Abstract
The packaging signal (Ψ) and Rev responsive element (RRE), enable unspliced HIV-1 RNAs’ export from the nucleus and packaging into virions. For some retroviruses, engrafting Ψ onto a heterologous RNA is sufficient to direct encapsidation. In contrast, HIV-1 RNA packaging requires 5′ leader Ψ elements plus poorly defined additional features. We previously defined minimal 5′ leader sequences competitive with intact Ψ for HIV-1 packaging, and here examined the potential roles of additional downstream elements. The findings confirmed that together, HIV-1 5′ leader Ψ sequences plus a nuclear export element are sufficient to specify packaging. However, RNAs trafficked using a heterologous export element did not compete well with RNAs using HIV-1’s RRE. Furthermore, some RNA additions to well-packaged minimal vectors rendered them packaging-defective. These defects were rescued by extending gag sequences in their native context. To understand these packaging defects’ causes, in vitro dimerization properties of RNAs containing minimal packaging elements were compared to RNAs with sequence extensions that were or were not compatible with packaging. In vitro dimerization was found to correlate with packaging phenotypes, suggesting that HIV-1 evolved to prevent 5′ leader residues’ base pairing with downstream residues and misfolding of the packaging signal. Our findings explain why gag sequences have been implicated in packaging and show that RRE’s packaging contributions appear more specific than nuclear export alone. Paired with recent work showing that sequences upstream of Ψ can dictate RNA folds, the current work explains how genetic context of minimal packaging elements contributes to HIV-1 RNA fate determination.
Keywords: RNA packaging, retrovirus, RNA dimerization, Mfold, Free energy calculation
Graphical Abstract
Introduction
During late phases of the HIV-1 replication cycle, unspliced RNAs transcribed from integrated proviruses play two broadly-defined roles: they serve as mRNAs that encode viral proteins and as genomic RNAs (gRNAs) that become encapsidated in progeny virions. How a single species of unspliced retroviral RNA can serve these two very different replication roles is a longstanding, incompletely answered question [1].
In the case of HIV-1, twenty-year-old literature suggests that RNA fates are governed by alternate RNA structures, formed by overlapping portions of HIV-1 RNA’s highly structured 5′ leader [2]. Consensus understanding of the key sequence determinants of packaging specificity have changed little in the ensuing years [3, 4]. Under physiologic-like salt conditions in vitro, an RNA consisting of the HIV-1 5′ untranslated region plus initial gag coding sequences can exist in an equilibrium mixture of monomeric and dimeric RNA species, and mutations that stabilize one fold or the other have been described [5] [6]. The packaging phenotypes of these mutations in the context of intact virus support a model in which the dimeric species is selected for packaging while the monomer preferentially contributes to mRNA functions [6].
The retroviral RNA 5′ leader elements required for packaging are called Ψ, for packaging signal [7], and the linkage between RNA dimerization and packaging has long suggested that formation of the RNA structure required for packaging requires RNA dimerization [8–10]. Despite considerable research effort involving both model RNAs studied in reconstituted systems and viral RNAs during HIV-1 replication, defining which portions of HIV-1’s 5′ leader constitute a minimal Ψ packaging signal has been challenging, in part due to the multitude of replication functions that are regulated by RNA elements within HIV-1 RNA’s 5′ leader [2]. Figure 1A presents one model for RNA secondary structure elements in the 5′ leader. Although most studies implicate the DIS through AUG hairpin regions as the core encapsidation signal [11–13], essentially all portions of the leader have been reported to play packaging roles or ruled unimportant in conflicting reports [3]. For example, the TAR hairpin has been implicated in packaging in some reports [14–16] as have the polyA hairpin [17, 18] portions of the PBS region [19], and gag coding sequences [20–22]. Initial studies suggested that the single hairpin between the major splice donor and the start of gag specified packaging, and thus this hairpin was designated Ψ, but subsequent work demonstrated this was insufficient [3, 4]. Even elements thousands of bases from the RNA’s 5′ end have been implicated in packaging [23, 24].
Figure 1.
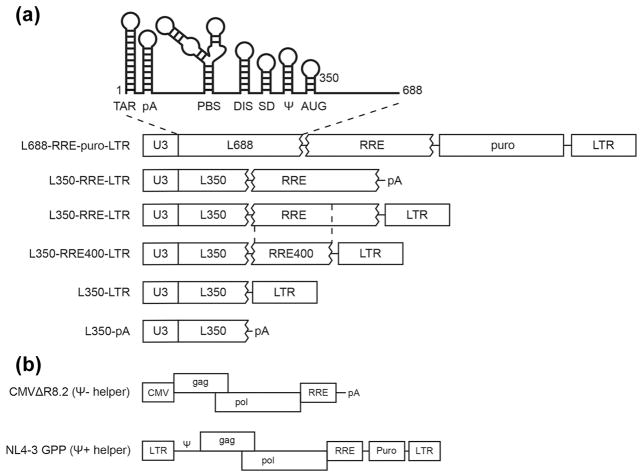
(A) Schematic representation of HIV-1 derived test vectors used to study minimal packaging determinants. At top: cartoon diagram of structural elements in HIV-1’s 5′ leader, including TAR, polyA, PBS, DIS, SD, Ψ and AUG stem-loops [2]. Below: Modular organization of test vectors. Transcription is controlled by the native HIV-1 promoter, U3. HIV-1 genome fragments are: L688: HIV-1 strain NL4-3 5′ leader from its 5′ end (RNA base 1) to base 688; L350: HIV-1 5′ leader sequence from base 1 to base 350; RRE: 858 base fragment of env (bases 7156–8014, using HXB2 numbering) that includes the RRE and downstream flanking sequences; RRE400: 400 base env fragment (bases 7156–7556) that includes the RRE; LTR: 3′ LTR. Non-retroviral sequences included in some of test vectors are puro: puromycin resistance expression cassette; and pA: minimal polyadenylation signal [see Materials and Methods]. (B) Schematic representation of HIV-1 derived helpers used in this study. CMVΔR8.2, an HIV-1 Ψ− helper and NL4-3-GPP, an HIV-1 Ψ+ helper, were described previously [38, 48].
Part of why the elements implicated in packaging have differed among published reports likely reflects the fact that disrupting one RNA structure can alter adjacent RNA folds. For example, destabilizing TAR ablates packaging [15, 25, 26]. However, it has been suggested that these TAR mutations exerted their effects by altering other elements, since the entire TAR hairpin could be removed without affecting packaging [25]. More recent evidence has suggested that initial folding events upon transcription initiation can dictate which folded structure downstream RNA elements will adopt [27].
Another challenge in defining minimal packaging sequences is HIV-1’s apparent promiscuity in packaging. For example, extensive alterations to the hairpin loop that contains the critical “dimer initiation signal” (DIS) palindrome believed to nucleate Ψ structure is surprisingly compatible with virus replication under some conditions [28], and even deletion of the entire region encompassing DIS through the AUG hairpin elements can reduce HIV-1 RNA packaging by as little as two-fold [29, 30]. When the difference between Ψ+ and Ψ− RNA packaging specificity is only two-fold, further delimitation of packaging elements can be compromised by experimental noise. Fortunately, however, when HIV-1 RNAs that lack 5′ leader packaging signals are co-expressed with RNAs that retain them, competition largely prevents leaderless RNA packaging, and thus packaging requirements can more accurately be defined under competition conditions [6].
