Summary
Cytoplasmic dynein-1 is a minus-end directed motor protein that transports cargo over long distances and organizes the intracellular microtubule (MT) network. How dynein motor activity is harnessed for these diverse functions remains unknown. Here, we have uncovered a mechanism for how processive dynein-dynactin complexes drive MT-MT sliding, reorganization, and focusing, activities required for mitotic spindle assembly. We find that motors cooperatively accumulate, in limited numbers, at MT minus-ends. Minus-end accumulations drive MT-MT sliding, independent of MT orientation, resulting in the clustering of MT minus-ends. At a mesoscale level, activated dynein-dynactin drives the formation and coalescence of MT asters. Macroscopically, dynein-dynactin activity leads to bulk contraction of millimeter-scale MT networks, suggesting that minus-end accumulations of motors produce network scale contractile stresses. Our data provide a model for how localized dynein activity is harnessed by cells to produce contractile stresses within the cytoskeleton, for example during mitotic spindle assembly.
Keywords: Dynein, Dynactin, Kinesin, Microtubule, Aster, Spindle, Minus-End
eTOC blurb
Tan et al. demonstrate how individual cytoplasmic dynein motors organize incoherent collections of microtubules into polarity-sorted structures at varying length scales. Dynein cooperatively accumulates into clusters at microtubule minus-ends; reorganizing microtubules exclusively via clusters, thus providing a molecular explanation for dynein’s structural role in mitotic spindle assembly.
Introduction
Cytoplasmic dynein-1 (dynein), is the only minus-end directed microtubule (MT) motor complex in animal cells capable of processive, long-distance transport (Allan, 2011; Cianfrocco et al., 2015; Vallee et al., 2012). The ~1.4 MDa, multi-subunit dynein complex is the largest and most complex of the MT motor proteins and belongs to the AAA+ family of ring-shaped molecular motors (Carter et al., 2016; Vale, 2000). It is responsible for a wide variety of cellular functions and transports many types of cargo, including membrane-bound organelles, mRNAs, stress granules, viruses, and misfolded proteins (Cianfrocco et al., 2015). In addition to canonical cargo transport, dynein has been implicated in intracellular MT organization. In neurons, dynein activity is necessary to maintain the correct polarity of axonal MTs (Zheng et al., 2008). During mitotic and meiotic spindle formation, dynein participates in coordinated large-scale MT reorganization (Clift and Schuh, 2015; Heald et al., 1996; Merdes et al., 2000; Rusan et al., 2002; Vaisberg et al., 1993). For example, dynein activity antagonizes kinesin-5-, and kinesin-12-dependent extensile MT-MT sliding (Ferenz et al., 2009; Mitchison et al., 2005; Tanenbaum et al., 2008; Tanenbaum et al., 2009; Vanneste et al., 2009). Indeed, inhibition of kinesin-5 results in dynein-driven spindle collapse into a monopolar structure (Ferenz et al., 2009; Tanenbaum et al., 2008). Dynein activity is also required to dynamically integrate and focus MT minus-ends into the spindle poles (Elting et al., 2014; Sikirzhytski et al., 2014), in both the presence and absence of centrosomes (Goshima et al., 2005; Heald et al., 1996). While dynein’s functions within the spindle are defined, the molecular mechanism for how it performs these activities remains to be fully understood.
How does dynein activity drive MT organization at the molecular level? Isolated dynein can generate forces to slide anti-parallel MTs in vitro and is sufficient to counteract kinesin-5 activity in vivo (Tanenbaum et al., 2013). Because this mechanism of dynein-driven sliding is restricted to anti-parallel MTs, how this observation is related to the focusing of parallel MT minus-ends at spindle poles is unclear (Goshima et al., 2005; Merdes et al., 2000; Morales-Mulia and Scholey, 2005). Furthermore, dynein’s multiple roles in spindle assembly likely require its accessory factor dynactin (Echeverri et al., 1996; Merdes et al., 2000; Mitchison et al., 2005; Wittmann and Hyman, 1999)(though see (Raaijmakers et al., 2013)), raising questions about the role of dynein mediated anti-parallel sliding versus functions that require dynactin. Thus, the molecular mechanism for how dynein exerts forces within the spindle remains unknown.
Isolated dynein is not strongly processive, but its motility is greatly stimulated by its association with the ~ 1 MDa multi-subunit activating complex dynactin (McKenney et al., 2014; Schlager et al., 2014; Torisawa et al., 2014). This molecular interaction requires a third, coiled-coil adapter protein to mediate dynein-dynactin binding, forming a tripartite dynein-dynactin-activator complex (McKenney et al., 2014; Schlager et al., 2014). The most well-characterized coiled-coil activator, BicD2, binds directly to Rab6 cargoes, is partially necessary for the maintenance of interphase MT arrays in cells, and strongly stimulates the formation of an ultra-processive dynein-dynactin-BicD2 co-complex (DDB) (Fumoto et al., 2006; Matanis et al., 2002; McKenney et al., 2014; Schlager et al., 2014; Urnavicius et al., 2015). Upon activation, DDB moves robustly towards MT minus-ends, and was previously observed to strongly accumulate at MT ends in vitro (McKenney et al., 2014). Similarly, accumulations of dynein and dynactin at MT minus-ends generated by laser severing of spindle fibers have also been reported in cells (Elting et al., 2014). During our studies of DDB motility in vitro, we observed that accumulations of motors at MT minus-ends were capable of generating robust forces along neighboring MTs. DDB accumulations were capable of simultaneously remaining bound to the minus-end of one MT, while generating pulling forces along another towards the minus-ends of a neighboring MT.
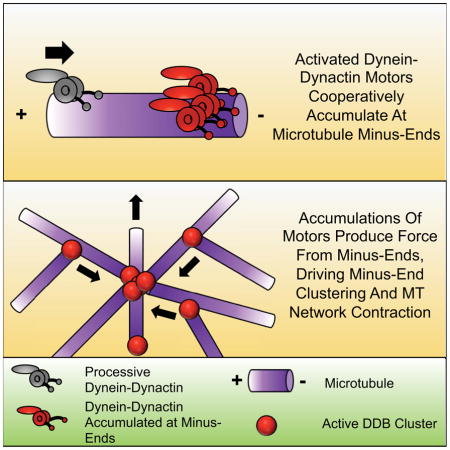
We have now studied this phenomenon in detail, and provide evidence that DDB accumulation at minus-ends is cooperative, suggesting an interaction between motors, and is likely limited by the protofilament number of the MT lattice. Strikingly, minus-end accumulations of DDB can drive the robust sliding of MTs, without a preference for initial MT orientation. Multiple sliding events lead to the dramatic rearrangement of MTs and the focusing of multiple MT minus-ends together, similar to dynein’s functions in spindle assembly. At larger length-scales, DDB drives the formation and coalescence of MT asters in vitro, recapitulating an activity previously shown to be dynein-dependent in cell extracts (Gaglio et al., 1995; Verde et al., 1991). This activity is sufficient to cause the bulk contraction of millimeter scale MT networks, a phenomenon previously observed in Xenopus laevis extract (Foster et al., 2015). Thus, the cooperative accumulation of activated dynein motors at MT minus-ends provides a simple and robust mechanism to drive large-scale reorganizations of MTs (Belmonte et al., 2017). Our data reveals a new molecular mechanism for how activated processive dynein fulfills its various roles during spindle assembly and in MT network reorganization in general, and also highlights the growing importance of cytoplasmic dynein’s ability to work together as small teams of motors (Rai et al., 2016; Rai et al., 2013).
Results
Processive Dynein-Dynactin Complexes Accumulate at MT Minus-Ends
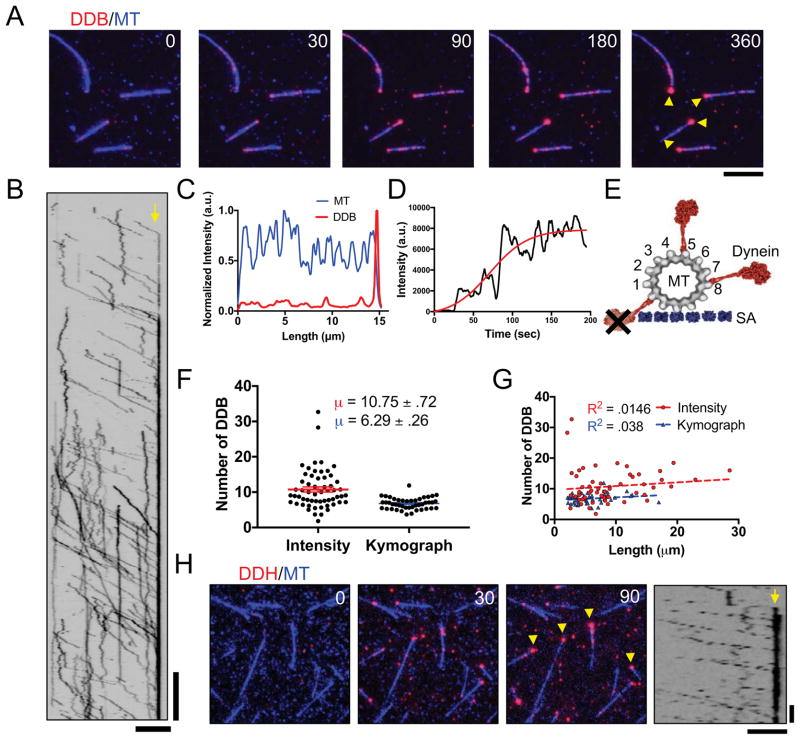
To explore the kinetics of DDB accumulation at MT minus-ends, we created a ‘drop-in’ assay where the motor complex is added to an observation chamber during continuous acquisition, allowing the observation of the accumulation of DDB on microtubules over time (Figure 1A, Movie S1). We used total internal reflection fluorescence (TIRF) microscopy to monitor DDB motility and accumulation along fluorescently-labeled MTs fixed to a coverglass. DDB molecules on the microtubule either moved processively along MTs toward the minus-ends or diffused on the MT lattice, as previously described (McKenney et al., 2014; Schlager et al., 2014) (Figure 1A, B). Notably, individual processive molecules often did not dissociate when reaching the minus-end of the MT, but rather remained bound, leading to the accumulation of multiple motors at the minus-end (Figure 1A, B, Movie S1). Quantification of the fluorescence signal in both the MT and DDB channels further confirmed that DDB accumulated at the ends of MTs (Figure 1C), and the DDB intensity at individual MT ends reached a steady state during our observation period of ~ 3 min. (Figure 1D).
Figure 1. Accumulation of Processive Dynein-Dynactin Complexes at MT Minus-ends.
(A) TIRF-M images of DDB (red) accumulating at MT (blue) minus-ends. DDB accumulations are marked by yellow arrows. Scale bar: 2 μm, time is given in sec. (B) Kymograph of DDB motility and accumulation on MT minus-ends. Minus-end denoted by yellow arrow. Scale bars: 2 μm and 30 sec. (C) Representative quantification of MT (blue) and DDB (red) average intensities over time per pixel along a MT, normalized to the maximum intensity along the same line in the respective channel. (D) Plot of moving average with 5 points on either side of minus-end DDB intensity over time. A 4-parameter logistic sigmoidal regression was fitted by least squares fit to the data (red), R2 = .795. (E) Scale diagram of dynein motor domains (red) bound to a 13 pf MT. The MT is attached to a surface via streptavidin molecules (SA, blue), as in our assays. Not all protofilaments are available for dynein binding on the surface-bound 13-protofilament microtubule due to dynein’s large size. Note that the entire DDB complex is much larger than the dynein motor domain depicted. (F) Quantification of the number of DDB complexes within minus-end accumulations using either fluorescent intensity analysis (red) or kinetic calculation methods (blue). N = 58 and 44 respectively, 2 independent experiments, see also Figure S1. Data represented as mean ± SEM. (G) Plot of number of DDB in minus-end accumulations vs MT length using either fluorescent intensity analysis at minus-ends (red), or using kinetic calculation methods (blue). N = 58 and 44 respectively, 2 independent experiments. (H) DDH complexes (red) also form accumulations at MT (blue) minus-ends, denoted by yellow arrows. Right, Kymograph of processive DDH complexes accumulating at to MT minus-ends (yellow arrow). Scale bars: 5 μm and 10 sec.
