Abstract
Background
Decision makers rely on meta-analytic estimates to trade off benefits and harms. Publication bias impairs the validity and generalizability of such estimates. The performance of various statistical tests for publication bias has been largely compared using simulation studies and has not been systematically evaluated in empirical data.
Methods
This study compares seven commonly used publication bias tests (i.e., Begg’s rank test, trim-and-fill, Egger’s, Tang’s, Macaskill’s, Deeks’, and Peters’ regression tests) based on 28,655 meta-analyses available in the Cochrane Library.
Results
Egger’s regression test detected publication bias more frequently than other tests (15.7% in meta-analyses of binary outcomes and 13.5% in meta-analyses of non-binary outcomes). The proportion of statistically significant publication bias tests was greater for larger meta-analyses, especially for Begg’s rank test and the trim-and-fill method. The agreement among Tang’s, Macaskill’s, Deeks’, and Peters’ regression tests for binary outcomes was moderately strong (most κ’s were around 0.6). Tang’s and Deeks’ tests had fairly similar performance (κ > 0.9). The agreement among Begg’s rank test, the trim-and-fill method, and Egger’s regression test was weak or moderate (κ < 0.5).
Conclusions
Given the relatively low agreement between many publication bias tests, meta-analysts should not rely on a single test and may apply multiple tests with various assumptions. Non-statistical approaches to evaluating publication bias (e.g., searching clinical trials registries, records of drug approving agencies, and scientific conference proceedings) remain essential.
Electronic supplementary material
The online version of this article (10.1007/s11606-018-4425-7) contains supplementary material, which is available to authorized users.
KEY WORDS: Cochrane Library, funnel plot, meta-analysis, publication bias, statistical test
INTRODUCTION
Systematic reviews and meta-analyses are an essential link in the chain of evidence translation and are frequently used to provide a single pooled estimate of the best available evidence for decision makers. Publication bias is recognized as a serious threat to the validity and generalizability of this pooled estimate. Studies with statistically significant findings are more likely to be published than those reporting statistically non-significant findings; thus, summary treatment effects may be under- or over-estimated.1–5 In one example, data on 74% of patients enrolled in the trials evaluating the antidepressant reboxetine remained unpublished.6 Published data overestimated the benefit of reboxetine vs. placebo by 115% and underestimated harm; the addition of unpublished data changed the non-significant difference between reboxetine and placebo shown in published data to an inferiority of reboxetine. Therefore, assessing publication bias has been recommended as a critical step in conducting systematic reviews and meta-analyses.7 Both non-statistical and statistical approaches have been widely accepted for this purpose.
Non-statistical approaches include searching for unpublished databases from clinical trials registries and drug or device approving agencies, and they provide a powerful tool to detect publication bias. In the reboxetine example, only few published studies were available to validate the benefit of reboxetine, and the majority (74%) of the data were unpublished. Statistical methods may not successfully detect publication bias when the number of available published studies is small as in this example.
However, identifying and accessing unpublished databases are not always possible. Therefore, statistical methods have been popular auxiliary tools to handle publication bias. Table 1 summarizes several statistical methods that are based on testing the asymmetry of the funnel plot, which is a plot that presents each study’s effect size against its precision or standard error.8,9 The trim-and-fill method not only detects but also adjusts for publication bias; nevertheless, it makes a rather strong assumption that the potentially unpublished studies have the most negative (or positive) treatment effects. Thus, it is generally recommended as a form of sensitivity analysis.10 Begg’s and Egger’s tests examine the association between the observed treatment effects and their standard errors; a strong association implies publication bias. The original Egger’s test regresses the standardized effect (i.e., the effect size divided by its standard error) on the corresponding precision (i.e., the inverse of the standard error).11 It is equivalent to a weighted regression of the treatment effect on its standard error, weighted by the inverse of its variance.12 The weighted regression is more familiar among meta-analysts, because it directly links the treatment effect to its precision without a standardization process. Several modifications of Egger’s test also use the technique of weighted regression: the dependent variable is also the treatment effect, but the independent variable differs. For example, Tang and Liu13 used the inverse of the square root of study-specific sample size as the regression independent variable, which was motivated by the sample-size-based funnel plot (effect size against sample size).
Table 1.
