Abstract
The importance of glutathione (GSH) in alternative cellular roles to the canonically proposed, were analyzed in a model unable to synthesize GSH. Gene expression analysis shows that the regulation of the actin cytoskeleton pathway is strongly impacted by the absence of GSH. To test this hypothesis, we evaluate the effect of GSH depletion via buthionine sulfoximine (5 and 12.5 mM) in human neuroblastoma MSN cells. In the present study, 70% of GSH reduction did not induce reactive oxygen species, lipoperoxidation, or cytotoxicity, which enabled us to evaluate the effect of glutathione in the absence of oxidative stress. The cells with decreasing GSH levels acquired morphology changes that depended on the actin cytoskeleton and not on tubulin. We evaluated the expression of three actin-binding proteins: thymosin β4, profilin and gelsolin, showing a reduced expression, both at gene and protein levels at 24 hours of treatment; however, this suppression disappears after 48 hours of treatment. These changes were sufficient to trigger the co-localization of the three proteins towards cytoplasmic projections. Our data confirm that a decrease in GSH in the absence of oxidative stress can transiently inhibit the actin binding proteins and that this stimulus is sufficient to induce changes in cellular morphology via the actin cytoskeleton.
Keywords: Glutathione, BSO, thymosin β4, gelsolin, profiling
Introduction
Gluthathion (GSH) is a tripeptide synthesized in two adenosine triphosphate-dependent steps: the glutamate cysteine ligase (GCL; rate-limiting enzyme in GSH synthesis) forms the dipeptide γ-L-glutamyl-L-cysteine, and then the glutathione synthetase binds a glycine to form GSH or γ-L-glutamyl-L-cysteinyl-glycine (Bains and Shaw, 1997; Dringen, 2000). GSH acts as a redox buffer due to its cysteine sulfhydryl group (-SH); GSH can therefore react directly with radicals through non-enzymatic reactions and constitutes the main barrier against oxidative damage caused by reactive oxygen species (ROS). GSH can also form xenobiotic conjugates through its enzymatic glutathione S-transferase activity and acts as an electron donor in the peroxide reduction catalyzed by the glutathione peroxidase enzyme (GPx) (Camera and Picardo, 2002).
Additionally, GSH and GSH metabolism have been implicated in cancer prevention, progression and treatment response. As a scavenger molecule, GSH can inhibit the action of different molecules through their interaction, including harmful molecules and anticancer drugs (Siddik, 2003; Wang and Lippard, 2005; Franco and Cidlowski, 2009; Chen and Tien Kuo, 2010). Also, GSH levels appear to be reduced after exposure to some xenobiotics (Becker and Soliman, 2009; Pierozan et al., 2016). In the nervous system, GSH is needed for defense against oxidative stress, and alterations in GSH metabolism have been reported in different pathologies including Parkinson’s disease and Alzheimer’s disease (Bains and Shaw, 1997; Ramaekers and Bosman, 2004). Thus, a reduced level of GSH appears to represent an important step in cell destabilization.
In addition to its several functions, data generated in our workgroup on a GCS-2 cell line (a GCL knockout cell line that is unable to synthesize GSH (Shi et al., 2000; Rojas et al., 2003; Valverde et al., 2006) suggested that the reduced level of intracellular GSH (2% of the wild type value of GSH) could generate changes in the expression of several genes. In addition, these cells can survive in the complete absence of GSH if N-acetyl cysteine is provided in the medium; thus, the reducing equivalents provided by GSH, and not GSH itself, protect cells from apoptosis. This means that GCS-2 cells with a severe reduction in GSH produce a metastable state compatible with survival. The results provided by this particular cell model could indicate that cell survival is compatible with low GSH intracellular levels without alterations of redox. In the present study, from the analysis of global expression of the GSC-2 model identifies the role of the absence of GSH in the pathways involved in the remodeling of the actin cytoskeleton and explores the hypothesis in a neuroblastoma model, under intracellular GSH modulation.
This study used MSN neuroblastoma cells to represent an early stage in neuronal development in which cells are pluripotent and retain the capability of expressing multiple neural crest-derived phenotypes (Abemayor and Sidellt, 1989). These cells also appear to be very sensitive to thiols depletion and have been used previously to observe changes in the cytoskeleton (Arias et al., 1993; Ko et al., 1997; Stabel-Burow et al., 1997).
The cytoskeleton is a dynamic system that consists of several filamentous networks that extend from the plasma membrane to the nuclear envelope and interconnect the cell nucleus to the extracellular matrix. Actin polymerizes to form filaments and participates in the generation and maintenance of cell morphology, polarity, endocytosis, intracellular trafficking, contractility, motility and cell division (Ramaekers and Bosman, 2004; Fu et al., 2015). However, actin filaments by themselves are not able to perform the processes involved. Instead, these processes require many proteins, including actin-binding proteins. Actin-binding proteins are responsible for orchestrating rounds of polymerization-depolymerization of the actin filaments. Our work focuses on three of the relevant functions of the actin binding proteins in disease development: thymosin β4, gelsolin and profilin (Winder and Ayscough, 2005; Lee et al., 2013; Cui et al., 2016).
