Abstract
In our previous work, we isolated Arbas Cashmere goat hair follicle stem cells (gHFSCs) and explored the pluripotency. In this study, we investigated the expression and putative role of Sox9 in the gHFSCs. Immunofluorescence staining showed that Sox9 is predominantly expressed in the bulge region of the Arbas Cashmere goat hair follicle, and also positively expressed in both nucleus and cytoplasm of the gHFSCs. When the cells were transfected using Sox9-shRNA, cell growth slowed down and the expression of related genes decreased significantly, cell cycle was abnormal, while the expression of terminal differentiation marker loricrin was markedly increased; cells lost the typical morphology of HFSCs; the mRNA and protein expression of gHFSCs markers and stem cell pluripotency associated factors were all significantly decreased; the expression of Wnt signaling pathway genes LEF1, TCF1,c-Myc were significantly changed. These results suggested that Sox9 plays important role in gHFSCs characteristics and pluripotency maintenance.
Electronic supplementary material
The online version of this article (10.1007/s10616-018-0206-8) contains supplementary material, which is available to authorized users.
Keywords: Arbas Cashmere goat hair follicle stem cells, Characteristic, Pluripotency, Sox9
Introduction
Sox9 is a transcription factor with a highly conservative HMG domain structure which plays different roles in the multiple biogenic activities and developmental defects such as embryo development, germ layer (ectoderm, endoderm, and mesoderm) specialization, cell fate determination, maintenance and differentiation of stem cells and precursor cells, adult tissue regeneration and damage repair, diseases caused by congenital hypoplasia, and acquired diseases (Jo et al. 2014). Current evidence has shown that the development of testicles, gristle, heart, lungs, pancreas, bile duct, hair follicle, kidney, inner ear, cornea, and central nervous system are associated with Sox9. In addition, Sox9 expression is still required in these organs in adults (Stolt et al. 2003; Chaboissier et al. 2004; Furuyama et al. 2011; Seymour et al. 2007). Hair morphogenesis and development in mammals is a rigorously organized signal regulation between epidermis and the underlying matrix (Liu et al. 2013), including currently recognized Wnt, Shh, Notch, BMP and other signaling pathways, and the Wnt pathway is considered to be the master regulator during hair follicle morphogenesis (Rishikaysh et al. 2014). In the past several decades, many molecules involved in the hair growth have been obtained by using gene editing technology (Schneider et al. 2009). Vidal et al. (2005) first found Sox9 expression in both the fetal as well as adult mice hair follicle through in situ hybridization. Nowak et al. (2008) further demonstrated that the generation and development of hair follicle stem cells in the early embryo stage development were dependent on Sox9. To date, mice have been used as the model for investigation of hair follicles, while studies about other mammals are very rare.
The Inner Mongolia Arbas Cashmere goat (Capra hircus) is local cashmere and meat dual-purpose breed, which produces fine, soft, quality cashmere, and the growth cycle of the hair follicle is relatively long and with an obvious demarcation line, and thus could help investigating the mechanisms involved in the cyclic growth of the hair follicles. However, few studies about the mechanism of Arbas Cashmere goat hair growth have been reported. In our previous studies, we isolated and identified Arbas Cashmere goat hair follicle stem cells (gHFSCs) (He et al. 2016b), and explored the pluripotency of gHFSCs (He et al. 2016a). In the present study, the expression and putative role of Sox9 in gHFSCs growth, differentiation, pluripotency, and feature maintenance were evaluated, and also Wnt signaling associated genes were detected aiming to provide experimental evidence for further mechanism investigation involved in the effects of Sox9 on gHFSCs.
Materials and methods
Reagent Paraformaldehyde, TritonX-100, DAPI were all purchased from Sigma (St. Louis, MO, USA). Antibodies of HFSCs markers and stemness markers were purchased from Abcam (Cambridge, MA, USA). Antibodies to LEF1, TCF1 and c-Myc were purchased from Cell Signaling Technology (Danvers, MA, USA). RNAiso Plus, PrimeScriptTMRT reagent kit and SYBR® Premix Ex Taq were purchased from Takara (Dalian, China). RIPA lysis buffer (Millipore, Darmstadt, Germany).
Cells The gHFSCs were cultured as previously mentioned (Cytotechnology. 2016 Dec; 68(6):2579–2588. 10.1007/s10616-016-9981-20) (He et al. 2016b). The goat keratinocytes (gKCs) were isolated and cultured by Nimantana, and safed in the laboratory animal research center of Inner Mongolia University.
Immunohistochemistry
Skin samples from adult goat dorsal were embedded in OCT, frozen sectioned for 6 µm thickness, and fixed in 4% paraformaldehyde for 30 min. They were permeabilized with 0.05% Triton X-100 in PBS for 15 min and then incubated for 1 h in blocking solution (10% goat serum) at room temperature. Blocking solution was replaced by the Sox9 primary antibody. Control was incubated with the blocking solution instead of primary antibody. Then samples were incubated with Cy3-conjugated anti-rabbit IgG for 45 min in the dark. Slides were counterstained by DAPI then visualized using a confocal microscope (Nikon, Tokyo, Japan).
