Abstract
We describe a new experimental system to study genome instability caused by fragile X (CGG)n repeats in mammalian cells. It is based on a selectable cassette carrying the HyTK gene under the control of the FMR1 promoter with (CGG)n repeats in its 5′-UTR, which was integrated into the unique RL5 site in murine erythroid leukemia cells. Carrier-size (CGG)n repeats dramatically elevate the frequency of the reporter’s inactivation making cells ganciclovir-resistant. These resistant clones have a unique mutational signature: a change in the repeat length concurrent with mutagenesis in the reporter gene. Inactivation of genes implicated in break-induced replication including POLD3, POLD4, RAD52, RAD51 and SMARCAL1, reduced the frequency of ganciclovir-resistant clones to the baseline level that was observed in the absence of (CGG)n repeats. We propose that replication fork collapse at carrier-size (CGG)n repeats can trigger break-induced replication, which result in simultaneous repeat length changes and mutagenesis at a distance.
Introduction
Genetic instability associated with DNA repeats is responsible for numerous hereditary disorders1. A sub-group of these diseases, including fragile X syndrome, Huntington’s disease, myotonic dystrophy, Friedreich’s ataxia and others are caused by expansions of trinucleotide DNA repeats in distinct human genes 2. Large-scale repeat expansions primarily occur during intergenerational transmissions 3. While in most somatic tissues these repeats are fairly stable, substantial length polymorphisms have been observed in dividing cells of peripheral blood and germline tissue 4–6. Expansions have also been demonstrated in non-dividing, post-mitotic cells in tissues implicated in disease 7. Thus, the mechanisms of repeat instability are of prime biological and biomedical importance. While initial breakthroughs in the field came from human pedigree analyses 3, the molecular mechanisms of repeat instability have been more intensively studied in model organisms, including bacteria, yeast, Drosophila and mice 8.
The development of yeast experimental systems has made it possible to carry out genetic analysis at the genome-wide level to identify genes affecting repeat expansions and contractions 9. Most of these gene products are involved in DNA replication and/or post-replication repair, indicating that problems arising during replication of long repetitive DNA are at the heart of the expansion process. Aside from expansion events, DNA repeats demonstrate three other types of genetic instabilities in yeast: chromosomal fragility 10, repeat-induced mutagenesis (RIM) 11 and complex genome rearrangements (CGRs) 12.
It remains unclear to what extent the data obtained in yeast is applicable to humans. Similarly to yeast, expandable repeats stall replication fork progression in mammalian cells 13–19, and the extent of fork stalling seems to correlate with repeat instability. Proteins involved in replication fork stability and post-replication repair have been implicated in repeat instability in mammalian cells as well 18, and repeats expand effectively in actively dividing ES cells 15,20 or during reprogramming of differentiated patient cells into iPSC 21. In the case of fragile X syndrome, replication delay at expanded (CGG)n repeats leads to X-chromosome fragility in humans 22, lending the disorder its name. Altogether, this reinforces the role of DNA replication in repeat instability.
With that said, processes other than DNA replication have also been implicated in repeat expansions in humans and mice. Most notably mismatch repair was first shown to promote repeat expansions in transgenic mouse models of multiple repeat expansion diseases 23–27. Subsequently, the role of mismatch repair in repeat expansions was confirmed in patient cell cultures and specifically designed human cell lines 28–30. Transcription and transcription-coupled repair were also shown to promote repeat instability 31–33. Finally, the DNA-helicases WRN and Rtel, which are involved in homologous recombination, appear to prevent expansions as well 34,35.
To establish the genetic control of repeat instability, it was important to generate a tractable system to select for repeat expansions in cultured mammalian cells. The first such system, based on chromosomally integrated APRT or HPRT reporters carrying (CAG)n repeats in their introns 36, was largely limited to studying repeat contractions and established the role of transcription-coupled repair 31,37,38 and re-replication 39 in the contraction process.
Here we describe and characterize a novel system for analyzing the instability of fragile X (CGG)n repeats in cultured mammalian cells. Fragile X syndrome is caused by expansions of (CGG)n repeats located in the 5′UTR of the human FMR1 gene. Normal individuals have 5 to 50 repeats, carriers possess 50-to-200 repeats, while affected individuals have more than 200 repeats 40. When the repeat length exceeds 200 copies, methylation of the FMR1 promoter leads to heterochromatin formation and gene silencing, which ultimately causes disease. In our system, a HyTK reporter gene was placed under the control of a human FMR1 promoter that contains carrier-size (CGG)n repeats in its 5′-UTR and was integrated into the RL5 site of murine erythroid leukemia cells 41. We reasoned that large-scale repeat expansions in our system would inactivate the HyTK reporter, making cells ganciclovir-resistant. Indeed, the presence of a (CGG)153 repeat in our cassette dramatically elevated the frequency of ganciclovir-resistant clones. While the majority (~70%) of these clones contained expanded or contracted (CGG)n repeats, the lengths of the resultant repetitive runs rarely exceeded the gene’s inactivation threshold 40. Instead, ganciclovir resistance was caused by the presence of mutations in the body of the HyTK reporter concordant with the repeat length changes. Only a small fraction of Gc-r clones contained large-scale expansions of (CGG)n repeats that silenced the reporter. Remarkably, an increase in the frequency of ganciclovir-resistant clones observed in the presence the (CGG)153 repeat disappeared when we knocked down genes implicated in break-induced replication (BIR) in mammalian cells 42. We propose that replication fork stalling caused by carrier-size (CGG)n repeats 43 triggers BIR resulting in repeat length changes concurrent with mutagenesis at a distance.
