Abstract
Mammalian Tolloid-like 1 (Tll-1) is a pleiotropic metalloprotease that is expressed by a small subset of cells within the precardiac mesoderm and is necessary for proper heart development. Following heart tube formation Tll-1 is expressed by the endocardium and regions of myocardium overlying the region of the muscular interventricular septum. Mutations in Tll-1 lead to embryonic lethality due to cardiac defects. We demonstrate that the Tll-1 promoter contains Nkx2–5 binding sites and that the Tll-1 promoter is activated by and directly binds Nkx2–5. Tll-1 expression is ablated by a dominant negative Nkx2–5 or by mutation of the Nkx2–5 binding sites within the Tll-1 promoter. In vivo, Tll-1 expression is decreased in the hearts of Nkx2–5 knockout embryos when compared with hemizygous and wild-type embryos. These results show that Nkx2–5 is a direct activator of Tll-1 expression and provide insight into the mechanism of the defects found in both the Tll-1 and Nkx2–5 knockout mice.
Keywords: endocardium, Nkx2–5, precardiac mesoderm, Tolloid-like 1
Introduction
The formation of the heart requires the precise migration, differentiation, and interaction of different embryonic cell types. The susceptibility of the heart to developmental anomalies underscores the complexity of these interactions. The heart initially forms from a region of anterior lateral plate mesoderm, called the precardiac mesoderm, specified during gastrulation (Olson and Srivastava 1996). The precardiac mesoderm contains cells that will give rise to myocardium and endocardium. The precardiac mesoderm forms bilaterally symmetric fields during gut formation that fuse at the midline to form the primitive cardiac tube. Within the initially straight cardiac tube, the outer myocardium and inner endocardium are separated by a region filled with extracellular matrix (ECM) known as the cardiac jelly (Olson and Srivastava 1996). The interaction between the inner endocardium and the outer myocardium is critical for the proper development of the mature heart.
The molecular pathways that are necessary for heart field formation are conserved from arthropods to mammals. In Drosophila, tinman is a homeodomain-containing gene that is necessary for the formation of the dorsal vessel, the Drosophila equivalent of a heart. Mutations in tinman result in the complete absence of the dorsal vessel (Bodmer 1993). A homolog of tinman, found in vertebrates, is Nkx2–5. This is the earliest known marker of the vertebrate heart field and defines the region of lateral plate mesoderm that will give rise to the heart (Komuro and Izumo 1993; Lints et al. 1993). Nkx2–5 functions as a transcriptional activator.
Mammalian Tolloid-like 1 (Tll-1) is an astacin-like metalloprotease that is a member of the Tolloid family of proteins. Tll-1 is a pleiotropic enzyme processing a variety of substrates including ECM proteins such as procollagen, laminin, and proteoglycans (Rattenholl et al. 2002; Veitch et al. 2003; Ge et al. 2004; Gonzalez et al. 2005). In addition, Tll-1 cleaves chordin, an inhibitor of bone morphogenetic proteins (BMPs), that is expressed by the notochord during early development (Scott et al. 1999). We have previously shown that Tll-1 is necessary for proper formation of the heart (Clark et al. 1999). Tll-1 knockout mice display severe cardiovascular defects, including atrial and ventricular septal defects, abnormal rotation of the heart, and displaced outflow tract. Tll-1 is expressed by a small subset of cells in the anterior precardiac mesoderm, within the cardiac crescent that is defined by the expression of Nkx2–5. Tll-1 continues to be expressed by the endocardium of the developing heart tube and by the myocardium overlying the location of the future muscular interventricular septum (MIVS) (Clark et al. 1999). The Tll-1 promoter contains three putative Nkx2.5 binding sites (Tamura et al. 2005). Two of these are located approximately 1500 basepairs upstream of the transcriptional start site, whereas the third is located ~ 140 basepairs upstream of the transcriptional start site. We therefore hypothesized that Tll-1 gene expression is regulated by Nkx2–5 within the precardiac mesoderm.
Materials and methods
Ribonuclease protection assay
Total RNA was isolated from mouse cerebellum tissues with a MELT Total RNA Isolation System (Ambion) according to the manufacturer’s instructions. The Ribonuclease Protection Assay Kit (RPA) antisense probe was synthesized using a MAXIscript In vitro Transcription Kit (Ambion) and labeled with α−32P-UTP (Amersham Biosciences). Then the probe was purified by electrophoresis in a 6% denaturing polyacrylamide gel. The RNase protection assay was carried out using a kit (RPA III; Ambion). For detection of protected fragments and calculation of the start site for Tll-1 transcription, a denaturing polyacrylamide sequencing gel was used. A sequencing reaction was used to measure the size of the protected fragment (Sequenase 2.0 DNA Sequencing Kit, USB Corporation). The gel was dried and exposed to film.