Next, we aimed to quantify the number of DDB complexes within the steady-state accumulations at MT minus-ends. Due to the large size of the DDB complex (Chowdhury et al., 2015), and geometric constraints of surface-attached MTs, we estimated that ~ 7–8 MT protofilaments are accessible to the DDB complex at minus-ends in our assays (Figure 1E). To quantify the numbers of DDB at minus-end accumulations, we utilized two methods to estimate the number of DDB molecules at steady state within the minus-end accumulations. First, we compared the integrated fluorescence intensity of the steady state minus-end accumulation to the intensity of single DDB molecules on the MT lattice, or nonspecifically bound to the glass near the MT (Figure S1). Second, we used kymograph analysis to calculate the flux of DDB molecules into the minus-ends, as well as the off-rate of single DDB molecules from minus-ends (Figure S1 and see below). Both methods yielded similar estimates of the number of individual DDB complexes within each minus-end accumulation, 10.75 ± 0.72 and 6.29 ± 0.25 complexes, respectively (Figure 1F). This equates to approximately one to two DDB complex per MT protofilament, assuming most taxol-stabilized MTs in our preparation contain 13 protofilaments, and not all protofilaments are accessible to the motor due to steric hindrance from PEG-biotin-streptavidin attachment to the glass surface (Figure 1E). Furthermore, the estimated number of DDB molecules in the minus-end accumulations did not correlate with MT length (Figure 1G). These results argue that DDB accumulations at MT minus ends are limited to relatively few motor complexes per MT end.
To examine if minus-end accumulation was specific to dynein-dynactin complexes formed with the BicD2N adapter protein, we performed similar experiments with an orthogonal adapter molecule, Hook3, which has also been previously shown to mediate dynein’s interaction with dynactin, and activate processive dynein motility (McKenney et al., 2014; Olenick et al., 2016; Schroeder and Vale, 2016). Isolated dynein-dynactin-Hook3 complexes (DDH) displayed robust processive movement, as previously described, and noticeably accumulated at MT minus-ends similar to DDB complexes (Figure 1H). Thus, we conclude that minus-end accumulation is an intrinsic property of activated, processive dynein-dynactin complexes, and does not depend on the type of adapter molecule that mediates dynein’s interaction with dynactin.
Mechanism of Minus-End Accumulation
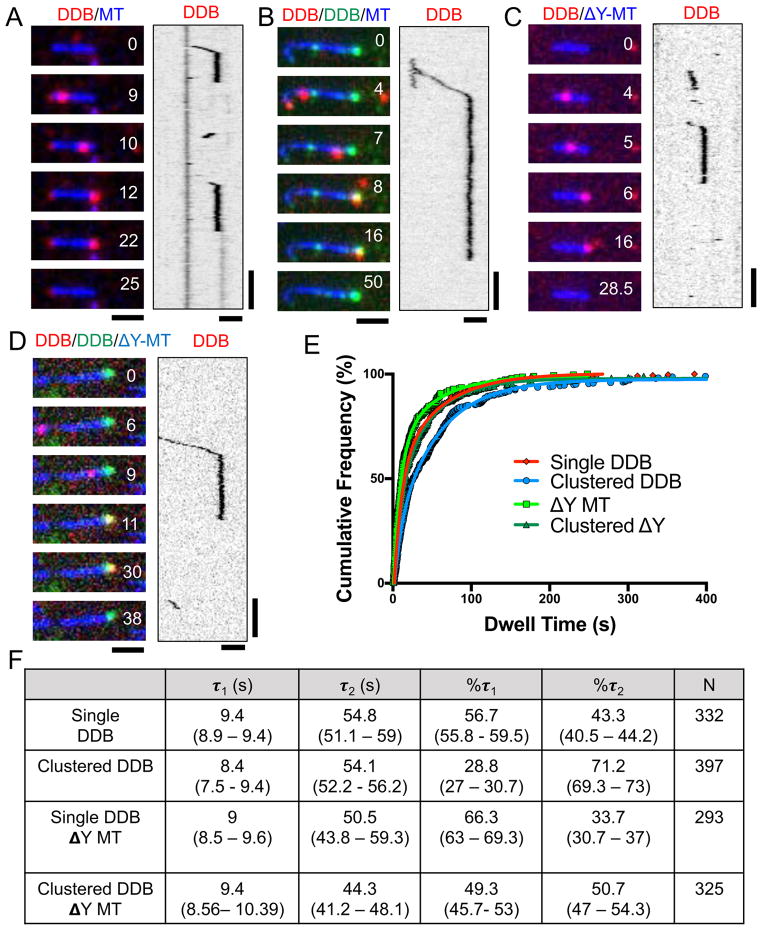
To explore the mechanism of minus-end accumulation, we first measured the rate of dissociation for individual DDB complexes at MT minus-ends (Figure 2A, E, F,). We found that the resulting dwell time distribution was best fit with a biphasic decay, suggesting two types of dissociation events at minus-ends (Figure 2E, F). The fit revealed a shorter dwell time of 9.4 seconds and a longer dwell time of 54.8 seconds, with 56.7% of the molecules displaying the faster dissociation time (Figure 2F). Thus, single molecules of DDB exhibit both fast and slow dissociation from MT minus-ends, with a preference for the former in our assay conditions.
Figure 2. Single Molecule Analysis of DDB Dwell Times at MT Minus-Ends.
(A) Representative TIRF-M images and associated kymographs of DDB (red) behavior at MT (blue) minus-ends. (B) Representative images and associated kymograph of single molecule spiking experiments at ~1 nM SNAP-TMR DDB (red) and ~30 nM SNAP-488 DDB (green) dwelling at MT minus-ends (blue). (C) Representative images and associated kymograph of DDB (red) dwelling at the ends of carboxypeptidase treated MTs (blue). (D) Representative images and associated kymograph of single molecule spiking experiments with ~1nM SNAP-TMR DDB (red) and ~30 nM SNAP-647 DDB (green) on carboxypeptidase treated MTs (blue). Time is given in sec. for A–D. Scale bars: 2 μm and 15 sec. (E) Cumulative frequency plots of DDB dwell times from the conditions in A–D. Solid lines are two-phase exponential regressions to the data. (F) Table summarizing the parameters of DDB cumulative frequency graphs, including the characteristic dwell time (τ) of short (τ1) and long (τ2) populations, percentage of molecules in each population, and number of molecules measured (N) are given. 95% confidence intervals given in parentheses. All regressions have a goodness of fit (R2) greater than .99. All data from 2–3 independent experiments.
To gain insight into how minus-end dissociation rates might be affected by other motors within a minus-end accumulation, we next performed single molecule spiking experiments with differentially labeled DDB. We mixed together a ratio of SNAP-488 and SNAP-TMR labeled DDB complexes such that all MTs in the chamber contained accumulations of SNAP-488 DDB at their minus-ends, while only individual molecules of SNAP-TMR DDB were observed to bind and move along MTs (Figure 2B). We then measured the dwell times of single SNAP-TMR DDBs within the context of the SNAP-488 minus-end accumulations. Strikingly, individual SNAP-TMR DDBs appeared to dwell much longer within minus-end accumulations of SNAP-488 DDB (Figure 2B, E, F). The same effect was observed when SNAP-647 DDB complexes were used to form minus-end accumulations (not shown), arguing against a non-specific effect of the SNAP dye used to label the DDB complexes. Quantitative analysis revealed that individual dwell times were not changed significantly, but there was nearly a two-fold shift in the population towards longer dwell times (Figure 2B, E, G). These results show that individual DDB molecules change their behavior at MT minus-ends depending on the presence of other DDB molecules at the MT end. Such a change in behavior suggests a cooperative interaction between DDB molecules at MT minus-ends that shifts equilibrium between two DDB dissociation rates towards longer dwell times.
DDB contains two distinct types of MT-binding domains (MTBD). Dynein s MTBD is located within the motor domain, and binds directly to the tubulin lattice (Carter et al., 2008; Redwine et al., 2012). The second MTBD, located at the N-terminus of the p150Glued subunit of dynactin, binds to the disordered tubulin tail domains (Culver-Hanlon et al., 2006; Lazarus et al., 2013; McKenney et al., 2016; Peris et al., 2006; Wang et al., 2014). In principle, the association of DDB to minus-ends could be mediated by one or both MTBDs. To distinguish between these possibilities, we treated taxol-stabilized MTs with carboxypeptidase A (CPA) to remove the C-terminal tyrosine residue of α-tubulin (Webster et al., 1987), which has been previously shown to greatly diminish the interaction of the p150Glued MTBD with the MT lattice (Figure S2A, B), but does not substantially effect on the interaction of dynein with MTs (McKenney et al., 2016). CPA treatment had little effect on the minus-end dwell time distribution of single DDB complexes as compared to WT MTs, suggesting that engagement of the p150Glued MTBD with the MT lattice is not strictly required for extended complex dwelling at MT minus-ends (Figure 2C, E, F). As with WT MTs, we observed a cooperative increase in the dwell time distribution of single DDB’s in the presence of minus-end DDB clusters on CPA MTs (Figure 2D, E, F). Removal of the α-tubulin tyrosine decreases the binding rate of DDB to MTs (McKenney et al., 2016), necessitating the use of higher (~ 2-fold) amounts of motor complex in these assays, possibly explaining the difference in magnitude of the cooperative effect observed on CPA versus WT MTs (Figure 2F). We conclude that DDB complexes accumulate cooperatively on MT minus-ends primarily through interactions of the dynein MTBD with MTs. While our data cannot exclude a minor role for the p150-MT interaction in this process, these results suggest it is not strictly required for cooperativity between DDB complexes at MT minus-ends.
Because of the observed change in dwell times at higher concentrations of DDB used in the CPA experiments above, we were curious how the concentration of motors affected the dwell time of individual complexes within minus-end clusters. In principle, if minus-ends are saturated with DDB complex, an incoming motor complex could either displace a pre-existing DDB from the minus-end cluster, dissociate immediately upon reaching the minus-end cluster, or stop moving and dwell behind the pre-existing minus-end cluster. The latter scenario would predict that minus-end clusters should grow in size over time as more and more incoming DDB’s accumulate behind the pre-existing cluster, a possibility that is excluded by our observation that minus-end clusters are limited in size (Figure 1), and minus-end clusters did not grow in size at higher concentrations of DDB (Figure S2C). We therefore measured single DDB dwell times within minus-end clusters at ~10-fold higher DDB concentration than was used in our previous assay (Figure 2B and Figure S2D, E). Strikingly, the high concentration of DDB abolished the cooperative effect on minus-end dwell times, and most of the population shifted to the faster dissociation time (Figure S2D, E). This observation suggests that incoming DDB complexes displace molecules from within the pre-existing cluster when minus-ends are saturated with DDB. Consistently, the propensity of incoming DDB complexes to immediately dissociate or to dwell at minus-ends was not effected by pre-existing minus-end clusters of molecules (Figure. S2F)
MT Reorganization by Minus-End Accumulations of DDB
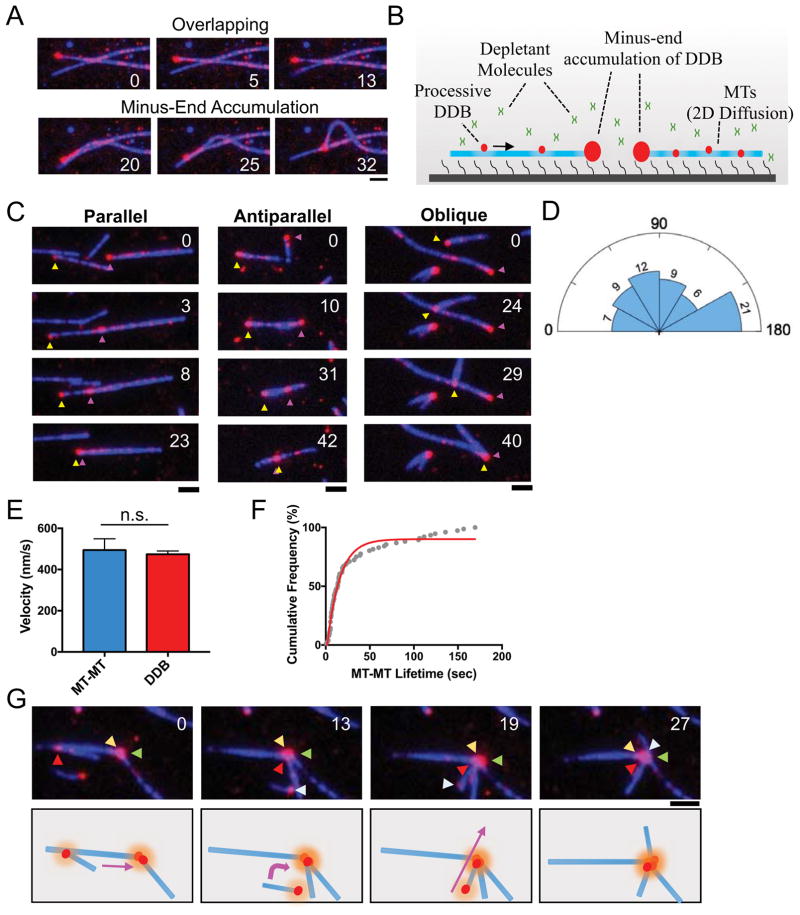
The ability to produce force along one MT while remaining bound to another is the basis for MT reorganization driven by molecular motors in cells. Isolated non-processive dimeric dynein can produce force from within MT-MT overlaps, possibly due to the low coordination and high flexibility between its two motor domains (Amos, 1989; Tanenbaum et al., 2013). Importantly, we observed that processive DDB did not obviously produce force between overlapping MTs in our assays (Figure 3A), reflecting a distinction between isolated dynein and dynein in complex with dynactin. Instead, forces between adjacent MTs were clearly visible once the minus-end DDB accumulation on one MT contacted an adjacent MT (Figure 3A).