Results
Experimental system to study genome instability mediated by (CGG)n repeats in cultured mammalian cells
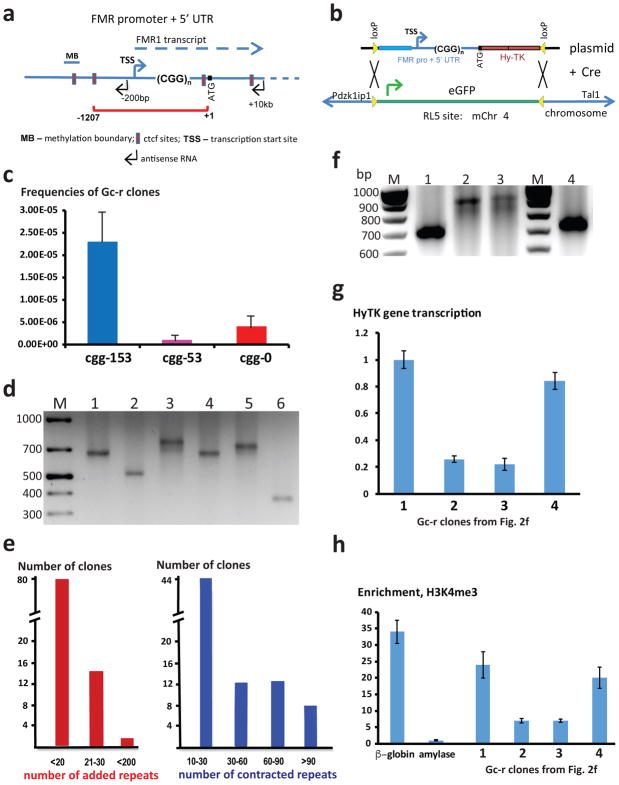
In humans, (CGG)n repeats expanded beyond 200 copies trigger heterochromatin formation - inhibiting the expression of FMR1 and, eventually, surrounding genes. We created a cassette in which the HyTK reporter (encoding an in-frame fusion of the hygromycin phosphotransferase with the herpes simplex virus thymidine kinase) was placed under the control of the FMR1 regulatory region containing its promoter with carrier-size (CGG)n repeats in its 5′-UTR (Fig 1a). This design places our reporter in a similar position with respect to the (CGG)n repeats as the coding part of the FMR1 gene. A single-copy of each FMR-CGGn-HyTK cassette containing 0, 53 or 153 (CGG)n repeats was inserted into the RL5 site in murine erythroid leukemia cells via Cre/loxP mediated exchange with a preexisting eGFP cassette 41 (Fig 1b). For all three repeat lengths (0, 53, 153), insertions in the same (A) orientation (transcribing reporter toward centromere) were isolated and compared. For the (CGG)153 repeat, insertions in the opposite (B) orientation of the cassette were also obtained. The RL5 locus was chosen, since it is not prone to spontaneous silencing 44, potentially minimizing the development of false-positive ganciclovir resistant clones resulting from the reporter’s silencing that is not related to the repeat. Our HyTK cell lines maintained a hygromycin resistant and ganciclovir sensitive phenotype for many months of culture. Based on the data from human studies, we expected that expansions beyond 200 copies of the (CGG)n repeat would inactivate the HyTK reporter and result in the accumulation of ganciclovir-resistant clones.
Figure 1. Experimental system to study genome instability caused by (CGG)n repeats.
a. FMR1 regulatory domain included in the selectable cassettes is shown by the red bracket; blue line represents human endogenous FMR 1 locus. b. FMR1-CGGn-HyTK cassettes were integrated into the RL5 locus in MEL cells via Cre-loxP recombination replacing the eGFP gene. A cassette can be integrated into the RL5 locus in two different orientations. c. Frequencies of Gc-r clones recovered. For clones with each experimental cassette, 96-well plates containing 106 cells were grown in the presence of ganciclovir, and Gc-r clones were counted. The number of analyzed plates were 17, 8 and 20 for CGG153-HyTK, CGG53-HyTK and CGG0-HyTK cassette, respectively. d. Sample PCR across the (CGG)153 run showing expanded (lanes 3 and 5), contracted (lanes 2 and 6) and unchanged repeats (lanes 1 and 4). e. Distribution of repeat expansions and contractions. f. PCR analysis of rare Gc-r clones containing large-scale repeats expansions (lanes 2–3); lane 1- starting repeat, lane 4 – small-scale expansion. g. qPCR analysis of the HyTK gene transcription in the clones shown in f. Means and standard deviations were calculated from three independent experiments. h. ChIP analysis of the H3K4me3 chromatin mark in the clones shown in f. The murine beta-major globin and the murine amylase genes were used as controls for open and condensed chromatin, respectively. Means and standard deviations were calculated from three independent experiments. Source data for d and f are available online.
PCR analysis of repeat length stability in the successfully targeted clones showed that even the longest (CGG)153 repeat in our cassette at the RL5 locus did not undergo length changes at a detectable frequency in cells cultured in the presence of hygromycin. Furthermore, the population doubling time in the hygromycin-containing media was not affected by the presence of the repeat.