Thymosin β4 is a monomer-sequestering protein that maintains a large pool of actin that allows rapid filament growth, clamps ATP-actin, and prevents its incorporation into filaments (Dedova et al., 2006). Gelsolin is a capping and severing protein that controls filament length by capping the barbed end, blocking the addition of new monomers and severing actin filaments to increase actin dynamics (Sun et al., 1999). Finally, profilin is also a monomer-sequestering protein but is involved in binding to ADP-actin to promote the nucleotide exchange (ADP for ATP) and facilitate new rounds of polymerization (Hertzog et al., 2004; Birbach, 2008).
Abnormalities in these essential cell components often result in disease (Ramaekers and Bosman, 2004). Studies have shown that the actin cytoskeleton and the actin-binding proteins participate in many processes related to carcinogenesis, such as invasion, metastasis and the epithelial-mesenchymal transition (Kedrin et al., 2007; Lorente et al., 2014; Cui et al., 2016). Moreover, alterations in cytoskeletal components have been implicated in the progression of some neurodegenerative disorders (McMurray, 2000; Ying et al., 2004).
Given the essential roles of the actin cytoskeleton and GSH in physiological and pathological processes, the aim of the present study was to confirm the hypothesis generated by the model GCS-2 in a scenario of GSH depletion in the regulation and remodeling of the actin cytoskeleton in human MSN neuroblastoma cells.
Materials and Methods
GCS-2 cell model
Cell culture
All studies used M15 complete medium, “knockout” DMEM supplemented with 15% embryonic stem cell-qualified FBS, 2 mM glutamine, 0.1 mM β-mercaptoethanol (BME), 100 units/mL of penicillin, and 100 μg/mL of streptomycin (all from GIBCO/BRL, Carlsbad, CA, USA, except BME, from Sigma-Aldrich, St. Louis, MO, USA). Mouse blastocyst cells derived from GCS -/- and the +/+ BDC1 mice were obtained. γGCS-deficient cells (GCS-2) were maintained in the above medium with 2.5 mM GSH (Sigma-Aldrich) and changed with fresh GSH containing medium daily. In the experiments involving GSH withdrawal, cells were washed twice with 1X PBS (GIBCO/BRL) and replaced with complete medium without supplemental GSH. Cultures were maintained at 37 °C in humidified incubators containing 5% CO2. Intracellular GSH levels in γGCS-deficient cells grown in the presence or absence of GSH were determined using HPLC/EC detection as described by Kleinman and Riche Jr (1995). Viability and cell number were determined using trypan blue exclusion staining (Sigma-Aldrich). Briefly, aliquots of 2.5 x 105 cells/mL were mixed with the trypan blue, and cells were examined by light microscopy. The results represent the average of three independent experiments with duplicate determinations.
cRNA preparation
We isolated total RNA using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and purified the RNeasy Total RNA Isolation Kit (Qiagen, Hilden, Germany). The SuperScript Choice system (Invitrogen) was used to synthesize double-stranded cDNA. Phase Lock Gels-phenol and chloroform extractions (Eppendorf, Hamburg, Germany) were used to clean up the cDNA template. We then generated biotin-labeled cRNA from this template using a BIOARRAY HIGHYIELD RNA Transcript Labeling Kit (Enzo Life Sciences, Farmingdale, NY, USA). In vitro transcription products were purified using RNeasy spin columns (Qiagen) and were quantified by spectrophotometric analysis. After the purification, the cRNA was fragmented using the standard procedure by Affymetrix to obtain a distribution of RNA fragments sized from approximately 35 to 200 bases. Fragmented RNA was checked with agarose gel electrophoresis.
Microarray analysis
A hybridization cocktail was prepared as recommended by Affymetrix, containing 0.05 μg/μL fragmented cRNA, 50 pM control oligonucleotide B2, 1.5, 5, 25 and 100 pM eukaryotic hybridization controls with bioB, bioC, bioD and cre genes, respectively, 0.1 mg/mL herring sperm DNA, 0.5 mg/ml acetylated BSA and 1X hybridization buffer. This hybridization cocktail was heated to 99 °C for 5 min and then used to fill the probe array cartridge. Hybridization was performed for 16 h with a rotation of 60 rpm in a rotisserie oven at 4 5°C.
After 16 h of hybridization, the hybridization cocktail was removed from the probe array, and the array was filled with non-stringent wash buffer. The GeneChip® Fluidics Station 400 (Affymetrix, Inc., Santa Clara, CA, USA) operated using Microarray Suite was used to wash and stain the probe arrays. We followed the manufacturer’s single stain protocol for eukaryotic targets. Arrays were washed twice and stained with a 10 μg/L streptavidin phycoerythrin solution. After staining, a final wash with non-stringent buffer was performed, and the arrays were scanned.
Data analysis
Image quantification, background subtraction and scaling were carried out with dChip software (Harvard, Boston, MA, USA) with 100% recall between control and lower GSH level chips and p<0.05 for the statistical algorithm (Li and Wong, 2001). DAVID Bioinformatics resources 6.8 was used to analyze the impacted pathways (Huang et al., 2009).