Figure 3. Minus-End Clustering Driven by Minus-End Accumulations of DDB.
(A) TIRF-M images of two separate events on the same set of loosely attached, overlapping microtubules (blue) without (top), and with (bottom), contact with the minus-end DDB cluster. Time is in sec., scale bar 2 μm. Note that no interactions between MTs are observed until the minus-end DDB cluster makes contact with the orthoganol MT. (B) Schematic of TIRF experiments involving microtubules held in the TIRF field non-covalently using depletion forces. In this system, MTs are free to move in two dimensions along the coverslip surface. (C) Representative TIRF-M images of parallel, antiparallel and oblique sliding by DDB minus-end clusters. MT minus-ends are inferred from DDB accumulation location, and designated by magenta and yellow arrows. (D) Rose diagram representing the relative distribution of initial angle of MT-MT sliding. Endpoints (0, 180°) were integrated into their nearest bins. The number of sliding events in each bin are indicated. Bin size = 30 degrees. N = 64. (E) Bar graph displaying the mean velocity of MT-MT sliding (blue) (474.1 ± 16.33 nm/s, N = 107, 2 independent experiments) in comparison to velocity of individual DDB molecules along MTs (red) (494.1 ± 55.09 nm/s, N = 43, 3 independent experiments). There is no statistical difference between the two conditions of DDB movement (P = 0.644, two-tailed T-test). Data represented as mean ± SEM. (F) Cumulative frequency plot of MT-MT foci life times. Solid red line is the single phase exponential decay fit to the data. Data composed of events where only two MT minus-ends are brought into contact by DDB motility and subsequently dissociate. τ = 14.44 s, N = 76, R2 = .979, 3 independent experiments. G) Top: TIRF-M images showing multiple MT-MT sliding events forming a mini-aster. Arrows label minus-ends of unique microtubules, inferred from DDB accumulation. Time is given in sec. Bottom: Schematic of MT movements observed in the movie. Colored arrows indicate direction of MT movement.
In the previous type of assay, MTs are bound tightly to the cover glass through near covalent biotin-streptavidin linkages between the MT lattice and the glass surface, greatly constraining the DDB-driven movement of MTs relative to one another. To fully explore the ability of DDB to drive MT-MT movements, we modified this assay to allow complete freedom of movement while keeping the MTs near the coverglass and within the TIRF illumination depth. We took advantage of the depletion force exerted by the addition of the inert polymer methylcellulose (Henkin et al., 2014), which prevents MTs from diffusing away from the coverglass, but still allows freedom of movement in two dimensions along the glass surface (Figure 3B, Movie S2).
Using this new assay, we observed that minus-end accumulations of DDB could robustly drive the sliding of MTs in nearly any orientation, including parallel, anti-parallel, and oblique angles (Figure 3C, D, Movie S2). Quantification revealed no obvious limitations on the angle of sliding driven by DDB, as sliding of all angles was observed (Figure 3D). MT-MT sliding velocity was identical to the speed of individual DDB complexes moving along the MT lattice (Figure 3E). Sliding always ended when the two minus-ends of the respective MTs (marked by high DDB signal) came within a diffraction-limited distance from one another, and quantification of the lifetime of the interaction between two minus-ends revealed a prolonged interaction lasting ~ 14 seconds (Figure 3F). We note that the data does not approach a definite plateau and further studies into the molecular interactions between DDB molecules at MT ends are required to understand the additional stability observed at longer lifetimes.
Multiple types of these sliding events often resulted in the accumulation of several individual MT minus-ends held together at the center of small asters (Figure 3G, Movie S3), which tended to accumulate over time during the assay. Similar sliding events and small aster formation was observed when the experiments were repeated with DDH complexes, indicating that this phenomenon is not unique to the BicD2N adapter protein, but rather a molecular property of activated, processive dynein-dynactin complexes (Figure S3B, C, D). Together, this data demonstrates that minus-end accumulations of dynein-dynactin complexes drive the robust sliding of MTs in any orientation resulting in two or more MT minus-ends brought into close proximity and held together for significant periods of time. Further, activated, processive dynein-dynactin complexes are sufficient to induce the formation of small MT asters.
Self-organization of MTs is Driven by Minus-end Accumulations of DDB
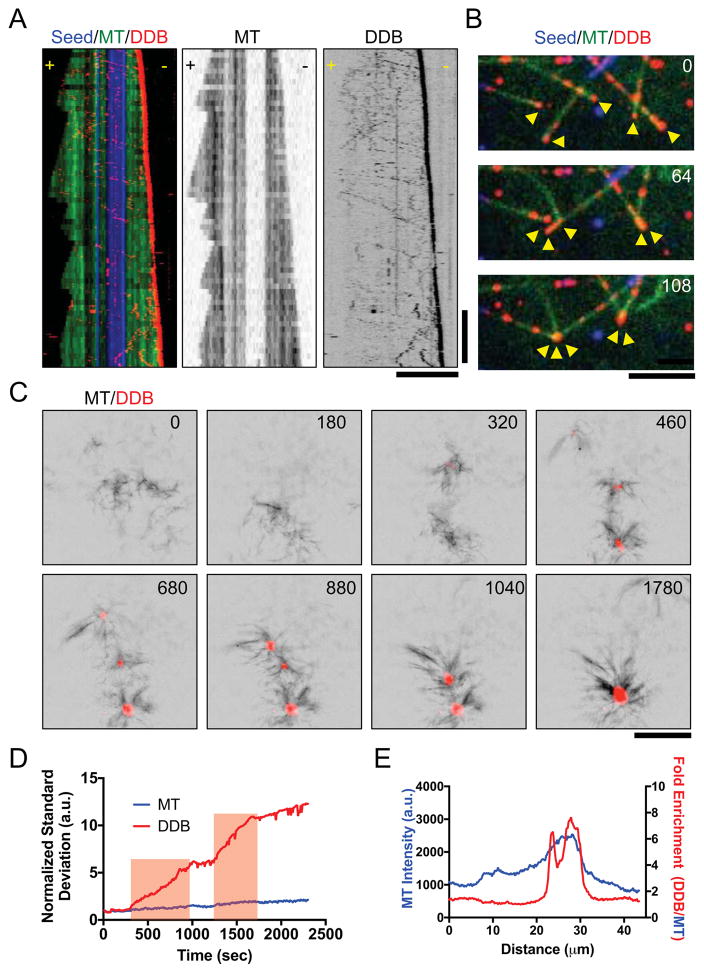
Because the previous analysis was performed on taxol-stabilized MTs, we wondered how DDB complexes interact with the minus-ends of dynamic MTs, as found in cells. We reconstituted dynamic MTs in vitro and observed DDB complexes moved along the full length of the lattice, and strongly accumulated at the dynamic minus-ends (Figure 4A, Movie S4). Further, accumulations of DDB on dynamic minus-ends were capable of driving MT-MT interactions (Figure 4B, Movie S4), similar to what we observed with taxol-stabilized MTs (Figure 3). These interactions often resulted in the focusing of two or more dynamic minus-ends together (Figure 4B, Movie S4), a function that we note is one of dynein’s key roles during mitotic spindle assembly (Goshima et al., 2005; Heald et al., 1996; Morales-Mulia and Scholey, 2005).
Figure 4. Minus-End Accumulations of DDB Drive Mesoscale MT Reorganization.
(A) Three-color kymograph of processive DDB (red) reveals strong accumulation at the growing minus-ends of dynamic MTs (green). Dynamic MT ends grown from GMPCPP-stabilized seeds (blue). Right: Individual fluorescent channels are reproduced for dynamic MTs and DDB for clarity. Scale Bars: 5 μm and 2 minutes. (B) Example of DDB accumulations (red) on growing MT minus-ends (green) driving MT-MT sliding to form minus-end clusters (arrows). Note that MT-MT sliding does not occur at MT crossover points not in contact with minus-end DDB clusters. Time in sec. Scale Bar: 5 μm. (C) Images of DDB-driven reorganization of growing microtubules (black) into asters with DDB (red) centers. Multiple asters coalesce into a single larger structure. Scale bar: 10 μm. Time in sec. (D) Plot of intensity contrast of both MT (blue) and DDB (red) channels expressed as standard deviation over time. Red bars indicate periods of merging between two asters. (E) Line scan plot of MT intensity (blue) across an aster and the fold increase of DDB fluorescence intensity divided by MT fluorescence intensity.
In cells, the collective action of MT motors drives the self-organization of the MT cytoskeleton. Our previous assays demonstrated that minus-end accumulations of DDB complexes are sufficient to sort and organize MTs by sliding until minus-ends are brought together. To understand how DDB accumulations at minus-ends could drive MT-MT interactions at a mesoscale level, on the order of hundreds of MTs, we studied the behavior of DDB in solutions of growing MTs. When 4 μM DDB was incubated in solutions of growing MTs, we observed the formation and coalescence of micron-size asters over time (Figure 4C, Movie S5). DDB signal accumulated strongly at the center of the MT asters during their formation (Figure 4C, Movie S5), indicating the asters formed by DDB-driven collection and focusing of MT minus-ends. Nearby asters often coalesced and fused into a single larger aster (Figure 4C, Movie S5), demonstrating a net contractile force produced by DDB in this system. Analysis of the image contrast, a measure of organization (Hentrich and Surrey, 2010), in the DDB channel showed the amount of contrast greatly increased at times corresponding to aster merging, and plateaued after a fusion event indicating DDB accumulation reaches steady state after the fusion event (Figure 4D). Line-scan analysis revealed that DDB intensity at the center of the asters was enriched relative to the MT signal (Figure 4E), indicating that the DDB foci accumulate specifically in the center of the asters, presumably at MT minus-ends. Cumulatively, these data demonstrate that processive DDB complexes cooperatively accumulate at growing MT minus-ends where they drive MT self-organization, focusing of MT minus-ends into asters, and the coalescence and fusion of MT asters.