The stability of the (CGG)n repeats at the lower or upper ends of the fragile X premutation range, (CGG)53 and (CGG)153, was then tested in the selectable system. Subclones of cell lines with the repeat-bearing cassettes were cultured in the absence of hygromycin for 6 days to remove selective pressure against expansions or other forms of repeat-mediated genetic instability followed by their redistribution to 96-well plates containing ganciclovir. Fig. 1c shows that the presence of 53 copies of (CGG)n repeat had no significant effect on the frequency of Gc-r clones, while 153 copies of the repeat increased their frequency by ~10-fold.
To establish the mechanisms leading to ganciclovir resistance, we carried out PCR analysis of repeat lengths in all Gc-r clones obtained. (CGG)n repeats in the Gc-r clones derived from the (CGG)53 bearing cell line showed no detectable length changes in all (~30) examined cases. By contrast, ~70% of ganciclovir resistant clones originating from the (CGG)153 bearing cell line contained expansions or contractions (Fig. 1d, Table 1). Note that there is a difference in the ratio of expansions to contractions between different clones with the CGG153-HyTK cassette in the A orientation (compare clones c2-29 and c3-1 in Table 1). We attribute these differences to clonal variations, possibly arising from changes in the replication and/or transcription profile of our cassette in those clones. Importantly, the ratio of expansions to contractions for the CGG153-HyTK cassette in the opposite (B) orientation is within the range of clonal variations for the cassette in the A orientation (Table 1). This indicates that repeat instability does not depend on repeat orientation in our system.
Table 1. Repeat instability in Gc-r clones originated from cell lines containing CGG153-HyTK cassettes in the RL5 locus.
71% of Gc-r clones had changes in the length of the CGG repeat across six experimental trials.
| N of Trials | CGG153-HyTK clone | Cassette orientation | Total number of Gc-r clones | Number and percentile of Gc-r clones with repeat instability | Repeat length changes in unstable clones |
|---|---|---|---|---|---|
| 2 | c2-29-1 | A | 58 | 31 (53%)* | 9 expansions (15%) 22 contractions (38%) |
| 2 | c2-29-2 | A | 57 | 43 (75%) | 15 expansions (26%) 28 contractions (49%) |
| 1 | c3-1 | A | 113 | 89 (80%) | 68 expansions (60%) 21 contractions (20%) |
| 1 | c2-38 | B | 15 | 9 (60%) | 3 expansions (20%) 6 contractions (40%) |
| Total: | 243 | 172 (71%) |
95 expansions (39%) 77 contractions (32%) |
||
The scale of repeat contractions amongst Gc-r clones varied widely - from less than 10 to more than 90 repeat units. In contrast, repeat expansions appear constrained: less than 30 repeats were added in the majority (98%) of the clones (Fig 1e). Only a small fraction (2%) of ganciclovir-resistant clones contained a repetitive run longer than 200 (CGG)n copies (Fig. 1f). We confirmed that the observed length changes were indeed repeat expansions and contractions by sequencing (CGG)n runs. Overall, the positive correlation observed in our system between the repeat length and its propensity to expand/contract is consistent with the behavior of repeats at disease loci and in many experimental systems 2.
Mechanisms leading to ganciclovir-resistance
As mentioned above, most of the ganciclovir-resistant clones obtained in our experimental system contained a repetitive run shorter than 200 (CGG)n copies. The expansions observed for the (CGG)153 array were less than 30 repeats in 98% of expanded clones, a length increase that does not cross the 200-repeat threshold for efficient heterochromatin formation. Moreover, ~30% of all Gc-r clones contained massive contractions in the starting (CGG)153 array. Altogether, these data made it very unlikely that ganciclovir resistance was the result of the repeat-mediated silencing of the HyTK cassette in the majority of our clones.
