Abstract
We have engineered the ecdysone-inducible mammalian expression system for general retroviral delivery to cultured mammalian cells. We inducibly expressed PTEN in the glioblastoma cell line, U87MG, lacking this gene. Because nearly all cells are recruited on induction, we find both up- and down-regulated genes by cDNA microarray analysis. The changes we see are similar to those observed after treatment with LY294002, an inhibitor of phosphatidylinositol 3-OH kinase, fully consistent with the model that PTEN antagonizes phosphatidylinositol 3-OH kinase. Both treatments result in suppressed expression of the transforming growth factor (TGF)-β gene and the genes of the cholesterol biosynthesis pathway. Our results illustrate the power of using a fully inducible expression system in conjunction with cDNA microarray analysis for exploring gene function.
Keywords: inducible expression‖glioblastoma‖DNA microarray
PTEN/MMAC1/TEP1 is a tumor suppressor on 10q22–23 that is found mutated in patients with Cowden's disease and in advanced cancers (1–5). PTEN was found to have sequence similarity to protein tyrosine phosphatases (1, 2, 6). The phosphatase activity of the PTEN gene product was clearly apparent only with highly negatively charged peptides poly(Glu-Tyr 4:1) (7). This observation led to the pursuit of other nonproteinaceous substrates of PTEN. Indeed, PTEN was demonstrated to have phosphoinositide-3-phosphatase activity capable of converting products of phosphatidylinositol 3-OH (PI3)-kinase to their unphosphorylated form at the D-3 position of the inositol ring (8, 9). Furthermore, forced expression of PTEN in cultured cells reduced the amount of phosphorylation of AKT-kinase, a downstream target of PI3-kinase (9–11). Significantly, a single germ-line mutation (G129E) in PTEN that caused loss of its lipid phosphatase activity, but not its ability to dephosphorylate poly(Glu-Tyr 4:1), was present in some Cowden's disease kindreds (9). This suggested that the main effect of loss of PTEN results from the unbridled activity of PI3-kinase. We desired to put this hypothesis to a more rigorous test by comparing the consequences of forced expression of PTEN to the chemical inhibition of PI3-kinase in a host lacking endogenous PTEN.
To conduct these experiments, we sought to use an inducible rather than a constitutive expression system. The acute responses of a host may be monitored by using inducible expression, whereas the constitutive expression of some genes may be toxic or deleterious to the host, leading to selection for tolerant subpopulations of cells after prolonged expression of the gene in the host. With constitutive expression systems, the gross overexpression of a gene may result in nonphysiological responses. By using inducible systems, in principle, levels of expression of the transgene can be adjusted to be within physiological ranges. Finally, the use of inducible systems allows the experimenter to explore the response of the same host cell to two different stimuli, thus controlling for clone-to-clone variability.
An ideal inducible system would have several features. First, it would be tightly regulated, with no expression in the uninduced state and a rapid inductive response. Second, there would be a dose dependence between the inducer added and transgene expression. Third, the components of the inducible system would be inert with respect to the host. Fourth, the proportion of host cells that respond when inducer is added (hereinafter referred to as recruitment) would be high. This last requirement is particularly important in two cases: when studying genes that confer growth disadvantage to the host cell, such as tumor suppressors; and when performing transcriptional profiling studies, wherein the presence of unresponsive cells would obscure the observation of gene repression caused by induction of the transgene.
The two most widely used inducible mammalian systems use tetracycline- or ecdysone-responsive transcriptional elements (12, 13). The chief drawback of the tetracycline-inducible system has been reported to be a relatively high background of expression in the uninduced state (12). The ecdysone-inducible system was developed to address this issue. The ecdysone-inducible system has a sensor that is made of two nuclear receptors/ transcription factors [modified ecdysone receptor (VgEcR) and retinoid X receptor (RXR)], which dimerize in the presence of inducer and activate transcription of a gene of interest from a cognate promoter. The gene of interest is placed under the control of this promoter. In a typical protocol, the sensor components are introduced by transfection into the cell line of interest, stable “host” clones are established by antibiotic selection, and the hosts are tested individually for their ability to induce a reporter gene introduced on a separate plasmid. A successful host then serves as a recipient of the inducible gene of interest (13).
In our experience, the existing methods for the construction of inducible cell lines had two important shortcomings: they were laborious, and the resulting inducible hosts showed considerable clonal variegation. Clonal variegation, resulting in fewer than 50% of the cells in a clone responding to inducer (poor recruitment), can be a significant obstacle to functional analysis. In this work we address these two problems associated with the plasmid-based systems. We have ameliorated the recruitment problem by re-engineering the system for retroviral delivery, thus assuring a more homogeneous expression of both sensor elements and inducible transgene. We describe a general strategy that uses fluorescence-activated cell sorting (FACS) to rapidly derive inducible cell lines. To date we have generated hosts from three human (U87MG, 293, and T47D) and two mouse cell lines (NIH 3T3 and NMuMG).