Constructs
The Tll-1/pGl-3 construct consists of the 2.2 kilobase Tll-1 promoter driving luciferase in the pGl-3 basic vector. The Nkx2–5 construct was made by cloning the full-length Nkx2–5 cDNA (a generous gift from Dr Gary Lyons) into the pcDNA3.1 vector (Invitrogen) at the BamHI restriction site. This results in Nkx2–5 expression driven by the Cytomegalovirus (CMV) promoter. The dominant negative Nkx2–5 consists of the Nkx2–5 DNA binding region with the transcriptional activation domain removed and replaced with the engrailed transcriptional repression domain (a generous gift from Dr Ilona Skerjanc).
Transfection assays
QCE-6 cells (a generous gift from Dr Carolyn Eisenberg) were transfected with Tll-1/pGl-3 with or without Nkx2–5 and Nkx2–5 DN. In the titration experiments, equal amounts of DNA were transfected using empty plasmid to make up the difference. In addition, transfection efficiency was monitored by cotransfection of pCMV- βgal. Transfection was accomplished by using either Lipofectin or Geneporter in 96 well plates. All transfections were completed in triplicate. Following transfection, cells were incubated at 37°C with 5% CO2 for 48 h after which the media was aspirated and cells were lyzed in Dual-Glo lysis buffer and assayed for both luciferase and β-galactosidase activity using the Dual-Glo system (Promega). Luciferase values were normalized to β-galactosidase values to account for transfection efficiency.
Mutational analysis
The Nkx2–5 binding sites within the Tll-1 promoter were mutated using the Quik-Change Site directed mutagenesis kit (Stratagene). The native Nkx2–5 proximal sequence was changed using the following primers: Forward 5’-CTGACACGTACCTCATCTATCGGCCGCTG-TTCGTGGACGCAAATGC-3’; Reverse 5’-GCATTTGCGT-CCACGAACAGCGGCCGATAGATGAGGTACGTGTCAG-3’. The native Nkx2–5 distal sequence was changed using the following primers: Forward 5’-GCAGACATCACTGTCT-ATTCACGGCGAGGCAGCTGTCCCCATATG-3’; Reverse 5’-CCATATGGGGACTGCCTCGCCGTGAATAGACAGT-GATGTCTG-3’. The native Nkx2–5 medial sequence was changed using the following primers: Forward 5’-CCTGCAGGAGCAACACCAGCATGGGCCATACAAAC-AGCTTTTCTGTTTGG-3’; Reverse 5’-CAAACAGAAAAG-CTGTTTGTATGGCCGATGCTGGTGTTGCTCCTGCAGGG-3’. Transfections were carried out as described above.
Nuclear extract and Nkx2–5 protein purification
Protein nuclear extracts were made by transfecting either the pcDNA3.1 plasmid containing the Nkx2–5 sequence or the pcDNA3.1 plasmid alone into COS-1 cells using Lipofectin (Invitrogen) in 100 mm plates. Following transfection, cells were incubated for 48 h, after which the cells were harvested and lyzed in a glass homogenizer. Following cell lysis the nuclei were isolated, lyzed and the protein extract dialyzed against 20 mM HEPES (4-(2-hydroxyethyl)-1 -piperazineethanesulfonic acid), pH 7.9, 0.25 m sucrose, 50 mM KCl, 200 μM ethylenediaminetetraacetic acid (EDTA), 500 μM dithio- threitol (DTT), 5 mM MgCl2, 100 μM ZnCl2, 500 nM Phenylmethylsulphonyl fluoride (PMSF), 10 μg leupeptin, and 2 ng aprotinin. Nkx2–5 protein was purified from nuclear lysates using 6% crosslinked, beaded agarose with attached Nkx2–5 polyclonal IgG (Carbolink kit, Pierce) according to the manufacturer’s recommendations and concentrated on Centriplus centrifugal filter devices (Millipore).
Electrophoretic mobility shift assay
Oligonucleotides with the same sequence as the proximal Nkx2–5 site, the mutated site (see above), or a nonsense sequence with the same base composition as the native sequence but randomized, were synthesized (IDTDNA). The sense and antisense oligonucleotides were hybridized by combining equimolar amounts of each oligonucleotide, heating to boiling, followed by slow cooling to room temperature. Following hybridization the double-stranded oligonucleotides were end-labeled with γ32P Adenosine 5’-triphosphate. 50 000 counts of labeled oligonucleotide, with or without competing oligonucleotides, were incubated with 0.6 μg purified Nkx2–5 protein or purified control protein extract for 30 min on ice after which they were electrophoresed on a 5% non-denaturing polyacrylamide gel. The gel was dried, exposed to a phosphor screen, and imaged on a Typhoon imager (GE).
