Abstract
Increased telomerase activity can be blocked by targeting the hTERT activity at both RNA and catalytic subunits. Various inhibitors had been used to regulate hTERT activity in glioblastoma cell lines and showed promising results. The present study hypothesized that the telomerase specific inhibitor BIBR1532 can effectively down-regulate the telomerase activity in LN18 glioblastoma cell line. LN18 glioblastoma cell line was treated with various concentrations of BIBR1532 at different time intervals. MTT assay was performed to determine cell viability after BIBR1532 treatment. hTERT mRNA and protein expression were determined by qRT-PCR and western blotting, respectively. Flow cytometry and TRAP assay was performed to detect the rate of apoptosis and telomerase activity in treated and control samples. One-way ANOVA was performed to compare the mean values of variables in control and BIBR1532 treated groups. LN18 cells showed a significant dose dependent cytotoxic effect after treatment with BIBR1532. hTERT mRNA expression in cells treated with 25, 100 and 200 μM BIBR1532 treated groups was decreased ~ 21, ~ 61.2, and ~ 77%, respectively (p < 0.05). We also observed that, BIBR1532 treatment reduced the expression of hTERT protein in LN18 cells in a dose dependent manner. The Flow cytometry data showed that, the drug induced significant increase in the total percentage of apoptotic cells with 200 μM concentration of BIBR1532 at all time points. BIBR1532 exhibited potent inhibition of telomerase activity in a dose-dependent manner in LN18 cells. BIBR1532 could induce apoptosis in LN18 cells through the downregulation of telomerase activity at transcriptional and translational level. We conclude that BIBR1532 may be a therapeutic agent to suppress telomerase activity, however, further efforts are necessary in order to explore this therapeutic strategy.
Electronic supplementary material
The online version of this article (10.1007/s10616-018-0205-9) contains supplementary material, which is available to authorized users.
Keywords: hTERT, BIBR1532, Glioblastoma, Regulation
Introduction
Gliomas are the most common heterogeneous primary brain tumors which arise from glial cells. In 2007, World Health Organization (WHO) classified gliomas into four different grades based on the cell type and severity (Louis et al. 2007). Among these glioblastoma (GB) is the most frequent and aggressive brain tumor in adults (Arvold and Reardon 2014). The hallmark features of GB are uncontrolled cellular proliferation, invasion, ability to form new blood vessels and evasion of apoptosis (Furnari et al. 2007). Even with the advancement in the field of surgery, chemotherapy and radiotherapy the prognosis of GB patients remains poor due to the presence of cancer stem cells that makes these tumors resistant to chemo and radio therapy (Singh et al. 2003; Yuan et al. 2004). Numerous molecular and genetic alterations which maintain molecular phenotypes of tumors are frequently seen in gliomas (Sathornsumetee et al. 2007; Zhang et al. 2012).
Human telomeres are unique protein-DNA complexes present at the ends of all eukaryotic chromosomes with a tandem repeats of TTAGGG sequences. They protect the chromosomes from end to end fusions and maintain genomic stability (Cong et al. 2002). Telomeres also play a major role in aging and cancer (Blasco 2005). Telomerase is a specialized ribonucleoprotein reverse transcriptase which maintains the telomeric TTAGGG repeats by adding DNA sequence repeats to the 3′ end of chromosomes (Cong et al. 2002; Osterhage and Friedman 2009). Telomerase holoenzyme consists of two major components: a catalytic protein component human telomerase reverse transcriptase (hTERT) and a RNA component (hTR) that acts as a template for adding hexanucleotide repeats (Artandi and DePinho 2010; Wai 2004). The expression and activity of telomerase has been observed in 80–90% of all cancer cells and involves in increased cell proliferation, cellular immortality and tumorigenesis (Kong et al. 2015). In contrast to that in most normal human cells the hTERT expression and telomerase activity are low or repressed (Counter et al. 1992; Harley et al. 1990). Hence the difference in hTERT expression and telomerase activity in cancer cells versus normal cells makes telomerase as an attractive target for the development of new cancer therapeutics. The expression and activity of telomerase correlated with the progression of malignant glioma (Falchetti et al. 1999; Komata et al. 2002). Therefore, telomerase is an essential target for improving the prognosis and treatment of gliomas.
In the recent years several therapeutic approaches have been evaluated to inhibit telomerase activity and block cancer cell growth. Previous studies demonstrated that inhibition of hTERT by RNA interference and pharmacological agents reduces telomerase activity and allows cells to undergo apoptosis (Lavanya et al. 2016; Lu et al. 2012; Mergny et al. 2002). In addition to that numerous telomerase inhibitors that block either the activity of the enzyme or recruitment of telomerase to telomeres have been developed. They are nucleoside analogs, catalytic inhibitors, hTR antisense oligonucleotides, ribozymes and G-quadruplex Ligands (Seimiya et al. 2002). Inhibitors such as imetelstat (GRN163L) have demonstrated a promising pharmacokinetic profile with fewer side effects and are currently in phase I and II clinical trials (Asai et al. 2003; Marian et al. 2010).
2-[(E)-3-naphtalen-2-yl-but-2-enoylamino]-benzoic acid BIBR1532, one of the most potent and specific inhibitors of hTERT, has been discovered recently (Damm et al. 2001; Kong et al. 2015). BIBR1532 is a nonpeptidic, non-nucleoside small molecule inhibitor that inhibits telomerase activity by specifically binding to the active site of hTERT (Damm et al. 2001). It has been observed that BIBR1532 has a strong ability to reduce growth across various cancer cells including breast, leukemia, lung, ovarian, chondrosarcoma and germ cell tumors (Bashash et al. 2013; Damm et al. 2001; Meng et al. 2012; Mueller et al. 2007; Parsch et al. 2008; Ruden and Puri 2013). In the present study we aim to investigate the effect of knockdown of hTERT and inhibition of telomerase activity by BIBR1532 in glioblastoma cells.
Materials and methods
Cell culture
The human glioblastoma cell line LN18 was purchased from the National Center for Cell Science (NCCS) (Pune, India). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum (Invitrogen, Life Technologies, Carlsbad, CA, USA) and antibiotics 100 U/ml penicillin and 100 µg/ml streptomycin (Life Technologies) at 37 °C in a humidified atmosphere with 5% CO2 and 95% air. The cells were grown in 6 well tissue culture plates and replated every 2–3 days to ensure log-phase growth.