We illustrate the effectiveness of the system by the analysis of the consequence of inducing PTEN expression in the glioblastoma host cell, U87MG, that lacks its own endogenous PTEN gene, and comparing that to the consequences of blocking PI3-kinase with LY294002.
Materials and Methods
Cells and Cell Culture Methods.
U87MG (human glioblastoma, ATTC no. HTB-14) was maintained in 10% FBS in DMEM. NIH 3T3 (mouse embryo fibroblast, ATCC no. CRL-1658) cell line was grow in 10% calf serum in DMEM. Other hosts were developed from the following cell lines: (i) 293 (human embryonal kidney, ATCC no. CRL-1573); (ii) T47D (human mammary gland ductal carcinoma, ATCC no. HTB133); and (iii) NMuMG (mouse mammary gland, ATCC no. CRL-1636). Retroviral-mediated gene transfer into recipient cell lines was carried out according to Wang et al. (14). Host cell lines expressing transgenes were maintained with hygromycin selection at 50 μg/ml. Muristerone A (Invitrogen) was reconstituted in absolute ethanol before adding to cells for induction. An equal amount of ethanol was added to plates that were uninduced.
Retroviral Vector Constructions.
The nuclear receptors/transcription factors RXR and VgEcR (Invitrogen) were cloned in pMAO Neo and pMAO Puro, respectively. RXR was amplified by PCR that used primers coding for EcoRI on the 5′ end and XhoI at the 3′ end and inserted as an EcoRI–XhoI fragment into pMAONeo, yielding RXR-pMAONeo (pRXR-MN). VgEcR was amplified with primers coding for EcoRI on both ends and inserted as an EcoRI fragment in pMAOPuro, yielding VgEcR-pMAOPuro (pVgEcR-MP). The MAO retroviruses are similar to the pBabe type of Moloney murine leukemia virus (MoMLV) retroviral vectors, but also carry the simian virus 40 origin of replication in the nonviral part of their backbone. The receptor-coding sequences were placed downstream of the 5′ long terminal repeat (LTR) promoters; the antibiotic resistance markers were placed downstream of PGK promoters as HindIII–ClaI inserts. Both transcripts terminated in the 3′ LTR poly(A) signal.
pI-TKHygro is a derivative of pSin (courtesy of David Beach), a self-inactivating retrovirus that lacks U3 enhancers in the 3′ LTR. The hybrid ecdysone response element (E/GRE) and polylinker (MCS) from pIND (Invitrogen) were amplified with BamHI–XhoI primers and cloned downstream of pSin's 5′LTR as an BglII/BamHI–XhoI fragment. This virus also carries a thymidine kinase enhancerless promoter (TKp) and the hygromycin-resistance gene (HYG) cassette as an XhoI–ClaI fragment between the polylinker and the 3′ LTR. The transcript orientation and structure are the same as for the MAO vectors with one important distinction, namely, the 3′ LTR carries a deletion of the U3 enhancers. On proviral integration, this deletion flanks the retroviral insert thereby removing the enhancers from both the 5′ and the 3′ LTRs. Therefore, the immediate vicinity of the glucocorticoid response elements is transcriptionally silent and assures tight induction control of the gene of interest. The pI-TKHygro is the parent vector for pI-EGFP, pI-LacZ and all PTEN inducible transgenes.
pI-LacZ was derived by subcloning LacZ into the polylinker of pI-TKHygro as an HindII–XhoI fragment.
pI-EGFP is a derivative of pI-TKHygro that has the TK-Hygro cassette deleted and the enhanced green fluorescent protein (EGFP; CLONTECH) reporter inserted as an EcoRI–SalI fragment.
Microscopy and Flow Cytometry.
β-Galactosidase expression was visualized by fixing cells in 0.5% glutaraldehyde, washing the cells, and then overlaying the cells with X-gal solution [0.2% X-Gal/2 mM MgCl2/5 mM K4Fe(CN)6/5 mM K3Fe(CN)6]. After sufficient blue color had developed, cells were washed and kept in PBS. Photographs of cells were taken with the Zeiss Axiovert 405 M fluorescence microscope with digital imaging capabilities. Cells were sorted by using the Coulter Elite ESP flow cytometer and carried out as described (15).
cDNA Microarray Analysis.
mRNA isolations were carried out according to the Poly(A) Pure kit (Ambion) protocol. Conversion of mRNA to double-stranded cDNA was performed by using the superscript cDNA synthesis kit (GIBCO/BRL) according to its protocol. High complexity representations (HCRs) of each sample were then prepared according to a protocol adapted from Lucito et al. (16). cDNA (400 ng) was digested with DpnII (four base-restriction endonucleases) for 8 h at 37°C. After phenol/chloroform extraction, the digested cDNA was precipitated by using 20 μg glycogen, 3 M sodium acetate, and absolute ethanol, then washed, resuspended, and ligated to the cohesive adaptor RBgl24 and RBgl12 (see Lucito et al., ref. 16). Primers complementary to adaptors were used to amplify desired amounts of amplicons for microarray hybridizations.
