Abstract
BACKGROUND
The Receptor for Advanced Glycation End Products (RAGE) is linked to cellular stress and inflammation during Alzheimer’s disease (AD). RAGE signals through Diaphanous-1 (DIAPH1), however the expression of DIAPH1 in the healthy and AD human brain is yet to be methodically addressed.
OBJECTIVE
To delineate the cell- and disease-state specific expression of DIAPH1 in the human medial temporal cortex during healthy aging and AD.
METHODS
We used semi-quantitative immunohistochemistry in the human medial temporal cortex paired with widefield and confocal microscopy and automated analyses to determine colocalization and relative expression of DIAPH1 with key cell markers and molecules in the brains of subjects with AD vs. age-matched controls.
RESULTS
We report robust colocalization of DIAPH1 with myeloid cells and increased expression during AD, which strongly correlated to increased neutral lipids and morphology of inflamed myeloid cells. DIAPH1 moderately colocalized with markers of endothelial cells, astrocytes, neurons, and oligodendrocytes.
DISCUSSION
Our findings localize DIAPH1 particularly to myeloid cells in the CNS, especially in AD in the locations of lipid droplet accumulation, thereby implicating RAGE-DIAPH1 signaling in dysregulated lipid metabolism and morphological changes of inflamed myeloid cells in this disorder.
Keywords: Alzheimer’s disease, RAGE, DIAPH1, Myeloid Cells, Microglia, Lipids, Inflammation
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder that currently has no cure and impacts millions of people worldwide. While the primary risk factor for AD is advanced age, recent innovations in genomic technology have implicated inflammatory lipid signaling in microglia, the myeloid cells of the Central Nervous System (CNS), as a possible contributing force to cognitive decline in this disorder [1–3]. However, despite these observations, the specific mechanisms by which myeloid cells become inflamed and putatively exacerbate neurotoxicity within the AD brain are unknown. Thus, it is critically important to develop our understanding of the molecules that govern the impact of myeloid cells on neuronal health and degeneration.
Mounting evidence suggests a potential role for the Receptor for Advanced Glycation Endproducts (RAGE) in regulating inflammation, oxidative stress, and neuronal stress during AD [4–8]. RAGE is an immunoglobulin-type, transmembrane receptor that is expressed on numerous cell types in the CNS and periphery, such as neurons, astrocytes, microglia, infiltrating myeloid cells, and endothelial cells (ECs). RAGE binds a diverse and unique repertoire of ligands, including nonenzymatically glycated proteins and lipids, S100/calgranulins, oligomeric amyloid aggregates such as pathological forms of Aβ and the pre-fibrillar toxic intermediates of islet amyloid polypeptide (IAPP) [9], high mobility group box 1 (HMGB1), and phosphatidylserine (PS). Extensive evidence has shown an increased burden of RAGE ligands in the AD brain, particularly AGEs, including glycated tau, and oligomeric Aβ. In addition, it is known that endothelial RAGE facilitates the transport of Aβ across the blood brain barrier (BBB), implicating RAGE and its ligand pools as putative drivers of disease progression [10, 11]. Furthermore, a recent structural analysis utilizing MRI technology revealed that region-specific atrophy of the right hippocampus substructure CA1 was significantly correlated to the AGER rs2070600 single nucleotide polymorphism (SNP) variant [12]. This variant has been previously associated with increased affinity to ligands and decreased circulating soluble RAGE (sRAGE). sRAGE, an isoform that possesses only the RAGE extracellular domains, is believed to function as a “decoy” receptor, which is predicted to be protective against inflammation and cellular stress by sequestering RAGE ligands and preventing their engagement of the full-length, transmembrane receptor. Thus, lowering of sRAGE may be accompanied by an increased ligand burden, subsequently driving increased RAGE-dependent cell signaling cascades that exacerbate cellular stress, impair lipid and cholesterol handling, and generate reactive oxygen species (ROS) production; processes that together may augur neuronal stress and death [13].
Our laboratory recently discovered that ligand binding to the extracellular domains of RAGE promotes binding of its cytoplasmic domain to the intracellular formin molecule, Diaphanous 1 (DIAPH1) [14]. This engagement of DIAPH1 is also required for RAGE-dependent signaling cascades that generate ROS, induce the upregulation of inflammatory cytokines, and, in myeloid cells, drive the downregulation of ATP binding cassette (ABC) cholesterol transporters, such as ABCA1 and ABCG1 [15, 16]. Besides its role in RAGE-DIAPH1-mediated inflammation, DIAPH1 is also important for actin cytoskeleton stability and rearrangement, and regulation of Rho-GTPase signaling and transcription factors [17, 18]. A recent study of human brain tumor cases suggested that DIAPH1 was expressed in human gliomas; however, the specific cell types expressing DIAPH1 in the brain in the absence of neoplasia were not elucidated [19]. Hence, very little is known about DIAPH1 expression pattern and functions within the CNS in the normal brain or in aging and dementia. Here, we aim to provide the first detailed analysis of DIAPH1 expression in distinct cell types in the human medial temporal cortex of aged non-demented (ND) control vs. demented AD human subjects.
Materials and Methods
2.1 Human Material
Human brain tissue (N=10/group) was obtained from the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. Human medial temporal cortex tissue was harvested 2–4 hours post-mortem and immediately fixed in paraffin. 40 μm thick coronal slices were preserved at −80°C in a 20% sucrose solution prior to immunohistochemistry. Non-demented age-controlled cases (ND) were defined through pathological analysis, indicating an absence of amyloid plaque and intraneuronal tau tangles expressed by a Braak and Braak score of ≤III and clinically, by a Mini Mental State Exam Score of ≥ 25. AD cases were defined by a Braak and Braak score of ≥V and Mini Mental State Exam Score of <25. Due to the known phenomena of increased AGEs in smokers, subjects were excluded if nicotine use was reported [20]. The study was approved by the Institutional Review Board for Sun Health Research Institute, with an exemption obtained by the New York University Institutional Review Board to use archived de-identified pathology specimens for research. Written informed consent was obtained from each subject by the Sun Health Research Institute prior to collection.
2.2 Mice
Wild-type (C57BL/6J) male mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and studies performed according to approved protocols of the Institutional Animal Care and Use Committee at New York University. Diaph1-deficient mice in the C57BL/6 background were generated as previously described[17]. All mice were humanely euthanized with ketamine and xylazine at age 3–6 months and brains were immediately harvested, washed in Dulbecco’s Phosphate-Buffered Saline (dPBS) solution, and fixed for 24 h in 4% paraformaldehyde and sectioned to 40 μm thickness for immunohistochemistry experiments.
2.3 Immunohistochemistry and Imaging
Indirect fluorescence immunohistochemistry was performed on 40 μm-thick coronal human temporal cortex and murine temporal cortex slices to detect colocalization of RAGE and DIAPH1 within various brain cell types including: microglia, astrocytes, endothelial cells (ECs), oligodendrocytes, and neurons (Supplementary Table 1). Free floating sections were washed 6 times with dPBS + 0.5% Tween and permeabilized for 24 h. Sections were blocked with Odyssey Blocking Buffer (Li-cor 927-4000) for 1 h, and all primary antibodies were diluted in blocking buffer and secondary antibodies were diluted in 1X dPBS. Primary antibodies were incubated at 4°C for 24 h, washed three times in dPBS, and secondary antibodies were incubated for 2 h at room temperature. Thioflavin S staining was conducted as previously described [21]. DIAPH1 antibody specificity was quantitatively validated on Diaph1-deficient mice and secondary antibody only negative controls were performed for all costaining combinations (Supplementary Figure 1).