Brief Descriptions for Various Publication Bias Tests and Summary of Test Results for the Cochrane Meta-Analyses
| Test | Designed for | Description | No. of meta-analyses with P value < 0.1 | |||
|---|---|---|---|---|---|---|
| Based on all eligible meta-analyses | Based on the restricted dataseta | |||||
| Non-binaryb | Binaryc | Non-binaryd | Binarye | |||
| Begg’s rank test | All outcomes | Use the rank correlation test to assess the association between standardized effect size and its standard error | 766 (7.2%) | 1479 (8.2%) | 108 (8.4%) | 165 (8.7%) |
| Trim-and-fill method | All outcomes | Estimate the number of suppressed studies, and calculate P value using its negative binomial distribution in the absence of publication bias | 706 (6.7%) | 1815 (10.1%) | 102 (7.9%) | 224 (11.8%) |
| Egger’s regression test | All outcomes | Weighted linear regression of y on s, with weights 1/s2 | 1426 (13.5%) | 2842 (15.7%) | 190 (14.7%) | 337 (17.7%) |
| Tang’s regression test | All outcomes | Weighted linear regression of y on , with weights N | 1045f (11.0%f) | 2064 (11.4%) | 128g (11.1%g) | 236 (12.4%) |
| Macaskill’s regression test | Binary outcomes | Weighted linear regression of y on N, with weights Ns × Nf/N | N/A | 2554 (14.1%) | N/A | 287 (15.1%) |
| Deeks’ regression test | Binary outcomes | Weighted linear regression of y on , with weights Ne | N/A | 2084 (11.5%) | N/A | 237 (12.4%) |
| Peters’ regression test | Binary outcomes | Weighted linear regression of y on 1/N, with weights Ns × Nf/N | N/A | 2135 (11.8%) | N/A | 249 (13.1%) |
y, effect size; s2, within-study variance; N, total no. of patients; Ns and Nf, no. of patients with and without events for binary outcomes respectively; Ne, effective sample size, defined as 4N0 × N1/N, where N0 and N1 are sample sizes the control and treatment groups respectively; N/A, not applicable
aThe restricted dataset consists of the meta-analyses with the largest numbers of studies in the corresponding Cochrane systematic reviews
bAmong 10,600 meta-analyses with non-binary outcomes
cAmong 18,055 meta-analyses with binary outcomes
dAmong 1291 meta-analyses with non-binary outcomes in the restricted dataset
eAmong 1906 meta-analyses with binary outcomes in the restricted dataset
fAmong 9530 meta-analyses whose total sample sizes are available
gAmong 1157 meta-analyses whose total sample sizes are available in the restricted dataset
When study outcomes are binary, the commonly used effect size odds ratio is mathematically associated with its standard error, even in the absence of publication bias.14,15 Because of this, Begg’s and Egger’s tests may have inflated false positive rates for binary outcomes, and alternative regression tests have been designed specifically to deal with this issue.15–17 For example, Macaskill et al.16 regressed log odds ratio on the study-specific total sample size. Deeks et al.15 used the “effective sample size” (defined in Table 1) as the regression independent variable, and Peters et al.17 modified Macaskill’s regression and used the inverse of the total sample size as the independent variable.
These various methods have been frequently applied to assess publication bias in systematic reviews, and some have been compared in simulation studies.17–19 It is generally recognized that Begg’s rank test has lower statistical power than others.12,14,16 However, comparison between these tests using empirical data, as opposed to simulation, is unavailable. Also, some simulation settings could be fairly unrealistic; for example, studies may be unpublished because of non-significant P values,20 or negative effect sizes,21 or some other obscure editorial criteria.22 Therefore, the exact mechanism of publication bias in a real meta-analysis cannot be reliably reproduced by simulation.
In this study, we apply seven commonly-used publication bias tests to a large collection of meta-analyses published in the Cochrane Library. We investigate the proportion of meta-analyses that have statistically significant publication bias detected by each test. We evaluate the agreement among the results produced by these tests and the effect of meta-analysis size on results. These empirical comparisons will aid researchers in properly assessing publication bias and interpreting test results in future systematic reviews.
METHODS
Data Source
The Cochrane Collaboration is a non-for-profit and non-governmental organization that produces systematic reviews on various healthcare-related topics. The Cochrane reviews are regularly updated, so a single review may have several versions. Also, some newly published reviews may be protocols that prepare data collection and analysis, so statistical data are not available from these protocols yet. Some early reviews have been withdrawn because they were merged into other reviews or were found to be flawed; their statistical data are also unavailable from the Cochrane Library.
We searched for all reviews in the Cochrane Library from 2003 Issue 1 to 2017 Issue 12. The issues before 2003 were not available online. All statistical data contained in each Cochrane review were downloaded at the link in the form of http://onlinelibrary.wiley.com/doi/10.1002/14651858.CDXXXXXX.pubY/downloadstats, where XXXXXX represents the Cochrane ID of the systematic review, and Y represents the review’s most current version. If a systematic review had only one version, the character string “.pubY” was removed from the foregoing link. We downloaded the data of all reviews iteratively using the R package “RCurl”23 on 6 December, 2017.
Analysis Approach
We classified the meta-analyses in the Cochrane reviews into those with non-binary or binary outcomes. For binary outcomes, regardless the analyses performed in the original reviews, we used the odds ratio as the effect size, because the methods of Macaskill’s, Deeks’, and Peters’ regressions were designed for the odds ratio. If the 2×2 table of a study contained zero data cell in one arm only, we added a continuity correction of 0.5 to all four cells so that the odds ratio and its variance can be properly estimated.24,25 Studies with zero data cells in both treatment and control arms were excluded because their odds ratios were not estimable.25–27 We considered meta-analyses containing at least five studies.