DDB Activity Drives Macroscale Contractile Stresses in MT Networks
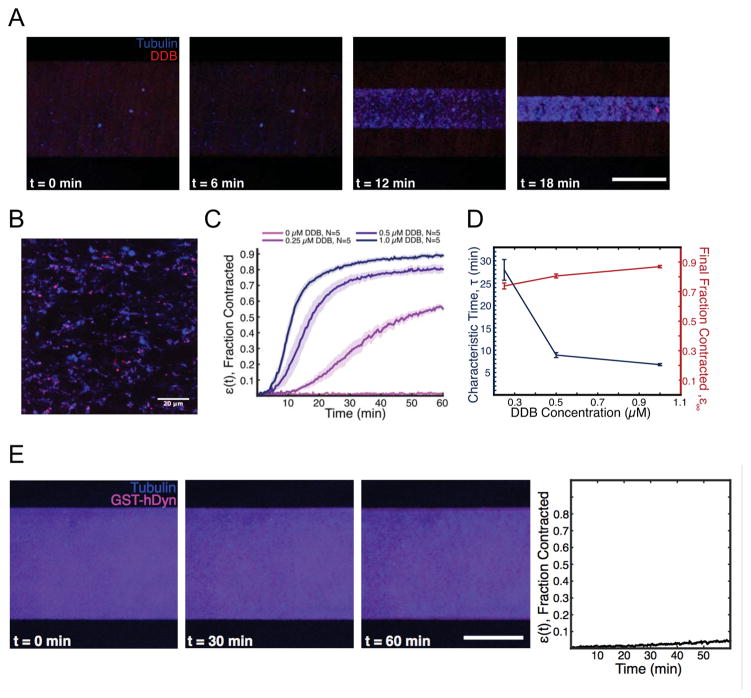
The previous data demonstrated that DDB motor activity localized at MT minus-ends drives MT reorganization by clustering minus-ends together into MT asters. The coalescence and fusion of these asters reveals a net contractile stress exerted by DDB motor activity within MT networks. Previous work in Xenopus egg extract revealed a dynein-dependent stress that drives the bulk contraction of millimeter-scale stabilized MT networks (Foster et al., 2015). These dynein-driven contractile stresses were hypothesized to be related to dynein’s role in mitotic spindle pole focusing, spindle healing, and spindle fusion. The bulk contraction observed in extracts was driven by MT aster formation and coalescence, similar to what we observe in purified solutions of DDB and MTs (Figure 4). We therefore set out to determine if purified DDB could recapitulate bulk MT network contraction, independent of other cellular factors found in Xenopus egg cytoplasm.
We combined various concentrations of purified, TMR-labeled DDB complexes with taxol stabilized, Atto- or Alexa-647-labeled MTs in microfluidic devices measuring 125 μm (H) × 0.9 mm (W) × 18 mm (L). In this system, the initially diffuse MT network exhibited spontaneous bulk contraction on the millimeter length scale, strongly resembling the bulk contraction previously observed in Xenopus extract (Foster et al., 2015)(Figure 5A, Movie S6). Imaging of the contracting network at higher magnification revealed the network was organized into aster-like structures with DDB fluorescence concentrated in the aster centers (Figure 5B, Movie S7), consistent with the network structures observed previously in Xenopus extracts (Foster et al., 2015), and the asters formed in our assays (Figure 4C). We quantified the contraction dynamics by measuring the width of the network as a function of time (Foster et al., 2015). When the concentration of DDB was increased, the contraction timescale decreased, with only a slight increase in the final fraction the network contracts (Figure 5C). This result is consistent with previous experiments in egg extract, and in qualitative agreement with the previously proposed active fluid model (Foster et al., 2015)(Figure 5C, D). The model predicts that the strength of contractile stresses, and thus the contraction timescale of the active fluid should depend on the concentration of motors, while the final density of the MT network should remain unchanged. Thus, a mixture of purified processive dynein-dynactin complexes and stabilized MTs is sufficient to recapitulate the bulk MT network contraction observed in Xenopus egg cytoplasm.
Figure 5. DDB Drives Bulk Contraction of Macroscale Networks of Microtubules.
(A) Representative images of a microfluidic containing a network of taxol-stabilized microtubules (blue). The MT network contracts over time due to forces exerted by DDB (red). Scale bar: 500 μm. (B) Higher magnification image of the contracting network of microtubules (blue), showing aster-like structures, with DDB (magenta) localizing towards the interior. Scale bar: 20 μm. (C) Average fractional network contraction as a function of time for varying DDB concentrations (mean ± s.e.m). (D) Characteristic time, τ, and final fraction contracted, ε∞, from fits to ε(t) curves as a function of DDB concentration. (E) Representative images of a microfluidic channel containing a network of taxol-stabilized microtubules (blue) and purified GST-hDyn (red). Note the network shows little contraction over much longer timescales than in (A). Right: Quantification of average fractional network contraction as a function of time.
A recombinant, minimal dimeric human dynein construct, GST-hDyn, was previously shown to be capable of bundling and sliding anti-parallel MTs in vitro, and antagonizing Eg5 activity within the mitotic spindle in vivo (Tanenbaum et al., 2013). Because this construct is not strongly processive (McKenney et al., 2014; Torisawa et al., 2014; Trokter et al., 2012), it cannot accumulate at MT minus-ends as we observe for DDB in vitro. Rather than generating forces at MT ends, GST-hDyn is thought to generate forces from within a MT-MT overlap using its two uncoordinated motor domains (DeWitt et al., 2012; Qiu et al., 2012; Tanenbaum et al., 2013). As a result, the sliding activity of GST-hDyn is restricted to anti-parallel MTs, whereas minus-end accumulations of DDB are not restricted by MT geometry (Figure 3C). To test if processivity and minus-end accumulation are important for dynein-driven MT network contraction, we incubated GST-hDyn with mixtures of MTs and observed minimal network contraction, even after much longer time scales than we investigated for DDB (Figure 5E, Movie S6). We conclude that cooperative accumulation of processive dynein-dynactin complexes at MT minus-ends provides a simple and robust mechanism to generate contractile stress within MT networks, and suggest this mechanism can account for dynein-dependent contractile stress generation within the mitotic spindle.
Discussion
Behavior of Individual Dynein-Dynactin Complexes at MT Minus-ends
Microtubule motors drive the self-organization of MT-based structures within cells. In addition to various kinesins, cytoplasmic dynein plays key roles in intracellular MT organization, and is critical for bipolar spindle assembly (Heald et al., 1996; Merdes et al., 1996; Raaijmakers et al., 2013; van Heesbeen et al., 2014). Dynein activity is necessary to collect, focus, and anchor parallel MT minus-ends into the spindle pole (Goshima et al., 2005; Maiato et al., 2004; Morales-Mulia and Scholey, 2005; Raaijmakers et al., 2013), as well as to antagonize kinesin forces acting on anti-parallel MTs (Cross and McAinsh, 2014; Drechsler and McAinsh, 2016; Drechsler et al., 2014; Kapitein et al., 2005; Sharp et al., 1999; Sturgill and Ohi, 2013; van den Wildenberg et al., 2008) in the spindle midzone. The molecular mechanism for how dynein fulfills these roles within the diverse spindle MT environment (parallel vs. anti-parallel MTs) remains unknown. Our results provide the first experimental support for a previously proposed hypothesis, derived from computational modeling, whereby a hypothetical minus-end motor with the ability to dwell at MT ends can be harnessed by the cell to drive MT organization in such diverse arrays of MTs (Burbank et al., 2007; Goshima et al., 2005; Nedelec, 2002).
Here, we have analyzed the single molecule and bulk behavior of activated, processive dynein-dynactin complexes at length-scales that span several orders of magnitude. At the nanoscale, we find that stable dynein-dynactin complexes, formed using two distinct adapter molecules, accumulate cooperatively at the minus-ends of single MTs, the natural endpoint of any dynein-based movement in cells. The cooperativity we observe in DDB accumulation likely reflects molecular interactions between individual DDB complexes at MT ends. The molecular basis of such an interaction remains to be determined, but we speculate that it could arise through motor domain interactions between adjacent DDB complexes in trans, akin to previously observed interactions between motor domains within a single dynein homodimer (Figure 6) (Amos, 1989; Kon et al., 2011; Torisawa et al., 2014; Toropova et al., 2017). It is also possible that DDB’s that reach the MT minus-end could impart structural changes to the MT lattice that favor further DDB retention. Indeed, dynein motor activity has been shown to affect MT plus-end dynamics in vitro (Laan et al., 2012) (Hendricks et al., 2012) and further work will be needed to determine if DDB affects minus-end dynamics.
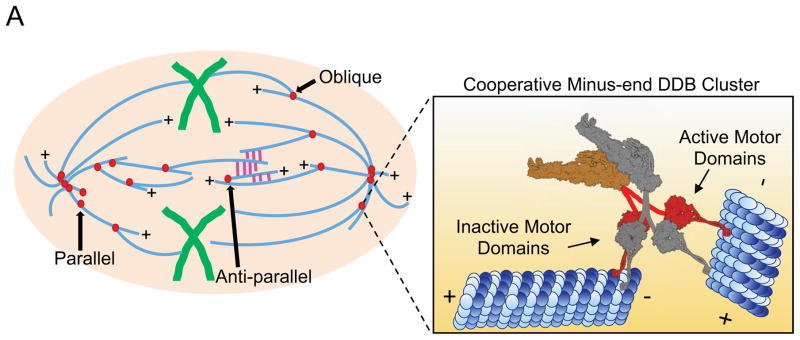
Figure 6. Model For Cooperative Accumulation of DDB Complexes At MT Minus-Ends That Drive MT Organization Within the Bipolar Mitotic Spindle.
Different orientations of MT-MT sliding are required at topologically unique locations within the mitotic spindle. Parallel sliding by minus-end DDB complexes (red) is important for spindle pole focusing while anti-parallel sliding resists kinesin forces (magenta) within the spindle midzone. Oblique interactions are likely important in integrating MTs into the spindle network (Elting et al., 2014; Redemann et al., 2017; Sikirzhytski et al., 2014). Inset: DDB complexes accumulate on MT minus-ends. We propose that individual complexes adopt an inactive confirmation, possibly through trans interactions with neighboring DDBs (gray), providing an anchor to the MT minus-end. Single motor domains from each DDB complex are free to bind, and produce force towards the minus-ends of adjacent MTs, thus, driving minus-end directed MT-MT sliding which produces net contractile force within MT networks.
The number of DDB complexes at MT minus-ends reaches steady state, even at high concentrations of DDB suggesting motor accumulation is intrinsically limited. Our measurements provide evidence that the number of DDB complexes within minus-end accumulations is constrained to approximately 6–11 DDB per minus end. Strikingly, this number is similar to the number of protofilament binding sites we estimate are available on a surface bound microtubule, suggesting that 1–2 DDB complexes may accumulate on each accessible protofilament at the MT minus-end. Further, the size of the accumulations do not change appreciably during periods of increased DDB flux into the minus-end, again suggesting limited binding sites at MT ends may limit accumulation growth (Figure S2C). These data are consistent with recent observations of limited dynein accumulations on the minus-ends of spindle fibers in cells (Elting et al., 2014; Hueschen et al., 2017). Our experiments further show that a larger flux of motors to the minus-end displaces resident complexes, counter-acting the cooperative accumulation of motors (Figure S2D, E, F). This result suggests that minus-end cooperativity could be inherently limited by the kinetics of motors that reach filament ends in cells, and cellular mechanisms such as post-translational modifications of tubulin that favor or disfavor dynein motion along particular subsets of microtubules (McKenney et al., 2016) could selectively tune the ability of the motor to cooperatively accumulate at specific filament ends and drive the reorganization of particular microtubules. Because DDB accumulation does not appear to require a strong interaction between p150Glued and the MT surface, we favor a model whereby individual DDB complexes remain bound to minus-ends via one or both of the dynein motor domains (Figure 6).