MSN cell model
Adherent human neuroblastoma MSN cells were grown in monolayers (Reynolds et al., 1986; Ramos-Espinosa et al., 2012). The cells were cultured in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Gibco, Life Technologies Corporation, Grand Island, NY, USA), 1% antibiotic-antimycotic (penicillin-streptomycin-amphotericin) (Gibco), 1% MEM non-essential amino acids (Gibco) and 1% QSN (glutamine-serine-asparagine) in tissue culture dishes in a humidified incubator under 95% air and a 5% CO2 atmosphere at 37 °C. Cells were subcultured at a density of 1x106 cells per dish and harvested by gently pipetting.
GSH depletion (BSO treatments)
A total of 1x106 MSN cells were seeded into a 100 mm tissue culture dish with 7 mL of supplemented RPMI 1640 medium. After 72 h, the culture was gently washed with PBS buffer, and 10 mL of supplemented RPMI 1640 medium was added. Buthionine sulfoximine (BSO, L-Buthionine-sulfoximine Sigma-Aldrich) treatments were administered at final concentrations of 0, 5, 12.5, 25 and 50 mM.
Cell viability
Cell viability was measured using the dual stain fluorescein diacetate (FDA)/ethidium bromide (BrEt) method as previously described (Hartmann and Speit, 1997). Briefly, the cells were mixed with a fluorochrome solution containing 0.02 μg/mL Et-Br and 0.015 μg/mL FDA (Sigma-Aldrich). The cells were then analyzed under an Olympus BX-60 fluorescence microscope with a UM61002 filter (Olympus, Tokyo, Japan). One hundred randomly chosen cells were evaluated per condition.
Reduced glutathione quantification
We used o-phthalaldehyde (OPT) (Sigma-Aldrich) as the fluorescent reagent to quantify the level of reduced glutathione due its specificity for GSH. A total of 100 μL of cells in PBS supplemented with protease inhibitors and 100 μL of meta-phosphoric acid (Sigma-Aldrich) precipitating reagent (1.67 g meta-phosphoric acid, 0.2 g EDTA (AMRESCO, Solon, OH, USA) and 30 g NaCl in 100 mL of distilled water) were added to a 0.6 mL Eppendorf tube, vortexed and centrifuged. The supernatant was decanted and frozen at -70 °C prior to further quantification. A total of 50 μL of the supernatant was added to a 1.5 mL Eppendorf tube with 1 mL of GSH buffer (0.1 M NaH2PO4 and 0.005 M EDTA, pH 8.0). Then, 50 μL of OPT (1mg/mL in methanol) was added to obtain a GSH-fluorescent conjugate. Next, 200 μL of the mixture from each Eppendorf tube was plated in an opaque 96-well plate and incubated for 15 min in the dark. The fluorescence was read in a BioTek FLx800 Fluorescence Microplate Reader (Winooski, VT, USA) with the emission set at 420 nm and the excitation set at 350 nm (Browne and Armstrong, 1998).
Reactive Oxygen Species (ROS)
ROS were determined with a modified fluorometric assay (Lee et al., 2003), which employs dihydrorhodamine 123 (DHR; Calbiochem-EMD Chemicals Inc. San Diego, CA, USA) as the probe. When DHR is oxidized by H2O2 in the presence of peroxidases, it produces the fluorescent compound rhodamine 123. Briefly, 100 μL of cells in PBS supplemented with protease inhibitors were centrifuged at 1,200 rpm for 5 min. Then, the supernatant was discarded, and 180 μL of buffer A (140 mM NaCl, 5 mM KCl, 0.8 mM MgSO4•7H2O, 1.8 mM CaCl2, 5 mM glucose and 15 mM HEPES, Sigma-Aldrich) and 20 μL of DHR (1 μM) were added and incubated at 37 °C for 2 min. The fluorescent product rhodamine 123 was measured using a spectrophotometer at 505 nm and interpolated in a curve of rhodamine 123.
Lipid Peroxidation (Lpx)
The thiobarbituric acid method was used to measure the concentration of malondialdehyde (MDA) (Bouaïcha and Maatouk, 2004). Briefly, 100 μL of cells in PBS supplemented with protease inhibitors and 100 μL of trichloroacetic acid (10% w/v) (Avantor Performance Materials Inc. Center Valley, PA, USA) were added to 1.5 mL Eppendorf tubes and centrifuged at 3,000 x g for 10 min. The supernatant was added to 1 mL of the thiobarbituric acid reagent (0.375%) (ICN Biomedicals Inc. Aurora, OH, USA), and the mixture was heated at 92 °C for 45 min. The absorbance of the thiobarbituric acid-MDA complex was measured at 532 nm using an ELISA spectrophotometer (Model 550 microplate reader, Bio-Rad, Hercules, Californa, USA). The data were interpolated onto a concentration curve of MDA (1,1,3,3-tetraethoxypropane) ranging from 0 to 10 nM.
Reverse transcriptase-polymerase chain reaction
Total RNA was isolated using TRIzol Reagent (Invitrogen) following the manufacturer’s protocol. The RNA quantity and purity were determined spectrophotometrically. The reverse transcriptase-polymerase chain reactions (RT-PCR) were performed using the Access RT-PCR System (Promega, MADISON, WI, USA) according to the manufacturer’s recommendations. The RT-PCR products were loaded onto a 3% agarose gel, and the mRNA levels were analyzed using the Kodak 1D v3.5.3 software. The following primers were used:
Actin (forward: catcatgaagtgtgacgtgg; reverse: atactcctgcttgctgatcc),
Thymosin β4 (forward: tgaacaggagaagcaagcag; reverse: tagacagatgggaaaggcag),
Gelsolin (forward: acggctgaaggacaagaaga; reverse: ttccaacccagacaaagacc),
Profilin (forward: ggaggcggattgaataagaag; reverse: ccatcaccctgcattgctaa) and
Ribosomal Protein L32 (forward: aagaagttcatccggcaccag; reverse: gcgatctcggcacagtaagat).