Immunocytochemistry
When the cells cultured on 24-well dishes with cover slips reached 70% of confluence, the gHFSCs were fixed in 4% paraformaldehyde at room temperature for 10 min and permeabilized with 0.05% Triton X-100 in PBS for 10 min. Antibody treatment and visualization were the same as Immunohistochemistry.
shRNA Transfection
Cells transfected with pGPU6/GFP/Neo shRNA vector (2 µg/µL) (GenePhama, Shanghai, China) targeting Sox9 mRNA were referred to as RNAi group, while cells transfected with non-specific shRNA to provide a negative control were referred to as NC group. The sequences of the sense and antisense strands of the goat sh-Sox9 were as follows: sense 5′-UCAACGGCUCGAGCAAGAATT-3′ and antisense 5′-UUCUUGCUCGAGCCGUUGATT-3′. The transfection was performed according to the manufacturer’s instructions. Briefly, 2 × 105 cells were seeded in 24-well culture plates without antibiotics and incubated with shRNA diluted with Opti-MEM (Invitrogen, Carlsbad, CA, USA) for 5 h, then replaced the MEM with fresh culture medium. After transfection, cells were screened by G418 (Sigma, St. Louis, MO, USA) for further analysis (Table S1).
Cell growth curve
The cells were seeded in 24-well plates at a density of 1 × 104 cells/mL and counts were performed every 24 h by hemocytometer. Three wells were counted for each time point, and the mean cell number was calculated to plot the cell growth curve.
Flow cytometry
After being harvested, gHFSCs were fixed in 70% alcohol for 2 h at 4 °C and treated with PI staining solution for 30 min at 37 °C in the dark. Flow cytometry was performed by BD FACS.
Real-time quantitive PCR
All primer sequences were determined using established GenBank sequences. Total RNA of the gHFSCs were extracted and reverse transcribed into cDNA with the PrimeScript™RT reagent kit (Takara) according to the manufacturer’s instructions. Q-PCR reaction system comprised 2 × SYBR Green Mix (10 μL), primer mix (1 μL), template (1 μL) and ddH2O (8 μL), and the parameters were as follows: 95 °C for 2 min followed by 40 cycles of 95 °C for 30 s and 60 °C for 30 s. To determine if there were multiple PCR amplicons, melting curves were constructed by heating final amplification reactions from 60 °C to 95 °C for 15 s, 60 °C for 30 s and 95 °C for 15 s in single degree steps. GAPDH was used as an internal reference gene. The relative mRNA expression level of each gene from triplicate experiments was calculated using the 2−ΔΔCt method (Schefe et al. 2006).
Western blot
Total cellular extracts of gHFSCs were obtained using RIPA lysis buffer (Millipore). Protein concentrations of the cell lysates were determined by spectrophotometry. Samples were electrophoretically separated by 12% SDS–polyacrylamide gels, and transferred onto nitrocellulose membranes and incubated with appropriate antibodies at 4 °C overnight. Blots were then incubated with peroxidase-conjugated secondary antibody, visualized by enhanced chemiluminescence. The intensity of each band was analyzed using ImageJ software. a-Tubulin was used as an internal reference protein.
Statistical analysis
All data are presented as a mean and standard error of the mean and analyzed using SPSS 17.0. A p value of less than 0.05 (p < 0.05) was considered statistically significant. Each experiment was repeated at least 3 times.
Results
Expression of Sox9 in gHF and gHFSCs
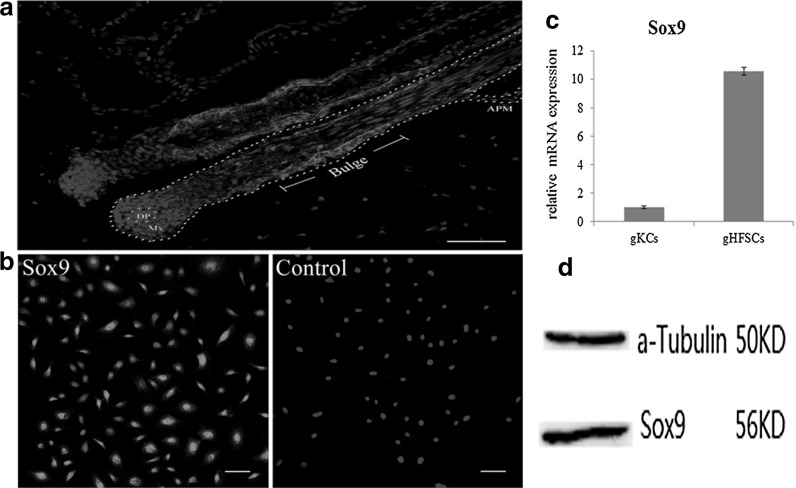
Immunohistochemistry of the frozen slices from the Arbas Cashmere goat skin showed expression of Sox9 in the whole ORS (outer root sheath) areas of gHF. The expression in the bulge region was evidently higher when compared to the other parts. Red fluorescence was also observed in the hair matrix (Fig. 1a). Cell immunohistochemistry showed that gHFSCs were Sox9 positive in both cytoplasm and nucleus (Fig. 1b). Q-PCR and western blot further confirmed the expression of Sox9 at transcription and protein levels in the gHFSCs (Fig. 1c, d).