CHME-3, an immortalized human microglial cell line (Nijaguna et al. 2015) was a kind gift from Assistant Professor Manjunatha M Venkataswamy (Department of Neurovirology, NIMHANS, Bangalore). CHME3 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 100 μg/ml penicillin–streptomycin (Invitrogen).
Drug treatment
The telomerase inhibitor BIBR1532 was obtained from (Genetix Biotech Asia Pvt Ltd., Bangalore, India). A stock solution of BIBR1532 at a concentration of 1 mM was prepared by dissolving the compound in sterile dimethylsulfoxide (DMSO), divided into aliquots and stored at − 20° C until use. LN18 cells were treated with relative amounts of BIBR1532 stock solution to attain concentrations of 25, 50, 100 and 200 µM.
MTT assay
The inhibitory effect of BIBR1532 on the metabolic activity of LN-18 cells was assessed by the uptake of thiazolyl blue tetrazolium bromide (Sigma Aldrich) by viable cells. Briefly, LN-18 cells were seeded at 5000/well in 96-well culture plates and incubated with desired concentrations of BIBR1532 for 24, 48 and 72 h. After removing the media, cells were further incubated with MTT solution [5 mg/ml in phosphate buffered saline (PBS)] at 37° C for 2 h and the untreated cells were defined as the control group. The MTT reaction was terminated through the addition of 100 μl of DMSO. The results were read by measuring absorption at 570 nm (620 nm as a reference) using plate reader (Tecan, infinite M200 Pro, A-5082, Austria). The effect of BIBR1532 was calculated as a percentage of control cell growth. Each experiment was performed in triplicate and repeated three times to assess for consistency of results. CHME3 cells were also incubated with different concentrations of BIBR1532 and cell viability was measured after 24 and 48 h.
RNA isolation and quantification
Total cellular RNA from LN18 cells was isolated at 48 h after treatment with BIBR1532 using a Pure LinkTM RNA Mini Kit (Ambion, Life technologies) according to the manufacturer’s instructions. The quantity of RNA samples was assessed spectrophotometrically using Nanodrop ND 2000c (Thermo Fisher Scientific, Waltham, MA, USA). RNA samples with a purity of 1.95–2.05 (A260/280) were used in this study. RNA stability was checked by formaldehyde agarose gel electrophoresis.
Quantitative real-time PCR
One microgram of total RNA was reverse transcribed into cDNA using MMLV reverse-transcriptase, oligo-dT, dNTPs and buffer, following the manufacturer’s instructions (Life Technologies). Power SYBR green universal master mix and 7500 fast real-time PCR system (Applied Biosystems, Foster City, CA, USA) were used for real-time PCR work. The details of primers are given in Table 1. Thermal cycling conditions including an initial activation step at 95 °C for 10 min, followed by 40 cycles, then each cycle at 95 °C for 30 s, 53 °C for 30 s, 72 °C for 30 s and a final extension step at 72 °C for 7 min. Each sample was assayed in triplicates. A housekeeping control gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control to correct for differences in the amount of RNA in each sample. Melt curve analysis was performed to confirm the specificity of the products, and the values for relative quantification were calculated based on the 2(−∆∆ CT) relative expression formula.
Table 1.
List of primers used for real time-PCR
| Sl. no | Primer name | Primer sequence 5′ to 3′ | No. of bases | Amplicon size (bp) |
|---|---|---|---|---|
| 1 | hTERT forward | GGAGCAAGTTGCAAAGCATTG | 21 | 182 |
| hTERT reverse | TCCCACGACGTAGTCCATGTT | 21 | ||
| 2 | GAPDH forward | GAAGGTGAAGGTCGGAGTCAAC | 22 | 71 |
| GAPDH reverse | CAGAGTTAAAAGCAGCCCTGGT | 22 |
Western blotting
After treatment with BIBR1532, the LN-18 cells were harvested and lysed with cold RIPA lysis buffer [50 mM Tris, 150 mMNaCl, 1 mmol/l EDTA, 0.1% sodium dodecyl sulphate, 1% Triton X-100, 1 mol/l phenyl methyl sulfonyl fluoride (pH 8.0): SRL Ltd, Bangalore, India]. The lysate was centrifuged at 8000×g for 20 min at 4 °C. The protein content in the lysates was measured using Bradford assay (Bio-Rad Laboratories Inc., Hercules, CA, USA). For western blot analysis 50 μg protein was resolved in 12% SDS-PAGE gels, transferred onto PVDF membranes (Bio-Rad) and subsequently incubated in blocking buffer (5% BSA, 1% Tween 20 in 20 mol/l Tris-buffered saline (pH 7.6); SRL Ltd) for 2 h. The membranes were incubated with monoclonal anti-hTERT antibody (#sc-377511; 1:1000) and anti-mouse secondary antibody conjugated to horseradish peroxidase (#sc-358914; 1:5000) for 1 h. The protein was detected with an enhanced chemiluminiscence detection kit (Bio-Rad Laboratories Inc.). Equal loading of protein was confirmed by stripping the blots and re-probing with monoclonal mouse anti-human β-tubulin (#sc-58882; 1:2000) antibody. Monoclonal antibodies were purchased from Santa Cruz Biotech Inc (Santa Cruz, Dallas, TX, USA). The bands were analyzed by NIH Image-J software (National Institutes of Health, Bethesda, MD, USA).
Flowcytometry
Flow cytometry assay was performed by Annexin V/PI double staining method (BD Pharmingen, San Diego, CA, USA) to evaluate the induction of apoptosis. LN-18 cells (2 × 105) were seeded in 6-well plates. The cells were treated with various concentrations of BIBR1532. The cells were harvested at different time points using trypsin, washed with 1X PBS and suspended in 100 µl of 1X binding buffer. The cells were incubated with 5 μl of fluorochrome-conjugated Annexin V and 5 μl of propidium iodide at room temperature in the dark for 15 min. Diverse labeling patterns allowed us to identify different cell populations: early apoptotic cells (PI−/Annexin V+); apoptosis/necrotic cells (PI+/Annexin V+) and viable cells (PI−/Annexin V−). The data were acquired by flow cytometry FACS Aria III (BD Biosciences). Apoptotic cells were detected and expressed as the percentage of total cells. The experiments were repeated three times.