Although 5′ leader Ψ elements of simple gammaretroviruses are sufficient to confer selective packaging onto heterologous RNAs, this is not the case for HIV-1 [31]. HIV-1 unspliced RNAs require both the viral protein Rev and the cis-acting Rev response element (RRE) to exit the nucleus, and active nuclear export plus cis-acting packaging sequences are required for an HIV-1 RNA’s packaging [32]. Previous studies have demonstrated that heterologous RNA trafficking mechanisms can substitute for the Rev/RRE interaction [12, 33, 34], but whether this is a vestigial function, like the packaging by HIV-1 of Ψ− RNAs in the absence of Ψ+ competition, or if heterologous RNA trafficking is fully equivalent to Rev-mediated RRE RNA export has not been addressed.
In our previous work delimiting minimal 5′ leader elements necessary for HIV-1 RNA packaging, nuclear export elements and other vector features were invariant [6]. The vector in those studies, which competed well with the intact native HIV-1 genomic RNA, contained the entire HIV-1 5′ leader, an 858 nt portion of the env gene that encompasses RRE, and sequences extending from upstream of the polypurine tract through the natural 3′ end of HIV-1 RNA (Figure 1). In the work here, we further dissect these elements to systematically address which HIV-1 sequences are required for competitive packaging.
Results
HIV-1 elements other than the 5′ leader and RRE are dispensable for competitive packaging
To define minimal HIV-1 sequences outside the core encapsidation signal that are required for packaging, the encapsidation of a series of test vector RNAs was monitored under competitive conditions (Figure 1A). Packaging of these was compared to L688-RRE-Puro-LTR, a vector previously shown to display competitive packaging levels indistinguishable from HIV-1 gRNA’s [6]. L688-RRE-Puro-LTR includes an extended 5′ leader (comprised of the entire 5′ untranslated region followed by the first 353 nt of gag, and here designated L688), 858 nt of env sequences containing the Rev response element (RRE), an expression cassette for puromycin resistance (Puro), and an HIV-1 3′ LTR with its preceding 70 nt, which includes the polypurine tract (LTR).
Variants of L688-RRE-Puro-LTR were designed to assess packaging roles of a) gag coding sequences downstream of core encasidation sequences, b) RRE sequences, and c) native HIV-1 3′ end sequences (Figure 1A). These derivatives, which all lack the puromycin resistance cassette, were: L350-RRE-LTR, in which all gag sequences except the first 15 nt were removed; L350-LTR, from which RRE-containing env sequences were deleted; L350-RRE-pA, in which the 3′ LTR fragment was replaced by a synthetic polyadenylation signal; and L350-pA, which also lacked RRE sequences.
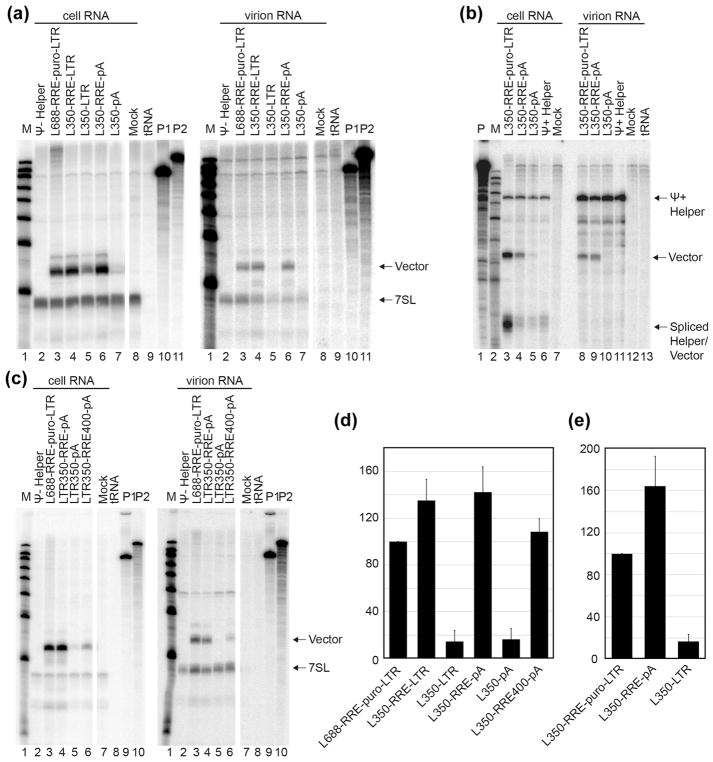
The packaging specificity of these derivatives was assessed by co-expressing them with an HIV-1 protein expressing “helper” construct (Figure 1B) in human 293T cells, and determining the levels of vector RNA in released virions. Figure 2A (and quantified in Figure 2D) shows results of RNAse protection assays that evaluated packaging of vectors mobilized using a Ψ− helper, which allowed virus-like particle formation and non-competitive test vector RNA packaging. Figure 2A cell RNA data evaluated the intracellular levels of the indicated vectors (Figure 1A), with products normalized to 7SL, a host RNA that is packaged into HIV-1 virions in proportion to Gag [35]. Figure 2A virion RNA data measured the virion incorporation of these vectors, again normalized to 7SL. Consistent with previous reports [32], RNAs lacking the RRE were packaged poorly if at all (Figure 2A virion RNA lanes 5 and 7) even after accounting for their reduced intracellular levels (Figure 2A cell RNA lanes 5 and 7), while the packaging of vectors lacking extended portions of gag or the 3′ LTR was indistinguishable from values for vectors retaining these HIV-1 sequences (Figure 2A lanes 3, 4 and 6). These results suggest gag sequences downstream of core encapsidation sequences as well as HIV-1’s natural 3′ end sequences are dispensable for HIV-1 packaging in the absence of competition.
Figure 2.
Cellular expression and packaging of HIV-1 based vectors RNA under non-competitive and competitive conditions, as monitored by RNase protection assay. (A) and (C) are RNA samples obtained from the co-expression of packaging test vectors with CMVΔR8.2, an HIV-1 Ψ− helper construct. Riboprobes A and B (see Materials and Methods) were used together in these experiments. (B) displays samples obtained from the co-expression test vectors with NL4-3-GPP, an HIV-1 Ψ+ helper construct. Riboprobe C was used in this RNase protection assay. Riboprobe C spans the splice donor and thus spliced products are observed in cells, but are not packaged. Within each panel, RNA samples extracted from cells are at the left and those from virus–containing media are on the right. Protected riboprobe fragments corresponding to test RNAs (Vector) and 7SL RNA (7SL) are labeled in panels A and C, and fragments corresponding to unspliced helper RNAs (Ψ+ Helper), test RNAs (Vector) and spliced test and helper RNAs (Spliced Helper/Test) are labeled in panel B. Lane designations indicate samples where indicated vectors were co-transfected with panel-specific helper; Ψ+ or Ψ− helper transfected alone; mock: mock transfected cells; tRNA: riboprobe protected with yeast tRNA; M: molecular weights marker; P: undigested riboprobe (P1 and P2 are shown separately in panels where two riboprobes were used together). RNase protection assays were performed as described in Materials and Methods. The closely-migrating multiple bands evident in some but not all lanes in Figure 2A and 3 were pooled during quantification, were not uniformly observed, and were an undetermined outcome of the digestion conditions. (D) Quantification of packaging efficiency of HIV-1 vectors under non-competitive conditions (panel A and its replicates). Packaging efficiency was calculated by dividing the ratio of vector RNA to 7SL RNA in the virion sample by the ratio of vector RNA to 7SL in cells as determined using RNase protection assays quantified by phosphorimager analysis. (E) Quantification of packaging efficiency of HIV-1 vectors under competitive conditions (panel C and its replicates). Packaging efficiency was calculated by dividing the ratio of vector RNA to helper RNA in the virion sample by the ratio of vector RNA to helper RNA in the cells as determined by RNase protection assay and quantified by phosphorimager analysis. For all quantifications, the results represent data from at least three independent transfection experiments.