Embryo collection
Heterozygous Nkx2–5 knockout males and females were crossed to obtain wild-type, null, and heterozygous progeny. Females were checked daily in the morning for a vaginal plug, which was considered embryonic day (E) 0.5. The pregnant dam was euthanized on E9.5. The embryos were collected and heart tubes extracted in ice-cold phosphate-buffered saline (PBS). The hearts were frozen in dry ice and then stored at −80°C. Polymerase chain reaction (PCR) genotyping of yolk sac DNA was done as previously described (Tanaka et al. 1999).
RNA isolation and real time PCR
Total RNA from embryo heart tissues was isolated using TRI REAGENT (Molecular Research Center, Inc.) according to the manufacturer’s instructions. Real time reverse transcription-polymerase chain reaction (RT-PCR) was carried out using 50 ng total RNA per reaction. RNA was combined with primer/probe sets and TaqMan Gold RT-PCR Master Mix (Applied Biosystems, Inc.). Gene-specific primers and probes were created for mouse Tll-1 and glyceraldehyde phosphate dehydrogenase (GAPDH) using the Primer Express Software (Applied Biosystems, Inc.) as shown in Table 1. Real time assays were run on an ABI 7000 (Applied Biosystems, Inc). The real-time PCR profile consisted of one cycle at 48°C for 30 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All reactions were repeated twice and normalized to GAPDH. Nkx2–5 was quantified using 15 ng cDNA and the primers sets shown in Table 1 and detection by iQ SYBR Green (Bio-Rad). The PCR profile consisted of one cycle at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All reactions were carried out twice and normalized to GAPDH. Statistics were carried out using ANOVA.
Table 1.
Primers and probes used for the analysis of mammalian Tolloid-like 1 (Tll-1) by real-time polymerase chain reaction
| Target | Species | Forward primer | Reverse primer | MGB probe |
|---|---|---|---|---|
| Tll-1 | Mouse | cgcccagaccgagacaac | gtactcttgacctggctggatgtt | atgtcaccatcattagag |
| Nkx2–5 | Mouse | attttacccgggagcctacggtgac | gctttgtccagctccactgccttct | |
| GAPDH | Mouse | gggaagcccatcaccatctt | cggcctcaccccatttg | agcgagaccccactaa |
GAPDH, glyceraldehyde phosphate dehydrogenase; MGB, 3’-Minor Groove Binder probe.
Results
Identification of the transcription start site in the mouse Tll-1 promoter
To facilitate the characterization of the mouse Tll-1 promoter we identified the start of transcription. We carried out a ribonuclease protection assay using a 32P end-labeled BamHI/EcoRI fragment corresponding to the 3’ end of the putative promoter region and including the most 5’ region of the previously published cDNA (Takahara et al. 1996). This probe was hybridized to total RNA isolated from mouse cerebellum in accordance with the RPA III kit protocol as described by the manufacturer (Ambion). We have previously shown that high levels of Tll-1 mRNA are found in the mouse cerebellum (Scott et al. 2000). The protected fragment was electrophoresed on a denaturing polyacrylamide gel. The probe protected a 226 bp fragment that indicates a start of transcription approximately 142 bases downstream of the proximal Nkx2–5 binding site (Fig. 1).
Fig. 1.
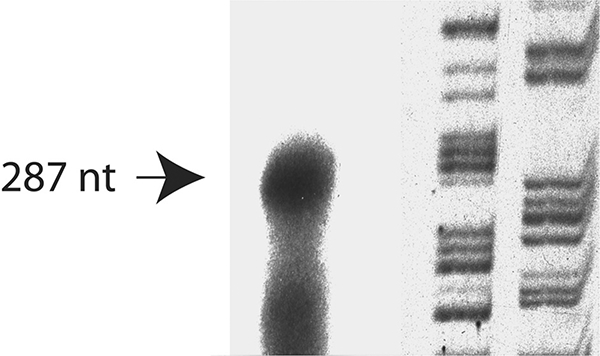
Identification of the transcription start site of the gene encoding Tolloid-like 1 by RNAse protection. The transcriptional start site was 750 bp upstream of the translation start site. Lane 1: RNAse protection of mouse cerebellum RNA; Lanes 2 & 3: Sequencing reaction (G, A).