Telomeric repeat amplification protocol (TRAP) assay
To investigate whether BIBR1532 treatment reduces telomerase activation, the enzymatic activity was measured by Telo TAGGG telomerase PCR ELISA kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer’s protocol. In brief, LN18 cells were collected 48 h after treatment with BIBR1532 at various concentrations. The cells were washed with cold PBS, lysed in 200 µl lysis buffer and incubated on ice for 30 min. The lysate was centrifuged at 16,000×g at 4 °C for 20 min. Later the supernatant was collected and the protein extracts were subjected to TRAP assay. For the TRAP reaction, 25 μl of reaction mixture, 10 μg of sample, and nuclease free water to make a final volume of 50 μl. PCR was performed and ELISA was carried out following the manufacturer’s guidelines. A total of 5 μl of PCR products was added to a streptavidin-coated 96-well plate and hybridized to a digoxigenin (DIG)-labeled telo-meric repeat-specific detection probe. The immobilized PCR products were detected with peroxidase-conjugated anti-DIG antibody. After addition of the stop reagent, the plate was assessed on a plate reader at a wavelength of 450 nm within 30 min. The cell extracts were heat inactivated for 10 min at 85 °C and used as negative controls.
Statistical analysis
Statistical analysis was performed with GraphPad Prism5.0 (GraphPad Software, San Diego, CA, USA). The data are expressed as mean ± standard deviation (SD) of three independent experiments. Difference in the mean values were evaluated using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. The least significant difference method was used to compare the mean values of different experimental groups. p < 0.05 was considered statistically significant.
Results
Effect of BIBR1532 on cell morphology
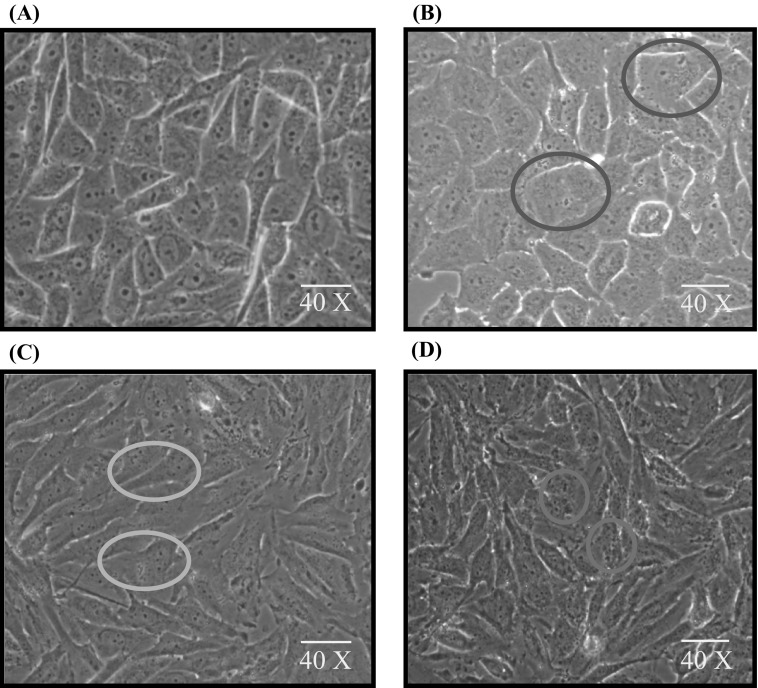
LN18 cells were cultured with or without BIBR1532 at different concentrations. When the cells were examined by phase contrast microscopy, LN18 cells treated with 25 µM BIBR1532 displayed flat and enlarged cell morphology. Cells treated with 100 and 200 µM BIBR1532 presented with narrow cell morphology and loss of nuclear membrane in each cell. LN18 cells at 200 µM showed black granule like structures inside the cells (Fig. 1).
Fig. 1.
Morphological changes induced by BIBR1532 in LN18 cells. Phase-contrast images (40× magnification) of LN18 cells. a, b Represent control cells and 25 µM concentration of BIBR1532 treated LN18 cells respectively. c, d Represent LN18 cells treated with 100 and 200 µM BIBR1532, respectively. Blue circle indicates enlarged cell morphology and red circle indicates the granule like structures inside the cells. Green and orange circle in (c) represents narrow cell morphology and loss of nuclear membrane in each cell. (Color figure online)
Effect of BIBR1532 on cell viability
The telomerase inhibitor BIBR1532 has been demonstrated to inhibit telomerase activity in many human cancer cell lines resulting in progressive shortening and dysfunction of telomeres. We tested whether BIBR1532 treatment could decrease the viability of LN-18 cells, by using the MTT assay.
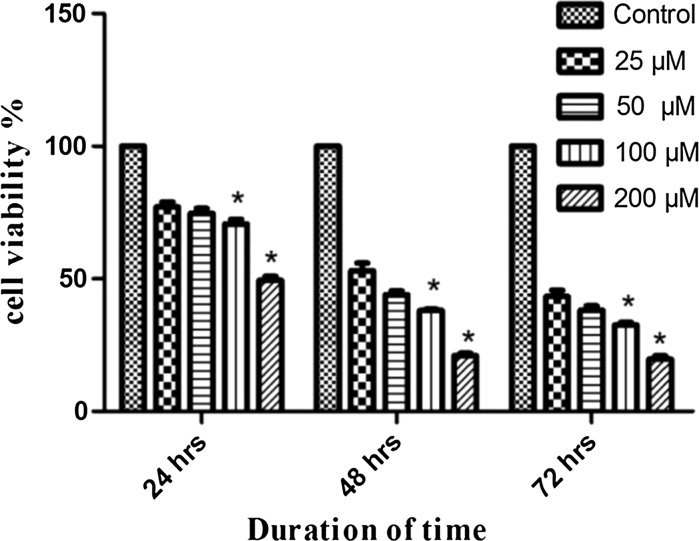
LN18 cells were incubated with (or without) BIBR1532 at various concentrations ranging from 10 to 200 µM and the cell viability was monitored for 24, 48 and 72 h. LN18 cells showed a significant dose dependent cytotoxic effect with IC50 value of 25 µM and 47, 56 and 62% reduction in cell viability after exposure to 25, 50 and 100 µM at 48 h. Treatment with 200 µM BIBR1532 caused more than 70% decrease in viability of LN18 cells at 48 and 72 h (Fig. 2). Taken together, these findings suggest that increasing concentrations of BIBR1532 above the concentration of 25 μM could exert a direct growth suppressive and cytotoxic effect, in contrast to the long-term cell cycle arrest mediated by substantial telomere erosion.