For meta-analyses with non-binary outcomes, we applied Begg’s rank test, the trim-and-fill method, and Egger’s and Tang’s regression tests to assess publication bias, as they were proposed for all types of outcomes.11,13,20,21 For meta-analyses with binary outcomes, we additionally considered Macaskill’s, Deeks’, and Peters’ regression tests, which were originally designed for binary outcomes to control false positive rates.15–17 The statistical significance level was set to 0.1 because the statistical power of the publication bias tests is generally low.11,16,20 Moreover, Cohen’s κ, a coefficient upper bounded by 1, was used to measure pairwise agreement among the publication bias tests.28 Typically, κ < 0 indicates no agreement; agreement is considered weak, moderate, and strong if κ lies in 0–0.4, 0.4–0.6, and 0.6–1, respectively.29
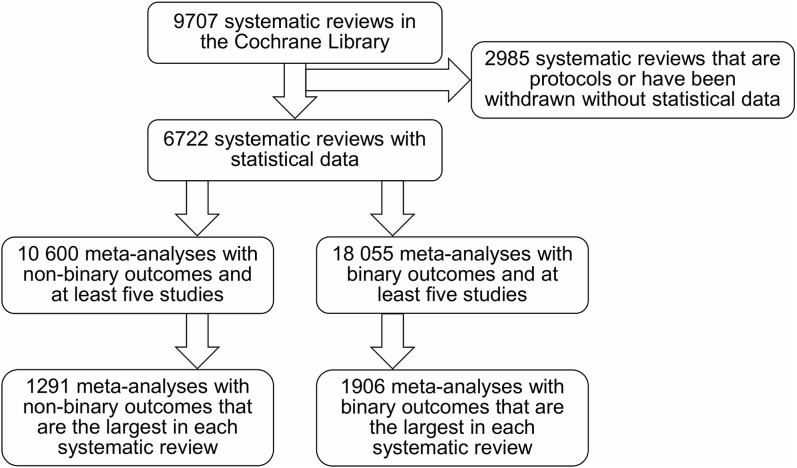
Multiple meta-analyses may be performed on different outcomes and treatment comparisons within a single review, but they probably used information from some common populations and thus may be dependent.30 To reduce the impact of such correlations, we also conducted the analysis using a restricted dataset. Specifically, the meta-analysis with the largest number of studies was chosen from each review. If a review contained more than one meta-analysis with the same largest number of studies, the meta-analysis with the largest total sample size was selected. If the total sample sizes were still equal, one meta-analysis was randomly chosen from those with the largest numbers of studies and total sample sizes. Figure 1 shows the process of meta-analysis selection.
Figure 1.
Flow chart of selecting the meta-analyses with non-binary and binary outcomes from the Cochrane Library.
RESULTS
A total of 9707 systematic reviews were collected for this empirical study. Among them, 2417 reviews had only one version, 4623, 1805, 656, 165, 33, 7 reviews had two, three, four, five, six, and seven versions respectively, and only one review had eight versions. In addition, 2985 reviews were protocols or had been withdrawn without statistical data in the Cochrane Library. After extracting the meta-analyses with at least five studies from the remaining 6722 reviews, we obtained a total of 28,655 meta-analyses; among them, 10,600 and 18,055 had non-binary and binary outcomes, respectively. Finally, for the restricted dataset, we obtained 1291 and 1906 unique meta-analyses with non-binary and binary outcomes respectively that were deemed independent.