Fig. 1.
Sox9 expression in the gHF and gHFSCs. a Immunohistochemistry identification of Sox9 in gHF. DP dermal papilla. Mx matrix. APM arrector pilli muscle. b Immunocytochemistry identification of Sox9 in gHFSCs. Scale bar 100 µm. c Relative mRNA expression of Sox9 in gHFSCs. gKCs: goat keratinocytes. d Protein expression of Sox9 in gHFSCs
Cell morphological changes
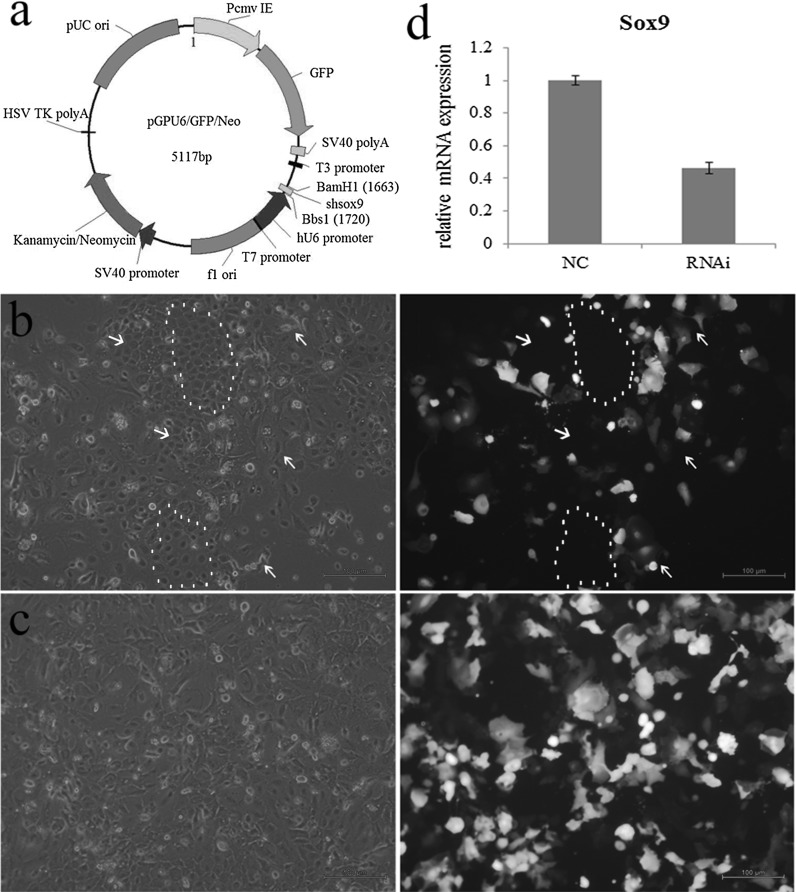
Microscopy at 24 h after Sox9-shRNA transfection showed that the cells with green fluorescence became flat and longer, cell adhesion, refractivity and junction between the cells were decreased, that is, the cell morphology has changed obviously. While the cells in the same visual field without green fluorescence still maintained the morphology of gHFSCs, which were relatively small and were connected closely with good adhesion and high refractivity (Fig. 2a). The cells transfected with Sox9-shRNA were referred to as RNAi group, and the cells tranfected with non-specific shRNA were referred to as NC group. The RNAi cells were screened by G418 which almost completely lost the morphological features of gHFSCs (Fig. 2b). Q-PCR showed that the level of Sox9 expression in the RNAi group was 0.4642-fold of the NC group, suggesting that the Sox9 expression was decreased by 53.58% (Fig. 2c).
Fig. 2.