A set of 14,000 human cDNA clones from the IMAGE consortium (purchased from Research Genetics, Huntsville, AL, and Invitrogen) were PCR amplified, purified, and robotically spotted on glass slides as described by Schena et al. (17). cDNA representations were labeled with fluorescent dyes (Cy3 or Cy5) by using the random priming method (http://nucleus.cshl.org/wigler/). Arrays were processed and hybridized according to protocols available on our web site (http://nucleus.cshl.org/wigler/).
After hybridization with samples labeled with Cy3 and Cy5, the arrays were scanned to measure fluorescence by using a GenePix 4000 laser scanner (Axon Instruments, Foster City, CA). Raw data files generated by the scanner software GENEPIX3 were imported into S-Plus (a mathematical/statistical package from StatSci Division, Mathsoft, Cambridge, MA) for analysis. For normalization, features for which R2 values were above 0.4, and 50% of feature pixels were 1 SD above background pixels in both channels, were considered. The data points were then median normalized and plotted. Experiments were performed in duplicate and color reversals (four hybridizations per sample pair).
RNase Protection Assays.
Total RNA was harvested from samples of induced and uninduced U87MG expressing wild-type PTEN, mutant PTEN, and LacZ. All RNase protection experiments were performed by using the RPA III kit from Ambion following the manufacturer's instructions.
Results
Retroviral-Based Ecdysone-Inducible System.
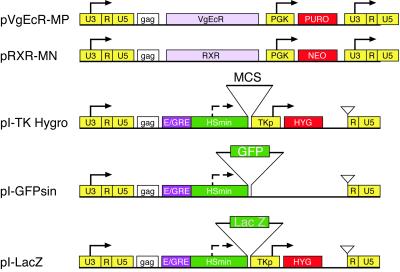
We placed the two receptors (VgEcR and RXR) on separate MoMLV-based viruses, expressed off the 5′ LTR (18, 19). Each receptor virus was marked with a different antibiotic resistance—puromycin for VgEcR and neomycin (G418) for RXR (Fig. 1). The gene of interest was introduced into a self-inactivating MoMLV-based retrovirus (I-TK-HYG), in which enhancers in the U3 region in the 3 ′ LTR are deleted so that, on proviral integration, no enhancers remain (20). This virus was marked with HYG, driven by a thymidine kinase enhancerless promoter (TKp). The gene of interest is subcloned into the multiple cloning site, which lies just downstream of the inducible cognate promoter (E/GRE/Hsmin). We produced amphotropic viruses by using high-titer packaging lines that were derived by Beach and Hannon (14).
Figure 1.
Vector descriptions. pVgEcR-MP, retroviral vector for constitutive expression of a modified Drosophila ecdysone receptor (containing the VP16 transactivation domain) from the LTR and marked with the puromycin-resistance gene; pRXR-MN, retroviral vector for constitutive expression of the RXR from the LTR and marked with the neomycin (G418)-resistance gene; pI-TK Hygro, self-inactivating retroviral ecdysone-inducible vector marked with HYG; pI-GFPsin, same vector as pI-TK Hygro but without hygromycin-selectable marker; pI-LacZ: pI-TK Hygro containing the β-galactosidase gene. U3-R-U5, LTR; gag, envelope gene; VgEcR, ecdysone receptor; PGK, phosphoglucokinase promoter; TKp, enhancerless thymidine kinase promoter; E/GRE, hybrid ecdysone response element (5×); Hsmin, minimal heat shock promoter; PURO, puromycin resistance gene; NEO, G418 resistance gene; MCS, multiple cloning site; GFP, green flourescent protein; LacZ, β-galactosidase gene.
Typically, the recipient cells were transduced with both receptor viruses in the same infection and selected for both puromycin and G418 resistance. Doubly resistant cell clones were isolated, expanded and tested for their ability to serve as a good “host.” To this end, each prospective host was infected with a β-galactosidase inducible virus, pI-LacZ (Fig. 1), which carries the HYG marker. Colonies were then selected for hygromycin resistance.
For each prospective host we determined the proportion of the hygromycin-resistant colonies that expressed any β-galactosidase on addition of inducer and the percentage of individual cells in each colony that express β-galactosidase. Thus we estimated how many clones expressing the gene of interest one needs to isolate to obtain, with high probability, a well-behaved inducible clone. Once a good host was established, it served as a recipient of the gene(s) of interest.