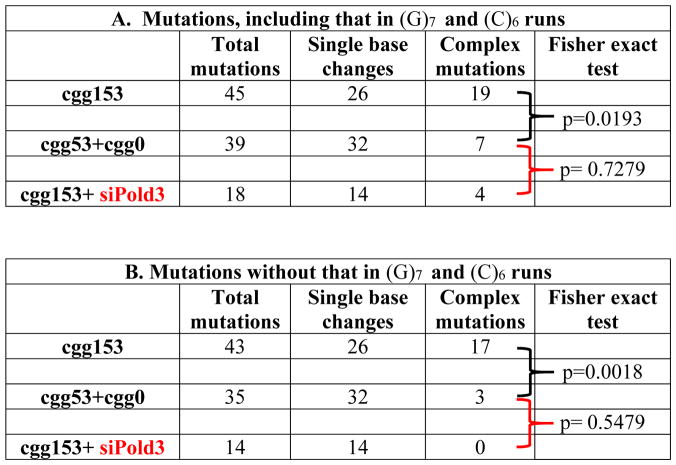
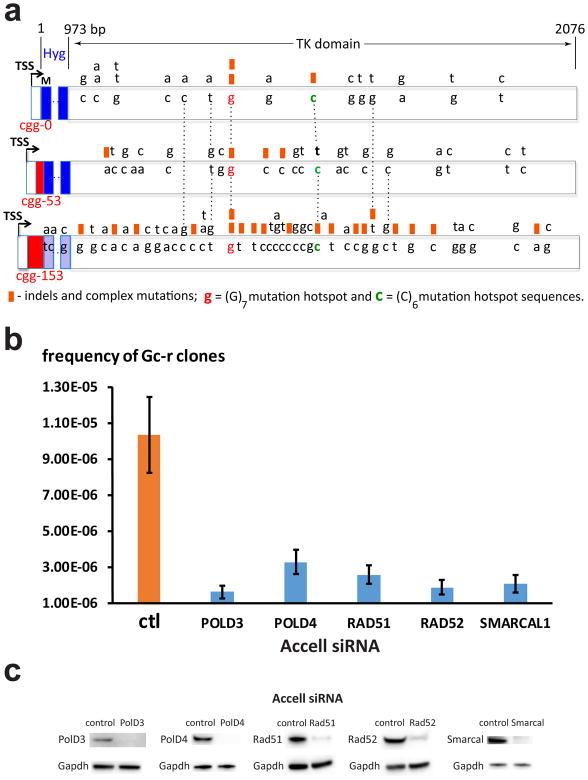
In a yeast experimental system, we have observed that unstable DNA repeats promote mutagenesis at a distance, a phenomenon we called RIM 11. We hypothesized, therefore, that mutations induced by the (CGG)153 repeat in the body of the TK domain of the HyTK gene could account for the ganciclovir resistance in our mammalian system as well. To test this hypothesis, we sequenced the reporter in a randomly selected fraction of Gc-r clones with small-scale expansions or contractions (66 out of 170 total). This sequencing analysis confirmed our hypothesis: two thirds of them (45 clones shown Table 2) contained mutations in the TK region of the HyTK gene that were likely causative, i.e. indels or complex mutations disrupting an open reading frame, as well as base substitutions in conserved regions of the thymidine kinase domain (Figs. 2a & S1). We conclude that a carrier-size (CGG)n repeat induces mutations at a distance in the TK domain of the reporter. Importantly, these mutations coincide with a change in the repeat length (an expansion or contraction). We believe, therefore, that both events result from a common source, such as a break within the repeat (see also Discussion).
Table 2. Repeat instability in Gc-r clones from cell lines with the CGG153-HyTK cassette is accompanied by an elevated frequency of complex mutations in the reporter as compared to CGG53- and CGG0-HyTK cassettes combined.
(A) Mutation patterns, including mutations of (G)7 and (C)6 runs in the TK domain in sequenced Gc-r clones; (B) mutation patterns without mutations of (G)7 and (C)6 runs in sequenced Gc-r clones. Statistical significance of the differences was verified by Fisher standard test. Treatment of the clone carrying CGG153-HyTK cassette with of Pold3 siRNA wipes out complex mutations.
Figure 2. Mechanisms of ganciclovir-resistance.
a. Loss of function mutations in the HyTK gene leading to Gc-r. DNA bases from the wild-type HyTK gene that underwent mutagenesis are shown inside boxed areas of different cassettes. Distances from the 3′ end of the (CGG)n run are shown at the top. Dashed vertical lines align identical positions. Black arrows show transcription start sites (TSS), red rectangles represent (CGG)n repeats, M stands for translation initiation codon, blue rectangles demarcate the Hy domain. Point substitutions are written above the wild-type DNA bases; orange squares above the wild-type DNA bases designate indels or complex mutations. b. Frequencies of Gc-r clones in the c3-1 cell line carrying FMR1-CGG153-HyTK cassette in the A orientation (Table 1) upon treatment with siRNA targeted against genes implicated in BIR. Means and standard deviations from 4 independent experiments are shown. c. Inactivation of mouse POLD3, POLD4, RAD51, RAD52 and SMARCAL1 genes by Accell siRNAs. Western blot analyses with specific antibodies show dramatic reductions in the expression levels of the corresponding proreins. Protein levels for GAPDH gene are shown as internal control. Source data for c are available online.
Note that small-scale expansions or contractions alone do not cause HyTK gene inactivation in our selectable system and, thus, their frequency cannot be assessed in our selectable system. Consequently, we cannot estimate the ratio of events in which repeat length changes are accompanied by mutagenesis versus that without mutagenesis in the HyTK gene. All we can say is that in our selective conditions, the most prevalent mutational event is a change in the repeat’s length combined with mutagenesis at a distance. Furthermore, as previously mentioned, repeats appeared stable in the original targeted cell lines under hygromycin selection, which suggests that small-scale changes are rare events overall.
We also sequenced 43 Gc-r clones that originated in cell lines containing cassettes with 0 or 53 (CGG)n repeats. 90% of them (39 clones) also contained mutations in the TK domain of the reporter. The spectra of mutations in the TK domain of GC-r clones originating from cells with the (CGG)153-, (CGG)53- and (CGG)0- bearing cassettes are compared in Figs. 2a and S1. The spectra were very similar for the cassettes with zero repeats and the shorter, (CGG)53 run, but spectrum was strikingly different for the (CGG)153 repeat. For statistical analysis in Table 2, we compared mutation spectra observed for the CGG153 cassette with that for CGG53 and CGG0 cassettes combined. In case of the CGG153 cassette, forty five percent of all mutations differed from simple point substitutions. Designated as complex in Table 2, 29% of them are indels and 16% are composite events including tandem base substitutions, short duplications or multiple mutational changes (Table S1). This is a significant difference (Fisher’s exact test, p=0.0193) with the other two cassettes where only 18% of observed mutations were indels or complex mutations (Table 2A). When we discounted mutations at the (G)7 and (C)6 runs within the HyTK reporter, which are putative mutation hotspots (highlighted in Fig. S1), this difference became even more profound (Fisher’s exact test, p=0.00187), as the fraction of complex mutations for (CGG)53 and (CGG)0 cassettes combined was reduced to 8.6% (Table 1B).