Multicolor wide-field micrographs were taken using the Nikon Eclipse NI-U upright microscope at 10x magnification and confocal photomicrographs were taken using the Zeiss 710 Confocal Laser Scanning microscope, with a 63x objective, scanning speed of 6 (6.3 μs dwell time / pixel) with averaging over 8 pictures, and all quantification analyses were applied to 2D micrographs, except for surface area to volume (SA:V) ratios of myeloid cells in which 20 μm Z-stacks were utilized. All images for each staining condition were generated by the matching microscope and laser settings, on the same day, with the experimenter naive to the tissue condition.
To determine the marker’s (M) and DIAPH1 (D) pixels of interest (M-POI), binary masks (segmentation) were generated by implementing a thresholding algorithm [22] where an unbiased python algorithm selected all pixels where the intensity values are greater than the threshold (th)[22].
We employed both intensity based and object based methods of analyzing DIAPH1 colocalization with (M-POI). We measured Cell Marker Area by the formula (M-POI / Field of View (FOV)). Overlap Area was used to determine the pixel x pixel density of co-expression of DIAPH1 Pixels above threshold (D-POI) and M-POI. We determined the expression of the proteins of interests in a semi-quantitative capacity by determining the Mean Intensity of DIAPH1 within M-POI. For SA:V ratio measurements, 20 μm z-stacks were subjected to the ImageJ 3D objects plugin and SA:V was calculated by dividing volume from surface area.
2.4 Statistics
Sample power calculations were performed by Power Analysis and Sample Size Software. To achieve 82% power to measure mean differences of at least 2-fold, with a standard deviation (SD) of 1.48, a minimum sample size of 8 subjects per group was required. We studied N=10 subjects per group. For all statistical analyses, data were collected via Python pipelines and organized in Graphpad Prism. Mann-Whitney-Wilcoxon test was used to assess if the two groups, AD and non-demented control (ND), differed in each of the 25 variables. The non-parametric method was used because the normality assumption of t-test may not be validated due to the small sample size. Histograms illustrating a dot for each patient display the mean +/− SD. For correlational analysis of Lipidtox and DIAPH1 area and Surface Area to Volume Ratios with DIAPH1 area, and for correlational analysis of DIAPH1 and Lipidtox area with 0 vs. 1 or 2 APOE4 alleles, a Pearson’s correlation was used. P value and estimated coefficients were reported to indicate significance and if the data were positively or negatively associated. Data were displayed with the line of best fit. All acquired data points were used in the analyses.
Results
3.1 Demographic Information for Subjects
We applied immunofluorescence methods to co-stain AD and ND brain sections from the medial temporal lobe of deceased human subjects. All subjects diagnosed with AD (mean age 82.3 +/− 6.5 years; see Table 1) had a Braak and Braak score of VI and clinical dementia, as determined by Mini Mental State Exam (MMSE) score (mean 9.6 +/− 9.87). Age-matched ND human subjects (mean age 81.6 +/− 10.3 years; see Table 1) had 0 or 1 alleles of APOE4, a Braak and Braak score ≤ III and excellent MMSE scores (mean 29.4 +/− .89), demonstrating normal, healthy characteristics of aging. All subjects had a post-mortem interval of 2–4 hours.
Table 1.
Demographic Information for the cases selected
| Non-Demented Control | Alzheimer’s disease | |
|---|---|---|
| Number of cases | 10 | 10 |
| Female : Male | 7:3 | 7:3 |
| Age at expiration (yrs) | 81.6 +/− 10.3 | 82.3 +/− 6.5 |
| Post-mortem interval (h) | 3.3 +/− 0.7 | 3.14 +/− 0.7 |
| APOE4 Frequency (0/1/2) | 8/2/0 | 3/5/2 |
| Braak Score (I/II/III/IV/V/VI) | 5/1/4/0/0/0 | 0/0/0/0/0/10 |
| MMSE | 29.4 +/− 0.89 | 9.6 +/− 9.87 |
| MMSE Interval to death (mo) | 27 +/− 29 | 17.5 +/− 10.7 |
| Cases with Diabetes | 5 | 2 |
3.2 Detection of DIAPH1 in Human Medial Temporal Cortex
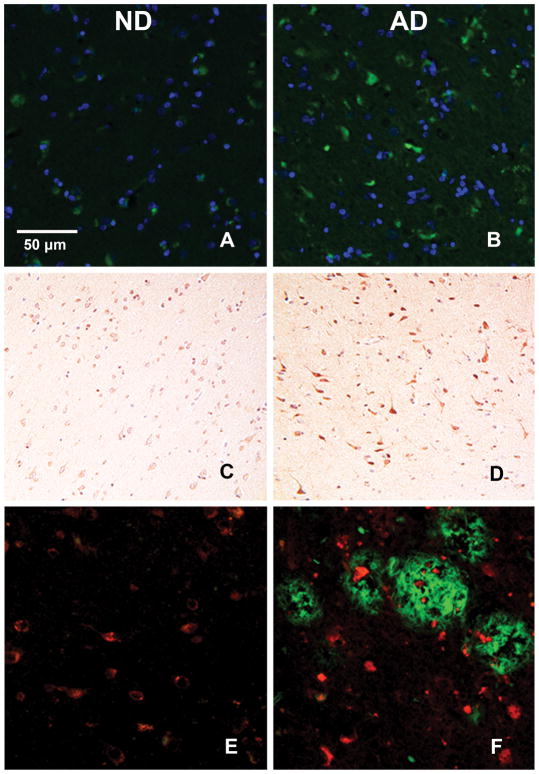
We first aimed to detect the presence or absence of DIAPH1 in the gray matter of human medial temporal cortex tissue. We conducted fluorescence immunohistochemistry and generated widefield epifluorescent micrograph images of ND and AD tissue (Figure 1A–B), where we first observed the abundance of DIAPH1 expression in the human brain and a qualitative increase in DIAPH1+ staining in the AD brain. In order to ensure the findings were not an artifact and to gain better insight into where DIAPH1 is expressed within the brain parenchyma, we conducted chromogenic immunohistochemistry for DIAPH1 and continued to observe a qualitative increase (Figure 1C–D); however, there was no apparent overlap with Thioflavin S (ThioS) positive plaques in AD (Figure 1E–F).
Figure 1. Increased DIAPH1 Expression in AD vs. ND human brain.
All panels contain representative human temporal cortex 4 μm thick slices stained as labeled and imaged at 20x by widefield microscopy. (A) and (B): DIAPH1 (green) and DAPI (blue) show a qualitative increase in immunofluorescence expression of DIAPH1 in the AD brain (B) when compared to aged matched control ND brain (A). (C) and (D): DIAPH1 (DAB) costained with Hematoxylin confirm staining patterns of DIAPH1 and relative qualitative increase in AD brain (D) vs. ND Control brain (C) under chromogenic immunohistochemistry conditions. (E) and (F): DIAPH1 expression (red) does not colocalize with ThioS+ plaques (green). ThioS+ staining is increased in AD brain (F) compared to ND brain (E). There are high intensity DIAPH1+ puncta near to and far from the ThioS+ plaques (F).