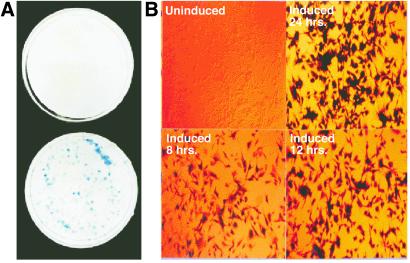
Our first experiments were performed with U87MG, a human glioblastoma cell line lacking endogenous PTEN. After double infection with receptor viruses (see Materials and Methods), and double selection, five candidate host clones were selected based on antibiotic resistance. They were infected with a β-galactosidase-inducible virus, split into two dishes at low density, and then hygromycin resistance was selected, resulting in ≈200 colonies per plate. One dish was induced with 0.5 μM muristerone A, an ecdysone analog, for 24 h and stained for β-galactosidase, and the proportion of blue colonies was determined. The second dish remained uninduced and was stained to determine background. For five of five host clones, all hygromycin resistant colonies were inducible (Fig. 2A Lower), and none showed detectable background (Fig. 2A Upper).
Figure 2.
(A) The retroviral ecdysone-inducible system yields virtually 100% inducible clones. Depiction of one of five candidate U87MG-inducible host clones, U87.23, infected with pI-LacZ and selected for hygromycin-resistant colonies. Each plate contained ≈200 colonies. The bottom dish was induced with 0.5 μM muristerone A for 24 h, fixed, and stained for β-galactosidase protein. The top dish was left uninduced, fixed, and also stained. Efficiency of LacZ expression was comparable for the remaining four clonal hosts (data not shown). (B) Complete recruitment by inducer at the single-cell level. The same U87.23 LacZ-inducible cell line was plated on coverslips and induced with 0.5 mM muristerone A. Cells were fixed and stained for β-galactosidase after 8 h (Lower Left), 12 h (Lower Right) and 24 h (Upper Right), or left uninduced (Upper Left). Note that even as early as 8 h all cells are recruited to express the inducible transgene.
One of the five hosts, U87.23 was selected to document the proportion of individual cells responding to inducer. To this end, individual hygromycin-resistant colonies were isolated, expanded, and plated onto coverslips for induction. Seven of seven colonies had greater than 99% inducibility after only 8 h in 0.5 μM muristerone A (Fig. 2B Lower Left). There was no detectable background (Fig. 2B Upper Left). Cells induced for 12 and 24 h displayed uniformly intense staining (Fig. 2B Lower Right and Upper Right, respectively). Liquid β-galactosidase activity assays demonstrated that the background β-galactosidase levels in the uninduced cell line were equal, within standard error, to the levels in the parental, nontransduced cell line (data not shown). Full induction was achieved with 1.0 μM muristerone A and was at least 200-fold above background.
The process of obtaining good inducible hosts for other cell lines was more laborious. Therefore, we modified the process for finding a good host by using sorting to select suitable hosts out of a large cell population. Cells were infected with three viruses: VgEcR-MP; RXR-MN, and GFP-Sin (Fig. 1). GFP-Sin is a self-inactivating virus identical to pI-TKHygro, but lacking the hygromycin-resistance marker and carrying ecdysone-responsive GFP. Cells expressing GFP in the absence of inducer are then removed by FACS. The remaining cells were expanded and induced with a very low dose of muristerone A (0.05 μM-0.2 μM). The top 1% of GFP-positive cells were collected. This step can be repeated if necessary to enrich for robustly inducing cells. Cells are plated for colony formation, induced, and examined for GFP fluorescence. Those showing full recruitment at the single-cell level were isolated, expanded, retested for inducibility, and used as hosts.
We tested this protocol with three cell lines: U87MG, NIH 3T3, and 293. To demonstrate that the inducible hosts produced by the FACS method can successfully serve as recipients of another inducible transgene, we infected several of the GFP-inducible hosts with pI-LacZ, then selected and isolated single clones. These clones were examined for their ability to induce both the GFP and the LacZ transgenes. Virtually all clones had 95% or better recruitment for inducible LacZ expression and fully retained their inducibility for GFP expression.
Effects of PTEN Induction on Growth, Morphology, and Transcription in U87MG Cells.
We sought to study the consequence of restoring PTEN in U87MG cells by using the inducible host, U87.23. To this end, the following host cell lines were generated: (i) U87.PTEN.WT, expressing wild-type PTEN; (ii) U87.PTEN.G129E, expressing the lipid phosphatase-defective mutant PTEN(G129E); (iii) U87.PTEN.G129R, expressing the catalytically inactive mutant PTEN(G129R); and (iv) U87.LacZ, expressing β-galactosidase.
