structurally homologous products which differ primarily in their regulatory regions. Hence, multiple genes permit a wide repertoire of regulatory responses resulting in differences in effective protein dosage and associated barrier quality. This increased capacity for modulating regulatory responses would permit subtle changes in barrier quality in response to environmental stimuli (9). A similar argument could account for multiple LEP genes. However, the alternative theory, that specific features of LEP group proteins confer distinct roles, is also tenable, and distinguishing between the two proposals depends on further experimentation.
XP5 and LEP are similar to known cornified envelope constituents loricrin, SPRRs, involucrin, and NICE-1 (12, 13, 15). The LEP N terminus resembles that of all of the above proteins and contains glutamine residues, identified in loricrin and SPRRs, as key sites for TGase activity (26–29). LEPs contain proline/lysine/cysteine (PKC) repeats in the N-terminal half, a feature of SPRRs, followed by serine/glycine/cysteine-rich regions (SGC), characteristic of loricrin. Family members differ with respect to the number of PKC and SGC repeats. An extreme example is the murine gene most similar to human LEP 18 (1110019L16Rik, Table 1), which is abnormally long as a result of extensively expanded SGC repeats. There is no human gene with these extended repeats, raising the possibility that the number of repeats confers no particular functional consequence.
The first member of this group of proteins, XP5, was detected by selection-cloning from a skin library with an EDC yeast artificial chromosome (YAC; ref. 12). Zhao and Elder (12) showed that XP5 expresses in skin but is absent from cultured keratinocytes. NICE-1, isolated from a screen of EDC members by using a calcium-induced keratinocyte library, is detected in cultured keratinocytes but not significantly in epidermis (13), suggesting that cultured keratinocytes undergo a variant form of skin differentiation. However, structurally, NICE-1 is distant from the LEP/XP5 group. Neither XP5 nor filaggrin transcripts were detected in this latter screen, suggesting that late differentiation markers were not induced in their library, consistent with our finding that LEP/XP5 members are not detected until very late in terminal differentiation of calcium-induced keratinocytes. NICE-1 function remains unknown; however, it has structural similarities to other TGase substrates, mouse homologues, and is probably another new cornified envelope constituent (13).
Eig3 is a murine member of the LEP group, detected by using the rapid analysis of gene expression technique to identify genes up-regulated in transgenic mice overexpressing E2F1 under the control of the basal keratinocyte-specific K5 promoter (15). Eig3 is strongly expressed in internal epithelia (forestomach), consistent with detection in internal epithelia in this work. Wang et al. (15) report weak skin expression; however, this may represent crossreaction to other family members, as skin expression is not detected here.
During development, barrier activity is acquired in steps or stages (25). Barrier activity is first detected at E16.5 of mouse development, determined by both colorimetric assays that measure the first stage in barrier formation and quantitative measurements of transepidermal water loss (TEWL). The TEWL assay shows that most barrier activity is acquired at E16.5 (25), whereas evaporimeter studies demonstrate that barrier activity continues to improve up to and beyond birth. There is a poor molecular appreciation of late changes during fetal barrier formation. We show here that LEP gene expression occurs after expression of other known cornified envelope proteins and very close to first acquisition of fetal barrier function. However, LEP protein is detected only after barrier formation has initiated. Mature filaggrin, a keratin filament-aggregating protein that forms the matrix of cornified cells or squames (30), is also detectable only after initial barrier formation (25). Filaggrin is also a minor cornified envelope component (31), showing that protein incorporation into the envelope continues after initial barrier formation. Because both proteins appear late during barrier development, an intriguing possibility is that LEPs play a role in envelope–matrix interaction during squame maturation.
Supplementary Material
Acknowledgments
This work was supported by Biotechnology and Biological Sciences Research Council Grants 97/A1/G/03357 (to D.M.) and S13352 (to K.N.) and by Medical Research Council (U.K.) Grant G9803920 (to M.J.H.).