Western blot analysis
MSN cells were lysed in RIPA buffer (150 mM NaCl, 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate and 50 mM Tris-HCl, pH 8.0) with a protease inhibitor mixture and then centrifuged. Total proteins were quantified using a bicinchoninic acid kit (Thermo-Fisher Scientific Inc., Rockford, IL, USA). Equal amounts of protein were loaded onto a polyacrylamide gel and then transferred to a nitrocellulose or polyvinylidene fluoride membrane as required (Schägger, 2006). The membrane was blocked with 5% nonfat milk and 1% albumin in TBS and then sequentially incubated with a primary antibody and horseradish peroxidase-conjugated secondary antibody. The primary antibodies employed were goat polyclonal anti-Thymosin β4 (1:2000), mouse monoclonal anti-Gelsolin (1:2000), rabbit polyclonal anti-Profilin 1/2 (1:2000), and mouse monoclonal anti-β Tubulin (1:2000) (all Santa Cruz Biotechnology, Santa Cruz, CA, USA); the antibodies were dissolved in blocking solution and incubated at 37 °C for 1 h. Then, the membranes were washed with TBS-0.1% Tween-20 and incubated with the secondary antibody. The secondary antibodies employed were HRP-conjugated goat anti-rabbit, HRP-conjugated rabbit anti-goat and HRP-conjugated goat anti-mouse, all supplied by Zymed (San Francisco, CA, USA). The antibodies were dissolved in blocking solution and incubated at 37 °C for 1 h. The bands were visualized using the Amersham ECL Western blotting detection reagents according to the manufacturer’s guidelines (GE Healthcare, Piscataway, NJ. USA).
Immunofluorescence
A total of 105 MSN cells were seeded onto coverslips coated with 0.1% gelatin in a 12-well plate and incubated for 72 h or until sub-confluent. Then, the culture was gently washed with PBS buffer, the medium was renewed and the 5 and 12.5 mM BSO treatments were added. After 24 or 48 h of BSO treatment, the media was removed and the MSN cells were fixed with 4% paraformaldehyde and permeabilized with PBS-0.2% Triton X-100. Prior to antibody incubation, the samples were blocked with 1% albumin and 5% fetal bovine serum in PBS. The primary antibodies employed were mouse anti-β-Tubulin (1:50; Invitrogen), goat polyclonal anti-Thymosin β4 (1:50, Santa Cruz Biotechnology), mouse monoclonal anti-Gelsolin (1:50, Santa Cruz Biotechnology) and rabbit polyclonal anti-Profilin 1/2 (1:50, Santa Cruz Biotechnology). The antibodies were dissolved in blocking solution and incubated at 37 °C for 1 h. Then, the coverslips were washed three times with PBS-0.2% Triton X-100 for 5 min. The secondary antibodies employed were FITC-goat anti-mouse (1:100, Invitrogen), FITC-goat anti-rabbit (1:100, Invitrogen) and FITC-bovine anti-goat (1:100, Santa Cruz Biotechnology). Alexa Fluor® 594 Phalloidin (1:50, Invitrogen) was also employed to stain the actin filament, and 300 nM 4’,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) was used to counterstain the cell nuclei. All of the secondary antibodies were dissolved in blocking solution. The cells were incubated with the secondary antibodies and Alexa Fluor® 594 Phalloidin at 37 °C for 1 h; DAPI was added during the last 5 min of the secondary antibody incubation. Finally, the coverslips were washed three times with PBS-0.2% Triton X-100 for 5 min, embedded in Dako Fluorescent Mounting Medium (Dako North America, Inc., Carpinteria, CA, USA) and mounted onto slides. The slides were examined using an Axio Observer inverted microscope (Carl Zeiss, Oberkochen, Germany) coupled to a confocal laser scanning LSM 710 DUO from Carl Zeiss (Plan Apochromat 40X/1.3 oil objective). Images were acquired using the ZEN 2008 software (Carl Zeiss) and analyzed using the Fiji image processing package, which is distributed by ImageJ (Schindelin et al., 2012).
Statistical analysis
The results are expressed as the percentage ± standard error (SE). Statistical significance was analyzed using one-way ANOVA and Student’s t-test, and p-values < 0.05 were considered statistically significant. All analyses were performed using the statistical software SigmaStat (Systat Software Inc., San Jose, CA, USA), and histograms were generated using the SigmaPlot software. We also performed a multiple linear regression using the SigmaStat software to determine whether the BSO treatments were related to the changes in cell viability, ROS and lipid peroxidation.
Results
Expression of actin cytoskeleton genes in cells unable to synthesize GSH-GCS-2
Global analysis of molecular functions in cells unable to synthesize GSH, obtained by the gene expression patterns and the number of genes involved with their respective p-value is presented in Table 1.