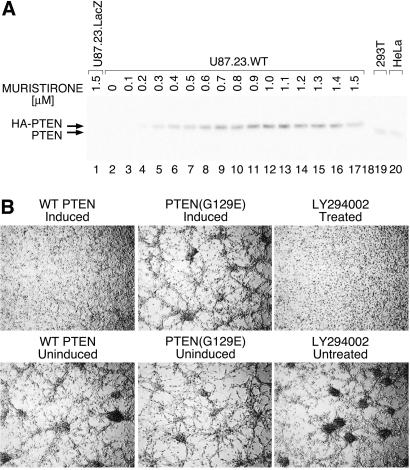
To ascertain the level of induction of PTEN, U87.PTEN.WT was induced with a range of concentrations (0.1 μM to 1.5 μM), and induced PTEN expression was compared with expression in HeLa and 293T cell lines by Western analysis (Fig. 3A). Inducer (0.5 μM) was sufficient to attain near physiological levels of PTEN expression (Fig. 3A, compare lane 7 to lanes 19 and 20). All of the other PTEN-expressing lines were also verified to be inducible by Western analysis.
Figure 3.
(A) Dose–response of PTEN-inducible line to muristerone A. Western blot analysis of dose–response for cell line, U87.23.WT, expressing hemagglutinin-tagged wild-type PTEN. Cells were induced with doses of muristerone A, in increments of 0.1 μM, from 0 (lane 2), to 1.5 μM (lane 17), for 30 h. For comparisons, lane 1 shows a cell line with inducible β-galactosidase (negative control) treated with 1.5 μM muristerone A, and lanes 19 and 20 show endogenous levels of PTEN in two PTEN-positive human cell lines, 293T, and HeLa respectively. The filter was probed by using anti-PTEN C terminus antibodies. Background expression is undetectable, even after very long exposures, and maximal expression is perhaps only a few fold elevated over endogenous levels. (B) PTEN and LY alter the cell growth pattern of U87MG cells in low serum in a similar fashion. U87MG cells carrying wild-type or mutant (G129E) inducible PTEN were seeded in 2% FBS DMEM at high density and either induced to express PTEN with 0.5 mM muristerone A (Upper Left and Upper Center), or left uninduced (Lower Left and Lower Center panels). U87MG cells were treated with 10 μM LY294002 for 24 h (Upper Right) or untreated (Lower Right). Nomarski photographs were taken 24 h after induction.
To assess the effect of PTEN on growth, we performed 4-day growth curves on U87.PTEN.WT, U87.PTEN.G129E, and U87.LacZ (negative control). Cells were grown in 2% FBS, with or without muristerone A at 0.5 μM. The induction of wild-type PTEN does not affect growth in U87MG cells when compared with induction of LacZ or PTEN(G129E) (data not shown). In addition, we have carried out the experiment at serum concentrations ranging from 0.5% to 10% FBS and by using both mass cultures and multiple independent clonal lines with consistent results.
Cells were examined for effects of PTEN induction on their morphology. Cell lines U87.PTEN.WT, U87.PTEN.G129E, and U87.PTEN.G129R were induced with 0.5 μM muristerone A. When grown in 2% FBS, uninduced cells appeared weakly attached to the plastic substrate and grew in a reticular formation. In the presence of inducer, the cultures expressing wild-type PTEN were flat and more adherent (Fig. 3B). Induction of G129E and the G129R mutant (data not shown) had no effect on morphology. All cultures were verified to be inducible by Western blotting (data not shown).
To establish whether the morphological effect was mediated by phosphatidylinositide-3, 4,5 phosphate (PIP3), we incubated U87.PTEN.WT cells with 10 μM LY294002, an inhibitor of PI3-kinase. This treatment produced a morphological change indistinguishable from the one caused by expression of PTEN (Fig. 3B), consistent with the hypothesis that the lipid phosphatase activity of PTEN is responsible for this phenotype.
To pursue further the similarities between PTEN induction and inhibition of the PI3-kinase pathway, we examined the transcriptional profiles of these two treatments by hybridization to cDNA microarrays. To generate fluorescently tagged samples for hybridization, we used high-complexity representations of cDNAs. The protocol for preparing cDNA representations is described in Materials and Methods. We prefer using cDNA representations over first-strand cDNA samples for hybridization to cDNA microarrays for two reasons: first, large amounts of DNA can be prepared from small amounts of biological samples; second, the resulting cDNA representations can be readily analyzed by Southern blotting for a probe of interest to confirm observations from the microarray data. To prove the validity of this method, we have shown by array hybridization experiments that the relative abundance of individual messages in the initial mRNA are preserved in the final cDNA representation (22).