To examine the packaging of vectors lacking extended gag and 3′ LTR sequences under competitive conditions, a subset of these vectors was co-expressed with Ψ+ helper (Figure 2B). The results confirmed that the core encapsidation sequences that reside within the first 350 nt of HIV-1 RNA’s 5′ end (L350) plus the 858 nt RRE fragment used here were sufficient for competitive packaging.
Figures 2C and E display data for vectors in which both 5′ leader and RRE fragments were trimmed. Based on vector RNA abundance relative to 7SL RNA, trimming of the 858 nt RRE fragment to generate a 400 nt fragment (RRE400; designed to retain the entire structured region and remove splice donor A7 [36]) demonstrated that this smaller portion of env also was sufficient to mediate competitive packaging (Figure 2C lanes 6).
Heterologous nuclear export signals can mediate packaging but do not compete well with RRE
Consistent with the hypothesis that RRE’s sole role in packaging is in the nuclear export of HIV-1 unspliced RNA, it has previously been reported that replacement of RRE sequences by the constitutive transport element (CTE) from the Mason-Pfizer monkey virus genome renders HIV-1 gene expression and RNA packaging Rev-independent [12, 34] [33]. RRE- and CTE-trafficked RNAs tap into different host RNA export pathways [37] and RNAs trafficked by these pathways do not freely intermix for co-packaging [33]. A recent report has suggested that RNA localization mediated by the CTE differs from that resulting from the interaction of Rev with RRE [38]. Thus, the work here examined CTE trafficked RNAs’ packaging under both competitive and non-competitive conditions. This was accomplished by generating a series of L688-RRE-LTR derivatives: one in which the RRE was replaced with CTE sequences, and a second in which CTE sequences were placed downstream of the RRE (L688-CTE-LTR and L688-RRE-CTE-LTR respectively; Figure 3).
Figure 3.
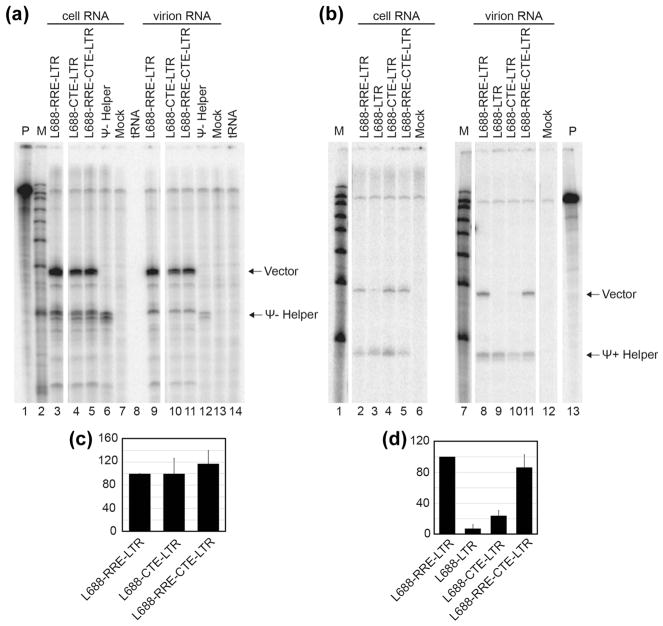
Packaging efficiency of vectors containing a heterologous RNA transport element under non-competitive or competitive conditions. RNase protection assays were performed after co-expression of test vectors with Ψ− (A) or Ψ+ (B) helper. Riboprobe D (see Materials and Methods) was used in these experiments. Protected riboprobe fragments corresponding to test (Vector) and helper (Ψ− or Ψ+ Helper) RNAs are labeled. Lane designations indicate co-expressed HIV-1 vectors, markers, and controls as in Figure 2. (C) and (D) Quantification of packaging efficiency of HIV-1 vectors under non-competitive (C) or competitive (D) conditions. The packaging efficiency was calculated by dividing the ratio of vector RNA to helper RNA in virion samples by the ratio of vector RNA to helper RNA in the cells as determined by RNase protection analysis and quantified by phosphorimager analysis. The results present data from at least three independent experiments.
The packaging of these vectors was first examined by RNAse protection assay under non-competitive conditions by co-expression of each test vector with a Ψ− helper (Figure 3A, with quantification in 3C). A comparison of vector and helper protected bands in cell samples showed that all the vectors tested here were expressed well and to similar levels. The ratios of these products in virus-derived RNA samples indicated that CTE-containing vectors were packaged as well as vectors containing RRE or RRE+CTE (Figure 3A).
Packaging was then examined under competitive conditions using a Ψ+ helper (Figure 3B, with quantification in 3D). A vector (L688-LTR) in possession of neither CTE nor RRE was included as a non-trafficked control. The results differed from those under non-competitive conditions. Consistent with previous reports and the findings above, L688-RRE-LTR competed effectively with Ψ+ helper, while packaging of the vector lacking nuclear export signals (L688-LTR) was below the level of detection (Figure 3B, compare lanes 8 and 9). However, in contrast to the findings in Figure 3A, where the CTE-containing vector L688-CTE-LTR was packaged well, L688-CTE-LTR packaging occurred at 5-fold reduced levels relative to RRE-containing vectors when tested under conditions of Ψ+ helper competition (Figure 3B lane 10 and Figure 3E). When packaging of a vector containing both CTE plus RRE sequences was examined (Figure 3B lane 11 and Figure 3E), competitive packaging was restored.