3.3 Quantitative Analysis Cell- and Disease State-specific DIAPH1 Expression
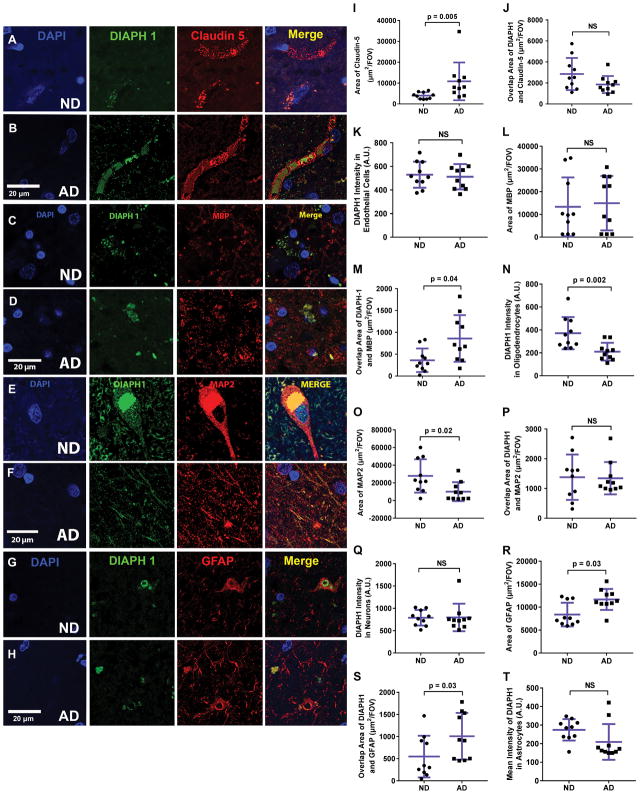
Thus, we continued this in depth analysis of DIAPH1 expression through a more quantitative and detailed approach in order to identify the distinct cell types that express DIAPH1, and to determine the relative expression level of DIAPH1 in each cell type in ND and AD brains. In these experiments, we used well-characterized antibodies to co-stain brain sections for DIAPH1 along with specific markers of the most prominent cell types in the brain: ECs, oligodendrocytes, neurons, astrocytes (Figure 2), and myeloid cells (Figures 3). To analyze these immunofluorescence images, we developed and employed a python computational pipeline for image quantification that utilizes the field standards of the ImageJ software to determine 1) Cell Marker Area, 2) Overlap Area, and 3) DIAPH1 Intensity, a semiquantitative assessment of relative DIAPH1 expression within the Overlap Area.
Figure 2. DIAPH1 Expression in Endothelial Cells, Oligodendrocytes, Neurons, and Astrocytes in the ND and AD human brain.
All panels contain representative human temporal cortex 40 μm thick slices stained as labeled and imaged at 63x by 2D and 3D confocal microscopy. (A) and (B): DIAPH1 (green) and Claudin5 (red) with DAPI (blue) immunohistochemistry to analyze DIAPH1 in ECs. (C) and (D): DIAPH1 (green) and MBP (red) with DAPI (blue) immunohistochemistry to analyze DIAPH1 in oligodendrocytes. (E) and (F): DIAPH1 (green) and MAP2 (red) with DAPI (blue) immunohistochemistry to analyze DIAPH1 in neurons. (G) and (H): DIAPH1 (green) and GFAP (red) with DAPI (blue) immunohistochemistry to analyze DIAPH1 in astrocytes. (I–T): Quantification of representative images of immunohistochemistry staining. (I): Area of Claudin5 is significantly increased in AD brain vs. ND brain. (J): Overlap Area of Claudin5 and DIAPH1 is high in both ND and AD conditions, but is unchanged between ND and AD brain. (K): DIAPH1 intensity in areas of overlap between DIAPH1 and Claudin5 is unchanged between ND and AD conditions. (L): Area of MBP is not different between ND and AD brain. (M): Overlap Area of MBP and DIAPH1 is increased in AD brain vs. ND brain. (N): DIAPH1 intensity in areas of overlap between DIAPH1 and MBP is reduced in AD brain vs. ND brain. (O) MAP2 Area is significantly decreased in AD brain vs. ND brain. (P): Overlap Area and (Q): Overlap Mean of DIAPH1 and MAP2 are unchanged in AD and ND conditions. (R): GFAP Area is significantly increased in AD brain vs. ND brains. (S): Overlap Area of GFAP and DIAPH1 is low, however, it is significantly increased in the AD brain vs. ND brain. DIAPH1 intensity is unchanged in areas of overlap between DIAPH1 and GFAP when comparing AD and ND brains. Data represented by mean +/− SD of n=10 cases per group (4–8 images analyzed per case). Mann-Whitney-Wilcoxon non-parametric tests with p values indicated. Scale bars = 20 μm for all representative images.
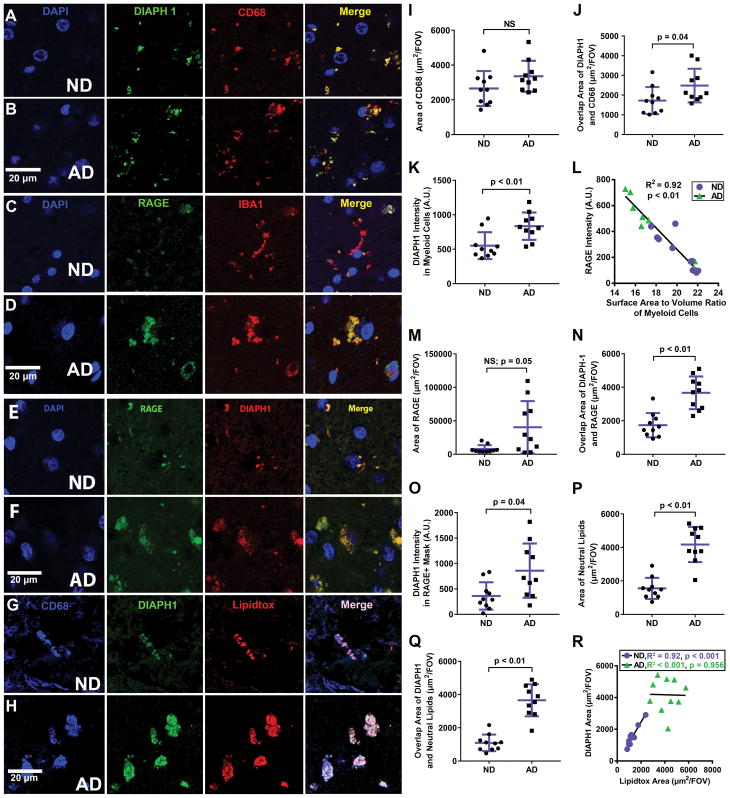
Figure 3. DIAPH1 and RAGE are Highly Expressed in Myeloid Cells in the Brain with Disease-State Specific Patterns.