We have shown hybridizations by using cDNA representations prepared from six sources: (i) U87.PTEN.WT untreated; (ii) U87.PTEN.WT treated with muristerone A to induce expression of wild-type PTEN; (iii) U87.PTEN.WT treated with LY294002; (iv) U87.PTEN.G129R untreated; (v) U87.PTEN.G129R treated with muristerone A to induce expression of the phosphatase defective mutant PTEN; and (vi) U87.PTEN.G129R treated with LY294002. We then performed a series of two-way, then four-way comparisons. For these comparisons, we established the ratio of hybridization intensities from cDNA of two sources.
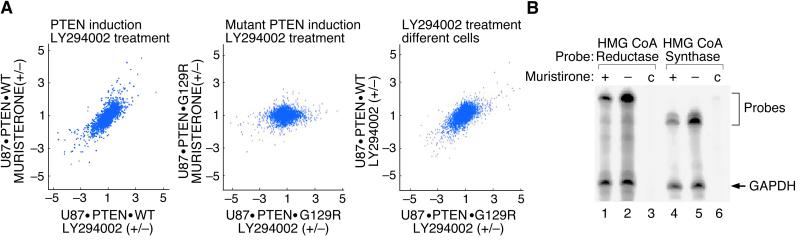
To compare PTEN expression and LY294002 treatment, we graphed the ratios obtained from U87.PTEN.WT treated with muristerone A or untreated with the ratios of U87.PTEN.WT treated with LY294002 or untreated. We observed a striking correlation in the transcriptional profiles (Fig. 4A Left). This effect was specific to the expression of wild-type PTEN, because ratios of U87.PTEN.G129R treated or untreated with muristerone A did not show any similarity to ratios of the same cells treated with LY294002 or untreated (Fig. 4A Center). Nevertheless, LY294002 induce similar changes in expression patterns in both U87.PTEN.WT and U87.PTEN.G129R cells (Fig. 4A Right).
Figure 4.
(A) Induction of PTEN and treatment with LY294002 affect a similar set of transcriptional targets. U87MG cell lines carrying inducible wild-type (U87.PTEN.WT) or mutant (U87.PTEN.G129R) PTEN were induced with 0.5 μM muristerone A for 36 h or treated with LY294002 for the same amount of time. mRNAs were isolated, converted to double-stranded cDNA. cDNAs were amplified to generate HCRs, labeled with fluorescent dyes, and hybridized to human cDNA microarrays carrying 14,000 probes. The normalized fluorescent ratios for individual array features were computed and then graphed. Scatter plots depict comparison of normalized ratios: U87.PTEN.WT treated (+) or untreated (−) with muristerone A versus the same cells treated or untreated with LY294002 (Left); U87.PTEN.G129R treated or untreated with muristerone A versus the same cells treated or untreated with LY294002 (Center); U87.PTEN.WT treated or untreated with LY294002 versus U87.PTEN.G129R treated or untreated with LY294002 (Right). Values on the axes are positive for ratios greater than 1 or negative reciprocals for ratios less than 1. The positive and negative signs represent induction or repression of gene expression, respectively. (B) RNase protection of HMG CoA reductase and HMG CoA synthase transcripts. U87MG cells carrying inducible PTEN were either induced with muristerone (lanes 1 and 4) or uninduced (lanes 2 and 5). Control (c) represents reactions carried out without total RNA (lanes 3 and 6). Total RNA was isolated and subjected to RNase protection assay by using anti-sense 32P-labeled probes representing the genes shown above the panel. Protected probes were purified and separated on a polyacrylamide gel as described in Sambrook et al. (21). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was used as a control to check integrity of mRNAs in the sample and to normalize gene-specific signal.
Microarray hybridization experiments are often plagued by “system noise” that generates variability. It is necessary to estimate variability to correctly interpret array data. To measure the extent of variability for each sample, we performed a “self-to-self” hybridization, where each sample was labeled separately with Cy3 and Cy5 and hybridized. Ideally the ratio of Cy3 to Cy5 for all probes would be 1. In our self-to-self experiments, we observed scatter about the ratio of 1, with very few ratios observed to be >2. Table 1 lists genes that over multiple experiments showed a 2-fold change when cells were induced to express PTEN and when cells were treated with LY294002. The expression of these genes were not altered when mutant PTEN was induced. It is significant that we saw down-regulated genes after induction of PTEN, because this is only possible with robust recruitment by our inducible system.
Table 1.