Unexpectedly, approximately one-third of the Gc-r clones with a change in length of the original (CGG)153 repeat contained no mutations within the TK domain. A priori, their phenotype could have resulted from: (1) a frameshift or nonsense mutation within the Hy domain; (2) heterochromatization of the FMR1 promoter somehow triggered by the carrier-size (CGG)n arrays in a subclonal mouse cell line; (3) stochastic gene inactivation by some unknown post-transcriptional mechanism in a subclonal cell line; or (4) lack of clonality, i.e., two or more subclones being mixed during selection in a 96-well plate. The first explanation implies that some Gc-resistant clones should be Hy-sensitive as well. We indeed detected twelve Hy-s clones with six of them containing mutations within the Hy domain (Fig. 2a), the remaining six carried no detectable mutations. To determine if reporter silencing occurred in mutation-free Gc-r Hy-s cell lines, five clones were tested for the relative mRNA levels from the reporter using semi-quantitative RT-PCR of the TK domain. Since all of them contained HyTK transcripts at a level similar to a cell line carrying the control cassette with no repeats, silencing triggered by the presence of carrier size (CGG)n repeats (up to n=175) did not appear to be the case. While we are not able to explain the Gc-r phenotype of the clones lacking mutations at present, we suspect that they may result from clonal variations in post-transcriptional RNA processing and/or nuclear retention associated with (CGG)n repeats.
In addition to clones described above, a small fraction (~2%) of Gc-r clones contained large-scale expansions reaching up to 240 (CGG)n repeats (Fig. 1f). Analysis of HyTK gene transcription by qPCR in these clones demonstrated a ~5-fold reduction in its mRNA level, as compared to the starting clone with the (CGG)153 repeat or a Gc-r clone with a small-scale expansion (Fig. 1g). Using ChIP (Fig. 1h), we found that the level of an active chromatin marker, H3K4me3, was decreased 3-to-4-fold in these clones. Furthermore, sequencing analysis revealed no mutations in the body of the reporter. Altogether, we conclude that the Gc-r phenotype of clones with large-scale expansions is due to the partial transcription silencing of the reporter in the presence of expanded repeats.
Role of break-induced replication in repeat-mediated genome instability
Altogether, the majority (70%) of Gc-r clones from the cell line with CGG153-HyTK cassette are characterized by a change in the repeat length (an expansion or contraction) concordant with the appearance of mutations, including complex mutations, in the body of the HyTK reporter. These data point to the involvement of an error-prone DNA repair pathway. Break-induced replication (BIR) was shown to be such a pathway in our yeast experimental system 45.
A key protein required for DNA synthesis during BIR is a small subunit of the lagging strand DNA polymerase δ, called Pol32 in yeast and PolD3 in mammalian cells 42. We thus looked at the role of PolD3 protein in repeat-mediated formation of Gc-r clones. To this end, we grew the cell line with the FMR-CGG153-HyTK cassette in the presence of either PolD3 siRNA or a non-targeted control followed by selection for ganciclovir resistance. The treatment with targeted siRNA led to practically complete loss of the PolD3 protein in our cell line (Fig. 2c). Strikingly, the frequency of Gc-resistant clones upon this treatment went down 6-fold, i.e. close to the level observed in the cell line with no repeat cassette (Fig. 2b). PCR analysis of repeat lengths in those clones showed repeats of unchanged length, some small-scale expansions, none of which could account for the ganciclovir resistance, and very few contractions (Fig. S2). We then looked at the pattern of reporter’s mutations in these clones (Fig. S1). Table 2 and Fig. S1 show that PolD3 inactivation resulted in the disappearance of complex mutations that were the hallmark of mutagenesis induced by the (CGG)153 repeat. This result indicated that complex mutations primarily occur in the course of BIR, as was previously suggested in 46.
We then looked at the role of other proteins implicated in BIR in mammalian cells using the same RNA interference approach. Fig. 2b shows that inactivation of another small subunit of DNA polymerase δ, PolD4 also decreased the frequency of Gc-r clones, albeit to a smaller extent than PolD3 inactivation (3-fold), which is consistent with its lesser role in BIR. Recombination proteins Rad52 and Rad51 have also been shown to function in mammalian BIR 42,47 and siRNA inhibition of these proteins in our selectable system resulted in a 4-to-5 fold decrease in the frequency of ganciclovir-resistant clones (Fig. 2B). siRNA inhibition of the annealing DNA-helicase SMARCAL1, which has been implicated in the replication fork-reversal and restart via BIR 48, caused a 5-fold decrease in Gc-resistance in our system (Fig. 2b). Altogether, these data strongly suggest that BIR might be responsible for the majority of mutational events mediated by long (CGG)n repeats.