Fig. 2.
Assessment of cell viability of LN18 cells following treatment with BIBR1532. The data are presented as mean ± SD from three independent experiments. *p < 0.05 versus control group
The effect of BIBR1532 on a normal human cell line (CHME3) was performed to confirm selectivity. CHME3 cells showed a significant dose dependent cytotoxic effect with IC50 value of 90 µM after 48 h. Cell viability was decreased to 56 and 49.6% after 24 and 48 h at a concentration of 90 µM (Supplementary Table 1).
Effect of BIBR1532 on hTERT expression in LN18 cells
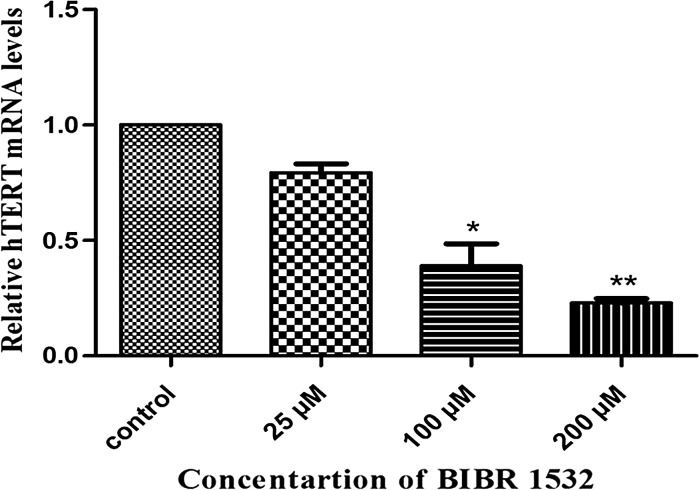
To verify the inhibitory effects of BIBR1532 on telomerase activity, we examined whether this agent could modulate the expression levels of hTERT mRNA. Real time PCR results showed that the level of hTERT mRNA expression in cells treated with 25, 100 and 200 µM BIBR1532 treated groups was decreased ~ 21, ~ 61.2, and ~ 77%, respectively, with a p < 0.05 (Fig. 3). These results also demonstrated that hTERT mRNA levels were decreased in a dose-dependent manner.
Fig. 3.
Quantitation of hTERT mRNA level in LN18 cells following BIBR1532 treatment. The relative mRNA expression of hTERT in control and BIBR1532 treated cells is shown. The values indicated the mean ± SD of 3 independent experiments in each group. *p < 0.05 and **p < 0.001 versus control group
Analysis of hTERT protein expression after treatment with various concentrations of BIBR1532 indicated significant down regulation of hTERT expression compared to untreated control. The level of hTERT protein was decreased to 44, 61, and 86%, at concentrations, of 25, 100, and 200 µM, respectively (Fig. 4).
Fig. 4.
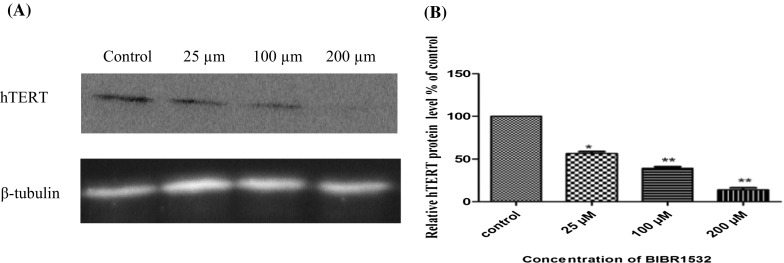
Quantitation of hTERT protein in control and BIBR1532 treated LN18 cells. a Western blot image of hTERT protein levels in the different groups. β-tubulin was used as an internal loading control. b Relative quantitation of hTERT protein plotted as hTERT/β-tubulin in each group. The data presented are mean ± SD of three independent experiments. *p < 0.005 and **p < 0.001 versus control group
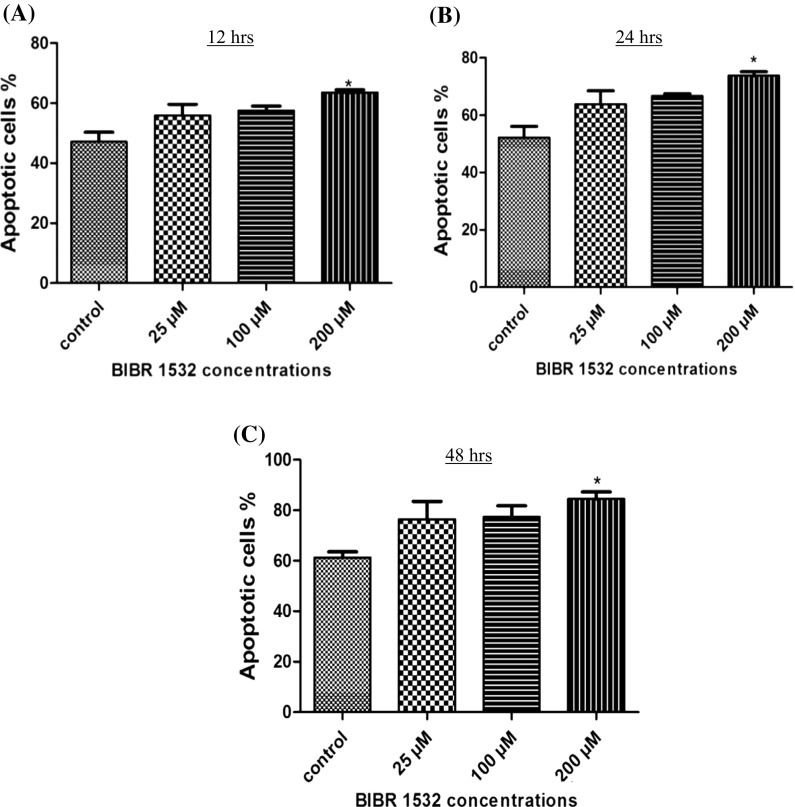
Effect of BIBR1532 on apoptosis in LN18 cells
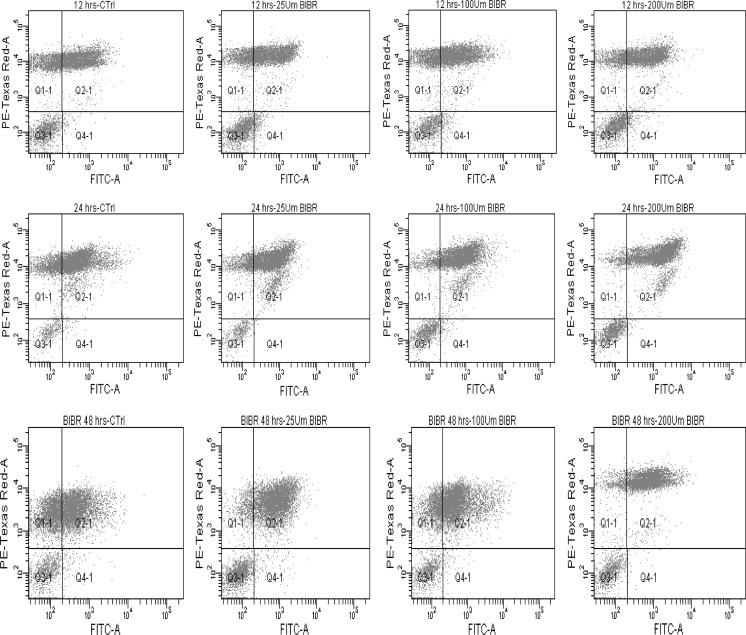