Table 1. Analysis of molecular functions impacted by the lack of GSH in mouse embryonic cells GCS-2 by DAVID Bioinformatics Resources 6.8.
| Term | Count | % | p value | Fold Enrichment | Benjamini | FDR |
|---|---|---|---|---|---|---|
| MF00107: Kinase | 86 | 5.77956989 | 6.68968E-07 | 1.72558967 | 0.00013980 | 0.00084452 |
| MF00108: Protein kinase | 68 | 4.56989247 | 9.29454E-06 | 1.73700140 | 0.00097081 | 0.01173303 |
| MF00126: Dehydrogenase | 32 | 2.15053763 | 0.00028276 | 2.00021197 | 0.01950907 | 0.35638051 |
| MF00123: Oxidoreductase | 71 | 4.77150537 | 0.00059677 | 1.49772870 | 0.02464379 | 0.75077381 |
| MF00261: Actin binding cytoskeletal protein | 40 | 2.68817204 | 0.00280201 | 1.62442364 | 0.07068307 | 3.48030055 |
| MF00091: Cytoskeletal protein | 73 | 4.90591397 | 0.00763658 | 1.34716657 | 0.10130483 | 9.22408705 |
| MF00071: Translation factor | 17 | 1.14247311 | 0.00281380 | 2.28303897 | 0.06333992 | 3.49470003 |
| MF00166: Isomerase | 21 | 1.41129032 | 0.00376944 | 2.00593441 | 0.07589561 | 4.65577076 |
DAVID Bioinformatics Resources 6.8;Term: Enrichment terms associated with the gene list; Count: Gene involved in the term; % : involved genes / total genes; p-value: modified Fisher exact p-value; Fold Enrichment: To globally correct enrichmentp-values of individual term members.; Benjamini and FDR: To globally correct enrichment p-values to control family-wide false discovery rate under certain rate.
Expression analysis showed that the actin cytoskeleton pathway is largely impacted by GSH absence and that actin binding proteins, such as cofilin 1 and 2, fascin homolog and gelsolin, were overexpressed. In contrast, the thymosin B4, profilin and capping protein muscle z-line beta alpha-2 were underexpressed (Table 2).
Table 2. Analysis of gene expression involved in actin cytoskeleton pathway in mouse embryonic cells GCS-2 by DAVID Bioinformatics Resources 6.8.
| Unigene ID | Gene Symbol | Name of the gene | Fold Change (log2) |
|---|---|---|---|
| Mm.261329 | Myl12a | Myosin | -7.7 |
| Mm.2647 | Pfn1 | Profilin | -5.16 |
| Mm.97858 | Kif1B | Kinesin family member 1B | -4.04 |
| Mm.142729 | Tmsb4 | Thymosin beta 4 | -2.9 |
| Mm.25321 | Nudcd3 | NudC domain containing 3 | -1.92 |
| Mm.21687 | Limd2 | Lim domain containing 2 | -1.91 |
| Mm.392504 | Capza2 | Capping protein muscle Z-line beta alpha 2 | -1.78 |
| Mm.253564 | Actn1 | Actinin, alpha 1 | -1.63 |
| Mm.441340 | Kif6 | Kinesin family member 6 | -1.55 |
| Mm.329322 | Fhod3 | Formin homolog 2 containing 3 | -1.41 |
| Mm.52297 | Fnbp1 | Formin binding protein 1 | -1.41 |
| Mm.428571 | Septin 11 | Septin II | -1.39 |
| Mm.272460 | Gabarap | Gamma.aminobutyric acid receptor associated protein | -1.28 |
| Mm.143877 | Mapre1 | Microtubule associated protein RP/EB family 1 | -1.25 |
| Mm.157770 | Cnn2 | Calponin 2 | 1.01 |