Discussion
Here we describe a mammalian experimental system designed to select for genetic instability mediated by the presence of fragile X (CGG)n repeats. We placed carrier size (CGG)n repeats in the 5′-UTR downstream of a synthetic genetic construct – downstream of the FMR1 promoter and upstream of a HyTK reporter. Expansions or other types of mutational events caused by these repeats inactivate the HyTK gene, making cells ganciclovir-resistant. We found that a longer, (CGG)153 repeat elevated the frequency of ganciclovir-resistance by roughly an order of magnitude as compared to a no-repeat control or a shorter (CGG)53 repeat. The majority (~70%) of these clones contained repeats that differed in length from the starting (CGG)153 repeat: contractions ranging from 10 to 90 repeats or expansions ranging from 10 to 30 repeats. Sequencing of these clones revealed indels or complex mutations in the HyTK reporter. Thus, the presence of the (CGG)153 run appeared to cause a simultaneous occurrence of two mutational events: a change in the repeat length concurrent with a mutation in the open-reading frame of the reporter.
Similar observations have previously been made for Friedreich’s ataxia patient fibroblasts, where changes in length of expanded (GAA)n repeats were concordant with mutagenesis in the flanking sequences 4. Other structure-prone DNA repeats were also found to induce mutagenesis at a distance in mammalian cells 49. In those studies, mutations were detected up to several kb away from the repeat, similarly to what we observe in our system.
We previously named such compound mutational event repeat-induced mutagenesis (RIM) 11 and hypothesized that they can result from replication fork stalling at the carrier-size repeats, which leads to fork breakage that is followed by an error-prone repair 11,49. This hypothesis seems highly applicable to our system, given that (CGG)n repeats stall replication fork progression in mammalian cells 16,43 and cause chromosomal fragility 22. A feasible mechanism for RIM is break-induced replication (BIR). This process is implicated in the repair of one-ended double-stranded breaks, was initially characterized in yeast and more recently documented in mammalian cells 42,47. Several replication and recombination proteins are required for BIR in both yeast and mammals. Most notably, the small subunit of DNA polymerase δ, called Pol32 in yeast and PolD3 in mammals 50 is essential for this process. We found that inactivation of PolD3 by siRNA led to a dramatic decrease in the frequency of Gc-r clones in our CGG153-HyTK cell line. Furthermore, sequencing of Gc-r clones recovered after PolD3 inactivation revealed the lack of complex mutations (Figs. S1&S2) characteristic of the wild-type cells. The role of another small subunit of DNA polymerase δ, PolD4 in BIR is less clear. While some data suggest that it may not be actively involved in BIR 42, other studies imply that it is essential for the process 51. In our system, PolD4 inactivation also reduced the frequency of Gc-r clones. The strand invasion step during BIR is promoted by recombination proteins Rad52 and sometimes Rad51 42,47. In our case, inactivation of either protein led to the elimination of repeat-induced Gc-r clones. Altogether, these data are consistent with the important role of BIR in RIM mediated by longer (CGG)n repeats.
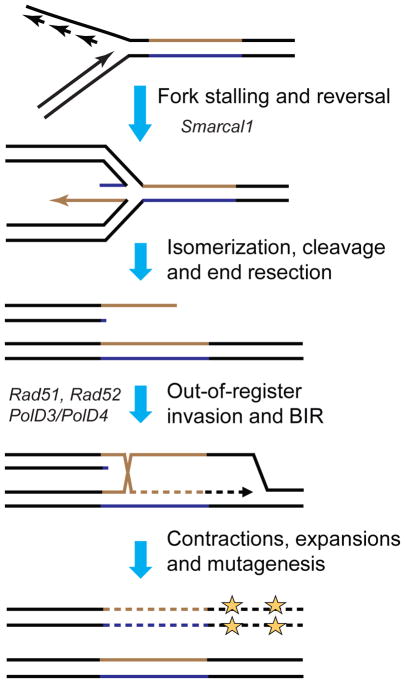
Our working model is presented in Fig. 3. The data from our and other labs show that carrier- and disease-size (CGG)n repeats stall replication fork progression in mammalian cells 16,43. Fork stalling at a long (CGG)n repeat in our CGG153-HyTK cell line might occasionally lead to fork reversal and isomerization of the reversed fork into a Holliday junction. In mammalian cells, SMARCAL1 protein is recruited to stalled replication forks promoting their regression 48. This role of SMARCAL1 is consistent with our model as its inactivation reduces the number of Gc-r clones in the CGG153-HyTK cell line. Resolution of this Holliday junction, presumably carried out by mammalian Mus81 protein 52, would then produce a one-ended DSB. Following end resection, a 3′-single stranded DNA containing repetitive runs is generated. This single-stranded tip can then invade the sister chromatid using Rad52 and Rad51 proteins to create a single D-loop. Subsequently, the D-loop is extended via conservative, PolD3- and/or PolD4-dependent DNA synthesis 42,47. Since the 3′-end of the invading strand contains a long repetitive run, this invasion might occur out-of-register, resulting in expansions or contractions. Alternatively, multiple template-switching events, which are known to occur at early stages of BIR 53 could be responsible for repeat length instability. BIR is a highly mutagenic form of DSB repair, owing to the conservative mode of replication combined with microhomology-mediated template switching 46. The latter explains our data on mutagenesis away from the repeat in the Hy-TK gene and accumulation of complex mutations, likely owing to template-switching events 46 (Table S2).
Fig. 3.
Proposed mechanism for mutational events triggered by carrier-size (CGG)n repeats.