All panels contain representative human temporal cortex 40 μm thick slices stained as labeled and imaged at 63x by 2D and 3D confocal microscopy. (A) and (B): DIAPH1 (green) and CD68 (red) with DAPI (blue) immunohistochemistry to analyze DIAPH1 in myeloid Cells. (C) and (D): RAGE (green) and IBA1 (red) with DAPI (blue) immunohistochemistry to analyze RAGE in myeloid cells. (E) and (F): DIAPH1 (green) and RAGE (red) with DAPI (blue) immunohistochemistry to illustrate high degree of overlap between the two molecules. (G) and (H): DIAPH1 (green) and Lipidtox (red) with DAPI (blue) immunohistochemistry to illuminate increased lipid burden in AD vs. ND brain and high degree of colocalization of DIAPH1 and neutral lipids in the AD and ND brains. (I–T): Quantification of representative images of immunohistochemistry staining. (I): Quantification of CD68 area shows a non-significant trend for increased CD68+ area in AD brain vs. ND brain. (J): Overlap Area of CD68 and DIAPH1 is high in both ND and AD conditions, and increased even further in AD brain vs. ND brain. (K): DIAPH1 intensity in areas of overlap between DIAPH1 and CD68 is significantly higher in AD vs. ND conditions. (L): Quantification of SA:V ratio in 3D Z-stacks displayed as 2D renderings in (E) and (F) reveals a strong, negative correlation between SA:V ratio and RAGE intensity in DIAPH1+ cells; purple circles = ND and green triangles = AD. (M): Quantification of RAGE area reveals a trend towards increased RAGE Area in AD brain vs. ND brain. (N): Overlap Area of RAGE and DIAPH1 is high in both ND and AD conditions, and increased even further in AD brain vs. ND brain (O): DIAPH1 intensity in areas of overlap between DIAPH1 and RAGE is significantly increased in AD brain vs. ND brain. (P): Area of Neutral Lipids is significantly decreased in AD brain vs. ND brain. (Q) Overlap Area of DIAPH1 and Neutral Lipids is increased in AD vs. ND conditions. (R): Linear Regression Analyses indicates a strong, positive relationship between DIAPH1 area and Lipidtox Area in the human brain for ND samples (purple circles; R2 =0.917, p < 0.001) and no correlation between DIAPH1 area and Neutral Lipid Area for AD samples (green triangles; R2 < .001, p = 0.956). In addition, AD cases have significantly higher Neutral Lipid and DIAPH1 content. Data represented by mean +/− SD of n=10 cases per group (4–8 images analyzed per case) for histograms. Mann-Whitney-Wilcoxon non-parametric tests with p values indicated. Scale bars = 20 μm for all representative images. Linear Regression analysis is displayed with one dot per case and a line of best fit.
Cell-specific DIAPH1 expression was low to moderate in ECs, oligodendrocytes, neurons and astrocytes (Figure 2A–H). Within the AD brain, the EC marker Claudin-5 was significantly higher than ND brain (p = 0.005) (Figure 2I). However, there was no concomitant change in Overlap Area or relative DIAPH1 Intensity within ECs (Figure 2J–K), indicating that while ECs are positive for DIAPH1, the changes observed in ECs within the AD brain are not associated with any other shift in DIAPH1 coexpression. Oligodendrocytes were marked by expression of myelin basic protein (MBP) and there were no changes observed for total MBP expression within the contexts of healthy aging and AD (Figure 2L). However, DIAPH1 expression was altered in the oligodendrocyte myelin, as observed by a relative increase in DIAPH1 and MBP Overlap Area (p = 0.04), but a decrease in relative DIAPH1 Intensity in MBP+ regions of the AD brain (p = 0.002). Taken together, these data indicate that DIAPH1 exhibits a more diffuse expression in oligodendrocytes (expanded area, low intensity) in the AD brain, as compared to oligodendrocyte DIAPH1 expression patterns (small punctate, high intensity) in the ND brain (Figure 2L–N). We selected microtubule-associated protein 2 (MAP2) as a marker for the perikarya and dendrites of neurons, and observed a relative decrease of MAP2 in the AD brain (p = 0.02) (Figure 2O). However, despite this change in cell marker expression in neurons, we did not see any associated shift in Overlap Area or DIAPH1 Intensity (Figure 2P–Q). In order to assess reactive astrocytes in the ND and AD brains, we utilized glial fibrillary acidic protein (GFAP). As previously reported, an increase in GFAP+ area was observed in the AD brain (p = 0.03) (Figure 2R). Interestingly, as the area of GFAP positivity increased, so did DIAPH1+ GFAP+ Overlap Area, indicating a concomitant increase in DIAPH1 expression, as astrocytic GFAP was upregulated in the AD brain (p = 0.03) (Figure 2S). However, there was no shift in DIAPH1 intensity within astrocytes; it remained low in the AD brain compared to other cell types (Figure 2T). We assessed pericyte colocalization with DIAPH1 through utilizing the α-Smooth Muscle Actin (α-SMA) antibody, but saw no colocalization in ND or AD (data not shown).
We next compared DIAPH1 and RAGE expression in the CD68+ myeloid cells of the ND and AD Medial Temporal Cortex (Figure 3A–H). While there was no significant difference in CD68+ areas between ND and AD subjects, a statistical trend (p = 0.05) towards increased CD68+ area was observed in AD brains (Figure 3I). This finding is consistent, given the ages of subjects within our study, with previous studies reporting microgliosis in the AD brain, which described peak levels of microglia-derived inflammatory markers early in the disease, which later wane as AD progresses. However, our results indicate a significant increase in Overlap Area between DIAPH1 and CD68 (p = 0.04), as well as a significant increase in DIAPH1 Intensity within CD68+ cells (p < 0.01) in the AD brain relative to the ND control brain (Figure 3J–K). This indicates that while the cell size and number of CD68+ myeloid cells in the brains were not significantly different, the DIAPH1 expression patterns within myeloid cells are significantly increased in AD.
3.3 DIAPH1 is co-expressed with RAGE and has strong correlations to lipid accumulation and morphology changes in myeloid cells during AD
Our laboratory has already published chromogenic immunohistochemistry findings that display RAGE expression in myeloid cells within the ND and AD brains and that indicate RAGE is increased in the context of AD, as evidenced by quantitative ELISA assays [8]. These data are replicated and further elucidated by this study. Not only did AD brains display a higher total area of RAGE expression when compared to ND brains, but there was also an increased extent of colocalization between RAGE and IBA1 within AD brains (Figure 3C–D). When SA:V assessments were applied to the 3D micrographs of IBA1 and RAGE expression, we determined a strong, inverse relationship between average RAGE intensity and the SA:V of myeloid cells within the brain (R2 = −0.092, p < 0.01). Specifically, the myeloid cells bearing the highest RAGE intensity display a more amoeboid morphology and lower SA:V, whereas the myeloid cells with the lowest RAGE intensity have higher SA:V ratios, indicative of a more homeostatic, ramified morphology (Figure 3L). In addition, the Overlap Area and relative DIAPH1 Intensity in areas of RAGE positivity are significantly increased in the AD brain (p < 0.01 and p = 0.04, respectively), indicating increased colocalization of these two signaling molecules within the AD brain (Figure 3N–O).
Having identified that myeloid cells are a primary source of DIAPH1 expression in both ND and AD human medial temporal cortex, and that the expression of DIAPH1 significantly increases in CD68+ myeloid cells in AD, we next set out to determine whether DIAPH1 colocalized with other key molecules in myeloid cells in AD brains. Substantial evidence indicates that besides proteins, lipids and cholesterol molecules also contribute to loss of homeostatic functions in myeloid cells during AD. Lipids regulate the trafficking and proteolytic activity of membrane bound proteins in microglia [23]. Several pathological features of AD include perturbed CNS membranes, lipid modulation of pathogenic Aβ production and degradation, and genetic variation of a number of lipid transport and signaling molecules, including TREM2, APOE4, and ABC transporters[24].