Differentially expressed genes under conditions of wild-type PTEN induction and inhibition by LY294002
| Gene name | Accession no. | WT | LY | Description |
|---|---|---|---|---|
| Down-regulated by treatment | ||||
| Stathmin | AA873060 | −4.8 | −3.5 | Phosphoprotein. Cell proliferation and differentiation |
| Transforming growth factor β-1 | R36467 | −3.1 | −3.9 | Cell proliferation and differentiation |
| ADP-ribosylation factor-4 | T71316 | −4.0 | −4.0 | GTP binding protein, activator of phospholipase |
| ADP-ribosylation factor-like 1 | N51280 | −2.8 | −5.0 | GTP binding protein |
| HMG-CoA synthase | X83618 | −3.5 | −4.7 | Cholesterol synthesis |
| HMG-CoA reductase | AI038284 | −2.5 | −2.7 | Cholesterol synthesis |
| Squalene synthase | AA018779 | −2.6 | −4.0 | Cholesterol synthesis |
| Human homolog of yeast IPP isomerase | H08899 | −2.3 | −3.1 | Cholesterol synthesis |
| Homo sapiens mRNA for epiregulin | N31585 | −2.7 | −3.0 | Epidermal growth factor; tumor growth-inhibitory factor |
| Interferon-related developmental regulator 1 | AA676598 | −3.0 | −2.8 | Muscle differentiation and signal transduction |
| Cytochrome C | R52654 | −2.5 | −3.3 | Mediates the electron transfer |
| H. sapiens mRNA for TFG protein | R60846 | −2.4 | −2.3 | Transmembrane receptor tyrosine kinase protooncogene |
| ER lumen protein retaining receptor 2 | AA626867 | −2.5 | −3.7 | Sorting of proteins |
| Adrenergic, beta-2, receptor | H90431 | −2.4 | −3.2 | Transmembrane G protein-coupled receptor |
| Homo sapiens insulin induced protein | H59620 | −2.5 | −3.4 | Growth response |
| Glycyl-tRNA synthetase | AA629909 | −2.6 | −5.0 | Protein translation |
| Alanyl-tRNA synthetase | AA156571 | −3.2 | −5.7 | Protein translation |
| Up-regulated by treatment | ||||
| Cartilage glycoprotein-39 precursor | AA434115 | 3.5 | 3.2 | Deposition and remodeling of cartilage matrix |
| Human sigma receptor mRNA, comp | W47484 | 3.8 | 3.1 | Interaction with antipsychotic drugs; involved in psychiatric disorders |
| ESTs, highly similar to J KAPPA | R19314 | 2.8 | 3.2 | Recombination signal binding protein |
| Fibronectin 1 | N26285 | 4.2 | 2.4 | Cell adhesion, morphology and surface architecture |
| Lactate dehydrogenase A | AA48961 | 3.2 | 2.6 | Converts L-lactate to pyruvate during anaerobic glycolysis |
Genes that consistently show similar patterns of expression on PTEN induction or LY294002 treatment and not on induction of mutant PTEN (G129R) are tabulated. Values in the WT (induction of wild-type PTEN) and LY (treatment with LY294002) columns denote fold changes in expression. A negative value is assigned for repression and a positive value is assigned for induction. Accession nos. are from GenBank. Descriptions of genes are from National Center for Biotechnology Information databases.
To investigate the reliability of the array data we used two techniques on several probes: Southern blots of cDNA representations and RNase protection assays (RPA) on total RNA. A representative RPA analysis of two genes, HMG-CoA synthase and HMG-CoA reductase, are shown in Fig. 4B. Both genes are suppressed on induction of wild-type PTEN and on treatment with LY294002. There was good correlation between these methods and the array data.
Discussion
We have developed and used an ecdysone-inducible gene expression system that utilizes retroviral delivery. The system appears to be close to ideal in many respects. It is relatively easy to create hosts. Gene induction is rapid and can be controlled in a dose-dependent manner, allowing us to study expression of a gene at physiological levels. There is negligible expression in the absence of inducer, which decreases the risk of acquisition of tolerance by the host. Recruitment of expression, that is the percentage of host cells that respond to inducer, is very high, as judged both by β-galactosidase and GFP expression studies. This conclusion is further strengthened by our transcriptional profiling, in which we observe both up-regulation and down-regulation of gene expression after addition of inducer. The down-regulation of gene expression consequent to induction of PTEN would not be observable were PTEN not induced in the great majority of cells.
In an ideal system, the inducer should be inert. Muristerone A is nearly inert, having no effects on cellular morphology or gene transcription that we have observed. However, the addition of muristerone leads to a slight inhibition of cell growth. This is not an intrinsic feature of the activation of the ecdysone receptor, because addition of muristerone to U87MG cells that lack the responsive system still results in slightly slower growth.
Our results show absence of growth effect of PTEN induction on U87MG cells. This observation is in marked contrast to the 6-fold increase in the length of the cell cycle observed by the group of Hong Sun (10). We think that this difference may be because of the expression of higher and nonphysiological levels of PTEN from the constitutive retroviral system used by Sun et al. (see figure 1C of that paper). It is our contention that our inducible U87MG cell line experiments carry more weight, because of the controlled PTEN expression. Moreover, it seems unlikely to us that physiological levels of PTEN inhibit the cell cycle severely because PTEN expression is found ubiquitously in proliferating cell lines and tissues.