Morphological changes of the cells. a Diagram of interference vector. b 48 h after transfection, white arrow shows the cells with green fluorescence with obvious morphological changes, red arrow and dashed line show cells without green fluorescence without obvious morphological changes. c Morphological changes of the cells after G418 screening. Scale bar 100 µm. d Changes of Sox9 relative mRNA expression. (Color figure online)
Cell proliferation was suppressed, and expression of terminal differentiation marker increased
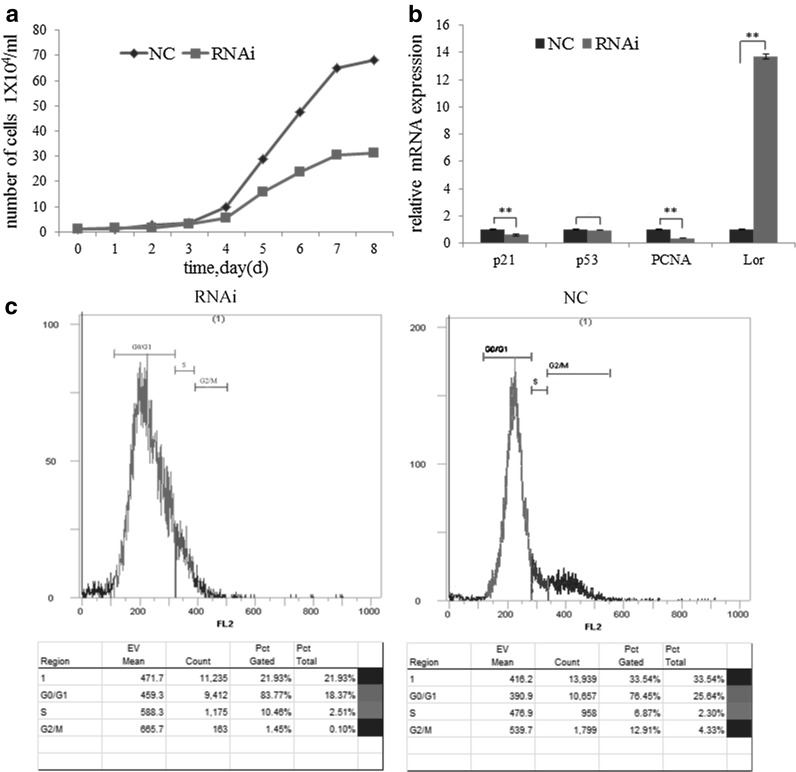
After the evaluation of the morphological changes of the cells, we further investigated the rate of cell growth and the effect on the cell cycle. In the RNAi group, cell growth was slower, number of cells in the logarithmic phase was less, and more dead cells occurred with the progression of cell cycle when compared with the NC group (Fig. 3a). Flow cytometry showed that in the NC group, 76.45% of the cells were in G0/G1 phase, 6.87% in S phase, and 12.91% in G2/M phase; while in the RNAi group, 83.77% of the cells were in G0/G1 phase, 10.46% in S phase, and 1.45% in G2/M phase (Fig. 3c).
Fig. 3.
Effects of Sox9 on gHFSCs growth. a Cell growth curve. c Cell cycle detection. b Differential expression of p21, p53, PCNA and loricrin. Mean and standard deviation are shown (n = 3). p values from t-test: *p < 0.05; **p < 0.01, relative to NC. NC: gHFSCs that transfected with non-specific shRNA to provide a negative control; RNAi: gHFSCs that transfected with sox9 shRNA
We further carried out Q-PCR to measure the expression of cell proliferation and apoptosis related genes including PCNA (proliferating cell nuclear antigen), p21, p53, and Lor (loricrin, a skin-derived cell differentiation marker), using the primers listed in Table 1. The results showed that p21, p53, PCNA, and Lor expression in the RNAi group was 0.6035 (reduced by 39.65%, p < 0.01), 0.9462 (p > 0.05), 0.3686 (reduced by 63.14%, p < 0.01), and 13.71 (p < 0.01) folds of the levels in the NC group (Fig. 3b).
Table 1.
Primer sequences of Sox9, p21, p53, PCNA, Lor, LEF1, c-Myc and GAPDH
| Gene name | NCBI accession | Sequence | Product (bp) | |
|---|---|---|---|---|
| Sox9 | XM_013972654.1 | Forward | 5′-CAAGTTCCCCGTCTGCATC-3′ | 112 |
| Reverse | 5′-GTGCGGCTTGTTCTTGCTC-3′ | |||
| p21 | XM_013973863.1 | Forward | 5′-ACTTGGACCTGTCGCTGTCC-3′ | 140 |
| Reverse | 5′-CCTGCGTTTGGAGTGGTAGAA-3′ | |||
| p53 | XM_005693530.2 | Forward | 5′-CCCATCCTCACCATCATCAC-3′ | 80 |
| Reverse | 5′-GCACAAACACGCACCTCAA-3′ | |||
| PCNA | XM_005688167.2 | Forward | 5′-GGCGTTCGTAATCGTGTTTTG-3′ | 81 |
| Reverse | 5′-CAGGTAGAAGGAGTGAGGAATGTGT -3′ | |||
| Lor | XM_010826802.1 | Forward | 5′-CATCTCCTCTCCTCACTCATCCTT-3′ | 117 |
| Reverse | 5′-GGTTTTCCCACAGCCCACT-3′ | |||
| GAPDH | XM_005680968.2 | Forward | 5′-TTGTGATGGGCGTGAACC-3′ | 127 |
| Reverse | 5′- CCCTCCACGATGCCAAA -3′ | |||
| LEF1 | NM_001285746.1 | Forward | 5′-CCCCACCTCTTGGCTGGTTTTCTCA-3′ | 177 |
| Reverse | 5′-TTGGCTCCTGCTCCTTTCTCTGTTC-3′ | |||
| c-Myc | XM_018058564.1 | Forward | 5′-GTGGTCTTCCCCTACCCGCTCAACGA-3′ | 322 |
| Reverse | 5′-ATCTCTTTAGGACCAACGGGCTGTGA-3′ |
Expression of gHFSCs specific markers were decreased
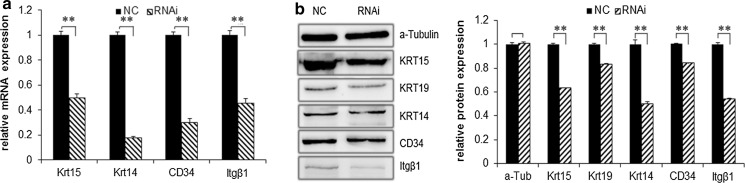
After Sox9 expression was interfered, the expression of the gHFSCs specific markers at transcription and protein levels were detected. The primer sequences are shown in Table 2. Q-PCR showed that the mRNA levels of Krt15, Krt14, CD34, and Itgβ1 in the RNAi group were 0.4963, 0.1763, 0.3024, and 0.4571 times of their expression in the NC group, respectively (all p < 0.01), namely the expressions of Krt15, Krt14, CD34, and Itgβ1 in the RNAi group were reduced by 50.37, 82.37, 69.76, and 54.29%, respectively (Fig. 4a).