Based on previous connections between AD and lipid/cholesterol dysfunction, we set out to determine whether there was a link between myeloid DIAPH1 expression and lipid content. Thus, we triple-stained brain sections for CD68, DIAPH1, and the neutral lipid dye, Lipidtox™ (Figure 3G–H). Our results show a highly significant increase in total neutral lipid area in AD brains compared to ND controls (p < 0.01) (Figure 3P). We identified a significant increase in Overlap Area between DIAPH1 and Lipidtox™ in the AD brain (Figure 3Q) and a significant linear relationship between Lipidtox™ area and DIAPH1 area in the ND brain (R2 = 0.917, p < 0.001), but no significant relationship between Lipidtox™ area and DIAPH1 area in the AD brain R2 < 0.001, p = 0.956) (Figure 3R). Importantly, both DIAPH1 and Lipidtox™ values were significantly increased in the AD brain when compared to the ND brain, however, the linear relationship between these two molecules in the context of progressive AD was lost. Collectively, these data suggest that myeloid cells are the primary cell types storing neutral lipid in the brain, and that there is a significant increase in lipid accumulation in AD brains that displays a strong, positive correlation with increased myeloid DIAPH1 expression. In conjunction with the strong colocalization of DIAPH1, RAGE, and CD68, we can extrapolate these findings to infer that the myeloid cells within the brain that have the highest lipid content, also possess the highest DIAPH1 and RAGE expression, but the lowest SA:V ratio, and thus the most amoeboid and inflamed morphology.
Finally, we performed correlational analyses to determine if there were any associations between the area of DIAPH1 or Lipidtox™ positivity and the presence of 0 versus 1 or 2 APOE4 alleles in the subjects. With respect to DIAPH1, we found a modest, but significant linear relationship between DIAPH1 intensity and APOE4 alleles (R2 = .213, p =.04). However, no significant relationship was observed between Lipidtox™ area and APO4 alleles (R2 = .188, p = .054).
1. Discussion
Collectively, these data provide the first quantitative evidence that DIAPH1 is expressed in the human medial temporal cortex and that its expression is increased in human AD, particularly in myeloid cells, as visualized by immunohistochemistry and microscopy. We determined cell-specific expression of DIAPH1 by quantifying colocalization with key CNS cell markers and molecules. The markers selected were chosen due to high degrees of specificity to a single cell type as opposed to other options that may be preferred due to illuminating the entire cell body, but may cross react to multiple cell populations. Thus, we optimized for diminishing Type 1 errors to the greatest extent possible. While neurons, ECs, oligodendrocytes, and astrocytes all express moderate amounts of DIAPH1, we discovered a robust expression of DIAPH1 in myeloid cells, that increased in the AD brain, which strongly correlated to lipid accumulation within these cells, in parallel with a concomitant decreases in ramified processes.
Previous studies have generated ample evidence to suggest a strong convergence between dysregulated lipid handling molecules in microglia and myeloid cells to an increased neuroinflammatory signature of the brain and robust increases in AD risk. First, genetic evidence from humans suggests a powerful link between inflammatory pathways (including complement and chemokines) and key lipid / cholesterol molecules (TREM2, ABCA7, APOE4, and beyond) with AD susceptibility [2, 3, 12, 25]. Second, subsequent analyses of mutations within animal models has illuminated major players from the innate immune system that increase or decrease AD progression, such as CCR2, CX3CR1, complement components, and CXCL8 and the previously mentioned lipid handlers [26–28]. In addition, burgeoning data in humans and mouse models indicate that systemic inflammation and transient infections can produce permanent worsening of ongoing neurodegenerative disease [29–31]. Finally, in vitro assays of microglia have shown that cholesterol content can alter and interfere with homeostatic functions [32]. However, despite these important findings, causal links of molecular mechanisms in myeloid cells to AD induction and progression remain largely inconclusive, and the specific myeloid cell signaling processes underlying cognitive decline remain elusive. Our data underscore the complexity of the lipid-inflammatory signaling nexus in healthy aging and AD, and how this corresponds to APOE4 variant expression. We report for the first time a modest, yet significant correlation between the area of DIAPH1 staining with the number of APOE4 alleles, however, no such significant association between LipidTox™ Area and APOE4 alleles was identified, indicating DIAPH1 positivity may be a more meaningful readout for a biomarker of disease state and AD risk than neutral lipid content directly. However, there is a strong, significant relationship between DIAPH1 and LipidTox™ Area in the ND brain, which implicates DIAPH1 upregulation as a candidate molecule to mediate aberrant lipid accumulation in healthy and prodromal states before full-blown AD presents, which may indicate a unique state of dysfunction in which both DIAPH1 and neutral lipids are high, but no correlation exists.
Decades of research have been dedicated to uncovering the key neuropathological characteristics of the AD brain, including the aggregation and accumulation of intracellular and extracellular proteins, particularly Aβ and hyperphosphorylated tau, which are believed to be detrimental to neuronal health and function. During this time, ample research illuminated that CNS myeloid cells display a more amoeboid, round shape in the settings of acute and chronic inflammation and that lipid accumulation within macrophages in the periphery is a potent sign of cellular stress, metabolic syndromes, and systemic pathology [16, 25]. From the foam cells during progressive atherosclerosis, to the lipid-laden adipose tissue macrophages during obesity, evidence from multiple organ niches have enhanced our understanding of the complex interface between immune regulation, bioenergetic phenotype, lipid accumulation, and cellular (dys)function. However, in the context of myeloid cells during AD, we have a relatively poor understanding how microglia and peripherally infiltrating myeloid cells may mediate cognitive decline, whether the differences observed in AD are due to a loss- or gain of critical functions, and which molecules are mediating molecular shifts in the context of non-inherited AD pathogenesis [33].
Importantly, when collating the literature, there are numerous connections between the pathophysiological phenotypes in AD and the RAGE-DIAPH1 signaling pathway, and further parsing may provide a meaningful approach to intervention strategies in AD. First, substantial evidence suggests that oligomeric forms of Aβ bind the variable domain of extracellular RAGE. It is known that RAGE is expressed in CNS myeloid cells, RAGE expression and ligand concentrations are increased in human and mouse model brains during AD, and overexpressing RAGE or expressing dominant negative RAGE (DN-RAGE) confers a detriment and benefit, respectively, to Aβ-dependent cognitive decline, plaque deposition, and mitochondrial-dependent cellular stress, strongly suggesting an exacerbating role of RAGE signaling in the context of cognitive decline [6, 8, 34, 35]. In addition, in conditions of systemic inflammation, such as sepsis models, RAGE and its ligands (Aβ, hyperphosphorylated tau, and AGEs) are elevated in rodent brains, and positively correlated to cognitive impairments[31, 36]. Based on our current findings, we postulate that studies examining potential roles for microglial DIAPH1 in the accumulation of Aβ, inflammatory lipids, and the development of cognitive decline in rodent models is warranted. These studies could unveil important mechanisms of disease that are unable to be tested in human post-mortem tissue.