Minimal Ψ packaging elements do not function in all sequence contexts
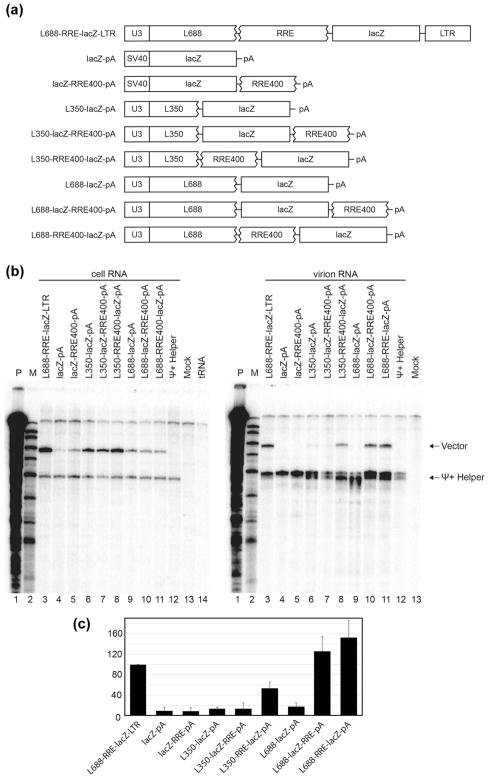
To address whether or not the minimal packaging sequences defined above were sufficient to drive the encapsidation of a foreign RNA into HIV-1 particles, subsets of these elements were incorporated into an RNA comprised of mammalian-expressed bacterial lacZ (Figure 4). Because the studies above indicated that the 3′ LTR sequences in a packagable vector could be replaced by a short synthetic polyadenylation signal (pA) and that a 400 nt RRE fragment (RRE400) was sufficient to support packaging, these minimal packaging elements were introduced into the lacZ expression construct one at a time, and the packaging of the resulting lacZ derived RNAs was monitored in the presence of Ψ+ helper competition. Figure 4 presents RNAse protection assay assessment of these RNAs in transfected cells and in the virions produced by them. Lanes 3 and 4 are RNase protection products of positive and negative control RNAs: one in which the puromycin resistance cassette within L688-RRE400-Puro-pA was replaced with lacZ sequences to generate L688-RRE-lacZ-LTR, and the second a lacZ RNA (lacZ-pA) that possessed no HIV-1 sequences and was expressed from a heterologous SV40 promoter. Comparing cell and virus RNAs by RNAse protection shows that as expected, competitive packaging was retained upon replacement of puro with lacZ in L688-RRE-lacZ-LTR, but that lacZ RNA lacking all HIV-1 sequences was not detectably packaged (Figure 4A and B lanes 3 and 4). Also as expected, appending RRE alone to the 3′ end of the Ψ− lacZ RNA failed to promote its packaging, as did inserting lacZ without RRE downstream of the minimal Ψ (Figures 4A and B lanes 5 and 6).
Figure 4.
Context effects of minimal packaging elements on the packaging of a heterologous RNA. (A) Schematic representation of HIV-1 derived test vectors for study of effects of lacZ sequence on packaging. (B) RNase protection assay of samples obtained from co-expression of test vectors with a Ψ+ helper. Cell RNA samples are at left and virus RNA is in the right panel. Riboprobe E, which is a chimeric riboprobe that includes sequences complementary to both gag and lacZ (see Materials and Methods), was used in these experiments. Protected riboprobe fragments corresponding to test (Vector) and helper (Ψ+ Helper) RNAs are indicated. Lane labels indicate the vectors that were co-expressed with Ψ+ helper. Ψ+ helper alone; mock: mock transfected cells; tRNA: riboprobe protected with yeast tRNA; M: molecular weights marker; P: undigested probe. (C) Quantification of packaging efficiency of HIV-1 vectors. The packaging efficiency was calculated by dividing the ratio of vector RNA to helper RNA in the virion by the ratio of vector RNA to helper RNA in the cells as determined by RNase protection analysis and quantified by phosphorimager analysis. The results represent data from at least three independent transfection experiments.
Surprisingly, however, addition of lacZ sequences between the minimal Ψ and RRE of the well-packaged L350-RRE400-pA vector yielded an RNA (L350-lacZ-RRE400-pA, Figures 4A and B lanes 7) that failed to be packaged. In contrast, re-ordering the genetic elements of this non-packaged vector (Figures 4A and B, lanes 8, L350-RRE400-lacZ-pA) fully restored vector packaging. When extended leader sequences were included upstream of lacZ plus RRE sequences (L688-lacZ-RRE400-pA and L688-RRE400-lacZ-pA; lanes 9 and 10), efficient vector packaging also was restored, as indicated by the quantification in panel C.
In vitro dimerization properties of packaging elements in altered contexts correlate with packaging properties
The above data suggested that lacZ sequences were not inherently inhibitory to packaging, but that proper functioning of minimal packaging elements was context-dependent. A possible explanation for this observation was that the gag sequences that naturally reside downstream of the core encapsidation signal serve as an insulator that allows upstream 5′ leader sequences to retain a packagable fold, whereas juxtaposition of the core encapsidation sequences within L350 with lacZ sequences interfered with formation of the required RNA structure.
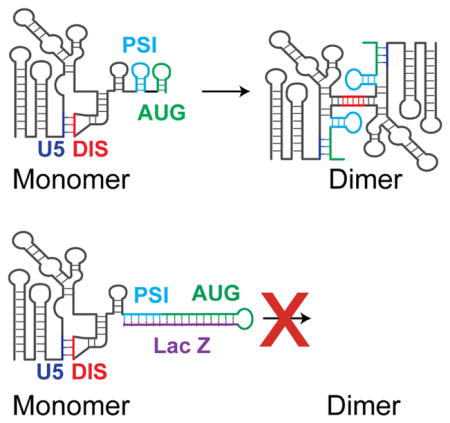
To test this notion and assess the influence of downstream nucleotides on 5′ leader structure, we measured the in vitro dimerization properties of RNAs composed of the 5′ ends of vectors that are either efficiently (L350-RRE-LTR) or poorly (L350-lacZ-RRE-pA) packaged on native agarose gels (Figure 5A). Previous NMR studies of HIV-1’s 5′-leader have shown that in its dimerization-competent form, the 5′ end of HIV-1 RNA adopts a structure in which residues of U5 are base paired with the region containing the gag start codon (indicated AUG in Figure 5), and that the palindromic sequence of DIS participates in a dimer interface (Figure 5B). In contrast, in the monomeric leader, residues in the AUG region form a hairpin and residues in U5 base pair with the DIS palindrome, thereby inhibiting intermolecular DIS:DIS base pairing [39–41]. Mutations that either stabilize U5:AUG element pairing or that destabilize the AUG hairpin promote dimerization, whereas mutations that enhance U5:DIS base pairing inhibit dimerization [39]. We therefore hypothesized that non-native residues downstream of AUG in vector RNAs that are not packaged might somehow interfere with U5:AUG base pair formation.
Figure 5.