To determine whether BIBR1532 induced apoptosis in LN18 cells Annexin V-FITC and PI staining was used to quantify apoptosis after treatment with BIBR1532 (Fig. 5). The results showed a significant increase in the total percentage of apoptotic cells at 200 µM of BIBR1532 with 63.6, 73.8, and 84.5% total apoptotic cells at 12, 24, and 48 h, respectively, compared to control (Fig. 6). These data suggest that the growth inhibitory effect of BIBR 1532 on LN18 cells is through apoptosis.
Fig. 5.
Flow cytometry profile of LN18 cells undergoing apoptosis following treatment with BIBR1532. Cells were treated with different concentrations of BIBR 1532 at different time points. Annexin V-FITC/PI double staining was performed on LN18 cells after treated with BIBR 1532 at 25, 100, and 200 μM for 12, 24 and 48 h. The graphs are representatives of three independent experiments. Apoptosis was measured using flowcytometry (X axis, Annexin V and Y axis PI). The Annexin V+/PI− cells represent apoptotic cells (Q4), and the Annexin V−/PI+ represent necrotic cells (Q1). Q3 represents viable cells and Q2 represents late apoptotic cells (Annexin V+/PI+ cells)
Fig. 6.
Quantitation of apoptosis in control and BIBR1532 treated LN18 cells at different time intervals. Bar diagram showing the total percentage of apoptotic cells after treatment with BIBR1532. The percentage of apoptotic cells increased with 200 µM concentration of BIBR1532 at different time points. The mean ± SD is taken from three independent experiments. *p < 0.05 was considered statistically significant
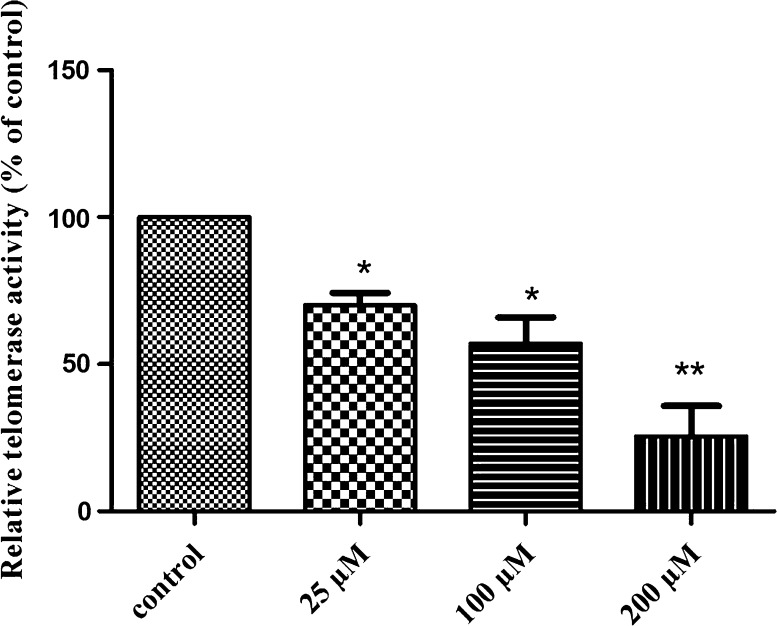
Effect of BIBR1532 on telomerase activity
To determine whether inhibition of hTERT mRNA and protein was correlated with inhibition of telomerase activity, we tested the effect of BIBR1532 telomerase activity in LN18 cells. TRAP assay was performed using partially purified telomerase from cells treated with different concentrations of BIBR1532 for 48 h indicated dose-dependent manner inhibition. While 25 µM of BIBR1532 resulted in a partial inhibition, treatment with 100 and 200 µM BIBR1532 inhibited the telomerase activity up to 42.9 and 74.4%, respectively (p < 0.05, Fig. 7).
Fig. 7.