Interestingly, RAGE ligand AGEs were also recently shown to regulate lipid accumulation in macrophages by regulating cholesterol uptake, esterification, and efflux [37]. However, all previous molecular studies of DIAPH1 and this formin molecule’s connections to RAGE and lipid regulation have been conducted in peripheral tissues. Thus, we have a limited understanding of how DIAPH1 may mediate these processes in the brain during health and disease.
We hypothesized that RAGE-DIAPH1 signaling potentiates neuroinflammation in the AD brain. However, before the consequences of RAGE-DIAPH1 signaling in the CNS could be investigated, we must first develop our understanding of where and when to look. Here, we provide the first salient evidence that DIAPH1 may potentially be involved in AD through unknown mechanisms regarding lipid handling in CNS myeloid cells, including microglia. Our data suggest pronounced DIAPH1 expression in CNS myeloid cells that is robustly increased in AD. Further, we corroborate our previous findings in peripheral smooth muscle cells, macrophages, and pancreatic β-cells to provide evidence here that RAGE and DIAPH1 display strong colocalization in the CNS. However, we also recognize that a limitation in extrapolating and generating more nuanced insights from the findings is low statistical power. Each case was specifically selected to minimize the post mortem interval as much as possible (<4h), because RAGE, DIAPH1, and RAGE ligands have previously been shown to robustly upregulate in the contexts of hypoxia and ischemia [38–42]. In future studies, with a more robust cohort size, more in-depth statistical analyses could prove to be potentially beneficial towards better understanding the mechanisms at play as well as specific patient subsets that may most or least benefit from RAGE-DIAPH1 pharmacological inhibition during neurodegeneration and if there is any sex effects of these findings. In addition, better understanding of how these data correlate to biomarkers from blood could also provide meaningful insight for treatment plans and the incorporation of a young cohort in order to understand RAGE-DIAPH1 changes during healthy aging could be instrumental as well.
In this study, our aim was to investigate how three seemingly disparate fields may integrate to provide evidence for understanding the contribution of RAGE and DIAPH1 signaling in neurodegeneration. While population-wide genomic studies had illuminated lipid and cholesterol dysregulation in the context of inflammatory neurodegeneration, these genetic variants and mutations only account for a small subset of the patient population impacted with AD and thus other mechanisms beyond those illuminated in GWAS must be at play. In fact, consistent with the complex nature of lipid- and inflammation-dependent factors, we were unable to observe a significant correlation between LipidTox™ area and the number of APOE4 alleles, albeit we acknowledge the limitations of the present study, in which we utilized a total of 10 AD subjects and 10 age-matched control individuals. Previous molecular biology studies of RAGE-DIAPH1 signaling driving cellular stress implicate RAGE and DIAPH1 in myeloid-cell dysfunction in inflammatory diseases, but this is the first detailed analysis of DIAPH1 in the central nervous system during aged health and AD. In addition, mouse models have implicated RAGE signaling abrogation through sRAGE or DN-RAGE to confer a benefit in AD-like mouse models, but studies addressing which specific cell types are contributing to this benefit, how, when, and to what extent aberrant lipid handling could be at the core of the dysfunction, have yet to be performed and reported. Here, we show that CNS myeloid cells that bear the highest RAGE and DIAPH1 expression concomitantly display the most amoeboid morphology and demonstrate a significantly increased lipid load and that all of these key endpoints are significantly altered in progressive AD. These data do not elucidate the specific mechanism(s) by which all of these processes are occurring, nor do they differentiate among correlation, causation, or time-specific consequences during AD progression. While there is no strong evidence to suggest that RAGE is an initiating factor in neurodegeneration, dementia, and AD, the data in mouse, human GWAS, and the present study all suggest there could be a role for RAGE-DIAPH1 signaling in exacerbating inflammatory phenotypes, particularly in myeloid cells, by contributing to lipid accumulation, aberrant signal transduction, and cellular dysfunction in the context of AD. These findings illustrate the importance of further investigation into the specific mechanisms of RAGE-DIAPH1 signaling and its impact on CNS myeloid cells during health and disease, specifically as a potential avenue of therapeutic intervention for halting or slowing neuroinflammation and lipid accumulation during AD.
Supplementary Material
All panels contain representative 4 μm thick slices of mouse (A) and (B) or 40 μm thick slices of human (C), (D), and (E): temporal cortex stained and imaged at 63x by 2D confocal microscopy. (A) and (B): DIAPH1 (green) with DAPI (blue) immunohistochemistry to analyze DIAPH1 in WT vs Diaph1-deficient mice. (C), (D), and (E): Secondary antibody only controls for all secondary combinations.
(A) Example of experimental image of GFAP, DIAPH1, and Merge staining. (B) Images of binary masks indicating the positive regions of staining (white) and negative/background regions of staining (black) as determined by automated thresholding techniques.
Supplementary Table 1. Antibodies used in staining.
Acknowledgments
The computational algorithm for this work is available at: https://github.com/FenyoLab/Confocal_Colocalization. We thank Dr. Richard Friedman (Columbia University) for his consultation on data analysis. We thank Ms. Latoya Woods for her expert assistance in the preparation of this manuscript. We are grateful to the Sun HealthResearch Institute Brain and Body Donation Program of Sun City, Arizona for the provision of human biological materials. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke [grant number: U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders], the National Institute on Aging [grant number: P30 AG19610 Arizona Alzheimer’s Disease Core Center], the Arizona Department of Health Services [contract 211002, Arizona Alzheimer’s Research Center], the Arizona Biomedical Research Commission [contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium] and the Michael J. Fox Foundation for Parkinson’s Research This work was supported by an NIH [grant numbers: 1F31AG054129-01 (NRSA for J.D.), T32CA009161 (Molecular Oncology and Immunology Training Grant for K.B.H.), and P32CA016087, S10OD01058 and S10OD018338 (NYU Experimental Pathology Histology and Immunohistochemistry Core Laboratory) and the Alzheimer’s Association [grant number: ZEN-17-440472] (Zenith Fellowship for AMS). This work was also supported in part by Research Funds, Diabetes Research Program, NYU Langone Medical Center (A.M.S.) and by the Laura and Isaac Perlmutter Cancer Center Support Grant. This work is inspired by and dedicated to C.L.S.
Abbreviations
- Aβ
Amyloid beta peptide
- ABC Transporters
Adenosine Binding Casette Transporters
- AD
Alzheimer’s disease
- APOE4
Apolipoprotein E variant 4
- BBB
Blood Brain Barrier
- CCR2
Chemokine Receptor 2 (Also known as cluster of differentiation 192)
- CD68
Cluster of Differentiation 68
- CX3CR1
CX3C Receptor 1 (Also known as G-protein coupled receptor 13)
- CNS
Central Nervous System
- DIAPH1
Diaphanous 1
- DN-RAGE
Dominant Negative RAGE
- ECs
Endothelial Cells
- ELISA
Enzyme Linked Immunoabsorbant Assay
- GFAP
Glial Fibrillary Acidic Protein
- HMGB1
High Mobility Group Box 1
- IAPP
islet amyloid polypeptide
- IBA1
Ionized Calcium-binding Adaptor Molecule 1
- MAP-2
Microtubule Associated Protein 2
- MBP
Myelin Basic Protein
- MMSE
Mini Mental State Exam
- ND
Non-demented
- POI
Pixel of Interest
- PS
Phosphatidyl Serine
- RAGE
Receptor for Advanced Glycation Endproducts
- ROS
Reactive Oxygen Species
- SA:V
Surface Area to Volume Ratio
- SNP
Single Nucleotide Polymorphism
- sRAGE
Soluable RAGE (RAGE lacking transmembrane domain)
- TREM2
Triggering Receptor Expressed on Myeloid Cells 2
Footnotes
Declarations of interest: none
Author contributions: J.D. designed the research, conducted experiments, analyzed data, consulted on algorithm creation, and wrote the manuscript. K.B.H. wrote the novel python code and contributed to writing the manuscript. M.R. generated all wide-field fluorescent and bright field images. M.H. was an independent observer for manual threshold selection. H.K. and H.L. analyzed data. A.A. assisted with experimental design and contributed to writing the manuscript. D.F. directed research. A.M.S. designed and directed research, analyzed data, and wrote the manuscript. The authors declare no competing financial interests.