Minus-End DDB Reorganizes MT Networks
Motor forces exerted from within overlaps drive most MT-MT sliding studied to date. Because of this, the geometry of sliding is typically limited by the intrinsic ability of the motor to move in one direction along the lattice. Here, we find that DDB produces force on adjacent MTs predominately from accumulations at MT minus-ends, allowing a much greater degree of freedom to drive MT-MT sliding regardless of MT orientation. The result of all DDB catalyzed sliding events, without a bias for initial MT orientation, is the coalescence of MT minus-ends together. This feature makes MT-MT sliding generated by DDB unique among MT motors reported, and may be a consequence of the relatively uncoupled mechanochemistry and flexibility of dynein’s two motor domains (DeWitt et al., 2012; Qiu et al., 2012). Additionally, we observe the ability of DDB to slide MTs is restricted to motors accumulated at minus-ends (Figure 3A), in contrast to strong lattice-lattice sliding observed for isolated yeast or mammalian dynein molecules (Tanenbaum et al., 2013), and most kinesin molecules studied to date (Fink et al., 2009; Hentrich and Surrey, 2010; Kapitein et al., 2005) This observation suggests that binding to dynactin may restrict dynein’s conformation flexibility, rendering the motor unable to bind adjacent MTs until it reaches the minus-end, providing an inherent self-limiting mechanism that we speculate may serve to separate dynein’s dual roles in transport of cargo along MT versus reorganization of the MT network.
The ability to slide parallel as well as anti-parallel MTs may provide insight into dynein’s known roles within different parts of the mitotic spindle (Figure 6). In the spindle mid-zone, enriched in anti-parallel MTs, dynein activity is thought to antagonize kinesin-5 and kinesin-12 dependent extensile sliding (Florian and Mayer, 2012; Mitchison et al., 2005; Tanenbaum et al., 2008; Tanenbaum et al., 2009; van Heesbeen et al., 2014). In contrast, at the spindle poles, where parallel MTs predominate, dynein activity is important to collect and focus MT minus-ends and anchor them to the spindle pole (Goshima et al., 2005; Maiato et al., 2004; Morales-Mulia and Scholey, 2005; Raaijmakers et al., 2013). Importantly, the mechanism of sliding described here is distinct from that previously reported for dimerized minimal dynein constructs, and likely explains why these constructs were unable to rescue focusing of parallel MTs at spindle poles (Tanenbaum et al., 2013). In addition, accumulations of dynein-dynactin at nascent MT minus-ends, produced by laser severing, have been demonstrated to capture neighboring MTs at oblique angles and drive poleward parallel sliding to repair spindle architecture (Elting et al., 2014; Sikirzhytski et al., 2014). The mechanism of dynein-dependent MT-MT sliding we present here provides a simple and robust model to explain how dynein activity can fulfill these diverse functions within the mitotic spindle.
How do accumulations of dynein-dynactin produce force along adjacent MTs, while maintaining attachment to the minus-end? The cooperative mechanism of accumulation may provide a clue, as it suggests molecular interactions among motors within the accumulation. We hypothesize that each motor may remain bound to the minus-end via a single motor domain, while its partner motor domain remains free to bind and produce force along adjacent MTs (Figure 6). Because the motor domains of dynein are not strongly coupled, and individual dynein dimers composed of a single active motor domain can still move processively in vitro (Cleary et al., 2014), we hypothesize that the force generated from minus-ends may be the sum of many individual motor domains working independently from each other. We propose that the minus-end bound motor domain may be held in a mechanically inactive, strongly MT bound state, through trans interactions with neighboring motor domains. Such a conformation could provide a stable tether to the minus-end of one MT, while allowing the free motor domain to produce force along an adjacent MT. However, our observation that DDBs remain accumulated at the growing minus-ends of dynamic MTs suggests that the motors within the accumulation are competent to rapidly switch between inactive, and actively processive states.
Because we observe two populations of off-rate for single DDBs, we suggest that the DDB complex samples at least two conformational states upon reaching the minus-end, and accumulation of more complexes favors one state over the other. The molecular basis for these two states remains to be determined, but we note that even the shorter dwell time that we observe at minus-ends is at least 20-fold longer than dwell times calculated in computer simulations to be effective in focusing of k-fiber minus-ends during spindle assembly (Goshima et al., 2005). This study concluded that the dwell time at minus-ends plays a critical role in determining if a molecular motor will perform well in focusing MT ends, an observation supported by our experimental data.
Minus-End Accumulations Provide Contractile Forces for MT Self-Organization
We have also examined the emergent behavior of molecular systems composed of MTs and DDB. At a mesoscale level, mixtures of growing MTs and DDB form micron-sized asters that fuse together via contractile forces exerted across tens of microns. Aster fusion is presumably driven by distal MT ends contacting the DDB core of another aster, and is reminiscent of dynein-driven aster and spindle fusion in Xenopus extracts (Gatlin et al., 2009). Previous in vitro studies have demonstrated similar aster formation using artificially multimerized kinesin motors (Nedelec et al., 1997; Surrey et al., 2001), or kinesin motors that naturally contain a second MT-binding site (Hentrich and Surrey, 2010). Aster formation in cell extracts has been shown to be dynein-dependent (Gaglio et al., 1995; Verde et al., 1991). Here we have shown that the native, dimeric DDB complex drives aster formation by generating force exclusively from small accumulations at MT ends, an important distinction from previous results with various kinesin motors. Further, DDB can slide and reorganize MTs in any orientation and possesses motor velocity and processivity large enough to overcome the speed of minus-end growth in vivo. The high velocity and extreme processivity of DDB may make the motor complex particularly well-suited to reach MT ends compared with much slower velocity and limited processivity of kinesin-5 (Kapitein et al., 2008; Kwok et al., 2006) or kinesin-14 (Fink et al., 2009; Hentrich and Surrey, 2010).
Remarkably, we find that purified DDB complex is sufficient to completely recapitulate the bulk contraction of MT networks, at millimeter length scales, observed previously in Xenopus extract (Foster et al., 2015). The contractile stresses exerted by dynein in mitotic extracts are likely to be related to the mechanisms used to cluster MT minus-ends during aster formation and spindle pole focusing in vivo. In cells, the mitotic molecules kinesin-14 (Goshima et al., 2005), NuMA (Merdes et al., 1996), and Lis1 (Moon et al., 2014; Raaijmakers et al., 2013; Tai et al., 2002) have been shown to be essential for the formation of asters and spindle poles, suggesting that dynein activity is likely tuned by other factors in cells. Kinesin-14 and dynein perform semi-redundant functions during spindle assembly (Goshima et al., 2005), and our work suggests that unbiased angle of sliding could be a key difference that differentiates dynein from kinesin-14 function.
In our assays, we used the well-characterized BicD2N adapter molecule to mediate the interaction between dynein and dynactin. Though BicD2 has be implicated in the organization of microtubules in interphase (Fumoto et al., 2006), BicD2 has no known role in mitosis in vivo. We hypothesize that NuMA could fill a similar role of mediating the dynein-dynactin interaction, leading to dynein motor activation and accumulation at MT minus-ends (Hueschen et al., 2017). NuMA’s own MT-binding domain (Haren and Merdes, 2002) could serve to enhance minus-end clustering by further augmenting motor dwell time at minus-ends or by providing another tether to the MT lattice during MT-MT sliding. Finally, the dynein regulatory factor LIS1, which is required for normal spindle assembly and locks dynein in a strongly MT-bound state (Huang et al., 2012; McKenney et al., 2010), also accumulates with DDB at minus-ends in vitro (Gutierrez et al., 2017). LIS1’s role at minus-ends remains to be examined, but it may further strengthen the DDB affinity for MT ends, leading to longer lasting interactions between MTs. The LIS1 binding partner NudEL also plays roles in mitotic spindle assembly and may further modulate dynein function at MT minus-ends (Wang et al., 2013). Further experiments reconstituting the mitotic environment in vitro will provide insight into the role of these molecules and processes.
In summary, our work reals a unique mechanism for dynein-driven contractile forces in a MT network and proposes a model for how dynein sorts and organizes MTs at spindle poles and in other cellular contexts. We propose that the contractile stresses involved in aster formation, poleward movement of k-fibers, and spindle pole focusing, are generated by small clusters of activated, processive dynein-dynactin complexes localized to MT minus-ends. We note that our molecular model for dynein-driven MT reorganization is more broadly applicable to other cellular contexts such as dynein-driven sorting of axonal microtubules (Rao et al., 2017; Zheng et al., 2008).
RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli (BL21DE3) | Agilent | 200131 |
| Escherichia Coli (XL10Gold) | Agilent | 200314 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SNAP-Cell TMR-Star | NEB | S9105S |
| SNAP-Surface Alexa Fluor 647 | NEB | S9136S |
| SNAP-Surface Alexa Fluor 488 | NEB | S9129S |
| Dylight 405 NHS Ester | ThermoFischer | 46400 |
| Alexa Fluor 647 NHS Ester | ThermoFischer | A37573 |
| Biotinylated poly(L-lysine)-[g]-poly(ethylene-glycol) (PLL-PEG-Biotin | SuSoS AG | PLL(20)-G[3.5]-PEG(2)/PEG(3.4)-biotin(50%) |
| Streptavidin | ThermoFischer | 21135 |
| Trolox (6-hydroxy-2,5,6,7,8-tetramethylchroman-2-carbonsaure, 97%) | Acros | AC218940050 |
| 3,4-Dihydroxybenzoic Acid (Protocatechuic acid) | Sigma-Aldrich | 37580 |
| Protocatachuate 3,4-Dioxygenase from Pseudomonas sp. | Sigma-Aldrich | P8279 |
| κ-caesin from bovine milk | Sigma-Aldrich | C0406 |
| Pierce Bovine Serum Albumin, Biotinylated | Thermo-Fischer | 209130 |
| GpCpp (guanosine-5′-[(α, β) – methyleno]triphosphate, Sodium salt | Jena Bioscience | Nu-405 |
| Paclitaxel | Sigma-Aldrich | T7402 |
| Pluronic F-157 | Sigma-Aldrich | P2443 |
| Glass cover slides (18×18-1.5) | Fischer | 12-541A |
| Superfrost Microscope slides | Fisher | 12-550-143 |
| Double Sided tape | 3M | 34-8517-0998-9 |
| Adenosine 5′-triphosphate disodium salt hydrate | Sigma-Aldrich | A2383 |
| Guanosine 5′-triphosphate sodium salt hydrate | Sigma-Aldrich | G8877 |
| Bovine Serum Albumin | Sigma-Aldrich | A2058 |
| Casein | Sigma-Aldrich | C7078 |
| Methyl Cellulose | Sigma-Aldrich | M0387 |
| Nonidet P 40 substitute (NP-40) | Sigma-Aldrich | 74385 |
| PDMS (polydimethylsiloxane, sylgard 184) | Sigma-Aldrich | 761036 |
| Dimethyl sulfoxide ACS Reagent | Sigma-Aldrich | 472301 |
| Atto 488 NHS ester | Sigma-Aldrich | 41698 |
| Recombinant DNA | ||
| Plasmid: pET28-StrepII-SNAPf-Hook3 | McKenney et al. Science 2014 | |
| Plasmid: pET28-StrepII-SNAPf-BicD2 | McKenney et al. Science 2014 | |
| Software and Algorithms | ||
| FIJI | Schindelin et al. 2012 | https://Fiji.sc/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| μManager | Edelstein et al. 2010 | https://micro-manager.org/ |
| Autocad 360 | Autodesk | https://client.autocad360.com/ |
| Silhouette Studio | Silhouette America | https://www.silhouetteamerica.com/software |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Richard James McKenney (RJMcKenney@ucdavis.edu).