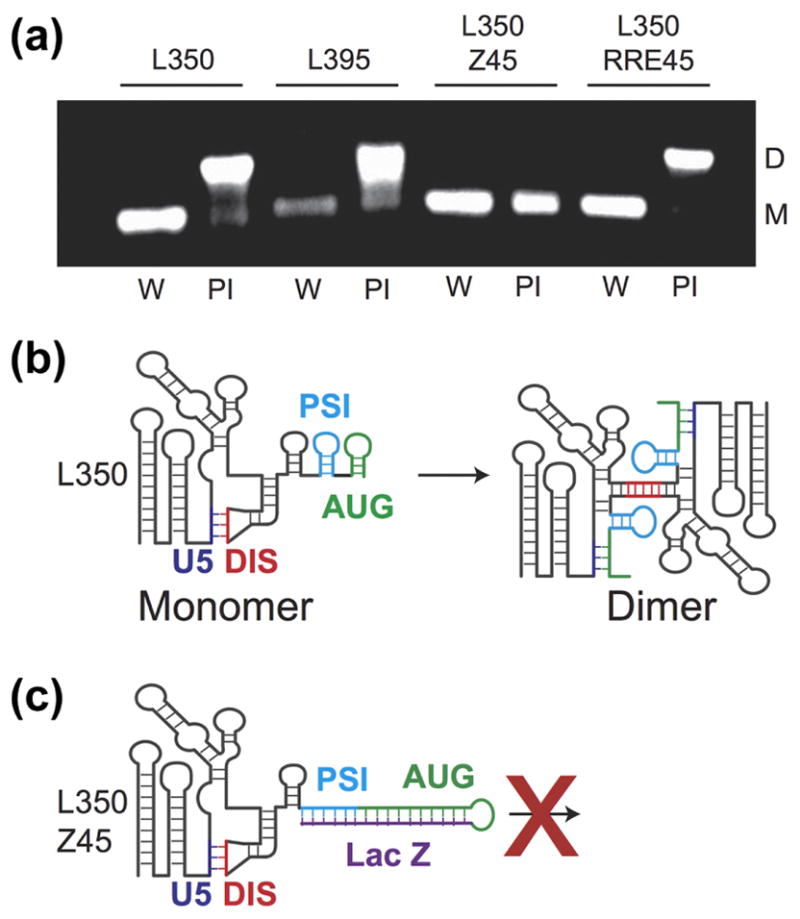
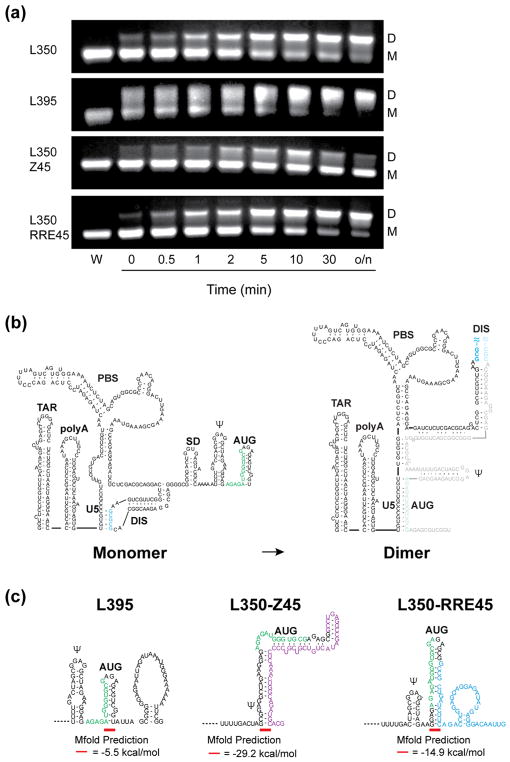
Influence of residues downstream of the 5′ UTR on the monomer-dimer RNA equilibrium. (A) Each construct was incubated overnight in water only (W) to form a monomer (M) or in physiological-like ionic strength (PI) buffer. L350 and L395 form a dimer (D) in PI buffer as expected. Addition of the non-native LacZ residues prevents dimerization in the L350-Z45 construct. L350-RRE retains dimerization behavior similar to the L350 and L395. (B) Cartoon representation of the 5′ UTR structure with the additional residues. In the monomer conformation, the region containing the AUG start codon forms a hairpin (green) and the dimer-promoting DIS hairpin (red) is sequestered by an intramolecular interaction with the U5 (blue). In the dimer conformation long-range base-pairing interactions between the AUG and U5 help expose the DIS hairpin and its palindromic sequence [39]. (C) The non-native residues in the L350-Z45 construct form a highly stable extended hairpin with AUG preventing the U5:AUG interaction, exposure of the DIS hairpin and thus formation of the dimer.
We have previously demonstrated that the L350 control RNA employed here, which contains the first 350 residues of the native 5′ leader, includes all 5′ leader sequences necessary for competitive packaging [6]. To address whether or not sequences downstream of L350 that differed among the above vectors affected RNA dimerization, the in vitro dimerization properties of additional RNAs were compared to L350. The other constructs studied here contained L350 plus either 45 native gag residues (5′-L395), 45 non-native residues from lacZ (5′-L350-Z45), or 45 residues of the 858 base RRE-containing env fragment (5′-L350-RRE45).
Under conditions of low ionic strength (10 mM KH2PO4 buffer, < 0.2 mM Na+), L350, L395, L350-Z45, and L350-RRE RNAs all exist predominantly as monomers, as expected (lanes designated “W” in Figure 5A). Overnight incubation of 5′-L350 in physiological-like ionic strength buffer solution (PI Buffer: 140 mM KCl, 10 mM NaCl, 1 mM MgCl2, and 10 mM HK2PO4, pH 7.4) led to an equilibrium shift that favored the dimer (L350 lane indicated PI in Figure 5A), as observed previously [41]. The dimerization behavior of L395, which includes additional native gag residues, was similar to that observed for 5′-L350 (L395 lanes). However, the 5′-L350-Z45 was unable to form a stable dimer under the physiological-like ionic conditions employed (L350-Z45; Figure 5A). Although 5′-L350-RRE also lacked gag sequences downstream of L350, this construct nevertheless exhibited dimerization properties similar to those of L350 and L395 (Figure 5A).
Varied incubation times were used to further investigate the thermodynamic equilibrium between structures for each construct (Figure 6A). 5′-L350 and 5′-L350-RRE45 showed a slow progression towards the dimer, whereas the L395 construct showed a much faster shift to the dimeric structure. In contrast, L350-Z45 exhibited a weak and transient dimer band after a few minutes of incubation in PI buffer, but was predominantly monomeric after reaching equilibrium (Figure 6A).
Figure 6.
Time dependent dimerization of in vitro RNAs, and structural predictions that correlate with RNA behaviors. (A) Time dependent equilibration of RNA monomer and dimer species. Time points of incubation in PI buffer are indicated in minutes or o/n for overnight. At equilibrium L350, L395, and L350-RRE form predominantly dimer conformations (77 %, 100 %, and 72 % dimers, respectively). L350-Z45 shows an initial dimerization behavior at 10 minutes but is predominantly monomer (73 % monomer) at equilibrium. (B) Secondary structure of the HIV-1NL4-3 5′-Leader in its monomeric and dimeric states. (C) Secondary structure prediction (MFold) of the downstream portion of the L359, L350-Z45, and L350-RRE45 constructs. L350-Z45 shows how non-native residues (purple) can remodel the Ψ and AUG hairpins and potentially prevent U5:AUG mediated dimerization.
Free energy calculations (MFold [42, 43]) of RNA secondary structures suggest a potential explanation for these findings (Figures 6B and C). Whereas L350, L395, and L350-RRE were predicted to adopt native-like secondary structures that include the Ψ and AUG hairpins, L350-Z45 adopted a dramatically altered low-energy structure in which the Ψ and AUG residues do not form hairpins but instead are base paired with downstream residues of Z45 (Figure 5 and 6, panels B and C; see Materials and Methods for information about the MFold prediction). These findings suggest that observed inhibition of RNA dimerization (and thus packaging) is due to the sequestration of AUG residues by non-native downstream elements, which prevents AUG from base pairing with U5 and exposing the DIS for dimerization (Figures 5C and 6C).