Table 2.
Primer sequences of the gHFSCs markers
| Gene name | NCBI accession | Sequence | Product (bp) | |
|---|---|---|---|---|
| GAPDH | XM_005680968 | Forward | 5′-TTGTGATGGGCGTGAACC-3′ | 127 |
| Reverse | 5′-CCCTCCACGATGCCAAA-3′ | |||
| Keratin15 | XM_005693823 | Forward | 5′-GCTTTGGTGGGGGTTTTGGC-3′ | 107 |
| Reverse | 5′-CAAGCGGTCATTGAGATTC-3′ | |||
| Integrinβ1 | XM_005701221 | Forward | 5′-AGTTCAGTTTGCTGTGTGTTTGG-3′ | 268 |
| Reverse | 5′-TTCCTTTGCTTCGGTTCGTT-3′ | |||
| CD34 | XM_005691045 | Forward | 5′-AGATGGTCTTGCAGCTTCCAC-3′ | 119 |
| Reverse | 5′-CACCTCTTGGGACGAAATCAC-3′ | |||
| Keratin14 | XM_005693820 | Forward | 5′-GCGTGGGTAGTGGTTTTGGT-3′ | 107 |
| Reverse | 5′-CAGGCGGTCATTCAGGTTC-3′ |
Fig. 4.
Differential expressions of gHFSCs specific markers. a Q-PCR and b Western blot detection of gHFSCs markers. α-Tubulin was used as an internal reference protein. Mean and standard deviation are shown (n = 3). p values from t-test: *p < 0.05;**p < 0.01, relative to NC. NC: gHFSCs transfected with non-specific shRNA to provide a negative control; RNAi: gHFSCs transfected with sox9 shRNA
Gray scale of protein bands from western blot showed that the protein levels of Krt15, Krt19, Krt14, CD34, and Itgβ1 were 0.6325, 0.8326, 0.5006, 0.8408, and 0.5390-folds of the levels in the NC group, respectively (all p < 0.01), that is, at protein level, the expressions of Krt15, Krt14, CD34, and Itgβ1 in RNAi group were reduced by 36.75, 16.74, 49.94, 15.92, and 46.10%, respectively (Fig. 4b).
Expression of pluripotency associated factors were sharply decreased
In this study, mRNA and protein expression of pluripotency and self-renewal associated factors Oct4, Nanog, Sox2, AKP, and TERT were measured after Sox9 expression was interfered, and investigated whether the transcription factor Sox9 could significantly affect the pluripotency of gHFSCs. The primer sequences used are shown in Table 3.
Table 3.
Primer sequences of the stemness markers
| Gene name | NCBI accession | Sequence | Product (bp) | |
|---|---|---|---|---|
| Oct4 | XM_013977063.1 | Forward | 5′-GCCAAGCTCCTAAAGCAGAAGA-3′ | 122 |
| Reverse | 5′-AAAGCCTCAAAACGGCAGATAG-3′ | |||
| Nanog | NM_001285576.1 | Forward | 5′-GTCTCTCCTCTTCCTTCCTCCA-3′ | 116 |
| Reverse | 5′-TCTTCCTTCTCTGTGCTCTCCTC-3′ | |||
| Sox2 | NM_001285672.1 | Forward | 5′-CATGATGGAGACGGAACTGG-3′ | 115 |
| Reverse | 5′-CGGGCTGTTCTTCTGGTTG-3′ | |||
| AKP | XM_013974604.1 | Forward | 5′-ATGGTCACCATGAAGGCAAAG-3′ | 125 |
| Reverse | 5′-ATGGTCTGCAGTGGCAAGGA-3′ | |||
| TERT | XM_013972889.1 | Forward | 5′-GTGCTGAACTACGAGCGAGC-3′ | 147 |
| Reverse | 5′-GTCCGCCTTGACGAAGTAGAG-3′ | |||
| GAPDH | XM_005680968.2 | Forward | 5′-TTGTGATGGGCGTGAACC-3′ | 127 |
| Reverse | 5′-CCCTCCACGATGCCAAA-3′ |
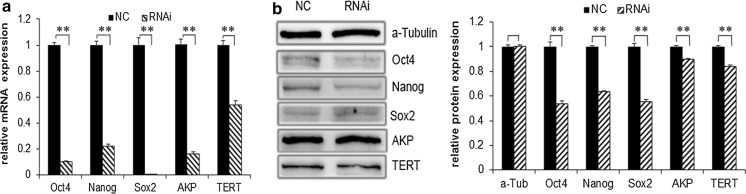
Q-PCR showed that the mRNA levels of Oct4, Nanog, Sox2, AKP, and TERT in RNAi group were 0.1066, 0.2221, 0.0082, 0.1652, and 0.5414-fold of the levels in the NC group, respectively (all p < 0.01), showing a significant reduction in the transcription levels of Oct4, Nanog, Sox2, AKP, and TERT in the RNAi group by 89.34, 77.79, 99.18, 83.48, and 45.86%, respectively (Fig. 5a). The protein levels of these factors in the RNAi group were 0.5425, 0.6401, 0.5591, 0.8975, and 0.8432-fold of the levels in the NC group, respectively (all p < 0.01), that is, the Oct4, Nanog, Sox2, AKP and TERT protein expressions were reduced by 45.75, 36.99, 44.09, 11.25, and 15.68%, respectively (Fig. 5b). These results showed that Sox9 interference could significantly reduce the expression of pluripotency associated factors at the transcription and protein levels.