Quantitation of telomerase activity in control and BIBR1532 treated LN18 cells. The data are presented as mean ± SD which is a representative of three independent experiments. *p < 0.05 and **p < 0.005 versus control
Discussion
Telomerase plays an important role in cancer development. It regulates the length of telomeres in tumor cells, maintains proliferative capacity and tumor progression in 90% of human malignancies (Shay and Wright 2011). hTERT, the catalytic component of telomerase is upregulated in several cancers including glioblastomas (Cong et al. 2002; Hahn et al. 1999). The expression and activity of telomerase was also correlated with poor survival in glioblastomas (Lotsch et al. 2013). Thus, studying telomerase has enormous potential in diagnostic, prognostic, and therapeutic applications in various cancers.
Several methods have been developed to inhibit telomerase activity. Previous studies demonstrated that telomerase has been inhibited or inactivated via gene knockout, antisense oligonucleotides, peptide nucleic acids, or nucleosidic reverse target inhibitors (Hahn et al. 1999; Herbert et al. 1999; Strahl and Blackburn 1996). But these methods showed limited success in vivo due to the inability of the compounds to cross cellular membranes under physiological conditions or due to their low efficacy. It has been reported that targeting either hTR or hTERT by antisense oligonucleotides reduced cell proliferation and tumor growth in human bladder and prostate cancer cells (Chen et al. 2003; Kraemer et al. 2004). However antisense strategies and gene therapy hold great potential for the treatment of several human diseases, but the possibility of gene therapy and clinical usefulness of most antisense compounds remain unclear. A previous study from our lab showed that, silencing of hTERT expression in glioblastoma cells by a specific siRNA resulted in decreased cell growth and increased cell apoptosis (Lavanya et al. 2016). However, in the past, several challenges have been encountered in the delivery of siRNA into solid tumors. Therefore, the clinical application of hTERT siRNA in the treatment of glioblastoma cancer may require other confirmed apoptosis-inducing agents. Hence, the use of small molecule inhibitors, may be considered as a potential anti-telomerase therapy in patients with glioblastoma (Ward and Autexier 2005).
Numerous telomerase inhibitors, that block either the activity of the telomerase or recruitment of the enzyme to telomeres, have been developed recently. One such compound is BIBR1532, which has opened the new therapeutic window of chemotherapy treatments. BIBR1532 is a non-nucleoside compound which binds to a site in the telomerase that is distinct from the site for deoxyribonucleotides and the DNA primer, and acts as a chain terminator during nucleotide polymerization, leading to inhibition of the catalytic activity of telomerase (Pascolo et al. 2002). It has been demonstrated that BIBR1532 does not cause chain termination events but rather prevents the formation of long reaction products. In particular, BIBR1532 leads to an overall decrease in the number of added TTAGGG repeats. This suggests that the inhibitor not only block the basic catalytic steps involved in template copying but also specifically impairs the elongation of the DNA substrate after its extension to the 5′ end of the template. Therefore, BIBR1532 may affect translocation of the enzyme DNA substrate complex or may promote dissociation between DNA substrate and the enzyme upon completion of template copying. As these steps are most likely unique to telomerase, this may project BIBR1532 a novel class of telomerase inhibitor (Pascolo et al. 2002).
Telomerase inhibition using BIBR1532 in human cancer cells of different histological origin leads to progressive shortening of telomeres down to a critical checkpoint (Damm et al. 2001) and inhibition of cell proliferation and also reduction in the tumorigenic potential following xeno-transplantation into nude mice (De Cian et al. 2008). BIBR1532 could selectively inhibit telomerase and induce apoptosis in human cancer cell lines derived from lung, breast, fibrosarcoma and prostate carcinoma through p21-induced senescence (Damm et al. 2001; Shi et al. 2015). In leukemia cells BIBR1532 directly induced a cytotoxic effect through the activation of p21 coupled with downregulation of c-Myc and hTERT transcription (Bashash et al. 2013). BIBR1532 inhibited the activity of telomerase through transcriptional suppression of survivin-mediated c-Myc and hTERT expression, increasing p73 and p21 expression, up-regulating the Bax/Bcl-2 molecular ratio and finally increasing P53-induced apoptosis (Bashash et al. 2012; Brassat et al. 2011). TP53 is the final executant of the telomerase inhibiting effect of BIBR1532. In P53-negative K562 cells, the telomere length was stabilized when it reached approximately 5 kb (Brassat et al. 2011).
Previous studies reported that long-term cultures of several human cancer cell lines in the presence of the telomerase inhibitor BIBR1532 at a concentration of 10 µM caused telomeric attrition and subsequent activation of the DNA damage response pathway, leading to cell cycle arrest and apoptosis (Damm et al. 2001). It has been demonstrated that a dose-dependent growth suppressive and antiproliferative effect has been shown in leukemia cell lines using increasing concentrations of BIBR1532 above 20 µM (El-Daly et al. 2005). Altogether, the results from previous studies demonstrated that BIBR1532 inhibits telomerase activity. In order to understand the role of BIBR1532 in altering telomerase activity, we first attempted to prove that BIBR1532 modulates telomerase activity and hTERT expression in glioblastoma cells.
In the current study, we have demonstrated that BIBR1532 represents a potent specific inhibitor of hTERT, which exhibits a selective cytotoxicity against the human glioblastoma cell LN18. This effect was observed when BIBR1532 was used at higher concentrations than at the previously reported concentration for inhibition of catalytic activity. This suggests that BIBR1532 exerts a direct cytotoxic potential, which may be separate from telomerase inhibition. The IC50 value for the purified telomerase enzyme has been determined to be 93 nM. In contrast, concentrations higher than 100 µM were more effective to inhibit RNA polymerases I to III (Damm et al. 2001).
We found that more than 70% decrease in viability when LN18 cells were exposed to the drug at a concentration of 200 µM of BIBR1532. Thus, an increase in the doses of BIBR1532 in cell culture results in a direct antiproliferative effect in contrast with delayed growth arrest. Evidence from previous studies showed that in endometrial cancer cells, inhibition of hTERT expression by BIBR1532 leads to an acute telomerase depletion and cell growth arrest (Kong et al. 2015). We also observed that BIBR1532 showed more cytotoxic effect on LN18 cells at a lower concentration (IC50 = 25 µM) when compared to CHME3 cells. These results suggest that LN18 cells are more sensitive to BIBR1532 at a low concentration than CHME3 cells.