Methods Details
elPorcine brain tubulin was isolated using the high-molarity PIPES procedure as described (Castoldi and Popov, 2003) and then labeled with biotin-, Dylight-405 NHS-ester, or Alexa647 NHS-ester as described (http://mitchison.hms.harvard.edu/files/mitchisonlab/files/labeling_tubulin_and_quantifying_labeling_stoichiometry.pdf). Microtubules were prepared by incubation of tubulin with 1mM GTP for 10 min. at 37°C, followed by dilution into 10 μM final taxol for an additional 20 min. For experiments using detyrosinated microtubules, 12 μg/mL carboxypeptidase was added at this step (McKenney et al., 2016). Microtubules were pelleted at 80K rpm in a TLA-100 rotor and the pellet was resuspended in 50 μL BRB80 with 10 μM taxol. StrepII-SNAPf-BicD2N and StepII-SNAPf-Hook3 was isolated from bacteria as described (McKenney et al., 2014). Briefly, a strepII-SNAPf-tagged adaptor constructs were expressed in BL21(DE3) cells (Agilent). The cells were grown at 36° C until OD600 of 0.6, then induced with .4 mM IPTG overnight at 18 ° C. Proteins were affinity-purified by streptactin beads, then further purified by size exclusion chromatography on a Superose 6 column in 60 mM Hepes pH 7.4, 50 mM K-acetate, 2 mM MgCl2, 1 mM EGTA, 10 % glycerol. Purified BicD2N and Hook3 was used to isolate DDB complexes from rat brain cytosol as previously described (McKenney et al., 2014). DDB complexes were labeled with in a ~4:1 ratio of dye:SNAPf-tagged protein at 2 μM SNAP-TMR, SNAP-Alexa647, or SNAP-Alexa488 dye (NEB) during the isolation procedure, and were frozen in small aliquots and stored at −80°C.
The stoichiometry of labeling and protein concentration was assessed using a Nanodrop One (ThermoFisher) and comparing the absorbance of total protein at 280nm to the absorbance at the SNAP-dye wavelength. Protein concentrations given are for the total amount of SNAP-labeled BicD2N used in each assay. Gel densitometry analysis of a typical DDB preparation reveals an approximate 25-fold excess of SNAPf-BicD2N over dynein heavy chain and therefore the amount of fully formed, active DDB complex in each assay is substantially lower than the reported value. We chose to report the measureable value of total labeled SNAPf-BicD2N due to possible variations in the final concentration of fully formed, active DDB from different preparations. All buffers and chemicals were from Sigma Aldrich.
TIRF Microscopy
All microscopy was performed on a custom built through the objective TIRF microscope (Technical Instruments, Burlingame CA) based on a Nikon Ti-E stand, motorized ASI stage, quad-band filter cube (Chroma), Andor laser launch (100 mW 405 nm, 150 mW 488 nm, 100 mW 560 nm, 100 mW 642 nm), EMCCD camera (iXon Ultra 897), and high-speed filter wheel (Finger Lakes Instruments). All imaging was performed using a 100X 1.45NA objective (Nikon) and the 1 or 1.5X tube lens setting on the Ti-E. Experiments were conducted at room temperature. The microscope was controlled with Micro-manager software (Edelstein et al., 2010).
TIRF chambers were assembled from acid washed coverslips (http://labs.bio.unc.edu/Salmon/protocolscoverslippreps.html) and double-sided sticky tape. Taxol-stabilized MTs were assembled with incorporation of ~ 10% Dylight-405-, and biotin-labeled tubulin. Chambers were first incubated with 0.5 mg/mL PLL-PEG-Biotin (Surface Solutions) for 10 min., followed by 0.5 mg/mL streptavidin for 5 min. Unbound streptavidin was washed away with 40 μL of BC buffer (80mM Pipes pH 6.8, 1mM MgCl2, 1mM EGTA, 1 mg/mL BSA, 1mg/mL casein, 10μMtaxol). MTs diluted into BC buffer were then incubated in the chamber and allowed to adhere to the streptavidin-coated surface. Unbound MTs were washed away with TIRF buffer (60 mM Hepes pH 7.4, 50 mM K-acetate, 2 mM MgCl2, 1 mM EGTA, 10 % glycerol, 0.5 % Pluronic F-127, 0.1 mg/mL Biotin-BSA, 0.2 mg/mL κ-casein, 10μM taxol). Unless otherwise stated, experiments were conducted in imaging buffer (60 mM Hepes pH 7.4, 50 mM K-acetate, 2 mM MgCl2, 1 mM EGTA, 10 % glycerol, 0.5 % Pluronic F-127, 0.1 mg/mL Biotin-BSA, 0.2 mg/mL κ-casein, 10μM taxol, 2 mM Trolox, 2 mM protocatechuic acid, ~50 nM protocatechuate-3,4-dioxygenase, and 2 mM ATP). The total ionic strength of this assay buffer, calculated according to established methods (Thiede et al., 2013), is 137.44 mM, which we note is very close to the ionic strength calculated inside living cells using FRET biosensors (Liu et al., 2017).
The resulting data was analyzed manually using kymograph analysis in ImageJ (FIJI). For velocity analysis, the velocity of an uninterrupted run segment from a kymograph was used. For images displayed in Figure, background was subtracted in FIJI using the ‘subtract background’ function with a rolling ball radius of 50 and brightness and contrast settings were modified linearly. In images where there was substantial drift, the “Descriptor-based series registration (2D/3D + T)” plug-in was used in FIJI with interactive brightness and size detections in the MT channel to stabilize images. Graphs were created using Graphpad Prism 7.0a and statistical tests were performed using this program. All variances given represent standard error of mean.
Continuous imaging (Drop-in) assay
Accumulation assays were conducted in Nunc Labtek II chambered coverglass system (Thermo Fischer Scientific, #155409). Fixation of microtubules were conducted as before with an adjusted protocol. 50 μL of PLL-PEG was added to the chamber for at least 10 minutes, then aspirated out. Streptavidin, and microtubules were added in the same fashion. Buffer exchanges were done as rapidly as possible to limit surface exposure to air. For imaging, 150 uL of imaging buffer was added in two stages. First, microtubules were aspirated out and 100 uL of imaging buffer was added to the chamber to begin imaging. Next, a 3x dilution of DDB was made in 50 uL of imaging buffer and carefully dropped into the chamber using a pipette during continuous imaging.
DDB minus-end cluster size quantification
We integrated the fluorescence intensity over a ROI around a MT minus-end cluster. Next 10–30 similar ROIs were drawn around surrounding individual molecules bound to the MT lattice or coverslip to provide an average signal intensity estimate for one DDB molecule. One large ROI was drawn in empty space near the microtubule to provide an accurate estimate for background fluorescence. The average intensity of the background fluorescence ROI was subtracted from the average intensity at minus-ends then integrated over the area of the ROI. This integrated intensity was then compared to the average integrated intensity for one DDB molecule to obtain numbers of DDB at ends.
The kymograph method of determining the number of DDB at minus-ends involved continuous imaging assay as described above. A line scan through the minus-end to obtain an intensity peak over time. The first maximum at saturation was used as a temporal fiducial for when steady state is achieved. We then drew a line scan further into the microtubule lattice and obtained intensity peaks corresponding to DDB traveling across the line to the minus-end. This is the flux of DDB into the minus-end until the time of saturation. This information along with the exponential decay resulting from DDB dwell times in a clustered environment, allowed us to determine the number of molecules at minus-ends at infinity seconds using the formula below.
Where N is the number of molecules at minus-ends. X is the rate at which DDB reach minus-ends. The rate constants k1 and k2 are for short and long dwelling DDB, respectively, and %k1 and %k2 are the percent of molecules with rate constants k1 and k2, respectively.
Single molecule experiments
Single molecule experiments with DDB and DDH were conducted using chamber slides. DDX at ~1 nM in imaging buffer was flowed into chambers containing fixed taxol-stabilized microtubules. Cluster dwell times were obtained by following the same conditions as above with DDB labeled with either Alexa647- or Alexa488-labled BicD2 at ~30 nM concentrations. Experiments with dynamic microtubules were conducted using coverslip-attached, biotin-labeled GMPCPP stabilized microtubule seeds. Next, imaging buffer containing ~1 nM DDB, 40 μM labeled tubulin (~5% Dylight-405), and 2 mM GTP was introduced into the chamber. In experiments involving carboxypeptidase treated microtubules, tubulin tail digestion was evaluated using the fluorescence intensity of bound TMR-labeled p150 on differentially labeled WT and CPA-treated microtubules in the same chamber.
Images were taken 0.5 seconds apart for 1500 frames. For experiments using dynamic microtubules, images were taken 1 second apart for 1500 frames. Single molecule dwell times were obtained using kymographs. Molecules that dwelled were defined as molecules that remained at the minus-end longer two frames. Molecules that did not dwell at minus-ends longer than two frames were considered to have fallen of immediately. Dwell times were defined as the time when a processive motor became immobile for at the end of a microtubule until it either became diffusive or signal was lost. Processive molecules that immediately dissociated (~20% of all molecules) when reaching the end of the microtubule, continued to dwell at the end of the microtubule at the last frame of capture, or overlapped with another molecule were not counted.
Sliding Assays and Data Analysis
Experiments were conducted in chamber slides. Fluorescently labeled microtubules diluted in 50 μl of BC+ buffer (BRB80, 1mg/mL casein, 0.2% methylcellulose) was flowed into the chamber. Microtubules not bound to the coverglass were washed out with 20 ul of BC+ buffer. Experiments were conducted in imaging+ buffer (imaging buffer with 0.2% methylcellulose) at 1 second per frame. All angles were defined as the smaller angle between positive ends of microtubules around a central pivot point joining two microtubules. Minus-end foci lifetimes began when minus-ends of two microtubules first contacted each other until they dissociated from one another. Foci that bundled or had another sliding event with a third microtubule were not counted.
Aster assays
Solutions for aster assays were prepared in 5 μL final volume on ice in TIRF assay buffer with 2 mM ATP, 2 mM GTP, 20 μM Dylight405-labeled tubulin, ~4 μM DDB, and 2 μM taxol. The solution deposited between a slide and coverslip then imaged in widefield at 25° C. DDB were prepared as described previously then concentrated to 10 μM using Amicon spin concentrators with a MW cutoff of 50 KDa (Millipore, UFC510096).
For analysis, background was subtracted in FIJI using the ‘subtract background’ function with a rolling ball radius of 50 and brightness and contrast settings were modified linearly. Standard deviation for the microtubule and DDB channel were obtained by selecting an ROI over the entire field of view. Normalized intensity was normalized to the initial intensity of the first pixel along the line scan.
Bulk Contraction Assay
Microfluidic devices were prepared as previously described (Foster et al., 2015). Briefly, channel of width 0.9mm and length 18mm were created using AutoCAD 360 (Autodesk) and Silhouette Studio (Silhouette America) software, cut from 125 μm thick tape (3M Scotchcal, St. Paul MN), and adhered to petri dishes. PDMS (Sylgard 184, Dow Corning, Midland, MI) was mixed at a 10:1 ratio, poured onto masters, degassed, and baked overnight at 60° C. Coverslips and PDMS devices were corona treated with air plasma for 1 minute each before bonding. Channels were loaded with a degassed blocking solution composed of 5mg/mL BSA (J.T. Baker, Center Valley, PA) and 2.5% Pluronic-F127 (Sigma, St. Louis, MO) and incubated overnight at 12° C. Channels were further incubated for at least 15 minutes with a solution of 2 mg/mL κ-casein before use. Taxol stabilized microtubules were made fresh daily as previously described (Nedelec et al., 1997) at a final tubulin concentration of 18.9 μM containing 1.7 μM Alexa-647 or Atto-647 labeled tubulin. Taxol microtubules were diluted 1:19 into a reaction mix such that the final mix was composed of 2.5mM ATP, 20 μM Taxol, DDB at the indicated concentration, and 20% DMSO, all in 1x DDB Buffer. This mix was loaded into a microfluidic channel, sealed using vacuum grease, and imaged immediately using a spinning disk confocal microscope (Nikon Ti2000, Yokugawa CSU-X1), an EMCCD camera (Hamamatsu), and a 2x objective using μManager acquisition software (Edelstein et al., 2010). Images were analyzed using ImageJ and custom written MATLAB software. To extract the contraction timescale and final fraction contracted, ε(t) curves were fit to a saturating exponential function using time points where ε(t) >0.1 as previously described (Foster et al., 2015). All conditions were repeated on at least 3 separate days, using DDB from 2 separate preparations. A similar protocol was used for GST-hDyn experiments, with Atto-488 labeled tubulin substituted as the labeled tubulin and hGST-Dyn1 buffer consisting of 50 mM Tris, pH 8.0 and 150 mM K-acetate substituted for DDB buffer. hGST-Dyn1 experiments were repeated on 3 separate days. We note that the concentrations of purified DDB used in these assays falls within the range of the measured concentrations of dynein (~1 μM) and dynactin (~0.7 μM) in Xenopus egg extract (Wang et al., 2013).