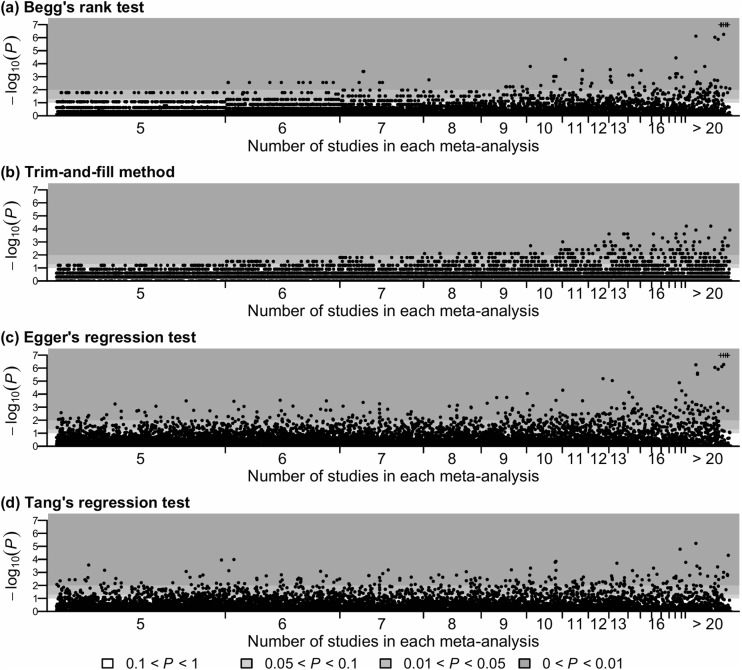
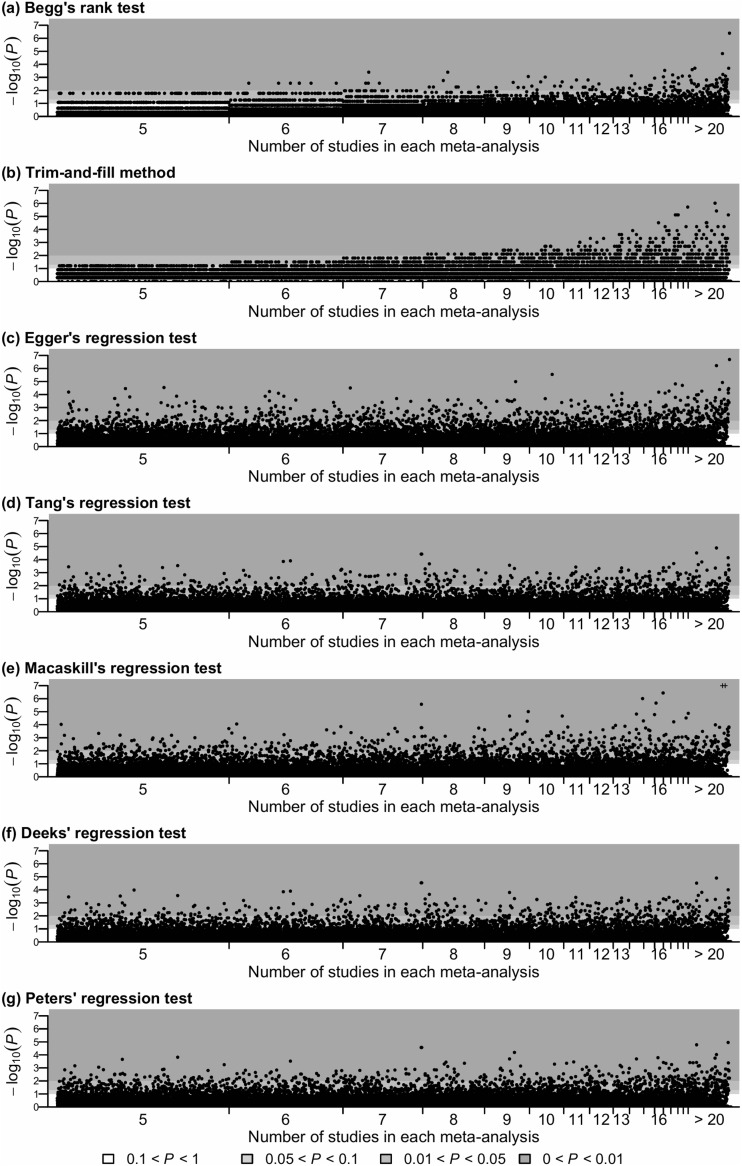
Figures 2 and 3 show the P values produced by the various publication bias tests for meta-analyses with non-binary and binary outcomes, respectively. The horizontal axis presents each meta-analysis sorted by its size (i.e., the number of included studies); the meta-analyses with the same size are sorted by their Cochrane IDs. The vertical axis shows the P values transformed by negative logarithm with base 10, and three statistical significance levels, 0.01, 0.05, and 0.1, are displayed. Both figures illustrate that the area representing small meta-analyses was much wider than that representing large meta-analyses, and most Cochrane meta-analyses contained less than 10 studies. Specifically, among the entire 28,655 meta-analyses with all types of outcomes, 7256 meta-analyses contained 5 studies, while only 191 meta-analyses contained 20 studies. The median number of studies was 7, and the lower and upper quartiles were 5 and 20, respectively.
Figure 2.
The P values produced by four publication bias tests for all 10,600 Cochrane meta-analyses with non-binary outcomes. Plus signs (+) indicate P values < 10−7. The total sample sizes were not reported in 1070 meta-analyses, so Tang’s test was not applicable for them, and panel (d) does not contain their results.
Figure 3.
The P values produced by seven publication bias tests for all 18,055 Cochrane meta-analyses with binary outcomes. Plus signs (+) indicate P values < 10−7.