Transcriptional regulation of hTERT and c-Myc is thought to be the major mechanism of human telomerase regulation (Cerni 2000; Wojtyla et al. 2011). c-Myc is believed to be a strong regulator of telomerase, and its ability to induce the transcriptional activation of hTERT has been shown to be increased in survivin gene transfectant cells (Endoh et al. 2005). To investigate the effect of BIBR1532 on transcription regulation of hTERT cells were subjected to treatment with different concentrations of BIBR1532. The mRNA and protein expression levels of hTERT were decreased in a dose-dependent manner following treatment with BIBR1532. Our data may define that high concentrations of BIBR1532 might delay cell proliferation and repress hTERT expression in LN18 cells through activation of c-Myc in glioblastoma cells. A previous study by Bashash et al. (2013) showed that decreased telomerase activity due to transcriptional suppression of survivin-mediated c-Myc and hTERT expression may be a postulated mechanism involved in BIBR1532-induced cell death in Nalm-6 leukemic cells (Bashash et al. 2013).
We have observed that BIBR1532 exposure led to a dramatic suppression of telomerase activity in glioblastoma cells. In LN18 cells the significant inhibition of telomerase activity was observed at 48 h. Treatment with 25 µM of BIBR1532 resulted in a partial inhibition of telomerase activity in LN18 cells, whereas treatment with higher concentrations 100 and 200 µM was associated with significant telomerase inhibition. BIBR1532 treatment caused a dose-dependent decrease in transcription of hTERT and in telomerase activity. hTERT is tightly regulated by various transcription factors and the hTERT core promoter is known to have two c-Myc, five Sp1, one Ets, and two Inr binding sites (Cong et al. 1999; Horikawa et al. 1999). In particular, the role of c-Myc in the regulation of hTERT transcription has been studied extensively (Kyo et al. 2000). c-Myc has been shown to bind to the c-Myc recognition sequence (E-box) at the promoter of hTERT and activate hTERT transcription (Wu et al. 1999). Our results may support the fact that c-Myc controls the transcription of hTERT and also suggest that the repression of telomerase activity could be via the decreased c-Myc DNA binding activity.
Apoptosis or programmed cell death is usually characterized by distinct morphological characteristics and energy-dependent biochemical mechanisms (Elmore 2007). The inhibition of telomerase activity has also been shown to induce apoptosis (Zhang et al. 2001). In our study, higher concentration BIBR1532 (200 µM) significantly resulted in an increase in apoptotic cells. However, BIBR1532 failed to catch early apoptosis response at all the time points.
Our results demonstrated that BIBR1532 induced apoptosis in human glioblastoma cells. This induction of apoptosis was associated with the down regulation of telomerase activity through transcriptional and posttranslational modification of hTERT. In view of accumulating evidence that BIBR1532 may be a therapeutic agent to suppress telomerase activity, further efforts are necessary in order to explore this therapeutic strategy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Lavanya C was supported by a UGC RGNF-Senior research fellowship (University Grants Commission-Rajiv Gandhi National Fellowship), New Delhi, India. The study was financially supported by DST-SERB (Department of Science & Technology-Science and Engineering research Board), Government of India. We thank Dr. Vijay Kumar Kalia for providing chemicals and reagents.
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvold ND, Reardon DA. Treatment options and outcomes for glioblastoma in the elderly patient. Clin Interv Aging. 2014;9:357–367. doi: 10.2147/CIA.S44259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai A, Oshima Y, Yamamoto Y, Uochi TA, Kusaka H, Akinaga S, Gryaznov S. A novel telomerase template antagonist (GRN163) as a potential anticancer agent. Cancer Res. 2003;63:3931–3939. [PubMed] [Google Scholar]
- Bashash D, Ghaffari SH, Zaker F, Hezave K, Kazerani M, Ghavamzadeh A, Vossough P. Direct short-term cytotoxic effects of BIBR 1532 on acute promyelocytic leukemia cells through induction of p21 coupled with downregulation of c-Myc and hTERT transcription. Cancer Investig. 2012;30:57–64. doi: 10.3109/07357907.2011.629378. [DOI] [PubMed] [Google Scholar]
- Bashash D, Ghaffari SH, Mirzaee R, Alimoghaddam K, Ghavamzadeh A. Telomerase inhibition by non-nucleosidic compound BIBR1532 causes rapid cell death in pre-B acute lymphoblastic leukemia cells. Leuk Lymphoma. 2013;54:561–568. doi: 10.3109/10428194.2012.704034. [DOI] [PubMed] [Google Scholar]
- Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- Brassat U, Balabanov S, Bali D, Dierlamm J, Braig M, Hartmann U, Brummendorf TH. Functional p53 is required for effective execution of telomerase inhibition in BCR-ABL-positive CML cells. Exp Hematol. 2011;39:66–76. doi: 10.1016/j.exphem.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Cerni C. Telomeres, telomerase, and myc. An update. Mutat Res. 2000;462:31–47. doi: 10.1016/S1383-5742(99)00091-5. [DOI] [PubMed] [Google Scholar]
- Chen Z, Koeneman KS, Corey DR. Consequences of telomerase inhibition and combination treatments for the proliferation of cancer cells. Cancer Res. 2003;63:5917–5925. [PubMed] [Google Scholar]
- Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet. 1999;8:137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm K, Hemmann U, Garin-Chesa P, Hauel N, Kauffmann I, Priepke H, Schnapp A. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- El-Daly H, Kull M, Zimmermann S, Pantic M, Waller CF, Martens UM. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood. 2005;105:1742–1749. doi: 10.1182/blood-2003-12-4322. [DOI] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh T, Tsuji N, Asanuma K, Yagihashi A, Watanabe N. Survivin enhances telomerase activity via up-regulation of specificity protein 1- and c-Myc-mediated human telomerase reverse transcriptase gene transcription. Exp Cell Res. 2005;305:300–311. doi: 10.1016/j.yexcr.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Falchetti ML, Pallini R, Larocca LM, Verna R, D’Ambrosio E. Telomerase expression in intracranial tumours: prognostic potential for malignant gliomas and meningiomas. J Clin Pathol. 1999;52:234–236. doi: 10.1136/jcp.52.3.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Herbert B, Pitts AE, Baker SI, Hamilton SE, Wright WE, Shay JW, Corey DR. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc Natl Acad Sci USA. 1999;96:14276–14281. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59:826–830. [PubMed] [Google Scholar]