The Tll-1 promoter is responsive to activation by Nkx2–5
The Tll-1 5’-flanking region contains putative Nkx2–5 transcription factor binding sites located 1741, 1438 and 142 bases upstream of the start of transcription (Fig. 2; see Tamura et al. 2005). These sites are conserved in both the mouse and human Tll-1 genes (data not shown). To determine if the Tll-1 promoter was responsive to Nkx2–5 we used the Tll-1/pGl-3 construct described previously (Tamura et al. 2005) which places the firefly luciferase gene under the control of the Tll-1 promoter. Further activation of the Tll-1 promoter was accomplished by cotransfection of the pcDNA3.1 vector containing the coding region of Nkx2–5 under the control of the CMV constitutive promoter. As a control, empty pGl-3 vector was used. The constructs were transfected in triplicate using GenePORTER transfection reagent (Gene Therapy Systems) into quail heart precardiac QCE-6 cells (a generous gift from Dr Carol Eisenberg) in 96 well plates. After 24 h, cells were harvested and luciferase activity was assayed as described (Tamura et al. 2005). QCE-6 cells were transfected with the Tll-1/pGl-3 construct and 1 ng, 10 ng, or 100 ng of pCDNA3.1 containing Nkx2–5. There was no increase in Tll-1 expression with 1 ng Nkx2–5 and only a slight increase in Tll-1 expression with 10 ng Nkx2–5 (Fig. 3). However the addition of 100 ng of Nkx2–5 containing plasmid resulted in a threefold increase in Tll-1 /pGl-3 expression over Tll-1 /pGl-3 without Nkx2–5. The construct containing the Tll-1 promoter displayed a 33-fold increase in luciferase activity over the empty vector (Fig. 3). These results indicate that the Tll-1 promoter is responsive to activation by Nkx2–5.
Fig. 2.

Map of Tolloid-like 1 (Tll-1) promoter.
Fig. 3.
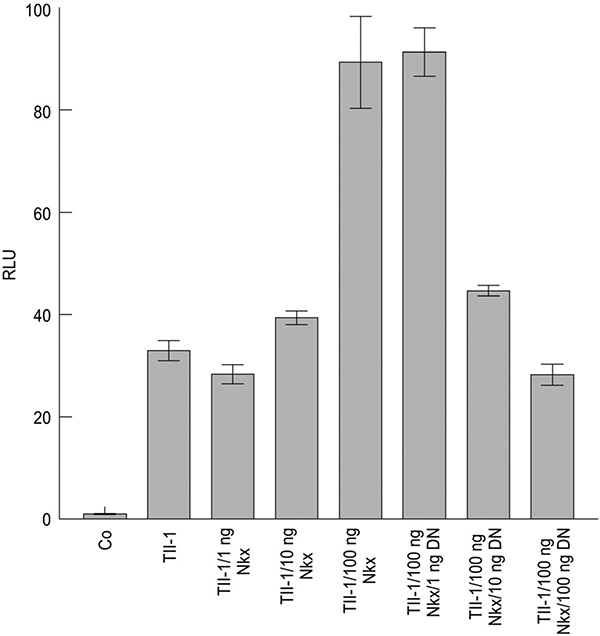
The 2.2kb Tolloid-like 1 (Tll-1) putative promoter functions as a promoter in vitro and is responsive to stimulation by Nkx2.5. The quail mesodermal cell line, QCE-6, was transfected with 0.1 μg of control plasmid (pGL-3), or pGL-3 with the mTll-1 promoter driving luciferase expression (Tll-1/pGL-3). Tll-1/pGL-3 was also cotransfected with constitutively expressed Nkx2.5 (mTll-1/pGL-3/Nkx) and/or a dominant negative construct (DN). Data are expressed as relative luciferase units (RLU) ± SEM.
To further characterize the responsiveness of the Tll-1 promoter to Nkx2–5 we used a dominant negative Nkx2–5 where the 60-aa homeobox DNA binding region of Nkx2–5 has been fused to the 196-aa repression domain of the mouse engrailed-2 protein (a generous gift from Dr Ilona Skerjanc). Addition of the construct containing the dominant negative Nkx2–5 had no effect at 1 ng, whereas addition of 10 ng and 100 ng reduced Tll-1/pGl-3 expression to the level of Tll-1 without Nkx2.5 (Fig. 3). These results indicate that the Tll-1 promoter is responsive to Nkx2–5 and that competition for an Nkx2–5 binding site by the dominant negative results in decreased expression of Tll-1.
We next sought to determine if the response was through direct transactivation of the Tll-1 promoter by Nkx2–5 or a different, indirect mechanism. We mutated the proximal Nkx2–5 (−142), medial Nkx2–5 (−1438), and distal Nkx2–5 (−1741) binding sites via site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene). This mutation resulted in the change of the core proximal Nkx2–5 binding site sequence from TAATA to CGGCC, medial Nkx2–5 binding site sequence from TAATT to CGGCC, and distal Nkx2–5 binding site sequence from TAATA to CGGCG. We then transfected these constructs as described above, with or without the addition of the pcDNA3.1 vector expressing Nkx2–5. Again, we observed a significant increase in Tll-1 expression when we compared the expression of luciferase driven by the wild-type Tll-1 promoter with the addition of Nkx2–5 to wild-type Tll-1 promoter without Nkx2–5 (Fig. 4). Mutation of all three Nkx2–5 sites results in a reduction of activation of the Tll-1 promoter to levels similar to that of the Tll-1 promoter without Nkx2–5 activation (Fig. 4). Mutation of individual Nkx2–5 sites resulted in either no decrease in activation of the Tll-1 promoter by Nkx2–5 (distal and medial Nkx2–5 sites) or a reduced level of activation by Nikx2–5 (proximal) (Fig. 4). Interestingly, mutation of two sites (medial/distal or proximal/medial) resulted in reduced activation of Tll-1 by Nkx2–5 but mutation of the proximal/distal sites had no effect on activation by Nkx2–5. Together, these studies indicate that the cis Nkx2–5 binding sites are necessary for activation of Tll-1 gene expression by the Nkx2–5 transcription factor.