Fig. 5.
Differential expressions of pluripotency associated genes in the gHFSCs. a Q-PCR and b Western blot detection of pluripotency associated genes in stem cells. α-Tubulin was used as an internal reference protein. Mean and standard deviation are shown (n = 3). p values from t-test: *p < 0.05; **p < 0.01, relative to NC. NC: gHFSCs transfected with non-specific shRNA to provide a negative control; RNAi: gHFSCs transfected with sox9 shRNA
Expression of LEF1, TCF1 and c-Myc were decreased
The mRNA and protein expression of Wnt signaling associated LEF1, TCF1 and c-Myc were measured after Sox9-shRNA transfection, and investigated whether Sox9 could significantly affect the pluripotency of gHFSCs. The primer sequences are shown in Table 1(no sequence of capra hircus TCF1 was found in NCBI).
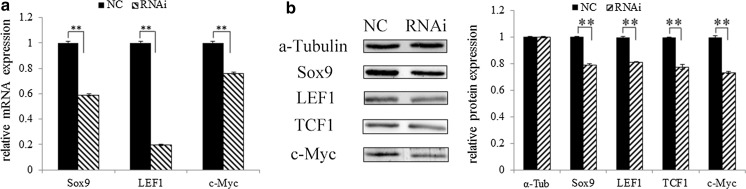
Q-PCR showed that the mRNA levels of Sox9, LEF1 and c-Myc in RNAi group were 0.5898, 0.1967, 0.7628-fold of the levels in the NC group, respectively (all p < 0.01) (Fig. 6a). The protein levels of Sox9, LEF1, TCF1 and c-Myc in RNAi group were 0.7931, 0.8112, 0.7745 and 0.7327 folds of the levels in the NC group, respectively (all p < 0.01), that is, these genes were reduced by 20.69, 18.88, 22.55 and 26.73%, respectively (Fig. 6b). It showed that Sox9 interference could significantly reduce the expression of LEF1, TCF1 and c-Myc at transcription and protein levels.
Fig. 6.
Differential expressions of LEF1, TCF1 and c-Myc in the gHFSCs. a Q-PCR and b Western blot detection. α-Tubulin was used as an internal reference protein. Mean and standard deviation are shown (n = 3). p values from t-test: *p < 0.05; **p < 0.01, relative to NC. NC: gHFSCs transfected with non-specific shRNA to provide a negative control; RNAi: gHFSCs transfected with sox9 shRNA
Discussion
Immunofluorescent staining showed that the expression pattern of Sox9 in the gHF bulge and gHFSCs was in agreement with the features that are similar in the mice and human hair follicles (Vidal et al. 2008).
Mice with Sox9 specifically knocked out from the skin showed several disadvantages which includes fragile hair, hair shaft atrophy, substantial reduction of the hair matrix cell number, decreased proliferation, outer root sheath cell apoptosis, hair bulb shortening, and lack or substantially reduced expression of HFSC markers (including CD34 and Krt15) at the bulge area (Vidal et al. 2005).
RNA interference is a sequence specific post-transcriptional gene silencing. Endogenous or exogenous double strand RNA (dsRNA) that has homologous complementary sequences with transcription products of target gene (mRNAs) could specifically degrade the corresponding mRNA in the cells, and thus effectively block the target gene (Bobbin and Rossi 2016). The application of RNA interference was most widely used in the investigation of gene functions. In the present study, microscopy at 6 and 12 h after Sox9-shRNA transfection found no suspension cells (dead cells), while the number of dead cells increased with time at 24 h. In addition, evident morphological changes of the cells were found after stable transfection.