- Komata T, Kanzawa T, Kondo Y, Kondo S. Telomerase as a therapeutic target for malignant gliomas. Oncogene. 2002;21:656–663. doi: 10.1038/sj.onc.1205072. [DOI] [PubMed] [Google Scholar]
- Kong W, Lv N, Wysham WZ, Roque DR, Zhang T, Jiao S, Zhou C. Knockdown of hTERT and treatment with BIBR1532 inhibit cell proliferation and invasion in endometrial cancer cells. J Cancer. 2015;6:1337–1345. doi: 10.7150/jca.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer K, Fuessel S, Kotzsch M, Ning S, Schmidt U, Wirth MP, Meye A. Chemosensitization of bladder cancer cell lines by human telomerase reverse transcriptase antisense treatment. J Urol. 2004;172:2023–2028. doi: 10.1097/01.ju.0000138157.46464.6e. [DOI] [PubMed] [Google Scholar]
- Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, Inoue M. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28:669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavanya C, Sibin MK, Srinivas Bharath MM, Manoj MJ, Venkataswamy MM, Bhat DI, Chetan GK. RNA interference mediated downregulation of human telomerase reverse transcriptase (hTERT) in LN18 cells. Cytotechnology. 2016;68:2311–2321. doi: 10.1007/s10616-016-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotsch D, Ghanim B, Laaber M, Wurm G, Weis S, Lenz S, Spiegl-Kreinecker S. Prognostic significance of telomerase-associated parameters in glioblastoma: effect of patient age. Neuro Oncol. 2013;15:423–432. doi: 10.1093/neuonc/nos329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MH, Liao ZL, Zhao XY, Fan YH, Lin XL, Fang DC, Yang SM. hTERT-based therapy: a universal anticancer approach (review) Oncol Rep. 2012;28:1945–1952. doi: 10.3892/or.2012.2036. [DOI] [PubMed] [Google Scholar]
- Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, Madden CJ, Bachoo RM. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res. 2010;16:154–163. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng E, Taylor B, Ray A, Shevde LA, Rocconi RP. Targeted inhibition of telomerase activity combined with chemotherapy demonstrates synergy in eliminating ovarian cancer spheroid-forming cells. Gynecol Oncol. 2012;124:598–605. doi: 10.1016/j.ygyno.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Hartmann U, Mayer F, Balabanov S, Hartmann JT, Brummendorf TH, Bokemeyer C. Targeting telomerase activity by BIBR1532 as a therapeutic approach in germ cell tumors. Invest New Drugs. 2007;25:519–524. doi: 10.1007/s10637-007-9063-6. [DOI] [PubMed] [Google Scholar]
- Nijaguna MB, Patil V, Urbach S, Shwetha SD, Sravani K, Hegde AS, Somasundaram K. Glioblastoma-derived macrophage colony-stimulating factor (MCSF) induces microglial release of insulin-like growth factor-binding Protein 1 (IGFBP1) to promote angiogenesis. J Biol Chem. 2015;290:23401–23415. doi: 10.1074/jbc.M115.664037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhage JL, Friedman KL. Chromosome end maintenance by telomerase. J Biol Chem. 2009;284:16061–16065. doi: 10.1074/jbc.R900011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch D, Brassat U, Brummendorf TH, Fellenberg J. Consequences of telomerase inhibition by BIBR1532 on proliferation and chemosensitivity of chondrosarcoma cell lines. Cancer Invest. 2008;26:590–596. doi: 10.1080/07357900802072905. [DOI] [PubMed] [Google Scholar]
- Pascolo E, Wenz C, Lingner J, Hauel N, Priepke H, Kauffmann I, Schnapp A. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J Biol Chem. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- Ruden M, Puri N. Novel anticancer therapeutics targeting telomerase. Cancer Treat Rev. 2013;39:444–456. doi: 10.1016/j.ctrv.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Sathornsumetee S, Reardon DA, Desjardins A, Quinn JA, Vredenburgh JJ, Rich JN. Molecularly targeted therapy for malignant glioma. Cancer. 2007;110:13–24. doi: 10.1002/cncr.22741. [DOI] [PubMed] [Google Scholar]
- Seimiya H, Oh-hara T, Suzuki T, Naasani I, Shimazaki T, Tsuchiya K, Tsuruo T. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-1991. Mol Cancer Ther. 2002;1:657–665. [PubMed] [Google Scholar]
- Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sun L, Chen G, Zheng D, Li L, Wei W. A combination of the telomerase inhibitor, BIBR1532, and paclitaxel synergistically inhibit cell proliferation in breast cancer cell lines. Target Oncol. 2015;10:565–573. doi: 10.1007/s11523-015-0364-y. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Strahl C, Blackburn EH. Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalized human cell lines. Mol Cell Biol. 1996;16:53–65. doi: 10.1128/MCB.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai LK. Telomeres, telomerase, and tumorigenesis: a review. Med Gen Med. 2004;6:19. [PMC free article] [PubMed] [Google Scholar]
- Ward RJ, Autexier C. Pharmacological telomerase inhibition can sensitize drug-resistant and drug-sensitive cells to chemotherapeutic treatment. Mol Pharmacol. 2005;68:779–786. doi: 10.1124/mol.105.011494. [DOI] [PubMed] [Google Scholar]
- Wojtyla A, Gladych M, Rubis B. Human telomerase activity regulation. Mol Biol Rep. 2011;38:3339–3349. doi: 10.1007/s11033-010-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, Yu JS. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- Zhang RG, Wang XW, Xie H. Telomerase activity inhibition and apoptosis induction of BEL-7404 human hepatoma cells by antisense oligonucleotides to telomerase RNA component. Shi Yan Sheng Wu Xue Bao. 2001;34:213–218. [PubMed] [Google Scholar]
- Zhang X, Zhang W, Cao WD, Cheng G, Zhang YQ. Glioblastoma multiforme: molecular characterization and current treatment strategy (review) Exp Ther Med. 2012;3:9–14. doi: 10.3892/etm.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.