References
- 1.Mao YF, Guo ZY, Pu JL, Chen YX, Zhang BR. Association of CD33 and MS4A cluster variants with Alzheimer’s disease in East Asian populations. Neurosci Lett. 2015;609:235–239. doi: 10.1016/j.neulet.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villegas-Llerena C, Phillips A, Garcia-Reitboeck P, Hardy J, Pocock JM. Microglial genes regulating neuroinflammation in the progression of Alzheimer’s disease. Curr Opin Neurobiol. 2015;36:74–81. doi: 10.1016/j.conb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Guglielmotto M, Aragno M, Tamagno E, Vercellinatto I, Visentin S, Medana C, Catalano MG, Smith MA, Perry G, Danni O, Boccuzzi G, Tabaton M. AGEs/RAGE complex upregulates BACE1 via NF-kappaB pathway activation. Neurobiol Aging. 2012;33:196.e113–127. doi: 10.1016/j.neurobiolaging.2010.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Hickman SE, Allison EK, Khoury JE. Microglial dysfunction and defective β-amyloid clearance pathways in aging Alzheimer’s disease mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang F, Lue LF, Yan S, Xu H, Luddy JS, Chen D, Walker DG, Stern DM, Yan S, Schmidt AM, Chen JX, Yan SS. RAGE-dependent signaling in microglia contributes to neuroinflammation, Abeta accumulation, and impaired learning/memory in a mouse model of Alzheimer’s disease. FASEB J. 2010;24:1043–1055. doi: 10.1096/fj.09-139634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giri R, Shen Y, Stins M, Du Yan S, Schmidt AM, Stern D, Kim KS, Zlokovic B, Kalra VK. beta-amyloid-induced migration of monocytes across human brain endothelial cells involves RAGE and PECAM-1. Am J Physiol Cell Physiol. 2000;279:C1772–1781. doi: 10.1152/ajpcell.2000.279.6.C1772. [DOI] [PubMed] [Google Scholar]
- 8.Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, Migheli A, Nawroth P, Stern D, Schmidt AM. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 9.Abedini A, Cao P, Plesner A, Zhang J, He M, Derk J, Patil SA, Rosario R, Lonier J, Song F, Koh H, Li H, Raleigh DP, Schmidt AM. RAGE binds preamyloid IAPP intermediates and mediates pancreatic β cell proteotoxicity. The Journal of Clinical Investigation. 2018;128:682–698. doi: 10.1172/JCI85210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi M, Yamagishi S. Possible involvement of advanced glycation end-products (AGEs) in the pathogenesis of Alzheimer’s disease. Curr Pharm Des. 2008;14:973–978. doi: 10.2174/138161208784139693. [DOI] [PubMed] [Google Scholar]
- 11.Deane R, Du Yan S, Submamaryan RK, LaRue B, Jovanovic S, Hogg E, Welch D, Manness L, Lin C, Yu J, Zhu H, Ghiso J, Frangione B, Stern A, Schmidt AM, Armstrong DL, Arnold B, Liliensiek B, Nawroth P, Hofman F, Kindy M, Stern D, Zlokovic B. RAGE mediates amyloid-β peptide transport across the blood-brain barrier and accumulation in brain. 2003;9:907. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z-X, Wan Y, Tan L, Liu J, Wang H-F, Sun F-R, Tan M-S, Tan C-C, Jiang T, Tan L, Yu J-T. Genetic Association of HLA Gene Variants with MRI Brain Structure in Alzheimer’s Disease. Molecular Neurobiology. 2017;54:3195–3204. doi: 10.1007/s12035-016-9889-z. [DOI] [PubMed] [Google Scholar]
- 13.Miller S, Henry AP, Hodge E, Kheirallah AK, Billington CK, Rimington TL, Bhaker SK, Obeidat Me, Melén E, Merid SK, Swan C, Gowland C, Nelson CP, Stewart CE, Bolton CE, Kilty I, Malarstig A, Parker SG, Moffatt MF, Wardlaw AJ, Hall IP, Sayers I. The Ser82 RAGE Variant Affects Lung Function and Serum RAGE in Smokers and sRAGE Production In Vitro. PLOS ONE. 2016;11:e0164041. doi: 10.1371/journal.pone.0164041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toure F, Fritz G, Li Q, Rai V, Daffu G, Zou YS, Rosario R, Ramasamy R, Alberts AS, Yan SF, Schmidt AM. Formin mDia1 mediates vascular remodeling via integration of oxidative and signal transduction pathways. Circ Res. 2012;110:1279–1293. doi: 10.1161/CIRCRESAHA.111.262519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson BI, Kalea AZ, Del Mar Arriero M, Harja E, Boulanger E, D’Agati V, Schmidt AM. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J Biol Chem. 2008;283:34457–34468. doi: 10.1074/jbc.M801465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daffu G, Shen X, Senatus L, Thiagarajan D, Abedini A, Hurtado Del Pozo C, Rosario R, Song F, Friedman RA, Ramasamy R, Schmidt AM. RAGE Suppresses ABCG1-Mediated Macrophage Cholesterol Efflux in Diabetes. Diabetes. 2015;64:4046–4060. doi: 10.2337/db15-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Zhang J, Mullin M, Dong B, Alberts AS, Siminovitch KA. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol. 2009;182:3837–3845. doi: 10.4049/jimmunol.0803838. [DOI] [PubMed] [Google Scholar]
- 18.Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Wang L, Chen J, Liang J, Xu Y, Li Z, Chen F, Du D. Knockdown of Diaph1 expression inhibits migration and decreases the expression of MMP2 and MMP9 in human glioma cells. Biomedicine & Pharmacotherapy. 2017;96:596–602. doi: 10.1016/j.biopha.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 20.Cerami C, Founds H, Nicholl I, Mitsuhashi T, Giordano D, Vanpatten S, Lee A, Al-Abed Y, Vlassara H, Bucala R, Cerami A. Tobacco smoke is a source of toxic reactive glycation products. Proceedings of the National Academy of Sciences. 1997;94:13915. doi: 10.1073/pnas.94.25.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajamohamedsait HB, Sigurdsson EM. Histological Staining of Amyloid and Pre-Amyloid Peptides and Proteins in Mouse Tissue. Methods in molecular biology (Clifton, NJ) 2012;849 doi: 10.1007/1978-1001-61779-61551-61770_61728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridler TW, CS Picture Thresholding Using an Iterative Selection Method. IEEE Transactions on Systems, Man, and Cybernetics. 1978;SMC-8:3. [Google Scholar]