Fig. 4.
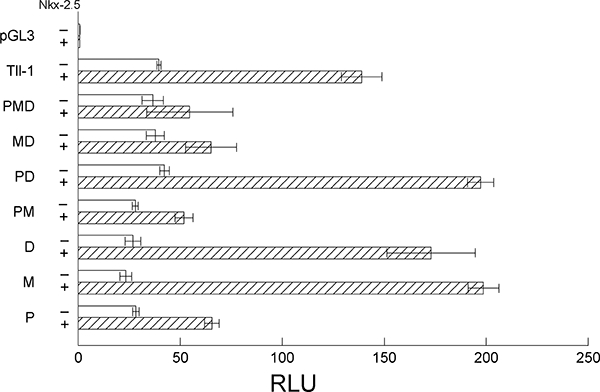
Mutation of the three Nkx2–5 sites ablates Tolloid-like 1 (Tll- 1) activation by Nkx2.5. QCE-6 cells were transfected with Tolloid- like 1 (Tll-1) with the proximal (P), medial (M), or distal (D) Nkx2–5 sites mutated with or without the cotransfection of Nkx2–5. Data are expressed as relative luciferase units (RLU) ± SEM.
Nkx2–5 protein directly binds to the Tll-1 promoter
Our previous experiments have shown that Nkx2–5 can activate the Tll-1 promoter and this activation appears to be through a direct interaction with the Tll-1 promoter. To show the direct binding of Nkx2–5 to the three Nkx2–5 binding sites within the Tll-1 promoter, we carried out electrophoretic mobility shift assays. Purified Nkx2–5 protein was incubated with double-stranded, end-labeled oligonucleotides corresponding to the proximal, medial and distal Nkx2–5 sites within the Tll-1 promoter, or with mutated forms of the sites (Fig. 5). In all three cases, binding of Nkx2–5 protein to the native Nkx2–5 sites within the Tll-1 promoter revealed bands that could be competed away with increasing concentrations of the unlabeled oligonucleotide (Fig. 5A-C). Furthermore, there is minimal direct binding of the Nkx2–5 protein to either the mutant Nkx2–5 sites or to nonsense sites indicating that Nkx2–5 binds specifically to the Nkx2–5 ci-elements in the Tll-1 promoter. We confirmed that the binding protein was Nkx2–5 by supershifting the complex for all three sites using an antibody directed against Nkx2–5 (Fig. 5A, right panel shows the proximal Nkx2–5 site. Medial and distal sites, data not shown).
Fig. 5.
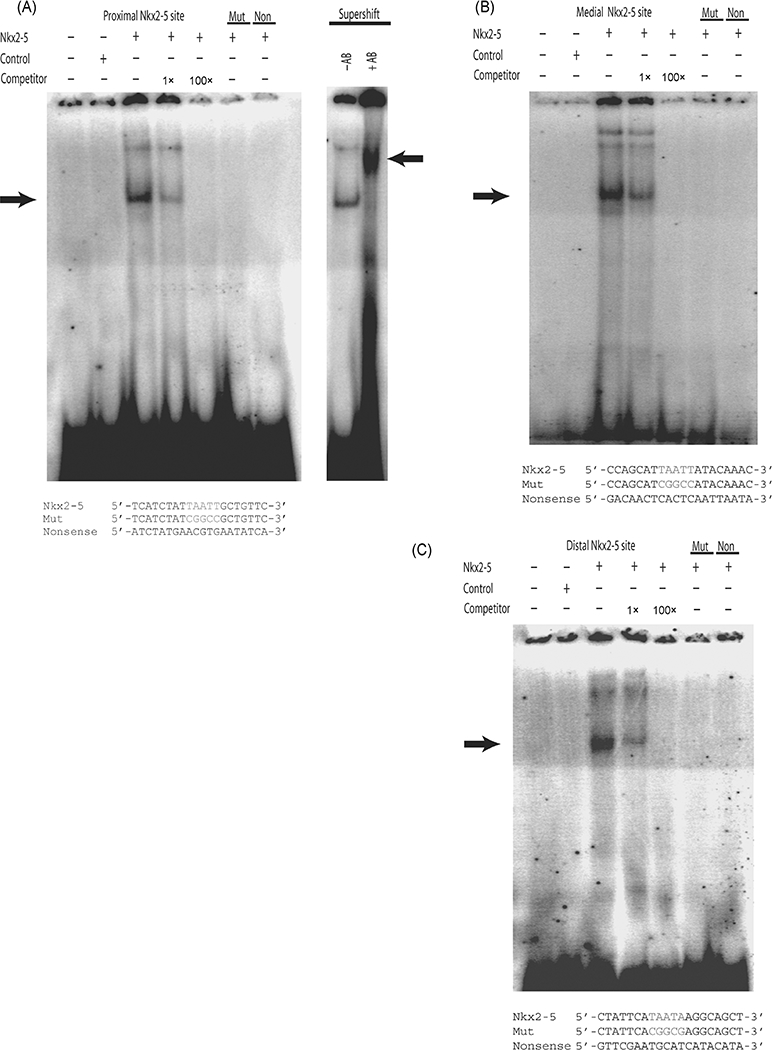
Purified Nkx2–5 protein binds to the Nkx2–5 ci-elements within the Tolloid-like 1 (Tll-1) promoter. (A) Electrophoretic mobility shift assay of the proximal Tll-1 Nkx2–5 site, the Mut site or a nonsense site with protein extracts containing Nkx2–5 grown in COS-1 cells. The sequences of the oligonucleotides are shown at the bottom of the figure. Changes in sequence from the mouse wild-type are indicated in red. Lane 1, Tll-1 proximal Nkx2–5 free probe. Lane 2, Tll-1 proximal Nkx2–5 site incubated with nuclear extract