Previous studies have shown that overexpression of Sox9 could increase the ability of clone formation and proliferation of ORS cells (Shi et al. 2013). In this study, the cell growth in the RNAi group was very slow after Sox9 expression was interfered; the cells could not form the normal cell clone and even stopped growing confirming that Sox9 could affect the proliferation of ORS derived cells. In vivo HFSCs without damage repair are mainly in the G0/G1 phase both in mice and human, and have the ability to form cell clones (Purba et al. 2014). In this study, flow cytometry showed that the number of cells in the quiescent state increased significantly in the RNAi group, percentage of cells in the G0/G1 and S phase increased while G2/M phase decreased significantly, suggesting that the cell division was abnormal and the cells could not progress to the G2/M phase. We speculated that these could be associated with the abnormal Sox9 expression which induced abnormalities in the DNA replication. The mean volume of the cells also increased, especially the cells in the S phase which was in accordance with the cell cycle results. p21 is involved in the regulation of transcription, apoptosis, DNA repair, as well as cell motility. It acts as a cell cycle repressor which is a well-known function. However, p21 appears to have a dual-face behavior, tumor suppressor or oncogene, depending on the cell type and cellular localization (Dutto et al. 2015). p21 interacts with PCNA at its carboxyl terminus. PCNA is a protein that plays a central role in DNA replication and repair. Lor is expressed in the stratum corneum which is a terminal differentiation marker of the skin-derived cells (Fuchs 1995). The expression of p21 and PCNA decreased significantly after Sox9 inhibition, and Lor expression was increased significantly which showed that the interfering Sox9 expression resulted in the suppression of gHFSCs proliferation, and induced terminal differentiation, which is consistent with the results found in the cell morphology, cell growth curve and flow cytometry. In the ESCs differentiation research, Sox9 directly regulated p21 expression, and p21 could bind to Sox2 and suppresses its expression (Yamamizu et al. 2014). The reduction of p21 expression in this study may also be affected by the same mechanism. As a tumor suppressor gene, p53 could promote cell death or persistently inhibit cell proliferation. Recent studies have shown that p53 could also exert effects in the cell survival (Chao 2015). Although p53 is not necessarily required in the normal cell proliferation and development, p53 plays a decisive role in DNA damage, anoxia, and cell apoptosis or survival that is caused by fluctuations in the nutrition. Post-translational modulations including phosphorylation, methylation, acetylation, and ubiquitination play important roles in the function of transcription factors. Latest studies have shown that ubiquitination could induce re-localization of p53, and decide p53 induced stress responses including cell proliferation, apoptosis, and treatment effects on cancers (Kruiswijk et al. 2015). In this study, cell death increased greatly while expression of p53 mRNA did not change significantly. This observation could be associated with the abnormal cell proliferation after Sox9 expression was suppressed, which may have induced the occurrence of phosphorylation and ubiquitination. Further studies are thus needed to investigate the post-transcriptional regulation of p53.
Both development and cyclic growth of hair follicles depend on the HFSCs, and Sox9 plays an important role in the development and maintenance of hair follicles (Christiano 2008). In this work, after Sox9 was inhibited, expression of gHFSCs specific markers Krt15, Krt19, Krt14, CD34, and Itgβ1 was reduced sharply at both transcription and protein levels. Combining with the obvious cell morphological changes, it indicates that Sox9 plays an important role in maintaining the characteristic of gHFSCs. Our previous studies have shown that gHFSCs could differentiate into osteogenic, chondrogenic, and myogenic lineages in vitro (He et al. 2016b), and the expression of pluripotency and self-renewal associated factors, including Oct4, Nanog, Sox2, AKP, and TERT, were detected at both transcription and protein levels (He et al. 2016a). In this study Q-PCR and western blot analysis showed that the expression of pluripotent factors reduced significantly at both transcription and protein levels after Sox9 inhibition in the gHFSCs, and the reduction of three core pluripotent regulatory factors, Oct4, Nanog, and Sox2 were more evident than AKP and TERT. As mentioned above, Sox9 could regulate Sox2 which affects the differentiation of ESCs. It has already been demonstrated that Sox2 and Oct4 could interact with each other in maintaining the pluripotency of stem cells (Masui et al. 2007), and Oct4 could also regulate the expression of Nanog (Loh et al. 2006). Therefore, the significant changes of the pluripotent factors in the gHFSCs demonstrated that Sox9 plays an important regulatory role in maintaining the pluripotency of gHFSCs. In vitro cultured skin stem cells are extremely sensitive to calcium concentration in the medium. Low level of calcium could maintain the features of the cells while high level of calcium could induce cell differentiation (Rinnerthaler et al. 2015). When cultured in high calcium medium, gHFSCs became longer, changed into fibroblast-like spindle shape morphology, Sox9 expression reduced, and Lor expression increased (Fig. s1).These findings indirectly demonstrated that Sox9 expression decreased after gHFSCs were differentiated.
Based on studies in human and mouse hair follicle, it shows that Wnt pathway induce hair follicle development, Shh is involved in morphogenesis and late stage differentiation, Notch signaling determines stem cell fate while BMP is involved in cellular differentiation. Hair placode formation depends on the activation of Wnt/β-catenin signaling. After placode forms, embryonic progenitor cells with attenuated Wnt/β-catenin signaling gain adult stem cell markers and become definitive long-term hair follicle stem cells at the end of organogenesis. Attenuation of Wnt/β-catenin signaling is a prerequisite for hair follicle stem cell specification since it suppresses Sox9, which is required for stem cell formation (Xu et al. 2015). In this study, the expression of LEF1, TCF1 and c-Myc was decreased both at transcription and protein level, however, the changes were not that obvious. So, it remains to be determined which, if any, other mechanisms lead to the maintenance of gHFSCs.