Abbreviations
- EDC
epidermal differentiation complex
- EST
expressed sequence tags
- ISH
in situ hybridization
- LEP
late envelope proteins
- NICE-1
newly identified component of the EDC
- SPRR
small proline-rich region protein
- TGase
transglutaminase
- GST
glutathione S-transferase
- En
embryonic day n
References
- 1.Nemes Z, Steinert P M. Exp Mol Med. 1999a;31:5–19. doi: 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- 2.Steinert P M, Marekov L N. Mol Cell Biol. 1999;10:4247–4261. doi: 10.1091/mbc.10.12.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch P, de Viragh P, Scharer E, Bundman D, Longley M, Bickenbach J, Kawachi Y, Suga Y, Zhou Z, Huber M, et al. J Cell Biol. 2000;151:389–400. doi: 10.1083/jcb.151.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djian P, Easley K, Green H. J Cell Biol. 2000;151:381–388. doi: 10.1083/jcb.151.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson N A, Lapic S, Welter J F, Eckert R L. J Biol Chem. 1997;272:12035–12046. doi: 10.1074/jbc.272.18.12035. [DOI] [PubMed] [Google Scholar]
- 6.Ruhrberg C, Watt F. Curr Opin Genet Dev. 1997;7:392–397. doi: 10.1016/s0959-437x(97)80154-2. [DOI] [PubMed] [Google Scholar]
- 7.Tesfaigzi J, Carlson D M. Cell Biochem Biophys. 1999;30:243–265. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- 8.Kartasova T, Darwiche N, Kohno Y, Koizumi H, Osada S, Huh N, Lichti U, Steinert P M, Kuroki T. J Invest Dermatol. 1996;106:294–304. doi: 10.1111/1523-1747.ep12340741. [DOI] [PubMed] [Google Scholar]
- 9.Cabral A, Voskamp P, Cleton-Jansen A M, South A, Nizetic D, Backendorf C. J Biol Chem. 2001;276:19231–19237. doi: 10.1074/jbc.M100336200. [DOI] [PubMed] [Google Scholar]
- 10.Steinert P M, Candi E, Kartasova T, Marekov L. J Struct Biol. 1998;122:76–85. doi: 10.1006/jsbi.1998.3957. [DOI] [PubMed] [Google Scholar]
- 11.Steinert P M, Kartasova T, Lyuben N M. J Biol Chem. 1998;19:11758–11769. doi: 10.1074/jbc.273.19.11758. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X P, Elder J T. Genomics. 1997;45:250–258. doi: 10.1006/geno.1997.4952. [DOI] [PubMed] [Google Scholar]
- 13.Marenholz I, Zirra M, Fisher D F, Backendorf C, Ziegler A, Mischke D. Genome Res. 2001;11:341–355. doi: 10.1101/gr.114801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The RIKEN Genome Exploration Research Group Phase II Team; the FANTOM Consortium. Nature (London) 2001;409:685–690. [Google Scholar]
- 15.Wang A, Johunson D G, MacLeod M C. Genomics. 2001;73:284–290. doi: 10.1006/geno.2001.6518. [DOI] [PubMed] [Google Scholar]
- 16.Mischke D. Subcell Biochem. 1998;31:71–104. [PubMed] [Google Scholar]
- 17.Marshall D, Hardman M J, Byrne C. J Invest Dermatol. 2000;114:967–975. doi: 10.1046/j.1523-1747.2000.00955.x. [DOI] [PubMed] [Google Scholar]
- 18.Lennon G G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel FM, Brent R, Kingston RE, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1998. [Google Scholar]
- 20.Raghunath M, Hennies H C, Velten F, Wiebe V, Steinert P M, Reis A, Traupe H. Arch Dermatol Res. 1998;290:621–627. doi: 10.1007/s004030050362. [DOI] [PubMed] [Google Scholar]
- 21.Byrne, C., Tainsky, M. & Fuchs, E. (1994) 120, 2369–2383. [DOI] [PubMed]
- 22.Jarnik M, Kartasova T, Steinert P M, Lichti U, Steven A C. J Cell Sci. 1996;109:1381–1391. doi: 10.1242/jcs.109.6.1381. [DOI] [PubMed] [Google Scholar]
- 23.Yuspa S H, Kilkenny A E, Steinert P M, Roop D R. J Cell Biol. 1989;109:1207–1217. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiColandrea T, Karashima T, Maatta A, Watt F M. J Cell Biol. 2000;151:573–586. doi: 10.1083/jcb.151.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardman M J, Sisi P, Banbury D N, Byrne C. Development (Cambridge, UK) 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- 26.Deng J, Pan R, Wu R. J Biol Chem. 2000;275:5739–5747. doi: 10.1074/jbc.275.8.5739. [DOI] [PubMed] [Google Scholar]
- 27.Candi E, Melino G, Mei G, Tarcsa E, Chung S-I, Marekov L N, Steinert P M. J Biol Chem. 1995;270:26382–26390. doi: 10.1074/jbc.270.44.26382. [DOI] [PubMed] [Google Scholar]
- 28.Tarcsa E, Candi E, Kartasova T, Idler W W, Marekov L N, Steinert P M. J Biol Chem. 1998;273:23297–23303. doi: 10.1074/jbc.273.36.23297. [DOI] [PubMed] [Google Scholar]
- 29.Candi E, Tarcsa E, Idler W, Kartasova T, Marekov L N, Steinert P M. J Biol Chem. 1999;274:7226–7237. doi: 10.1074/jbc.274.11.7226. [DOI] [PubMed] [Google Scholar]
- 30.Dale B A, Resing K A, Presland R B. In: The Keratinocyte Handbook. Leigh I M, Birgitte Lane E, Watt F M, editors. Cambridge, U.K.: Cambridge Univ. Press; 1994. pp. 323–350. [Google Scholar]
- 31.Simon M, Haftek M, Sebbag M, Montezin M, Girbal-Neuhauser E, Schmitt D, Serre G. Biochem J. 1996;317:173–177. doi: 10.1042/bj3170173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.