| Mm.28357 | Map1lc3b | Microtubule associated protein 1 light chain 3 beta | 1.2 |
| Mm.99996 | Kif1c | Kinesin Family member 1c | 1.23 |
| Mm.478285 | Dcin5 | Dynactin 5 | 1.26 |
| Mm.278357 | Klc1 | Kinesin light chain 1 | 1.26 |
| Mm.276826 | Cfl2 | Cofilin 2 | 1.27 |
| Mm.295284 | Stom | Stomatin | 1.27 |
| Mm.276504 | Nudcd2 | NudC domain containing 2 | 1.28 |
| Mm.40068 | Tubb3 | Tubulin beta 3 Class III | 1.3 |
| Mm.7688 | Kif3c | Kinesin Family member 3c | 1.32 |
| Mm.258986 | Mark2 | Microtubule affinity regulating kinase 2 | 1.34 |
| Mm.299774 | Jup | Junction plakoglobin | 1.34 |
| Mm.272368 | Crip1 | Cystein-Rich protein 1 | 1.38 |
| Mm.249479 | Dync1i2 | Dynein cytoplasmic 1 intermediate chain2 | 1.38 |
| Mm.5567 | Palim1 | PDZ and Lim domain 1 (elfin) | 1.42 |
| Mm.27063 | Trip6 | Thyroid hormone receptor interactor 6 | 1.44 |
| Mm.205601 | Cttn | Cortactin | 1.48 |
| Mm.30010 | Arpc1b | Actin related protein 2/3 complex subunit 1B | 1.49 |
| Mm.273538 | Tubb5 | Tubulin beta 5 class1 | 1.49 |
| Mm.392113 | Tuba1b | Tubulin alpha 1B | 1.56 |
| Mm.20829 | Emp3 | Epithelial membrane protein3 | 1.6 |
| Mm.271967 | Lasp1 | Lim and SH3 protein 1 | 1.61 |
| Mm.208601 | Tln1 | Talin 1 | 1.62 |
| Mm.289306 | Arpc4 | Actin related protein 2/3 complex subunit 4 | 1.64 |
| Mm.289106 | Add1 | Aduccin 1 (alpha) | 1.64 |
| Mm.6919 | Dctn1 | Dynactin 1 | 1.65 |
| Mm.238285 | Ketd10 | Potassium channel tetramerisation | 1.74 |
| Mm.38450 | Sept 9 | Septin 9 | 1.88 |
| Mm.21109 | Gsn | Gelsolin | 1.93 |
| Mm.271711 | Tagln2 | Transgelin 2 | 2.11 |
| Mm.329655 | Cfl1 | Cofilin 1 | 2.27 |
| Mm.289707 | Fscn | Fascin homolog1 | 2.44 |
| Mm.288974 | Arpc5 | Actin related protein 2/3 complex subunit 5 | 2.91 |
| Mm.1287 | Mapt | Microtubule-associated protein TAU | 3.08 |
| Mm.275648 | Pdlim7 | PDZ and Lim domain 7 | 4.18 |
| Mm.441431 | Syn2 | Synapsin II | 4.64 |
| Mm.371777 | Pmp2 | Peripheral myelin protein 2 | 5.34 |
| Mm.218624 | Sh3yl1 | SH3 domain YSC-like 1 | 13.25 |
Depletion of intracellular GSH levels
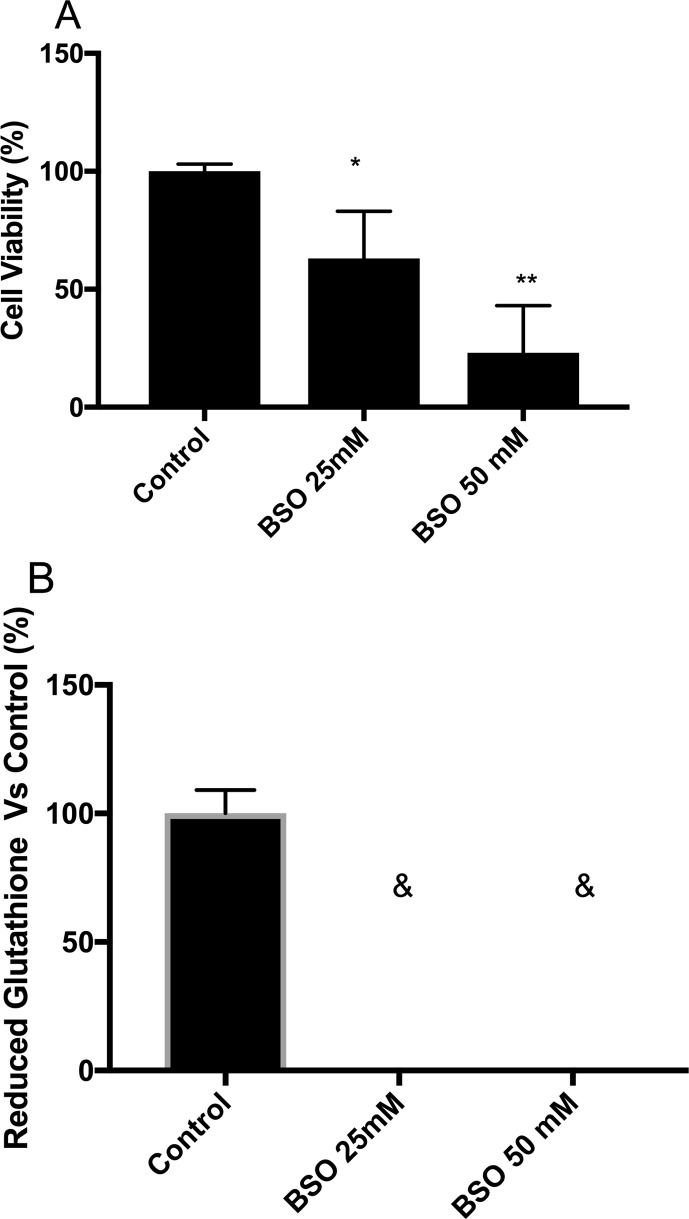
To achieve the maximum GSH depletion in the shortest amount of time, MSN cells were treated with 25 and 50 mM BSO for 24 h. We found significantly decreased cell viability and GSH levels at both concentrations (Figure 1A and 1B, respectively). Therefore, we tested lower BSO concentrations (5 and 12.5 mM) without a cell viability effect (Table 3). The 5 and 12.5 mM BSO treatments for 24 h could decrease the GSH level to approximately 70% compared with the control (Table 3). Therefore, we used these conditions for the assays. After we set up the working BSO concentrations, we evaluated the cell viability and GSH level following treatment for 48 h. The BSO treatments did not affect the cell viability of the MSN cells (Table 3), and the intracellular GSH level was decreased to approximately 80% compared with the control (Table 3).