Overall, Table 1 shows that Begg’s rank test and the trim-and-fill method detected statistically significant publication bias in much fewer meta-analyses than regression-based tests. In particular, for small meta-analyses, Figures 2 and 3 indicate that the P values produced by Begg’s rank test and the trim-and-fill method were generally larger than those produced by regression tests. For example, among the meta-analyses containing 5 studies, most P values produced by Begg’s rank test and all P values produced by the trim-and-fill method were greater than 0.05, while the regression tests implied substantial publication bias with P values much less than 0.01 in some meta-analyses. In addition, Begg’s rank test and the trim-and-fill method were more likely to detect publication bias in large meta-analyses than in small ones. Furthermore, note that all P values of the trim-and-fill method were discontinuous and massed at several specific values, because this method used the negative binomial distribution, which was discrete, to calculate P value.21 Many P values of Begg’s rank test were also massed at several specific values. This is because the rank test calculated an exact P value, taking some discontinuous values, when the number of studies was small and the treatment effects had no ties; otherwise, the P value was calculated using the normal approximation of the rank statistic’s distribution.
Compared with Begg’s rank test and the trim-and-fill method, the significance of publication bias assessed by regression-based tests seemed to be less dependent on the size of meta-analysis. Table 1 shows that Egger’s test detected statistically significant publication bias in 13.5% of meta-analyses with non-binary outcomes and 15.7% of those with binary outcomes. These proportions were higher than the other regression tests. The numbers of meta-analyses with statistically significant publication bias detected by Tang’s, Deeks’, and Peters’ tests were similar for binary outcomes. Moreover, the P value plots of Tang’s and Deeks’ tests in Figure 3 were fairly similar. However, the plots of the other regression tests were noticeably different: one test may not detect statistically significant publication bias for a meta-analysis, while another test could lead to an extremely small P value for the same meta-analysis.
Table 2 quantifies the agreement among the tests using Cohen’s κ coefficient. The upper panel analyzes all extracted Cochrane meta-analyses, and the lower one is based on the restricted dataset that consisted of the largest meta-analysis from each Cochrane review. Results were in general consistent between the two analyses. In the lower table, in which the meta-analyses were from different reviews and may be deemed independent, Begg’s rank test and the trim-and-fill method had a rather weak agreement (κ ≤ 0.40), and their agreement with the regression tests was also weak. Egger’s test had moderate agreement with Tang’s, Deeks’, and Peters’ regression tests. Most Cohen’s κ coefficients between Tang’s, Macaskill’s, Deeks’, and Peters’ tests were close to 0.60, which implied moderate agreement. The Cohen’s κ coefficient between Tang’s and Deeks’ tests was close to 1, implying a near perfect agreement; this confirms the original observation in Figure 3.
Table 2.
Cohen’s κ Coefficients for the Agreement Among Seven Publication Bias Tests. Within Each Sub-Table, the Results in the Upper and Lower Triangular Are Based on the Cochrane Meta-Analyses with Non-Binary and Binary Outcomes, Respectively
| Based on all Cochrane meta-analyses with at least five studies: | ||||||
| Begg | 0.22 | 0.45 | 0.30 | N/A | N/A | N/A |
| 0.26 | T & F | 0.35 | 0.21 | N/A | N/A | N/A |
| 0.45 | 0.42 | Egger | 0.48 | N/A | N/A | N/A |
| 0.25 | 0.27 | 0.41 | Tang | N/A | N/A | N/A |
| 0.13 | 0.21 | 0.34 | 0.54 | Macaskill | N/A | N/A |
| 0.25 | 0.28 | 0.42 | 0.93 | 0.52 | Deeks | N/A |
| 0.24 | 0.24 | 0.38 | 0.65 | 0.45 | 0.64 | Peters |
| Based on the meta-analyses that are the largest in their corresponding Cochrane systematic reviews: | ||||||
| Begg | 0.40 | 0.50 | 0.35 | N/A | N/A | N/A |
| 0.29 | T & F | 0.46 | 0.27 | N/A | N/A | N/A |
| 0.46 | 0.43 | Egger | 0.47 | N/A | N/A | N/A |
| 0.25 | 0.27 | 0.43 | Tang | N/A | N/A | N/A |
| 0.14 | 0.21 | 0.38 | 0.56 | Macaskill | N/A | N/A |
| 0.25 | 0.27 | 0.44 | 0.94 | 0.54 | Deeks | N/A |
| 0.23 | 0.25 | 0.43 | 0.68 | 0.50 | 0.67 | Peters |
Cohen’s κ coefficients ≥ 0.60 are in italics
Begg, the rank test; Egger, Tang, Macaskill, Deeks, and Peters, the regression tests; T & F, the trim-and-fill method; N/A, not applicable
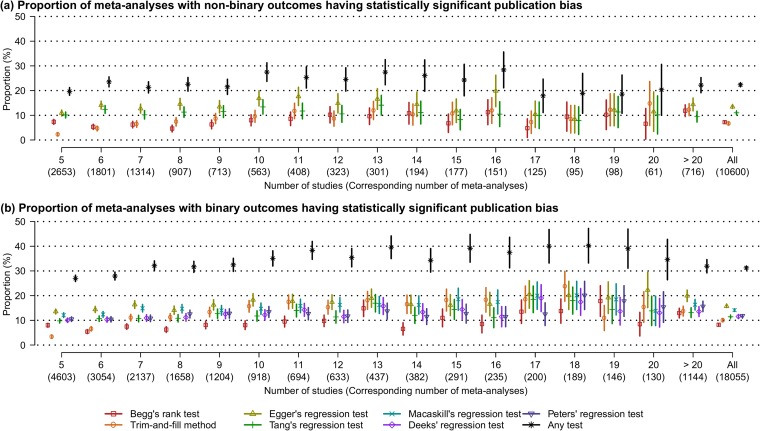
Categorized by the number of studies, Figure 4 describes the proportions of meta-analyses having statistically significant publication bias based on the various tests and the Wald-type 95% confidence intervals of these proportions. The lower panel indicates that the proportion tended to be greater for larger meta-analyses with binary outcomes. Also, the proportions of the Cochrane meta-analyses having statistically significant publication bias were approximately between 10 and 30% for most sizes of meta-analyses. Publication bias was detected by at least one test in more than 20% of meta-analyses with non-binary outcomes and in more than 30% of meta-analyses with binary outcomes.