Discussion
RNA elements involved in HIV-1 packaging have been studied extensively over the decades, with attempts to precisely define the required sequences often yielding contradictory results [44]. This is in part due to the multitude of functional information in the 5′ leader, which complicates its genetic dissection. The high level of genetic conservation in HIV-1’s 5′ leader likely reflects how sensitive 5′ leader structures are to genetic changes [5]. At least some of the disagreements in defining minimal packaging elements may reflect differences in experimental approach. For example, assayed in the absence of competition, HIV-1 RNA packaging is promiscuous, with most 5′ leader elements dispensable for virus replication under certain experimental conditions [28].
Unlike simple retroviruses, HIV-1’s Ψ is not sufficient to mediate heterologous RNA packaging. It has long been recognized that HIV-1 RNA packaging requires a mechanism for RNA nuclear export in addition to 5′ leader packaging sequences [31, 34], and the competitive packaging approaches used here allowed a closer examination of this requirement. One novel finding of the current report is that whereas RNAs trafficked by heterologous nuclear export signals like the Mason Pfizer monkey virus constitutive transport element (CTE) can complement the defect of a Rev plus RRE-deficient HIV-1, CTE and RRE-mediated trafficking are not functionally equivalent and RRE-trafficked RNAs are preferentially packaged under competitive conditions. Although less straightforward phenomenology cannot be ruled out, the fact that our CTE-trafficked vectors were packaged well in the absence of Ψ+ competition while vectors that possessed neither CTE nor RRE were not detectably packaged under non-competitive conditions suggests both that the packaging signal was folded correctly and that nuclear export occurred for L688-CTE-LTR.
On initial examination, these observations appear to conflict with a previous report’s, in which CTE and RRE expressed vectors were observed to co-package, albeit at reduced rates [33]. We are not certain why our two reports differed, but it is important to note that the other authors’ study did not monitor intracellular RNA levels and that they used 4X CTE vectors, which are known to maximize CTE function. Using multimerized CTEs was appropriate for a study intended to maximize packaging and where packaging competition per se was not addressed. However, these differences in experimental approach may have contributed to why our observations of CTE-containing RNA packaging were more similar to the three-fold reduced levels previously reported by McBride et al, who also used both transport systems co-expressed together, than to those of Moore et al [33].
Some of the conflicting literature about what portions of HIV-1 RNA are required for packaging may reflect the fact that alterations to HIV-1 vector RNAs can drastically change their intracellular abundance. This phenomenon is vividly evident from much of the cell RNA data in the current work, such as the differences among vectors in intracellular vector: 7SL RNA ratios that are evident in the cell RNA panel of Figure 2A. A confounding factor in early HIV-1 packaging studies was apparent cell-type dependent differences, which may have reflected differing levels of RNA expression [45]. Retroviral vector packaging is often measured as the amount of viral RNA in virions without considering the RNAs’ levels within cells, and thus studies employing this approach may erroneously conclude that packaging is defective for a vector that is instead unstable or expressed at a low level. Some evidence of the extent to which altering vector expression plasmids can affect intracellular RNA levels can be seen for vectors lacking nuclear export signals in the current report’s figures (eg: Figure 2 constructs lacking RRE). In some cases, not measuring intracellular RNA levels relative to a well-packaged but poorly expressed standard likely overestimates a vector’s packaging efficiency. Virus-like particles generated by Ψ− helpers contain enhanced levels of host mRNAs [46], and reduced intracellular viral RNA levels may relax packaging specificity and increase the frequency of alternate RNA packaging.
The key findings of this report relate to how genetic context can influence the folding behavior of minimal RNA elements required for packaging. While this article was in preparation, Hu et al [22] published results of their experiments where they found that varying the sequences downstream of the first 362 nt of HIV-1 affected packaging efficiency. Those studies showed that vectors containing the native 5′-UTR (untranslated region) followed by codon optimize gag sequences (syngag) were packaged with relatively poor efficiency, and it was concluded that residues in gag play an indirect but mechanistically unknown role in packaging [22].
Here, the roles of gag sequences on core encapsidation sequence structure were examined more directly, using agarose gel studies to test the role downstream sequences play in the monomer-dimer equilibrium. Overnight and time dependent incubations demonstrated that constructs with native HIV sequence (L350, L395, and L350-RRE45) favored the dimer, as opposed to the L350-Z45 construct that favored the monomer. These results argue for three conclusions. First, gag sequence downstream of the core encapsidation region can stabilize the dimer conformation as evidenced in faster rate of dimer formation for the L395 construct than the L350 construct. This is consistent with the cell culture-based work described above, which showed that increasing the length of gag sequences increases packaging [22]. Second, downstream, non-gag HIV-1 sequences that can form a local, stable hairpin structure do not interfere with dimer formation, as evident from the ability of L350-RRE45 to readily dimerize. Third, non-native downstream sequences that, according to free energy-base predictions, are capable of sequestering AUG and preventing formation of the U5:AUG structure (e.g., lacZ), inhibit dimerization and thereby inhibit dimerization-dependent RNA packaging. Our MFold calculations suggest that the previously-reported syngag constructs may also misfold in a manner that sequesters residues of AUG and prevents U5:AUG formation [22]. It thus appears that the gag sequences of HIV genomes have evolved to allow appropriate coding of amino acids without interfering with formation of 5′ leader structures that regulate genome function. Similarly, previous mutagenesis experiments aimed at identifying packaging determinants in gag may have been confounded by mutation-induced misfolding of the 5′-UTR.
Although the current work enhances our understanding of how a single species of unspliced RNA can contribute to many HIV-1 replication roles, understanding of HIV-1’s RNA packaging selectivity remains incomplete. For example, it is still unclear what determinants contribute to observations that spliced HIV-1 RNAs and HIV-derived RNAs gutted of known packaging signals remain better-packaged than most cellular mRNAs [29, 47]. Nonetheless, by advancing understanding of the regulation and structural complexity of the HIV-1 5′ leader, and by demonstrating that RRE-trafficked RNAs outcompeted CTE-trafficked RNAs for packaging, the results here indicate that the RRE may contribute more to packaging specificity than merely by transporting RNAs to the cytoplasm,
In summary, our findings are consistent with proposals that the primary determinants of packaging reside within the 5′-leader of the genome. The ability of downstream RNA modifications to interfere with both dimerization and packaging is likely due to their ability to induce misfolding of the upstream dimerization and packaging signals, and their contributions to authentic RNA trafficking within cells. Future experiments aimed at determining if misfolding occurs in the manner predicted by free energy calculations or other types of long-range interactions are now being planned.
Materials and methods
Plasmid construction
All HIV-1 vectors and RNAs analyzed in this report are derivatives of HIV-1 strain NL4-3. The Ψ+ helper used here (NL4-3 GPP) has been described [39] and contains HIV-1 NL4-3 sequences with env replaced by a puromycin-resistance expression cassette. CMVΔR8.2 [48] which contains a CMV promoter in place of the HIV-1 5′ leader, served as the Ψ− helper. In the absence of Ψ+ competition, the unspliced Ψ− transcript of CMVΔR8.2 is encapsidated in virus-like particles at levels approximately 20% the level of Ψ+ RNAs [6].