In this study, we investigated the Sox9 functions in the in vitro cultured gHFSCs, and the results were in agreement with the Sox9 functions in other stem cells, and in addition it also provided evidence to support that Sox9 acts as a factor regulating the pluripotency and plasticity of stem cells in the domestic animals.
Conclusions
From the above experimental results and analysis we summarized below
Sox9 was predominantly focused in the bulge region of adult gHF, where gHFSCs exist, and Sox9 could be detected at both transcription and protein levels in the cultured gHFSCs. After Sox9 expression was inhibited, gHFSCs growth was greatly suppressed, and terminal differentiation markers increased significantly; the cells lost the morphological features of HFSCs, and the expression of gHFSCs specific markers decreased sharply; expressions of stem cell pluripotency factors decreased significantly; expression of Wnt signaling genes LEF1, TCF1 and c-Myc decreased. These findings together demonstrated that Sox9 plays an important regulatory role in maintaining the characteristics and pluripotency of the gHFSCs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank all the staff participating in this research. This work was supported by a grant of the Key Special Projects in Breeding New Varieties of Genetically Engineered Organisms (2014ZX08008002) from Ministry of Science and Technology of the People’s Republic of China.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no competing interests.
References
- Bobbin ML, Rossi JJ. RNA interference (RNAi)-based therapeutics: delivering on the promise? Annu Rev Pharmacol Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- Chaboissier M-C, et al. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- Chao CCK. Mechanisms of p53 degradation. Clin Chim Acta. 2015;438:139–147. doi: 10.1016/j.cca.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Christiano AM. Hair follicle epithelial stem cells get their sox on. Cell Stem Cell. 2008;3:3–4. doi: 10.1016/j.stem.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Dutto I, Tillhon M, Cazzalini O, Stivala LA, Prosperi E. Biology of the cell cycle inhibitor p21CDKN1A: molecular mechanisms and relevance in chemical toxicology. Arch Toxicol. 2015;89:155–178. doi: 10.1007/s00204-014-1430-4. [DOI] [PubMed] [Google Scholar]
- Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol. 1995;11:123–154. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- He N, Dong Z, Zhu B, Nuo M, Bou S, Liu D. Expression of pluripotency markers in Arbas Cashmere goat hair follicle stem cells. In Vitro Cell Dev Biol-Anim. 2016;52:782–788. doi: 10.1007/s11626-016-0023-3. [DOI] [PubMed] [Google Scholar]
- He N, Dong Z, Tao L, Zhao S, Bou S, Liu D. Isolation and characterization of hair follicle stem cells from Arbas Cashmere goat. Cytotechnology. 2016;68:2579–2588. doi: 10.1007/s10616-016-9981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, Kang R, Shi LL, Mok J, Lee MJ, Haydon RC. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1:149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- Liu S, Zhang H, Duan E. Epidermal development in mammals: key regulators, signals from beneath, and stem cells. Int J Mol Sci. 2013;14:10869–10895. doi: 10.3390/ijms140610869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y-H, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Masui S, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purba TS, Haslam IS, Poblet E, Jiménez F, Gandarillas A, Izeta A, Paus R. Human epithelial hair follicle stem cells and their progeny: current state of knowledge, the widening gap in translational research and future challenges. Bioessays. 2014;36:513–525. doi: 10.1002/bies.201300166. [DOI] [PubMed] [Google Scholar]
- Rinnerthaler M, Streubel MK, Bischof J, Richter K. Skin aging, gene expression and calcium. Exp Gerontol. 2015;68:59–65. doi: 10.1016/j.exger.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Rishikaysh P, Dev K, Diaz D, Qureshi WMS, Filip S, Mokry J. Signaling involved in hair follicle morphogenesis and development. Int J Mol Sci. 2014;15:1647–1670. doi: 10.3390/ijms15011647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s C T difference” formula. J Mol Med. 2006;84:901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- Schneider MR, Schmidt-Ullrich R, Paus R. The hair follicle as a dynamic miniorgan. Curr Biol. 2009;19:R132–R142. doi: 10.1016/j.cub.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, et al. Expression and functional role of Sox9 in human epidermal keratinocytes. PLoS ONE. 2013;8:e54355. doi: 10.1371/journal.pone.0054355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier M-C, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal VP, et al. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- Vidal VPI, Ortonne N, Schedl A. SOX9 expression is a general marker of basal cell carcinoma and adnexal-related neoplasms. J Cutan Pathol. 2008;35:373–379. doi: 10.1111/j.1600-0560.2007.00815.x. [DOI] [PubMed] [Google Scholar]
- Xu Z, et al. Embryonic attenuated Wnt/b-catenin signaling defines niche location and longterm stem cell fate in hair follicle eLife. 2015;4:e10567. doi: 10.7554/eLife.10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizu K, Schlessinger D, Ko MS. SOX9 accelerates ESC differentiation to three germ layer lineages by repressing SOX2 expression through P21 (WAF1/CIP1) Development. 2014;141:4254–4266. doi: 10.1242/dev.115436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.