Figure 1. Maximum depletion of intracellular GSH levels generate effects on survival. (A) Cell viability of MSN cells treated with high concentrations of buthionine sulfoximine, BSO, (25 and 50 mM) for 24 h assessed by fluorescein diacetate/ethidium bromide (FDA/EtBr) dual fluorochrome staining. (B) Corresponding intracellular GSH levels of MSN cells treated with BSO (&: not detected) determined fluorometrically (estimated limit of detection for the fluorometric method was 0.31 nmol of GSH/mg of protein). Statistical significance was determined by Student’s t-test. *p<0.05, **p<0.01. Experiments were performed three times.
Table 3. Absence of oxidative stress under GSH depletion.
| Hoursa | Treatment | Cell viabilityb,c | GSH b,d | ROSb,e | LPxb,f |
|---|---|---|---|---|---|
| Control | 100.00 ± 4.82 | 100.00 ± 9.05 | 100.00 ± 6.95 | 100.00 ± 14.02 | |
| 24 | BSO 5 mM | 105.40 ± 2.88 | 31.33 ± 6.03* | 106.75 ± 10.03 | 82.21 ± 22.06 |
| BSO 12.5 mM | 102.35 ± 0.36 | 23.21 ± 6.23* | 100.91 ± 12.87 | 104.26 ± 19.08 | |
| Control | 100.00 ± 0.74 | 100.00 ± 6.05 | 100.00 ± 6.95 | 100.00 ± 14.02 | |
| 48 | BSO 5 mM | 92.21 ± 5.07 | 15.45 ± 6.10* | 109.50 ± 9.35 | 103.61 ± 19.59 |
| BSO 12.5 mM | 89.14 ± 0.36 | 18.21 ± 5.09* | 113.06 ± 13.96 | 128.81 ± 19.74 |
Length of treatment in hours.
Data are expressed as the percentage with respect to controls ± standard error.
Cell viability after BSO treatments assessed by FDA/EtBr dual fluorochrome stain.
GSH values determined by o-phthalaldehyde (OPT) method after BSO treatment.
ROS: reactive oxygen species; ROS were measured by the generation of rhodamine 123 in MSN cells after BSO treatment. ROS positive control (Cadmium chloride 50 μM, 2 h): 138.29 ± 13.55.
LPx: Lipid peroxidation; LPx level assessed with the TBARS method using a MDA curve in MSN cells after treatment with BSO. Experiments were performed three to seven times.
Student’s t-test; p<0.01 versus control.
Absence of oxidative stress under GSH depletion
The redox state evaluation of MSN cells after BSO treatment was determined by measuring ROS levels and the end products of Lpx. It is important to emphasize that our aim was to decrease intracellular GSH levels without initiating an oxidative stress state to establish the effect trigger by GSH depletion on the regulation of the actin cytoskeleton. ROS were measured by the generation of rhodamine 123 in MSN cells treated with 5 and 12.5 mM BSO for 24 and 48 h. No changes in the ROS level were found compared to the control condition, suggesting that the depletion of the intracellular GSH level did not induce ROS generation (Table 3). Additionally, we measured Lpx end products and found that MSN cells treated with 5 and 12.5 mM BSO for 24 h were unable to generate Lpx end products; in contrast, a slight increase was observed after the 48 h treatment with 12.5 mM BSO. To ensure that the loss of GSH did not induce oxidative stress, we conducted a multiple linear regression analysis to evaluate the influence of ROS and Lpx due to GSH depletion by BSO treatment for cell viability (Table 4). Neither ROS nor lipid peroxidation appeared to be necessary to predict cell viability. Thus, the detected ROS and Lpx levels did not affect cell viability, and non-oxidative stress generation could be inferred.
Table 4. Multivariate analysis of oxidative stress markers with cell viability.
| Multiple linear regression | Coefficient | Standard Error | p | VIFa |
|---|---|---|---|---|
| Constant | 139.162 | 50.107 | 0.069 | |
| ROSb | -0.222 | 0.568 | 0.722 | 2.425 |
| LPxc | -0.153 | 0.118 | 0.284 | 2.425 |
VIF: variance inflation factor;
ROS: reactive oxygen species;
LPx: lipid peroxidation. Multiple linear regression model uses a set of independent variables (ROS and LPx) to explain influences on the dependent variable (cell viability). In this case, the analysis shows that none of the independent variables appear necessary to predict cell viability, which means that the detected ROS and Lpx levels do not affect cell viability.