purified from COS-1 cells transfected with pc DNA 3.1 control plasmid. Lane 3, Tll-1 proximal Nkx2–5 site incubated with Nkx2–5 protein. Lane 4, Tll-1 proximal Nkx2–5 site incubated with Nkx2–5 protein and 1 × cold Tll-1 proximal Nkx2–5 site competitor. Lane 5, same as Lane 4 except with 100× cold competitor. Lane 6, Mut sequence incubated with Nkx2–5 protein. Lane 7, Nonsense sequence incubated with Nkx2–5 protein. Lane 8, Same as Lane 3 control antibody. Lane 8 Same as Lane 3 incubated with anti-Nkx- 2–5 antibody showing supershift. (B) Electrophoretic mobility shift assay of the medial Tll-1 Nkx2–5 site, the Mut site or a nonsense site with protein extracts containing Nkx2–5 grown in COS-1 cells. The sequences of the oligonucleotides are shown at the bottom of the figure. Changes in sequence from the mouse wild-type are indicated in red. Lane 1, Tll-1 medial Nkx2–5 free probe. Lane 2, Tll-1 medial Nkx2–5 site incubated with nuclear extract purified from COS-1 cells transfected with pc DNA 3.1 control plasmid. Lane 3, Tll-1 medial Nkx2–5 site incubated with Nkx2–5 protein. Lane 4, Tll-1 medial Nkx2–5 site incubated with Nkx2–5 protein and 1 × cold Tll-1 medial Nkx2–5 site competitor. Lane 5, same as Lane 4 except with 100 × cold competitor. Lane 6, Mut sequence incubated with Nkx2–5 protein. Lane 7, Nonsense sequence incubated with Nkx2–5 protein. (C) Electrophoretic mobility shift assay of the distal Tll-1 Nkx2–5 site, the Mut site or a nonsense site with protein extracts containing Nkx2–5 grown in COS-1 cells. The sequences of the oligonucleotides are shown at the bottom of the figure. Changes in sequence from the mouse wild-type are indicated in red. Lane 1, Tll-1 distal Nkx2–5 free probe. Lane 2, Tll-1 distal Nkx2–5 site incubated with nuclear extract purified from COS-1 cells transfected with pc DNA 3.1 control plasmid. Lane 3, Tll-1 distal Nkx2–5 site incubated with Nkx2–5 protein. Lane 4, Tll- 1 distal Nkx2–5 site incubated with Nkx2–5 protein and 1 × cold Tll-1 distal Nkx2–5 site competitor. Lane 5, same as Lane 4 except with 100 × cold competitor. Lane 6, Mut sequence incubated with Nkx2–5 protein. Lane 7, Nonsense sequence incubated with Nkx2–5 protein.