The test vector L688-RRE-puro-LTR was previously described [49], where it was designated “Native”, with its construction described in [6], where it was called 5′-LNL4-3 (aka “Native vector”; or pSRK1428-4). Derivatives used in the current study were constructed by combining fragments of HIV-1 sequences generated using overlap PCR, inserting them into pGEM-T easy (Promega), and sequencing them prior to use. The minimal cleavage/polyadenylation signal was originally derived from Promega vector psiCheck1, and was provided by Aaron Goldstrohm (University of Minnesota). The CTE fragment used here was amplified from NL4-3ΔenvΔrevCTE ([50] kindly provided by Paul Bieniasz, The Rockefeller University). The lacZ sequences used in the Figure 4 vectors were amplified from pHR’CMVlacZ (GenBank accession # AF105229). The vector designated lacZ-pA in Figure 4 was pSVβ-Galactosidase (Promega). The riboprobe expression constructs were generated by subcloning PCR products into pBluescript KS as described below.
Cell culture and transfection
Human embryonic kidney 293T cells were maintained in Dulbecco’s Modified Eagle Medium (Invitrogen) supplemented with 10% FBS (Gemini) and penicillin-streptomycin (Invitrogen) at 37°C with 5% CO2. A fresh monolayer of cells, approximately 70% confluent, was transfected with HIV-1 helper and vector plasmids using polyethylenimine (Polysciences) with a ratio of 1 μg total plasmid DNA to 4 μg polyethylenimine in 800 μl of 150 mM NaCl [51]. All transfections included 5 μg of either Ψ+ or Ψ− helper plasmids, and test vector plasmids were added at a molar ratio of 1:1 (Ψ+ helper:test) or 2:1 (Ψ− helper:test). pBluescriptKS plasmid DNA was added where needed to ensure each transfection contained equal amounts of DNA. Transfection mixtures were incubated 15 min. at room temperature, then added to 8 ml of media in 10 cm2 plates of cells. Media was replaced 24 hrs. post-transfection and virus-containing media and cells harvested 48 hrs. post-transfection.
Virus isolation & quantification
Virus particles were collected 48 hrs. after transfection by filtering cell culture media through a 0.22 μm filter and centrifuging through a 20% sucrose cushion at 25,000 rpm for 2 hrs. A real-time reverse-transcription PCR assay modified from Vermeire J., et al [52] was performed to quantify virus content. Briefly, 2 μl of diluted virus lysate was transferred to Light Cycler 480 96-well plates (Roche) containing PCR mix (10mM Tris, pH 8.3, 50mM KCL, 0.15% Triton X-100, 2mM MgCl2, 0.2mM dNTPs, 1× EvaGreen (Biotium), 1mM DTT, 0.08 μg/ul MS2 RNA, 0.3μM of forward and reverse MS2 primers, 4 u RNasin (Promega) and 0.5 u Go Taq (Promega)). PCR was performed using the Light Cycler 96 (Roche) and the data analyzed with Light Cycler 96 SoftWare 1.1.
Cell and virus RNA isolation and RNase protection assays
Cellular and viral RNA was isolated using TRIzol according to the manufacturer’s protocol (Ambion) and DNase treated. RNase protection assays (RPAs) were conducted as previously described [53] using cell RNA from approximately 1/60th of one plate of transfected cells and viral RNA normalized to RT activity. Riboprobes were transcribed from linearized plasmid templates using either T7 or T3 polymerase (Promega) and [α-32P]-rCTP (Perkin-Elmer) as previously described [53]. The riboprobes used in this study are: Riboprobe A, aka pSKh112, which protects 124nt of HIV-1 RNA upstream of splicing donor site 1 (bases 165–288); Riboprobe B, aka pSKh78, which protects 100nt of human 7SL RNA; Riboprobe C, aka pJTC171-4, which is a chimeric riboprobe with one portion complementary to the HIV-1 strain NL4-3 5′ leader (bases 218–501) and a second portion that was derived from the helper and is complementary to 285nt of unspliced Ψ+helper RNA,133nt of unspliced test vector RNA and 74nt of spliced RNA; Riboprobe D, aka pSKh212-1, which is complementary to the 3′ ends of the CTE containing set of HIV-1 vectors used here and protects 145nt of test vector RNA, 97nt from Ψ− helper RNAs, and 93nt of Ψ+ helper RNAs; and Riboprobe E, aka pSKh90, which is a chimeric riboprobe that protects 201nt of gag (bases 1356–1556) from the helper RNA and 289nt of lacZ contained in test Figure 4 test vectors.
In vitro RNA template preparation and transcription
In vitro transcription templates were generated by PCR amplification (EconoTaq PLUS 2X Master Mix, Lucigen) of pUC19 derivatives encoding the different RNAs using a forward primer 20–30 nucleotides upstream to the T7 promoter site and a reverse primer with the first two nucleotides containing 2′ O-methyl modifications to optimize transcription termination [54]. RNAs were synthesized using 7.5 mL reactions, each containing 80 mM Tris·HCl (pH 9.0), 2 mM DTT, 20% (vol/vol) DMSO, 2 mM spermidine, 20 mM MgCl2, 3–6 mM NTPs, 12 mg of PCR-amplified DNA template, and 0.15 mg T7 RNA polymerase. Reactions were quenched after a 6-h incubation at 37 °C by addition of 250 mM EDTA and RNA was purified by electrophoresis on denaturing polyacrylamide denaturing gels (SequaGel, National Diagnostics) at 20 W overnight. RNA bands were visualized by UV-shadowing, excised, and eluted using the Elutrap electroelution system (Whatman) at 150 V overnight. The eluted RNAs were washed with 2 M NaCl and then desalted using a 30-kDa MWCO Amicon Ultra-4 Centrifugal Filter Device (Millipore) at 5000 rpm at 16 °C. The concentration of each sample was determined by optical absorbance at 260 nm, and sample homogeneity was confirmed by denaturing polyacrylamide gel electrophoresis (PAGE).
Native agarose gel electrophoresis
In vitro RNA samples were incubated at 37 °C in either RNase free Millipore filtered water or PI Buffer (10 mM KH2PO4, pH 7.4, 1 mM MgCl2, 122 mM KCl, 10 mM NaCl) overnight. Time dependent experiments had samples incubated for the times indicated such that all incubation periods would finish simultaneously. Samples were immediately placed on ice when removed from the incubator. Native agarose gel loading solution containing 0.17% Bromophenol Blue and 40% (vol/vol) Sucrose was added to each sample and mixed. 250 ng of each sample was loaded onto a 1% (vol/vol) agarose TBM gel containing 0.2 mM MgCl2 and 0.5 μg/mL ethidium bromide in the gel and running buffer. Gels were run at room temperature at 115 V for 90 min and visualized by UV illumination. Percent monomer versus dimer populations were measured from photographed gels (Kodak Gel Logic 200 Imaging System) using ImageJ software [55]. To understand the effect the 3′ residues had on the stability of the AUG region, the RNA secondary structures were predicted with Mfold using sequence from U306 to the end of each corresponding construct (L350, L395, L350-Z45, L350-RRE45) [42, 43]. Relative free energies for predicted hairpins that sequester the AUG element were calculated separately for each individual hairpin. All structures were calculated de novo without inclusion of any experimentally derived restraints.