Figure 4.
Proportions of the Cochrane meta-analyses having statistically significant publication bias (P value < 0.1) based on various tests and their 95% confidence intervals. “Any test” implies the proportion of the meta-analyses having statistically significant publication bias detected by at least one test. The label “All” on the horizontal axis represents all the extracted meta-analyses with non-binary (upper panel) or binary (lower panel) outcomes.
Figures S1–S3 in the Supplementary Materials online show the P value plots and the plot of proportions of having publication bias based on the restricted dataset. The trends in these plots were similar with those in Figures 2, 3, and 4, although the 95% confidence intervals in Figure S3 were wider than those in Figure 4 because the restricted dataset contained much fewer meta-analyses.
DISCUSSION
Main Findings
Using a large collection of meta-analyses, this empirical study has illustrated that publication bias is frequently found using standard tests in meta-analyses conducted in the Cochrane systematic reviews. This finding underscores the need to routinely assess publication bias in future evidence synthesis research. Egger’s regression test detected statistically significant publication bias in more meta-analyses than others.
Tang’s and Deeks’ regression tests were shown to have almost identical performance. Tang’s method was motivated by examining the asymmetry of the sample-size-based funnel plot for all types of outcomes, and the regression independent variable is the total sample size within each study.13 Deeks’ method was originally developed for meta-analysis of diagnostic tests, and the regression independent variable is the “effective sample size” (Table 1).15 If the allocation ratio for the treatment and control groups is close to 1:1, which is common in randomized controlled trials, then the “effective sample size” is close to the total sample size. Therefore, it is not surprising to obtain similar results using Tang’s and Deeks’ tests.
Limitations and Strengths
This study has several limitations. For example, the Cochrane Library contains meta-analyses only in healthcare-related specialties; therefore, the results may not be generalized to other fields. In addition, due to the lack of a gold standard test for publication bias, we never know whether the results of this study directly imply statistical power or true comparison of the accuracy of these tests. For example, Egger’s test detected publication bias in more meta-analyses than others possibly because it was more sensitive or had a higher risk of false positive.17
All seven tests considered in this study were based on the funnel plot; however, the funnel plot’s asymmetry needs to be interpreted from various perspectives. For example, since small studies may be biased due to poor methodological quality (e.g., design flaws such as inadequate allocation concealment) and they commonly enroll high-risk individuals, the funnel plot can be viewed as an approach to evaluating small study effects in general, rather than publication bias in particular.14,31,32 In addition, the P value plots in Figures 2 and 3 indicate that some publication bias tests tended to detect more statistically significant publication bias in larger meta-analyses. As the number of studies increases, a meta-analysis likely collects more heterogeneous or outlying studies, which can be sources of causing the funnel plot’s asymmetry other than publication bias. Outliers may appear in meta-analysis due to several reasons. For example, some study results could be extreme because of errors in the process of recording, analyzing, or reporting data.33 Also, if a review did not strictly follow pre-specified inclusion and exclusion criteria, some studies may be improperly included showing extreme results (compared to other studies with proper inclusion criteria). Outliers may lead to a heavy tail at one side of the treatment effect distribution; thus, the funnel plot may look asymmetric, but it is not caused by publication bias.
Heterogeneity between studies caused by differences in patient selection, baseline disease severity, study location, and other factors affects the interpretation of funnel plot’s asymmetry. A random-effects meta-analysis is usually applied to account for the heterogeneity; a normal distribution is conventionally specified to model study-specific underlying treatment effects.34,35 This model is appropriate if the heterogeneity permeates the entire collection of studies; however, the heterogeneity may be mostly limited to several subgroups, and the studies within each subgroup share a common overall treatment effect. In the presence of such multiple subgroups, even if the funnel plot within each subgroup is fairly symmetric, the funnel plot based on the entire collection of studies can be asymmetric. This asymmetry is induced by heterogeneity, not publication bias.36,37 Performing separate analysis within each subgroup is more appropriate for such data than pooling the results of all studies.