The combination of inducible systems with transcriptional profiling is especially powerful. The analysis of PTEN expression in U87MG cells confirms the major hypothesis of PTEN action, that its effects result from its PIP3 phosphatase activity. The addition of an inhibitor of PI3-kinase, LY294002, produces a change in the transcriptional profile of U87MG that is very similar to that produced by the induction of wild-type PTEN. Induction of the PTEN(G129R) mutant, which is catalytically inert, does not produce these changes. Our arrays do not, however, contain all of the transcription units of the cell, so major differences may have been missed.
The transcriptional profiling did produce some unexpected findings. First, the genes of the cholesterol biosynthesis pathway, namely HMG-CoA synthase, squalene synthase, and HMG-CoA reductase were down-regulated by PTEN and LY294002. If generally true, this would suggest that certain hormones and growth factors, such as insulin, that activate the PI3-kinase pathway, may also increase cholesterol biosynthesis. We have not seen this connection made in the literature. Second, one of the most strongly down-regulated genes, by both LY294002 and PTEN, was transforming growth factor (TGF) β (Table 1). Interestingly, TGF-β is known to down-regulate PTEN (6). Thus there appears to be a mutual antagonism of these two genes.
Transcriptional profiling of the response of cells to a stimulus does not distinguish primary from secondary responses. It is possible, for example, that the wide-ranging similarity of PTEN induction and LY294002 treatment results from the secondary response of cells to down-regulation of TGF-β. Additional experiments would be needed to resolve this question.
Acknowledgments
We thank Raju Kucherlapati and Geoffery Childs (Albert Einstein School of Medicine) for sharing their expertise in microarray technology; Lewis Cantley (Harvard Medical School) and Ron Evans (Salk Institute) for useful technical discussions; and Ramon Parsons (Columbia University), Ron Evans, and Raju Kucherlapati (Harvard Medical School) for their helpful comments regarding the manuscript. This work was supported by grants to M.W. from the National Institutes of Health and National Cancer Institute (5R01-CA78544), Tularik Inc.; 1:9, The Long Island Breast Cancer Action Coalition; Lillian Goldman and the Breast Cancer Research Foundation. M.W. is an American Cancer Society Research Professor. J.S. was a graduate student in the Department of Genetics and Development, Columbia University. K.C. is a graduate student in the Department of Microbiology, State University of New York, Stony Brook.
Abbreviations
- PI3-kinase
phosphatidylinositol 3-OH kinase
- HCR
high complexity representation
- PIP3
phosphatidylinositide-3, 4,5-phosphate
- HYG
hygromycin-resistance gene
- FACS
fluorescence-activated cell sorting
- LTR
long terminal repeat
- MoMLV
Moloney murine leukemia virus
- RXR
retinoid X receptor
- VgEcR
modified ecdysone receptor
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S, Puc J, Miliarcsis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck P A, Pershouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Longford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Liaw D, Marsh D J, Li J, Dahia P L, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, et al. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 4.Nelen M R, van Staveren W C, Peeters E A, Hassel M B, Gorlin R J, Hamm H, Lindboe C F, Fryns J P, Sijmons R H, Woods D G, et al. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 5.Lynch E D, Ostermeyer E A, Lee M K, Arena J F, Ji H, Dann J, Swisshelm K, Suchard D, MacLeod P M, Kvinnsland S, et al. Am J Hum Genet. 1997;61:1254–1260. doi: 10.1086/301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D-M, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 7.Myers M P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 9.Myers M, Pass I, Batty I H, Van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D-M, Sun H. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stambolic V, Suzuki A, de la Pompa J L, Brothers G M, Mirtsos C, Sasaki T, Ruland J, Penninger J M, Siderovski D P, Mak T W. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 12.No D, Yao T-P, Evans R M. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saez E, No D, West A, Evans R M. Curr Opin Biotechnol. 1997;8:608–616. doi: 10.1016/s0958-1669(97)80037-7. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Xie L Y, Allan S, Beach D, Hannon G J. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lybarger L, Chervenak R. Methods Enzymol. 1999;302:189–199. doi: 10.1016/s0076-6879(99)02018-2. [DOI] [PubMed] [Google Scholar]
- 16.Lucito R, Nakamura M, West J A, Han Y, Chin K, Jensen K, McCombie R, Gray J W, Wigler M. Proc Natl Acad Sci USA. 1998;95:4487–4492. doi: 10.1073/pnas.95.8.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 18.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:1068–1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu S F, von Ruden T, Kantoff P W, Garber C, Seiberg M, Ruther U, Anderson W F, Wagner E F, Gilboa E. Proc Natl Acad Sci USA. 1986;83:3194–3198. doi: 10.1073/pnas.83.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Stolarov J. Ph.D. thesis. New York: Columbia University; 2000. [Google Scholar]