Quantification and Statistical Analysis
Unless otherwise stated, all data was analyzed manually using ImageJ (FIJI). Graphs were created using Graphpad Prism 7.0a and statistical tests were performed using this program. All variances given represent standard error of mean.
Images for bulk contraction assays were analyzed using ImageJ and custom written MATLAB software. To extract the contraction timescale and final fraction contracted, ε(t) curves were fit to a saturating exponential function using time points where ε(t) >0.1 as previously described (Foster et al., 2015).
Supplementary Material
Figure S1: Determination of Numbers of DDB Molecules at MT Minus-Ends, related to Figure 1. (A) A kymograph of a microtubule accumulating DDB at its minus-end (reproduced from Figure 1B). A line scan was drawn along the region corresponding to the MT minus-end (red). The earliest peak of the fluorescence intensity plateau along this line scan was used as a fiducial for when the minus-end accumulation reaches saturation. A second line scan (pink) extending as far as the DDB intensity plateau fiducial in the Y-axis was drawn further into the MT as close to the minus-end as possible without interference from the growing minus-end accumulation. (B) Intensity plot of the microtubule minus-end (red line in kymograph (A)). The earliest peak of the fluorescence intensity plateau as indicated by the red highlight was used as a fiducial for when a minus-end reaches saturation. (C) Intensity plot in the microtubule lattice used to count the number of DDB that enter the minus-end accumulation until saturation. Every peak intensity on this new line scan corresponds to a single DDB crossing the point in space. Red circles indicate peaks and correspond to individual DDB. (D) Example of intensity analysis for numbers of DDB at MT ends. Yellow, white, and green circles indicate ROIs for minus-end accumulations, background fluorescence normalization, and single molecules, respectively. Scale bar: 2 μm. (E) Plot of the rate at which DDB enter the minus-end. μ = .1658 ± .04. N = 44.
Figure S2: The p150-MT Interaction is Not Required for Cooperative Accumulation of DDB at Minus-ends, related to Figure 2. (A) Representative image of TMR-labeled p150-SNAP (red) binding affinity for WT MTs (blue) compared to carboxypeptidase A (CPA)-treated MT (cyan). Scale bars: 5 μm, 30 sec. (B) Quantification of integrated p150 intensity on either WT or CPA-treated MT. Data represented as intensity minus background per pixel of p150 signal along a microtubule. Average WT and CPA-treated intensity are 2950 a.u. ± 370 and 1100 a.u. ± 176, respectively. P=.0001 by Student’s T-test. Error bars represent SEM. N = 90 and 64 MTs, from two independent experiments. (C) Kymograph showing that DDB accumulation at a MT minus-end (red arrow) is constant in size even at much higher flux of incoming DDB molecules. Scale bars: 5 μm, 30 sec. (D) Plot of cumulative frequency comparing single DDB dwell times within minus-end DDB clusters at 1X and 10X DDB concentration. Note 1X DDB concentration data is reproduced from Figure 2E. (E) Table summarizing the parameters of cumulative frequency graphs comparing 1X and 10X DDB concentrations, including the characteristic dwell time (τ) of short (τ1) and long (τ2) populations, percentage of molecules in each population, and number of molecules measured (N) are given. 95% confidence intervals given in parentheses. All regressions have a goodness of fit (R2) greater than .99. All data from 2–3 independent experiments. (F) Plot of the percent of incoming DDB molecules to the MT minus-end that either dwell (filled) or fall off immediately (empty) on bare minus-ends (red) and minus-ends with DDB clusters (blue). Percent of molecules that dwell are 77% and 83% for vacant and clustered minus ends, respectively. P = .2734 by Fischer’s exact test.
Figure S3: Minus-End Accumulations of DDH Drive MT Reorganization Similar To DDB, related to Figure 3. (A) TIRF-M image of DDH (red) forming clusters (yellow arrows) at MT minus-ends. Scale bar: 5 μm. (B) Top: Image frames showing steps during MT reorganization into a mini-aster. Arrows highlight minus-end accumulations that drive sliding in that frame. Bottom: Schematic depicting the DDB-driven movements of MTs. Red dots indicate DDB clusters at minus-ends. Arrows highlight the direction of sliding MTs. Scale bar: 2 μm, time is in sec. (C) Examples of parallel, anti-parallel, and oblique sliding driven by minus-end accumulations of DDH. Yellow and green arrows indicate minus-ends of sliding microtubules inferred from DDH accumulation. Scale bar: 5 μm, time is in sec. (D) Examples of mini-asters formed through DDH-driven MT-MT sliding. Scale bars: 5 μm.
Corresponds to Figure 1A. Taxol-stabilized microtubules (blue) were fixed to coverglass. The microtubules were then continuously imaged by TIRF-M while ~30 nM DDB (Red) was added to the system.
Corresponds to Figure 3C. Taxol-stabilized microtubules (blue) were fixed within the TIRF-M field by depletion forces, and are free to diffuse in the xy-plane. Under these conditions, we observe DDB minus end clusters (red) driving parallel (left), antiparallel (middle), and perpendicular (right) MT-MT sliding. Time is labeled in seconds, scale bar: 2 μm.
Taxol-stabilized microtubules (blue) were fixed to the TIRF-M field by depletion forces and are free to diffuse in the xy-plane. Under these conditions, multiple DDB-driven (red) sliding events lead to a stable topologically organized mini-aster. Time is labeled in seconds, scale bar: 2 μm.
Corresponds to Figure 4B. 20 μM soluble tubulin (green) was added to GMP-CPP stabilized MT seeds (blue) in the presence of 2mM GTP and ATP. DDB complexes (red) form clusters on growing MT minus ends. MT-MT sliding events driven by minus end clusters result in MT-MT foci (yellow arrows).
Corresponds to Figure 4C. 4 μM DDB (red) was added to 40 μM free tubulin (black) in the presence of 2 mM ATP and GTP. MTs gather into asters with strong accumulation of DDB at their cores, and individual asters coalesce over time.
Upper: Corresponds to Figure 5A. 1 μM DDB (red) was added to a microfluidic channel filled with 1 μM taxol-stabilized microtubules (blue). Over time, the network of MTs contracts, driven by DDB forces. Network tearing is observed near the end of the moving, resulting in lateral network movement (Foster et al. 2015). Lower: Corresponds to Figure 5E. 1 μM of GST-hdynein (red) mixed with 1 μM taxol-stabilized microtubules (blue) does not lead to bulk network contraction, suggesting that processivity and minus end accumulation are necessary for this activity. Time is shown in minutes.
Corresponds to Figure 5B. Higher magnification imaging of a contracting network of 1 μM taxol-stabilized microtubules (blue) by 1 μM DDB (red) shows that DDB localize to the centers of aster like MT structures. Time is shown in minutes.
Highlights.
Dynein-dynactin cooperatively form limited-sized motor clusters at MT minus-ends.
End clusters provide spatial restriction of force production to MT minus-ends.
End clusters slide MTs without orientation bias, leading to minus-end focusing.
End clusters produce contractile forces on millimeter-scale MT networks.
Acknowledgments
Thanks to Elizabeth Christine Paz for generating the original macros for image analysis. We thank members of the McKenney and Ori-McKenney labs for critical input during the project and Christina Hueschen and Sophie Dumont for valuable advice and feedback. We acknowledge Dan W. Nowakowski (N Molecular Systems, Inc.) for molecular graphics help. This work was supported by the Kavli Institute for Bionano Science and Technology at Harvard University, National Science Foundation grant DMR-0820484 to DJN, National Institutes of Health grant R35GM124889 to RJM, and startup funding provided by UCD to RJM.
Footnotes
Author Contributions: RJM and DJN conceived the project. RT and PJF performed the experiments and analyzed the data. All authors wrote the manuscript and assembled the figures.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- Amos LA. Brain dynein crossbridges microtubles into bundles. J Cell Sci. 1989;93:19–28. doi: 10.1242/jcs.93.1.19. [DOI] [PubMed] [Google Scholar]
- Belmonte J, Leptin M, Nedelec F. A Theory That Predicts Behaviors Of Disordered Cytoskeletal Networks. bioRxiv. 2017 doi: 10.15252/msb.20177796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Carter AP, Diamant AG, Urnavicius L. How dynein and dynactin transport cargos: a structural perspective. Curr Opin Struct Biol. 2016;37:62–70. doi: 10.1016/j.sbi.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein expression and purification. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Ketcham SA, Schroer TA, Lander GC. Structural organization of the dynein-dynactin complex bound to microtubules. Nat Struct Mol Biol. 2015;22:345–347. doi: 10.1038/nsmb.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco MA, DeSantis ME, Leschziner AE, Reck-Peterson SL. Mechanism and regulation of cytoplasmic dynein. Annu Rev Cell Dev Biol. 2015;31:83–108. doi: 10.1146/annurev-cellbio-100814-125438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary FB, Dewitt MA, Bilyard T, Htet ZM, Belyy V, Chan DD, Chang AY, Yildiz A. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nature communications. 2014;5:4587. doi: 10.1038/ncomms5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, Schuh M. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nature communications. 2015;6:7217. doi: 10.1038/ncomms8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross RA, McAinsh A. Prime movers: the mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Biol. 2014;15:257–271. doi: 10.1038/nrm3768. [DOI] [PubMed] [Google Scholar]
- Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nature Cell Biology. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McAinsh AD. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci U S A. 2016;113:E1635–1644. doi: 10.1073/pnas.1516370113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD. The Kinesin-12 Kif15 is a processive track-switching tetramer. Elife. 2014;3:e01724. doi: 10.7554/eLife.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr Protoc Mol Biol. 2010;Chapter 14(Unit14):20. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting MW, Hueschen CL, Udy DB, Dumont S. Force on spindle microtubule minus ends moves chromosomes. J Cell Biol. 2014;206:245–256. doi: 10.1083/jcb.201401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P. Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol. 2009;19:1833–1838. doi: 10.1016/j.cub.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- Florian S, Mayer TU. The functional antagonism between Eg5 and dynein in spindle bipolarization is not compatible with a simple push-pull model. Cell reports. 2012;1:408–416. doi: 10.1016/j.celrep.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Foster PJ, Furthauer S, Shelley MJ, Needleman DJ. Active contraction of microtubule networks. Elife. 2015;4 doi: 10.7554/eLife.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumoto K, Hoogenraad CC, Kikuchi A. GSK-3beta-regulated interaction of BICD with dynein is involved in microtubule anchorage at centrosome. EMBO J. 2006;25:5670–5682. doi: 10.1038/sj.emboj.7601459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin JC, Matov A, Groen AC, Needleman DJ, Maresca TJ, Danuser G, Mitchison TJ, Salmon ED. Spindle fusion requires dynein-mediated sliding of oppositely oriented microtubules. Curr Biol. 2009;19:287–296. doi: 10.1016/j.cub.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez PA, Ackermann BE, Vershinin M, McKenney RJ. Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. J Biol Chem. 2017 doi: 10.1074/jbc.M117.790048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Merdes A. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J Cell Sci. 2002;115:1815–1824. doi: 10.1242/jcs.115.9.1815. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial bipolar spindles around artifical chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur EL. Dynein tethers and stabilizes dynamic microtubule plus ends. Current biology: CB. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin G, DeCamp SJ, Chen DT, Sanchez T, Dogic Z. Tunable dynamics of microtubule-based active isotropic gels. Philos Trans A Math Phys Eng Sci. 2014;372 doi: 10.1098/rsta.2014.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich C, Surrey T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J Cell Biol. 2010;189:465–480. doi: 10.1083/jcb.200910125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueschen CL, Kenny SJ, Xu K, Dumont S. NuMA recruits dynein activity to microtubule minus-ends at mitosis. Elife. 