Highlights.
Ψ plus nuclear export signals are the only HIV-1 elements required for specific RNA packaging
RNAs trafficked using non-HIV nuclear export signals do not efficiently compete with HIV RNAs
Sequences downstream of Ψ can disrupt Ψ structure and RNA dimerization in vitro
HIV-1 sequences evolved to concertedly maintain RNA folding and biologic functions
Acknowledgments
We thank Cleo Burnett for help preparing the figures. Support from the NIH (F31 GM123803 to J.D.B. and R01 GM42561 to A.T. and M.F.S.) is gratefully acknowledged.
Footnotes
Declarations of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leblanc J, Weil J, Beemon K. Posttranscriptional regulation of retroviral gene expression: primary RNA transcripts play three roles as pre-mRNA, mRNA, and genomic RNA. Wiley Interdiscip Rev RNA. 2013;4:567–80. doi: 10.1002/wrna.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 3.Mailler E, Bernacchi S, Marquet R, Paillart JC, Vivet-Boudou V, Smyth RP. The Life-Cycle of the HIV-1 Gag-RNA Complex. Viruses. 2016:8. doi: 10.3390/v8090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lever AM. HIV-1 RNA packaging. Adv Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 5.van Bel N, Ghabri A, Das AT, Berkhout B. The HIV-1 leader RNA is exquisitely sensitive to structural changes. Virology. 2015;483:236–52. doi: 10.1016/j.virol.2015.03.050. [DOI] [PubMed] [Google Scholar]
- 6.Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, et al. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417:224–39. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann R, Mulligan RC, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–9. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 8.Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SH, Soheilian F, et al. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013;9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat Rev Microbiol. 2004;2:461–72. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 10.Johnson SF, Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 2010;6:e1001007. doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–9. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride MS, Schwartz MD, Panganiban AT. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–54. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci U S A. 1994;91:4945–9. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem. 2004;279:22243–9. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 15.Helga-Maria C, Hammarskjold ML, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73:4127–35. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jalalirad M, Saadatmand J, Laughrea M. Dominant role of the 5′ TAR bulge in dimerization of HIV-1 genomic RNA, but no evidence of TAR-TAR kissing during in vivo virus assembly. Biochemistry. 2012;51:3744–58. doi: 10.1021/bi300111p. [DOI] [PubMed] [Google Scholar]
- 17.Clever JL, Eckstein DA, Parslow TG. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J Virol. 1999;73:101–9. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the u5 RNA sequences. Virology. 2002;303:152–63. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 19.Clever JL, Miranda D, Jr, Parslow TG. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J Virol. 2002;76:12381–7. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luban J, Goff SP. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–93. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchschacher GL, Jr, Panganiban AT. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–9. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Nikolaitchik OA, Rahman SA, Chen J, Pathak VK, Hu WS. HIV-1 Sequence Necessary and Sufficient to Package Non-viral RNAs into HIV-1 Particles. J Mol Biol. 2017;429:2542–55. doi: 10.1016/j.jmb.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chamanian M, Purzycka KJ, Wille PT, Ha JS, McDonald D, Gao Y, et al. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe. 2013;13:181–92. doi: 10.1016/j.chom.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolaitchik OA, Hu WS. Deciphering the role of the Gag-Pol ribosomal frameshift signal in HIV-1 RNA genome packaging. J Virol. 2014;88:4040–6. doi: 10.1128/JVI.03745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das AT, Vrolijk MM, Harwig A, Berkhout B. Opening of the TAR hairpin in the HIV-1 genome causes aberrant RNA dimerization and packaging. Retrovirology. 2012;9:59. doi: 10.1186/1742-4690-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vrolijk MM, Ooms M, Harwig A, Das AT, Berkhout B. Destabilization of the TAR hairpin affects the structure and function of the HIV-1 leader RNA. Nucleic Acids Res. 2008;36:4352–63. doi: 10.1093/nar/gkn364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharytonchyk S, Monti S, Smaldino PJ, Van V, Bolden NC, Brown JD, et al. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci U S A. 2016;113:13378–83. doi: 10.1073/pnas.1616627113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill MK, Shehu-Xhilaga M, Campbell SM, Poumbourios P, Crowe SM, Mak J. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J Virol. 2003;77:8329–35. doi: 10.1128/JVI.77.15.8329-8335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laham-Karam N, Bacharach E. Transduction of human immunodeficiency virus type 1 vectors lacking encapsidation and dimerization signals. J Virol. 2007;81:10687–98. doi: 10.1128/JVI.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berkhout B, van Wamel JL. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–32. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkowitz RD, Hammarskjold ML, Helga-Maria C, Rekosh D, Goff SP. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–23. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 32.Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J Virol. 2010;84:6598–604. doi: 10.1128/JVI.02264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5:e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, et al. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci U S A. 1994;91:1256–60. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onafuwa-Nuga AA, Telesnitsky A, King SR. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. Rna-a Publication of the Rna Society. 2006;12:542–6. doi: 10.1261/rna.2306306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann DA, Mikaelian I, Zemmel RW, Green SM, Lowe AD, Kimura T, et al. A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J Mol Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 37.Rekosh D, Hammarskjold ML. Intron retention in viruses and cellular genes: Detention, border controls and passports. Wiley Interdiscip Rev RNA. 2018 doi: 10.1002/wrna.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker JT, Sherer NM. Subcellular Localization of HIV-1 gag-pol mRNAs Regulates Sites of Virion Assembly. Journal of Virology. 2017:91. doi: 10.1128/JVI.02315-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–5. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran T, Liu Y, Marchant J, Monti S, Seu M, Zaki J, et al. Conserved determinants of lentiviral genome dimerization. Retrovirology. 2015;12:83. doi: 10.1186/s12977-015-0209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keane SC, Van V, Frank HM, Sciandra CA, McCowin S, Santos J, et al. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc Natl Acad Sci U S A. 2016;113:13033–8. doi: 10.1073/pnas.1614785113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA (New York, NY) 2014;20:846–54. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keane SC, Summers MF. NMR Studies of the Structure and Function of the HIV-1 5′-Leader. Viruses. 2016:8. doi: 10.3390/v8120338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 46.Rulli SJ, Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol. 2007;81:6623–31. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, et al. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007;35:2695–704. doi: 10.1093/nar/gkm153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 49.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–21. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, et al. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keene SE, King SR, Telesnitsky A. 7SL RNA is retained in HIV-1 minimal virus-like particles as an S-domain fragment. J Virol. 2010;84:9070–7. doi: 10.1128/JVI.00714-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, et al. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One. 2012;7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyazaki Y, Garcia EL, King SR, Iyalla K, Loeliger K, Starck P, et al. An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus. J Mol Biol. 2010;396:141–52. doi: 10.1016/j.jmb.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA (New York, NY) 1999;5:1268–72. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasband WS. ImageJ. Bethesda, MD: U. S. National Institutes of Health; 1997–2016. [Google Scholar]