Because heterogeneity is common in meta-analyses,38–40 researchers need to carefully assess heterogeneity before making conclusions about publication bias. For example, Ioannidis and Trikalinos30 advised that it may not be appropriate to use the publication bias tests if I2 statistic38,41 is greater than 50% or Q statistic42,43 is significant with P value < 0.1. Although these criteria may not be rigorous for determining whether the publication bias tests are appropriate, a fairly large heterogeneity measure should alert researchers to interpret the funnel plot’s asymmetry with great cautions.
Each Cochrane meta-analysis conducted a subgroup test to identify potential subgroups; if the test indicated the presence of multiple subgroups, our study extracted the meta-analysis within each subgroup. Therefore, although it was infeasible to examine whether a funnel plot’s asymmetry was caused by publication bias or subgroup effect for each of the 28,655 Cochrane meta-analyses, extracting meta-analyses within subgroups has allowed us to reduce the subgroup effect on the funnel plots.
Practical Implications
Decision makers rely on meta-analytic estimates to trade off benefits and harms. If such estimates were erroneous because of publication bias, “Evidence to Decision” frameworks44 can be misled by skewed balance of benefits and harms and the resulting recommendations may be erroneous or detrimental to patient care. Because the agreement among most publication bias tests is weak or moderate, researchers need to carefully interpret the result produced by a single test. As publication bias tests usually have low statistical power,11,16,20 a single test that has a non-significant P value may lead to a false-negative conclusion. Instead of relying on the conclusion from a single test, researchers should assess publication bias using a variety of methods because different tests make different assumptions on the association between the treatment effects and precision measures. Lastly, considering the importance of publication bias and the challenges in statistically ascertaining its presence, systematic reviewers should resort to non-statistical approaches. These approaches include comparing published evidence to data available in clinical trials registries, records of drug or device approving agencies such as the Food and Drug Administration, and scientific conference proceedings.
Electronic Supplementary Material
(PDF 1104 kb).
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they do not have a conflict of interest.
REFERENCES
- 1.Begg CB, Berlin JA. Publication bias: a problem in interpreting medical data. J R Stat Soc Ser A (Stat Soc). 1988;151(3):419–63. doi: 10.2307/2982993. [DOI] [Google Scholar]
- 2.Sutton AJ, Duval SJ, Tweedie RL, et al. Empirical assessment of effect of publication bias on meta-analyses. BMJ. 2000;320(7249):1574–77. doi: 10.1136/bmj.320.7249.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thornton A, Lee P. Publication bias in meta-analysis: its causes and consequences. J Clin Epidemiol. 2000;53(2):207–16. doi: 10.1016/S0895-4356(99)00161-4. [DOI] [PubMed] [Google Scholar]
- 4.Kicinski M. Publication bias in recent meta-analyses. PLoS ONE. 2013;8(11):e81823. doi: 10.1371/journal.pone.0081823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin L, Chu H. Quantifying publication bias in meta-analysis. Biometrics. 2017. 10.1111/biom.12817. [DOI] [PMC free article] [PubMed]
- 6.Eyding D, Lelgemann M, Grouven U, et al. Reboxetine for acute treatment of major depression: systematic review and meta-analysis of published and unpublished placebo and selective serotonin reuptake inhibitor controlled trials. BMJ. 2010;341:c4737. doi: 10.1136/bmj.c4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murad MH, Montori VM, Ioannidis JPA, et al. How to read a systematic review and meta-analysis and apply the results to patient care: users’ guides to the medical literature. JAMA. 2014;312(2):171–79. doi: 10.1001/jama.2014.5559. [DOI] [PubMed] [Google Scholar]
- 8.Light RJ, Pillemer DB. Summing Up: The Science of Reviewing Research. Cambridge: Harvard University Press; 1984. [Google Scholar]
- 9.Sterne JAC, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol. 2001;54(10):1046–55. doi: 10.1016/S0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- 10.Peters JL, Sutton AJ, Jones DR, et al. Performance of the trim and fill method in the presence of publication bias and between-study heterogeneity. Stat Med. 2007;26(25):4544–62. doi: 10.1002/sim.2889. [DOI] [PubMed] [Google Scholar]
- 11.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothstein HR, Sutton AJ, Borenstein M. Publication Bias in Meta-Analysis: Prevention, Assessment and Adjustments. Chichester: Wiley; 2005. [Google Scholar]
- 13.Tang J-L, Liu JLY. Misleading funnel plot for detection of bias in meta-analysis. J Clin Epidemiol. 2000;53(5):477–84. doi: 10.1016/S0895-4356(99)00204-8. [DOI] [PubMed] [Google Scholar]
- 14.Sterne JAC, Gavaghan D, Egger M. Publication and related bias in meta-analysis: power of statistical tests and prevalence in the literature. J Clin Epidemiol. 2000;53(11):1119–29. doi: 10.1016/S0895-4356(00)00242-0. [DOI] [PubMed] [Google Scholar]
- 15.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–93. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Stat Med. 2001;20(4):641–54. doi: 10.1002/sim.698. [DOI] [PubMed] [Google Scholar]
- 17.Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295(6):676–80. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 18.Moreno SG, Sutton AJ, Ades AE, et al. Assessment of regression-based methods to adjust for publication bias through a comprehensive simulation study. BMC Med Res Methodol. 2009;9:2. doi: 10.1186/1471-2288-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bürkner PC, Doebler P. Testing for publication bias in diagnostic meta-analysis: a simulation study. Stat Med. 2014;33(18):3061–77. doi: 10.1002/sim.6177. [DOI] [PubMed] [Google Scholar]
- 20.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 21.Duval S, Tweedie R. A nonparametric “trim and fill” method of accounting for publication bias in meta-analysis. J Am Stat Assoc. 2000;95(449):89–98. [Google Scholar]
- 22.Hedges LV. Modeling publication selection effects in meta-analysis. Stat Sci. 1992;7(2):246–55. doi: 10.1214/ss/1177011364. [DOI] [Google Scholar]
- 23.Temple LD. RCurl: General Network (HTTP/FTP/...) Client Interface for R. R package version 1.95-4.8, 2016.