Fork stalling and reversal can lead to the formation of the one-ended DSB. Repair of this DSB via BIR can result in repeat expansion or contractions concordant with mutagenesis at a distance (see text for details). Strands of the repeat are shown in brown and blue; flanking DNA is in black. Proteins involved at various steps of the process are indicated.
Repeat-induced mutagenesis was previously documented in the case of Friedreich’s ataxia 4, but has not been observed for FX-associated syndromes or other repeat expansion diseases in humans. A small number of fragile X-affected males have a deletion hot-spot in the vicinity of the (CGG)n repeat in several fragile X patients 54, which could be frecapitulated by DNA replication through the repeat 55. In light of our data, it could be of interest to estimate mutational load at genomic segments surrounding the (CGG)n repeat in somatic tissues of carriers with premutation-size FMR1 alleles. Carrier-size repeats have been linked to three late-onset diseases called fragile X-associated tremor/ataxia syndrome (FXTAS) 56, fragile X-associated primary ovarian insufficiency (FXPOI) 57 and fragile-X-associated diminished ovarian reserve (FXDOR) 58. These diseases are believed to be triggered by an elevated level of the “toxic”, repeat-bearing mRNA 59. The fact that mRNA levels are increased in these patients may argue against the accumulation of mutations in the FMR1 gene under these circumstances, as these mutations could lead to RNA decay. Note, however, that the frequency of RIM in our system is only 5·10−6, which could not possibly affect the overall level of the FMR1 mRNA in FXTAS or FXPOI patients.
A small fraction (2%) of our Gc-r clones contained repeats that exceeded the threshold length for gene inactivation (>200 repeats) established in fragile X studies. We did not observe any mutations in the reporter cassette in these clones, and its inactivation seemed to be due to heterochomatin formation triggered by expanded (CGG)n repeats. Obviously, our candidate gene analysis is not applicable to these clones, given their strong underrepresentation among all GC-r clones. Thus, we cannot assess whether the observed large-scale expansions occur by the same mechanism as small-scale repeat length changes that are accompanied by mutagenesis at a distance.
Methods
Methods, including references, are available at:
Supplementary Material
Acknowledgments
We thank Karen Usdin for the generous gift of the p32.9 plasmid, Eric Bouhassira for providing us with the RL5 cell line, David Gennert for technical assistance and Alexander Neil for his invaluable editorial help. Supported by the NIH grants R01GM60987 and P01GM105473 to SMM and R01CA093729 to KMV.
Footnotes
Accession Codes
Not applicable.
Data Availability Statement
Source data for Figures are available at:
References
- 1.Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 3.Ashley C, Jr, Warren ST. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- 4.Bidichandani SI, et al. Somatic sequence variation at the Friedreich ataxia locus includes complete contraction of the expanded GAA triplet repeat, significant length variation in serially passaged lymphoblasts and enhanced mutagenesis in the flanking sequence. Hum Mol Genet. 1999;8:2425–2436. doi: 10.1093/hmg/8.13.2425. [DOI] [PubMed] [Google Scholar]
- 5.Chong SS, et al. Gametic and somatic tissue-specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1995;10:344–350. doi: 10.1038/ng0795-344. [DOI] [PubMed] [Google Scholar]
- 6.Monckton DG, Wong LJ, Ashizawa T, Caskey CT. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum Mol Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- 7.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usdin K, House NCM, Freudenreich CH. Repeat instability during DNA repair: Insights from model systems. Crit Rev Bioch Mol Biol. 2015;50:142–167. doi: 10.3109/10409238.2014.999192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JC, Mirkin SM. The balancing act of DNA repeat expansions. Curr Opin Genet Dev. 2013;23:280–288. doi: 10.1016/j.gde.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 11.Shah KA, Mirkin SM. The hidden side of unstable DNA repeats: Mutagenesis at a distance. DNA Repair. 2015;32:106–112. doi: 10.1016/j.dnarep.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGinty RJ, et al. Nanopore sequencing of complex genomic rearrangements in yeast reveals mechanisms of repeat-mediated double-strand break repair. Genome Res. 2017;27:2072–2082. doi: 10.1101/gr.228148.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandok GS, Patel MP, Mirkin SM, Krasilnikova MM. Effects of Friedreich’s ataxia GAA repeats on DNA replication in mammalian cells. Nucleic Acids Res. 2012;40:3964–3974. doi: 10.1093/nar/gks021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Follonier C, Oehler J, Herrador R, Lopes M. Friedreich’s ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nat Struct Mol Biol. 2013;20:486–494. doi: 10.1038/nsmb.2520. [DOI] [PubMed] [Google Scholar]
- 15.Gerhardt J, et al. Stalled DNA Replication Forks at the Endogenous GAA Repeats Drive Repeat Expansion in Friedreich’s Ataxia Cells. Cell Rep. 2016;16:1218–1227. doi: 10.1016/j.celrep.2016.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerhardt J, et al. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat Chem Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, et al. Altered replication in human cells promotes DMPK (CTG)(n) · (CAG)(n) repeat instability. Mol Cell Biol. 2012;32:1618–1632. doi: 10.1128/MCB.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rindler MP, Clark RM, Pollard LM, De Biase I, Bidichandani SI. Replication in mammalian cells recapitulates the locus-specific differences in somatic instability of genomic GAA triplet-repeats. Nucleic Acids Res. 2006;34:6352–6361. doi: 10.1093/nar/gkl846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seriola A, et al. Huntington’s and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum Mol Genet. 2011;20:176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 21.Ku S, et al. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAA·TTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen RS, Canfield TK, Lamb MM, Gartler SM, Laird CD. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 23.Manley K, Shirley TL, LF, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 24.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 25.Savouret C, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ezzatizadeh V, et al. The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol Dis. 2012;46:165–171. doi: 10.1016/j.nbd.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao XN, et al. Mutsbeta generates both expansions and contractions in a mouse model of the Fragile X-associated disorders. Hum Mol Genet. 2015;24:7087–7096. doi: 10.1093/hmg/ddv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wohrle D, et al. Heterogeneity of DM kinase repeat expansion in different fetal tissues and further expansion during cell proliferation in vitro: evidence for a casual involvement of methyl-directed DNA mismatch repair in triplet repeat stability. Hum Mol Genet. 1995;4:1147–1153. doi: 10.1093/hmg/4.7.1147. [DOI] [PubMed] [Google Scholar]
- 29.Du J, et al. Role of mismatch repair enzymes in GAA.TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J Biol Chem. 2012;287:29861–29872. doi: 10.1074/jbc.M112.391961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halabi A, Ditch S, Wang J, Grabczyk E. DNA mismatch repair complex MutSbeta promotes GAA.TTC repeat expansion in human cells. J Biol Chem. 2012;287:29958–29967. doi: 10.1074/jbc.M112.356758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamori M, Pearson CE, Thornton CA. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum Mol Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao XN, Usdin K. Gender and cell-type-specific effects of the transcription-coupled repair protein, ERCC6/CSB, on repeat expansion in a mouse model of the fragile X-related disorders. Hum Mutat. 2014;35:341–349. doi: 10.1002/humu.22495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan NL, et al. The Werner syndrome protein promotes CAG/CTG repeat stability by resolving large (CAG)(n)/(CTG)(n) hairpins. J Biol Chem. 2012;287:30151–30156. doi: 10.1074/jbc.M112.389791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frizzell A, et al. RTEL1 inhibits trinucleotide repeat expansions and fragility. Cell Rep. 2014;6:827–835. doi: 10.1016/j.celrep.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorbunova V, et al. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol Cell Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 38.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee N, Lin Y, Santillan BA, Yotnda P, Wilson JH. Environmental stress induces trinucleotide repeat mutagenesis in human cells. Proc Natl Acad Sci USA. 2015;112:3764–3769. doi: 10.1073/pnas.1421917112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu YH, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 41.Feng YQ, Lorincz MC, Fiering S, Greally JM, Bouhassira EE. Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. Mol Cell Biol. 2001;21:298–309. doi: 10.1128/MCB.21.1.298-309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol. 2009;16:226–228. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebersole T, et al. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10:2779–2791. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JC, Harris ST, Dinter T, Shah KA, Mirkin SM. The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat Struct Mol Biol. 2017;24:55–60. doi: 10.1038/nsmb.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakofsky CJ, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sotiriou SK, et al. Mammalian RAD52 Functions in break-induced replication repair of collapsed DNA replication forks. Mol Cell. 2016;64:1127–1134. doi: 10.1016/j.molcel.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lugli N, Sotiriou SK, Halazonetis TD. The role of SMARCAL1 in replication fork stability and telomere maintenance. DNA Repair. 2017;56:129–134. doi: 10.1016/j.dnarep.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Vasquez KM. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair. 2014;19:143–51. doi: 10.1016/j.dnarep.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roumelioti FM, et al. Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep. 2016;17:1731–1737. doi: 10.15252/embr.201643169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasero P, Vindigni A. Nucleases Acting at Stalled Forks: How to Reboot the Replication Program with a Few Shortcuts. Annu Rev Genet. 2017;51:477–499. doi: 10.1146/annurev-genet-120116-024745. [DOI] [PubMed] [Google Scholar]
- 53.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 54.de Graaff E, et al. Hotspot for deletions in the CGG repeat region of FMR1 in fragile X patients. Hum Mol Genet. 1995;4:45–49. doi: 10.1093/hmg/4.1.45. [DOI] [PubMed] [Google Scholar]
- 55.Edamura NK, Pearson CE. DNA methylation and replication: implications for the “deletion hotspot” region of FMR1. Hum Genet. 2005;118:301–304. doi: 10.1007/s00439-005-0037-5. [DOI] [PubMed] [Google Scholar]
- 56.Hagerman RJ, Hagerman PJ. The fragile X premutation: into the phenotypic fold. Curr Opin Genet Dev. 2002;12:278–283. doi: 10.1016/s0959-437x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- 57.Sherman SL. Premature ovarian failure among fragile X premutation carriers: parent-of-origin effect? Am J Hum Genet. 2000;67:11–13. doi: 10.1086/302985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Man L, Lekovich J, Rosenwaks Z, Gerhardt J. Fragile X-Associated Diminished Ovarian Reserve and Primary Ovarian Insufficiency from Molecular Mechanisms to Clinical Manifestations. Front Mol Neurosci. 2017;10:e290. doi: 10.3389/fnmol.2017.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagerman R, Au J, Hagerman P. FMR1 premutation and full mutation molecular mechanisms related to autism. J Neurodev Dis. 2011;3:211–224. doi: 10.1007/s11689-011-9084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.