Expression of Tll-1 is decreased in the hearts of Nkx2–5 knockout mouse embryos
To investigate expression of Tll-1 in vivo, we assayed Tll-1 mRNA levels by quantitative real-time PCR in the hearts of Nkx2–5 knockout, heterozygotes and wild type embryos. Heterozygous Nkx2–5 knockout males and females were crossed to obtain wild-type, null, and heterozygous progeny. At 9.5 days’ gestation, the embryos were harvested and the hearts isolated and processed for RNA isolation. Nkx2–5 is expressed by the myocardium throughout the heart tube in E9.5 mouse embryos (Lints et al. 1993). As mentioned above, Tll-1 is expressed by the endocardium as well as regions of the myocardium overlying the future MIVS. Real-time PCR revealed a nearly twofold decrease in Tll-1 mRNA in knockouts when compared with wild- type embryos. Heterozygotes displayed an intermediate level of expression (Fig. 6). Analysis of the Nkx2–5 confirmed the absence of Nkx2–5 expression in the knockout embryos. Since at E9.5 the only cell types in which Nkx2–5 is coexpressed with Tll-1 is in the myocardium, Tll-1 endocardial expression could mask a more significant decrease in myocardial expression in the knockout. We cannot exclude the possibility, however, that Nkx2–5 gene dosage indirectly affects Tll-1 expression in the endocardium. Nevertheless, these data suggest that Nkx2–5 activation of Tll-1 gene expression is important in vivo during cardiac development.
Fig. 6.
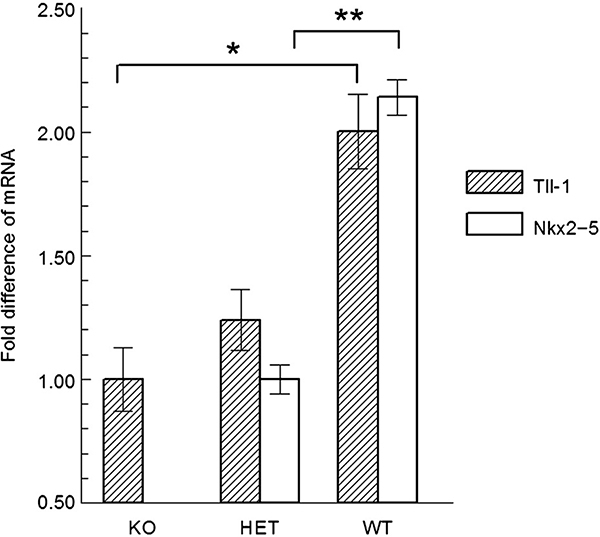
Real-time polymerase chain reaction (PCR) analysis of Tll-1 and Nkx2–5 mRNA expression in the hearts of Nkx2–5 knockout (KO) mouse embryos, heterozygous (HET) Nkx2–5 knockout mouse embryos, and wild type (WT) mouse embryos. Asterisks indicate statistical significance by anova. * P < 0.05; ** P < 0.001.
Discussion
The studies presented here show that Tll-1 is directly activated by Nkx2–5. There are three Nkx2–5 binding sites within the Tll-1 promoter and the Tll-1 promoter is responsive to the addition of Nkx2–5. This activation can be ablated by either mutation of the binding site or blockage of transcription activation by a dominant negative construct. We have also shown that Nkx2–5 protein directly binds to the Nkx2–5 binding sites in the Tll-1 promoter. Finally, in the hearts of embryonic Nkx2–5 knockout mice, Tll-1 mRNA expression is significantly decreased when compared with wild-type embryos.
Our finding that Nkx2–5 directly activates Tll-1 provides a mechanism for the expression observed in the precardiac mesoderm. Tll-1 is expressed by a subset of cells in the anterior precardiac mesoderm (Clark et al. 1999). Other unknown spatially restricted factors must also regulate or suppress Tll-1 expression, given that not all Nkx2–5 expressing cells in the precardiac mesoderm express Tll-1. In the mouse, deletion of Nkx2–5 results in the death of embryos at approximately 9–10 days postfertilization as a result of arrested cardiac development (Lyons et al. 1995; Tanaka et al. 1999). These defects include a lack of looping morphogenesis at the linear heart tube stage (8.25–8.50 days p.c) and no expression of the myosin light-chain 2 V gene (MLC2V). In contrast, Tll-1 ablation in mice results in embryonic death at ~13.5 days of gestation from valve and septal defects. Interestingly, Tll-1 mutant embryos also have defects in the position of the heart, which indicates a possible defect in looping. This indicates that the lack of Nkx2–5 activation of Tll-1 may be responsible for some but not all of the defects found in the Nkx2–5 knockout mice (Clark et al. 1999).
The myocardial expression of Tll-1 mRNA at the location of the future MIVS is particularly interesting in light of the recent identification of a secondary heart field (Mjaatvedt et al. 2001). The secondary heart field is a population of cells originating from the splanchnic and pharyngeal mesoderm that give rise to the outflow tract, right ventricle and ventricular septum and express Nkx2–5 (Verzi et al. 2005; Prall et al. 2007). Future studies will determine if Tll-1 plays a similar role in the primary and secondary heart fields in response to Nkx2–5 activation.