2017;6 doi: 10.7554/eLife.29328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol. 2006;2:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus JE, Moughamian AJ, Tokito MK, Holzbaur EL. Dynactin subunit p150(Glued) is a neuron-specific anti-catastrophe factor. PLoS Biol. 2013;11:e1001611. doi: 10.1371/journal.pbio.1001611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Poolman B, Boersma AJ. Ionic Strength Sensing in Living Cells. ACS Chem Biol. 2017;12:2510–2514. doi: 10.1021/acschembio.7b00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Huynh W, Vale RD, Sirajuddin M. Tyrosination of alpha-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J. 2016 doi: 10.15252/embj.201593071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE Induce a Persistent Dynein Force-Producing State. Cell. 2010;141:304–316. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HM, Youn YH, Pemble H, Yingling J, Wittmann T, Wynshaw-Boris A. LIS1 controls mitosis and mitotic spindle organization via the LIS1-NDEL1-dynein complex. Hum Mol Genet. 2014;23:449–466. doi: 10.1093/hmg/ddt436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol Biol Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec F. Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J Cell Biol. 2002;158:1005–1015. doi: 10.1083/jcb.200202051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec FJ, Surrey T, Maggs AC, Leibler S. Self-organization of microtubules and motors. Nature. 1997;389:305–308. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- Olenick MA, Tokito M, Boczkowska M, Dominguez R, Holzbaur EL. Hook Adaptors Induce Unidirectional Processive Motility by Enhancing the Dynein-Dynactin Interaction. J Biol Chem. 2016;291:18239–18251. doi: 10.1074/jbc.M116.738211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, et al. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174:839–849. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Derr ND, Goodman BS, Villa E, Wu D, Shih W, Reck-Peterson SL. Dynein achieves processive motion using both stochastic and coordinated stepping. Nature structural & molecular biology. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Medema RH. Systematic dissection of dynein regulators in mitosis. J Cell Biol. 2013;201:201–215. doi: 10.1083/jcb.201208098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A, Pathak D, Thakur S, Singh S, Dubey AK, Mallik R. Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell. 2016;164:722–734. doi: 10.1016/j.cell.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai AK, Rai A, Ramaiya AJ, Jha R, Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–182. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- Rao AN, Patil A, Black MM, Craig EM, Myers KA, Yeung HT, Baas PW. Cytoplasmic Dynein Transports Axonal Microtubules in a Polarity-Sorting Manner. Cell reports. 2017;19:2210–2219. doi: 10.1016/j.celrep.2017.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Baumgart J, Lindow N, Shelley M, Nazockdast E, Kratz A, Prohaska S, Brugues J, Furthauer S, Muller-Reichert T. C. elegans chromosomes connect to centrosomes by anchoring into the spindle network. Nature communications. 2017;8:15288. doi: 10.1038/ncomms15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwine WB, Hernandez-Lopez R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Tulu US, Fagerstrom C, Wadsworth P. Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J Cell Biol. 2002;158:997–1003. doi: 10.1083/jcb.200204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder CM, Vale RD. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J Cell Biol. 2016;214:309–318. doi: 10.1083/jcb.201604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikirzhytski V, Magidson V, Steinman JB, He J, Le Berre M, Tikhonenko I, Ault JG, McEwen BF, Chen JK, Sui H, et al. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J Cell Biol. 2014;206:231–243. doi: 10.1083/jcb.201401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol. 2013;23:1280–1290. doi: 10.1016/j.cub.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E. Physical properties determining self-organization of motors and microtubules. Science. 2001;292:1167–1171. doi: 10.1126/science.1059758. [DOI] [PubMed] [Google Scholar]
- Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Vale RD, McKenney RJ. Cytoplasmic dynein crosslinks and slides anti-parallel microtubules using its two motor domains. Elife. 2013;2:e00943. doi: 10.7554/eLife.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C, Lakamper S, Wessel AD, Kramer S, Schmidt CF. A chimeric kinesin-1 head/kinesin-5 tail motor switches between diffusive and processive motility. Biophys J. 2013;104:432–441. doi: 10.1016/j.bpj.2012.11.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisawa T, Ichikawa M, Furuta A, Saito K, Oiwa K, Kojima H, Toyoshima YY, Furuta K. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol. 2014;16:1118–1124. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- Toropova K, Mladenov M, Roberts AJ. Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat Struct Mol Biol. 2017;24:461–468. doi: 10.1038/nsmb.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokter M, Mucke N, Surrey T. Reconstitution of the human cytoplasmic dynein complex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20895–20900. doi: 10.1073/pnas.1210573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nature Cell Biology. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesbeen RG, Tanenbaum ME, Medema RH. Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell reports. 2014;8:948–956. doi: 10.1016/j.celrep.2014.07.015. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Verde F, Berrez JM, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Crevenna AH, Kunze I, Mizuno N. Structural basis for the extended CAP-Gly domains of p150(glued) binding to microtubules and the implication for tubulin dynamics. Proc Natl Acad Sci U S A. 2014;111:11347–11352. doi: 10.1073/pnas.1403135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ketcham SA, Schon A, Goodman B, Wang Y, Yates J, 3rd, Freire E, Schroer TA, Zheng Y. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell. 2013;24:3522–3533. doi: 10.1091/mbc.E13-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster DR, Gundersen GG, Bulinski JC, Borisy GG. Differential turnover of tyrosinated and detyrosinated microtubules. Proc Natl Acad Sci U S A. 1987;84:9040–9044. doi: 10.1073/pnas.84.24.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Hyman T. Recombinant p50/dynamitin as a tool to examine the role of dynactin in intracellular processes. Methods Cell Biol. 1999;61:137–143. doi: 10.1016/s0091-679x(08)61978-0. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, Younger SH, Zimmerman S, Jan LY, Jan YN. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nature Cell Biology. 2008;10:1172–1180. doi: 10.1038/ncb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Determination of Numbers of DDB Molecules at MT Minus-Ends, related to Figure 1. (A) A kymograph of a microtubule accumulating DDB at its minus-end (reproduced from Figure 1B). A line scan was drawn along the region corresponding to the MT minus-end (red). The earliest peak of the fluorescence intensity plateau along this line scan was used as a fiducial for when the minus-end accumulation reaches saturation. A second line scan (pink) extending as far as the DDB intensity plateau fiducial in the Y-axis was drawn further into the MT as close to the minus-end as possible without interference from the growing minus-end accumulation. (B) Intensity plot of the microtubule minus-end (red line in kymograph (A)). The earliest peak of the fluorescence intensity plateau as indicated by the red highlight was used as a fiducial for when a minus-end reaches saturation. (C) Intensity plot in the microtubule lattice used to count the number of DDB that enter the minus-end accumulation until saturation. Every peak intensity on this new line scan corresponds to a single DDB crossing the point in space. Red circles indicate peaks and correspond to individual DDB. (D) Example of intensity analysis for numbers of DDB at MT ends. Yellow, white, and green circles indicate ROIs for minus-end accumulations, background fluorescence normalization, and single molecules, respectively. Scale bar: 2 μm. (E) Plot of the rate at which DDB enter the minus-end. μ = .1658 ± .04. N = 44.
Figure S2: The p150-MT Interaction is Not Required for Cooperative Accumulation of DDB at Minus-ends, related to Figure 2. (A) Representative image of TMR-labeled p150-SNAP (red) binding affinity for WT MTs (blue) compared to carboxypeptidase A (CPA)-treated MT (cyan). Scale bars: 5 μm, 30 sec. (B) Quantification of integrated p150 intensity on either WT or CPA-treated MT. Data represented as intensity minus background per pixel of p150 signal along a microtubule. Average WT and CPA-treated intensity are 2950 a.u. ± 370 and 1100 a.u. ± 176, respectively. P=.0001 by Student’s T-test. Error bars represent SEM. N = 90 and 64 MTs, from two independent experiments. (C) Kymograph showing that DDB accumulation at a MT minus-end (red arrow) is constant in size even at much higher flux of incoming DDB molecules. Scale bars: 5 μm, 30 sec. (D) Plot of cumulative frequency comparing single DDB dwell times within minus-end DDB clusters at 1X and 10X DDB concentration. Note 1X DDB concentration data is reproduced from Figure 2E. (E) Table summarizing the parameters of cumulative frequency graphs comparing 1X and 10X DDB concentrations, including the characteristic dwell time (τ) of short (τ1) and long (τ2) populations, percentage of molecules in each population, and number of molecules measured (N) are given. 95% confidence intervals given in parentheses. All regressions have a goodness of fit (R2) greater than .99. All data from 2–3 independent experiments. (F) Plot of the percent of incoming DDB molecules to the MT minus-end that either dwell (filled) or fall off immediately (empty) on bare minus-ends (red) and minus-ends with DDB clusters (blue). Percent of molecules that dwell are 77% and 83% for vacant and clustered minus ends, respectively. P = .2734 by Fischer’s exact test.
Figure S3: Minus-End Accumulations of DDH Drive MT Reorganization Similar To DDB, related to Figure 3. (A) TIRF-M image of DDH (red) forming clusters (yellow arrows) at MT minus-ends. Scale bar: 5 μm. (B) Top: Image frames showing steps during MT reorganization into a mini-aster. Arrows highlight minus-end accumulations that drive sliding in that frame. Bottom: Schematic depicting the DDB-driven movements of MTs. Red dots indicate DDB clusters at minus-ends. Arrows highlight the direction of sliding MTs. Scale bar: 2 μm, time is in sec. (C) Examples of parallel, anti-parallel, and oblique sliding driven by minus-end accumulations of DDH. Yellow and green arrows indicate minus-ends of sliding microtubules inferred from DDH accumulation. Scale bar: 5 μm, time is in sec. (D) Examples of mini-asters formed through DDH-driven MT-MT sliding. Scale bars: 5 μm.
Corresponds to Figure 1A. Taxol-stabilized microtubules (blue) were fixed to coverglass. The microtubules were then continuously imaged by TIRF-M while ~30 nM DDB (Red) was added to the system.
Corresponds to Figure 3C. Taxol-stabilized microtubules (blue) were fixed within the TIRF-M field by depletion forces, and are free to diffuse in the xy-plane. Under these conditions, we observe DDB minus end clusters (red) driving parallel (left), antiparallel (middle), and perpendicular (right) MT-MT sliding. Time is labeled in seconds, scale bar: 2 μm.
Taxol-stabilized microtubules (blue) were fixed to the TIRF-M field by depletion forces and are free to diffuse in the xy-plane. Under these conditions, multiple DDB-driven (red) sliding events lead to a stable topologically organized mini-aster. Time is labeled in seconds, scale bar: 2 μm.
Corresponds to Figure 4B. 20 μM soluble tubulin (green) was added to GMP-CPP stabilized MT seeds (blue) in the presence of 2mM GTP and ATP. DDB complexes (red) form clusters on growing MT minus ends. MT-MT sliding events driven by minus end clusters result in MT-MT foci (yellow arrows).
Corresponds to Figure 4C. 4 μM DDB (red) was added to 40 μM free tubulin (black) in the presence of 2 mM ATP and GTP. MTs gather into asters with strong accumulation of DDB at their cores, and individual asters coalesce over time.
Upper: Corresponds to Figure 5A. 1 μM DDB (red) was added to a microfluidic channel filled with 1 μM taxol-stabilized microtubules (blue). Over time, the network of MTs contracts, driven by DDB forces. Network tearing is observed near the end of the moving, resulting in lateral network movement (Foster et al. 2015). Lower: Corresponds to Figure 5E. 1 μM of GST-hdynein (red) mixed with 1 μM taxol-stabilized microtubules (blue) does not lead to bulk network contraction, suggesting that processivity and minus end accumulation are necessary for this activity. Time is shown in minutes.
Corresponds to Figure 5B. Higher magnification imaging of a contracting network of 1 μM taxol-stabilized microtubules (blue) by 1 μM DDB (red) shows that DDB localize to the centers of aster like MT structures. Time is shown in minutes.