- 23.Tansley GH, Burgess BL, Bryan MT, Su Y, Hirsch-Reinshagen V, Pearce J, Chan JY, Wilkinson A, Evans J, Naus KE, McIsaac S, Bromley K, Song W, Yang HC, Wang N, DeMattos RB, Wellington CL. The cholesterol transporter ABCG1 modulates the subcellular distribution and proteolytic processing of beta-amyloid precursor protein. J Lipid Res. 2007;48:1022–1034. doi: 10.1194/jlr.M600542-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Mandas A, Abete C, Putzu PF, la Colla P, Dessi S, Pani A. Changes in cholesterol metabolism-related gene expression in peripheral blood mononuclear cells from Alzheimer patients. Lipids Health Dis. 2012;11:39. doi: 10.1186/1476-511X-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerterp M, Murphy AJ, Wang M, Pagler TA, Vengrenyuk Y, Kappus MS, Gorman DJ, Nagareddy PR, Zhu X, Abramowicz S, Parks JS, Welch C, Fisher EA, Wang N, Yvan-Charvet L, Tall AR. Deficiency of ABCA1 and ABCG1 in Macrophages Increases Inflammation and Accelerates Atherosclerosis in Mice. Circulation research. 2013;112 doi: 10.1161/CIRCRESAHA.1113.301086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, Paul SM, Holtzman DM. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickman SE, El Khoury J. TREM2 and the neuroimmunology of Alzheimer’s disease. Biochem Pharmacol. 2014;88:495–498. doi: 10.1016/j.bcp.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T, Vitorica J, Ransohoff RM, Herrup K, Frautschy SA, Finsen B, Brown GC, Verkhratsky A, Yamanaka K, Koistinaho J, Latz E, Halle A, Petzold GC, Town T, Morgan D, Shinohara ML, Perry VH, Holmes C, Bazan NG, Brooks DJ, Hunot S, Joseph B, Deigendesch N, Garaschuk O, Boddeke E, Dinarello CA, Breitner JC, Cole GM, Golenbock DT, Kummer MP. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes C, Cunningham C, Zotova E, Woolford J, Dean C, Kerr S, Culliford D, Perry VH. Systemic inflammation and disease progression in Alzheimer disease. Neurology. 2009;73:768–774. doi: 10.1212/WNL.0b013e3181b6bb95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de la Monte SM. Type 3 diabetes is sporadic Alzheimers disease: Mini-review. Eur Neuropsychopharmacol. 2014;24:1954–1960. doi: 10.1016/j.euroneuro.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasparotto J, Girardi CS, Somensi N, Ribeiro CT, Moreira JCF, Michels M, Sonai B, Rocha M, Steckert AV, Barichello T, Quevedo J, Dal-Pizzol F, Gelain DP. Receptor for advanced glycation endproducts mediates sepsis-triggered amyloid-β accumulation, tau phosphorylation, and cognitive impairment. Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.M117.786756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CYD, Tse W, Smith JD, Landreth GE. Apolipoprotein E Promotes β-Amyloid Trafficking and Degradation by Modulating Microglial Cholesterol Levels. Journal of Biological Chemistry. 2012;287:2032–2044. doi: 10.1074/jbc.M111.295451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nature Neuroscience. 2011;14:1227. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- 34.Lue LF, Walker DG, Brachova L, Beach TG, Rogers J, Schmidt AM, Stern DM, Yan SD. Involvement of microglial receptor for advanced glycation endproducts (RAGE) in Alzheimer’s disease: identification of a cellular activation mechanism. Exp Neurol. 2001;171:29–45. doi: 10.1006/exnr.2001.7732. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki N, Fukatsu R, Tsuzuki K, Hayashi Y, Yoshida T, Fujii N, Koike T, Wakayama I, Yanagihara R, Garruto R, Amano N, Makita Z. Advanced Glycation End Products in Alzheimer’s Disease and Other Neurodegenerative Diseases. The American Journal of Pathology. 1998;153:1149–1155. doi: 10.1016/S0002-9440(10)65659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liliensiek B, Weigand MA, Bierhaus A, Nicklas W, Kasper M, Hofer S, Plachky J, Grone HJ, Kurschus FC, Schmidt AM, Yan SD, Martin E, Schleicher E, Stern DM, Hammerling GG, Nawroth PP, Arnold B. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J Clin Invest. 2004;113:1641–1650. doi: 10.1172/JCI18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L, Wang Y-R, Li P-C, Feng B. Advanced glycation end products increase lipids accumulation in macrophages through upregulation of receptor of advanced glycation end products: increasing uptake, esterification and decreasing efflux of cholesterol. Lipids in Health and Disease. 2016;15:161. doi: 10.1186/s12944-016-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Toure F, Qu W, Lin L, Song F, Shen X, Rosario R, Garcia J, Schmidt AM, Yan SF. Advanced glycation end product (AGE)-receptor for AGE (RAGE) signaling and up-regulation of Egr-1 in hypoxic macrophages. J Biol Chem. 2010;285:23233–23240. doi: 10.1074/jbc.M110.117457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liesz A, Dalpke A, Mracsko E, Antoine DJ, Roth S, Zhou W, Yang H, Na S-Y, Akhisaroglu M, Fleming T, Eigenbrod T, Nawroth PP, Tracey KJ, Veltkamp R. DAMP Signaling is a Key Pathway Inducing Immune Modulation after Brain Injury. The Journal of Neuroscience. 2015;35:583. doi: 10.1523/JNEUROSCI.2439-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bucciarelli LG, Ananthakrishnan R, Hwang YC, Kaneko M, Song F, Sell DR, Strauch C, Monnier VM, Yan SF, Schmidt AM, Ramasamy R. RAGE and modulation of ischemic injury in the diabetic myocardium. Diabetes. 2008;57:1941–1951. doi: 10.2337/db07-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greco R, Demartini C, Zanaboni AM, Blandini F, Amantea D, Tassorelli C. Modulation of cerebral RAGE expression following nitric oxide synthase inhibition in rats subjected to focal cerebral ischemia. European Journal of Pharmacology. 2017;800:16–22. doi: 10.1016/j.ejphar.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Origlia N, Criscuolo C, Arancio O, Yan SS, Domenici L. RAGE Inhibition in Microglia Prevents Ischemia-Dependent Synaptic Dysfunction in an Amyloid-Enriched Environment. The Journal of Neuroscience. 2014;34:8749. doi: 10.1523/JNEUROSCI.0141-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All panels contain representative 4 μm thick slices of mouse (A) and (B) or 40 μm thick slices of human (C), (D), and (E): temporal cortex stained and imaged at 63x by 2D confocal microscopy. (A) and (B): DIAPH1 (green) with DAPI (blue) immunohistochemistry to analyze DIAPH1 in WT vs Diaph1-deficient mice. (C), (D), and (E): Secondary antibody only controls for all secondary combinations.
(A) Example of experimental image of GFAP, DIAPH1, and Merge staining. (B) Images of binary masks indicating the positive regions of staining (white) and negative/background regions of staining (black) as determined by automated thresholding techniques.
Supplementary Table 1. Antibodies used in staining.