The activation of Tll-1 by Nkx2–5 provides for a novel transcriptional pathway during embryonic cardiac development. Nkx2–5 is activated by a number of factors including bone morphogenetic protein 2 (BMP-2) (Andree et al. 1998). In this work we show that Nkx2–5 is capable of activating Tll-1 gene expression. Interestingly, Tll-1 protein has a protease activity that not only processes components of the ECM, but also cleaves chordin, a potent inhibitor of the BMPs (Scott et al. 1999). Thus, BMP-2 activation of Nkx2–5 may result in increased expression of Tll-1, which has the ability to cleave chordin liberating bound BMPs, which are then able to further activate the system. Whether a positive feedback loop functions in vivo remains to be determined.
Acknowledgments
The following grants supported this study: American Heart Association Northland Affiliate Scientist Development Grant 0235534Z to T.G.C., National Center of Research Resources Grant P20 RR15567 to T.G.C. and the National Institute of Child Health and Human Development Grant K12-HD001487 to P.Y.J.
References
- Andree B, Duprez D, Vorbusch B, Arnold HH & Brand T 1998. BMP-2 induces ectopic expression of cardiac lineage markers and interferes with somite formation in chicken embryos. Mech. Dev 70, 119–131. [DOI] [PubMed] [Google Scholar]
- Bodmer R 1993. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118, 719–729. [DOI] [PubMed] [Google Scholar]
- Clark TG, Conway SJ, Scott IC, Labosky PA, Winnier G, Bundy J, Hogan BL, Greenspan DS 1999. The mammalian Tolloid-like 1 gene, Tll1, is necessary for normal septation and positioning of the heart. Development 126, 2631–2642. [DOI] [PubMed] [Google Scholar]
- Ge G, Seo NS, Liang X, Hopkins DR, Hook M & Greenspan DS 2004. Bone morphogenetic protein-1/ tolloid-related metalloproteinases process osteoglycin and enhance its ability to regulate collagen fibrillogenesis. J. Biol. Chem 279, 41626–41633. [DOI] [PubMed] [Google Scholar]
- Gonzalez EM, Reed CC, Bix G, Fu J, Zhang Y, Gopalakrishnan B, Greenspan DS, lozzo RV 2005. BMP-1/Tolloid-like metalloproteases process endorepellin, the angiostatic C-terminal fragment of perlecan. J. Biol. Chem 280, 7080–7087. [DOI] [PubMed] [Google Scholar]
- Komuro I & Izumo S 1993. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl Acad. Sci. USA 90, 8145–8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I & Harvey RP 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 419–431. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP 1995. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 9, 1654–1666. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR 2001. The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol 238, 97–109. [DOI] [PubMed] [Google Scholar]
- Olson EN & Srivastava D 1996. Molecular pathways controlling heart development. Science 272, 671–676. [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP 2007. An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenholl A, Pappano WN, Koch M, Keene DR, Kadler KE, Sasaki T, Timpl R, Burgeson RE, Greenspan DS, Bruckner-Tuderman L 2002. Proteinases of the bone morphogenetic protein-1 family convert procollagen VII to mature anchoring fibril collagen. J. Biol. Chem 277, 26372–26378. [DOI] [PubMed] [Google Scholar]
- Scott IC, Blitz IL, Pappano WN, Imamura Y, Clark TG, Steiglitz BM, Thomas CL, Maas SA, Takahara K, Cho KW, Greenspan DS 1999. Mammalian BMP-1/ Tolloid-related metalloproteinases, including novel family member mammalian Tolloid-like 2, have differential enzymatic activities and distributions of expression relevant to patterning and skeletogenesis. Dev. Biol 213, 283–300. [DOI] [PubMed] [Google Scholar]
- Scott IC, Steiglitz BM, Clark TG, Pappano WN & Greenspan DS 2000. Spatiotemporal expression patterns of mammalian chordin during postgastrulation embryogenesis and in postnatal brain. Dev. Dynamics 217, 449–456. [DOI] [PubMed] [Google Scholar]
- Takahara K, Brevard R, Hoffman GG, Suzuki N & Greenspan DS 1996. Characterization of a novel gene product (mammalian tolloid-like) with high sequence similarity to mammalian tolloid/bone morphogenetic protein-1. Genomics 34, 157–165. [DOI] [PubMed] [Google Scholar]
- Tamura G, Olson D, Miron J & Clark TG 2005. Tolloid-like 1 is negatively regulated by stress and glucocorticoids. Brain Res. Mol. Brain Res 142, 81–90. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N & Izumo S 1999. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126, 1269–1280. [DOI] [PubMed] [Google Scholar]
- Veitch DP, Nokelainen P, McGowan KA, Nguyen TT, Nguyen NE, Stephenson R, Pappano WN, Keene DR, Spong SM, Greenspan DS, Findell PR, Marinkovich MP 2003. Mammalian tolloid metalloproteinase, and not matrix metalloprotease 2 or membrane type 1 metalloprotease, processes laminin-5 in keratinocytes and skin. J. Biol. Chem 278, 15661–15668. [DOI] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E & Black BL 2005. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol 287, 134–145. [DOI] [PubMed] [Google Scholar]