- 24.Walter SD, Cook RJ. A comparison of several point estimators of the odds ratio in a single 2 × 2 contingency table. Biometrics. 1991;47(3):795–811. doi: 10.2307/2532640. [DOI] [PubMed] [Google Scholar]
- 25.Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions. Chichester: Wiley; 2008. [Google Scholar]
- 26.Sweeting MJ, Sutton AJ, Paul LC. What to add to nothing? Use and avoidance of continuity corrections in meta-analysis of sparse data. Stat Med. 2004;23(9):1351–75. doi: 10.1002/sim.1761. [DOI] [PubMed] [Google Scholar]
- 27.Bradburn MJ, Deeks JJ, Berlin JA, et al. Much ado about nothing: a comparison of the performance of meta-analytical methods with rare events. Stat Med. 2007;26(1):53–77. doi: 10.1002/sim.2528. [DOI] [PubMed] [Google Scholar]
- 28.Cohen J. A coefficient of agreement for nominal scales. Educ Psychosoc Meas. 1960;20(1):37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 29.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–74. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 30.Ioannidis JPA, Trikalinos TA. The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey. Can Med Assoc J. 2007;176(8):1091–96. doi: 10.1503/cmaj.060410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sterne JAC, Egger M, Davey Smith G. Investigating and dealing with publication and other biases in meta-analysis. BMJ. 2001;323(7304):101–05. doi: 10.1136/bmj.323.7304.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbord RM, Egger M, Sterne JAC. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med. 2006;25(20):3443–57. doi: 10.1002/sim.2380. [DOI] [PubMed] [Google Scholar]
- 33.Lin L, Chu H, Hodges JS. Alternative measures of between-study heterogeneity in meta-analysis: reducing the impact of outlying studies. Biometrics. 2017;73(1):156–66. doi: 10.1111/biom.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S-LT N. Meta-analysis: formulating, evaluating, combining, and reporting. Stat Med. 1999;18(3):321–59. doi: 10.1002/(SICI)1097-0258(19990215)18:3<321::AID-SIM28>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 35.Borenstein M, Hedges LV, Higgins JPT, et al. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods. 2010;1(2):97–111. doi: 10.1002/jrsm.12. [DOI] [PubMed] [Google Scholar]
- 36.Sterne JAC, Sutton AJ, Ioannidis JPA, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 37.Peters JL, Sutton AJ, Jones DR, et al. Assessing publication bias in meta-analyses in the presence of between-study heterogeneity. J R Stat Soc Ser A (Stat Soc). 2010;173(3):575–91. doi: 10.1111/j.1467-985X.2009.00629.x. [DOI] [Google Scholar]
- 38.Higgins JPT, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgins JPT. Commentary: heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2008;37(5):1158–60. doi: 10.1093/ije/dyn204. [DOI] [PubMed] [Google Scholar]
- 40.Ioannidis JPA, Patsopoulos NA, Rothstein HR. Reasons or excuses for avoiding meta-analysis in forest plots. BMJ. 2008;336(7658):1413–15. doi: 10.1136/bmj.a117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 42.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10(1):101–29. doi: 10.2307/3001666. [DOI] [Google Scholar]
- 43.Whitehead A, Whitehead J. A general parametric approach to the meta-analysis of randomized clinical trials. Stat Med. 1991;10(11):1665–77. doi: 10.1002/sim.4780101105. [DOI] [PubMed] [Google Scholar]
- 44.Alonso-Coello P, Schünemann HJ, Moberg J, et al. GRADE Evidence to Decision (EtD) frameworks: a systematic and transparent approach to making well informed healthcare choices. 1: Introduction. BMJ. 2016;353:i2016. doi: 10.1136/bmj.i2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1104 kb).