Glutathione depletion triggers changes in the cell shape
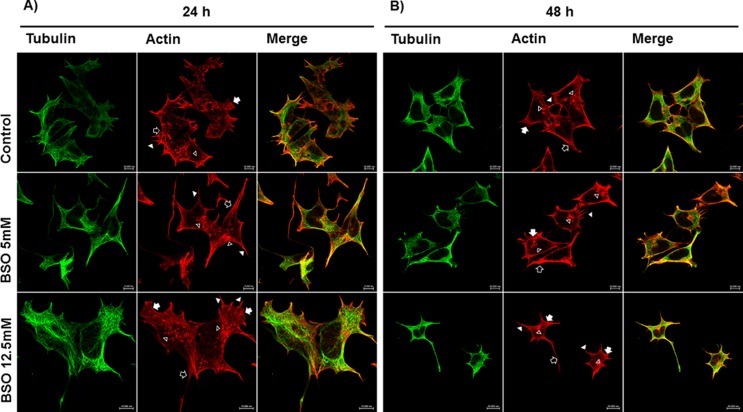
Confocal microscopy images showed drastic morphological changes in the MSN cells when GSH was depleted by the BSO treatments (Figure 2). After 24 h of GSH depletion, the actin confocal images showed that the control cells displayed the characteristic cell shape and were polarized. Lamellipodia (arrow) with some cytoplasmic projections or filopodia (arrowhead) were observed at one end, whereas a cone shape (empty arrow) with a few cytoplasmic projections and dot-shaped structures (empty arrowhead) that appeared to be focal adhesions was observed at the other end. In the confocal images of control cells the microtubules were clearly resolved as long fibers that were distributed throughout the cytoplasm to the cell periphery and delimited the space occupied by the nucleus. Actin confocal images following the 5 mM BSO treatment showed the loss of the characteristic cell shape and changes in cell polarity, including large cytoplasmic projections along the cell surface, the presence of both lamellipodia (arrow) and filopodia (arrowhead) and dot-shaped structures (empty arrowhead) near the projections. In contrast, the microtubules showed the same characteristics and distribution as the control treatment. Following the 12.5 mM BSO treatment, the actin confocal images showed drastic changes in cell polarity, with the presence of lamellipodia (arrow), many filopodia (arrowhead) and several dot-shaped structures (empty arrowhead). A large filopodium that resembled a neurite (in this case an axon) was observed at one end of the cell. Conversely, the microtubules remained unchanged in structure and organization. The microtubules in both the control and GSH-depleted conditions showed changes in distribution that corresponded to the changes in cell shape due to the actin cytoskeletal rearrangements (Figure 2A).
Figure 2. Comparative morphological changes induced by 24 and 48 h BSO treatments. Confocal microscopy images of MSN cells showing that the 48 h BSO treatments caused more drastic morphological changes compared to the 24 h BSO treatments. Panel A corresponds to Control, BSO 5 mM and BSO 12.5 mM 24 h treatments; Panel B corresponds to Control, BSO 5 mM and BSO 12.5 mM 48 h treatments. Microtubules were visualized by staining with FITC-labeled β-Tubulin (mouse anti-β-tubulin/FITC-goat anti-mouse), and actin filaments were visualized by staining with Alexa Fluor® 594 Phalloidin-labeled F-actin. The bar represents 10 μm 40X. Arrow: lamellipodia; arrowhead: filopodia; empty arrow: cone shape; empty arrowhead: cytoplasmic projections and dot-shaped structures.
After the 48 h treatments, the actin filaments and microtubules in the MSN control cells showed a similar pattern to that described following the 24 h control treatment. In the confocal images of the 48 h 5 mM BSO treatment, the loss of the characteristic cell shape present in the MSN control cells was evident, with dramatic polarity changes, retraction of the cytoplasm resulting in a round shape, cytoplasmic projections (filopodia, arrowhead) that extended beyond the leading edge of the membrane and the marked presence of dot-shaped structures (empty arrowhead). No changes were observed in the microtubules. After the 48 h 12.5 mM BSO treatment, the MSN cells showed changes similar to those observed after the 48 h 5 mM BSO treatment: the cytoplasm retracted and became rounded, filopodia (arrowhead) were present along the cell periphery and in some cells a neurite axon-like structure was very evident and gave the cells a neuron-like shape (empty arrow). No important changes were evident in the structure or the organization of the microtubules, however they exhibited changes in distribution corresponding to the actin cytoskeletal rearrangements and the changes in cell shape (Figure 2B).
Comparison of cell shapes
To visualize the changes in cell shape in the confocal images we used the image processing package Fiji (distributed by ImageJ) to isolate 25 images of the predominant cell shapes in the control and BSO treatments. For this, we used the actin confocal images. Figure 3 shows the predominant cell shapes in the control conditions (24 and 48 h, upper panel). In these images, the cells are polarized, one end has a lamellipodium (arrow) with some cytoplasmic projections or filopodia (arrowhead), and the far end has a cone shape (empty arrow) with a few filopodia (arrowhead). In contrast, the different BSO treatments (lower panel) resulted in the constant (approximately 70% of the cells) and very evident loss of the characteristic cell shape, dramatic changes in polarity, more evident cytoplasmic projections, either lamellipodia (arrow) or filopodia (arrowhead) and in some cases the filopodia resembles an axon (empty arrow) and gives the cell a neuron-like cell shape (lower panel).
Figure 3. Comparative chart of the predominant cell shapes in 25 cells in the control vs. BSO treatments. Using the image processing package Fiji (distributed by ImageJ), we obtained representative images of the predominant cell shapes in the control conditions (upper panel) and BSO treatments (lower panel). Arrow: lamellipodia; arrowhead: filopodia; empty arrowhead: cytoplasmic projections and dot-shaped structures.
Gene expression levels of actin-binding proteins under GSH depletion
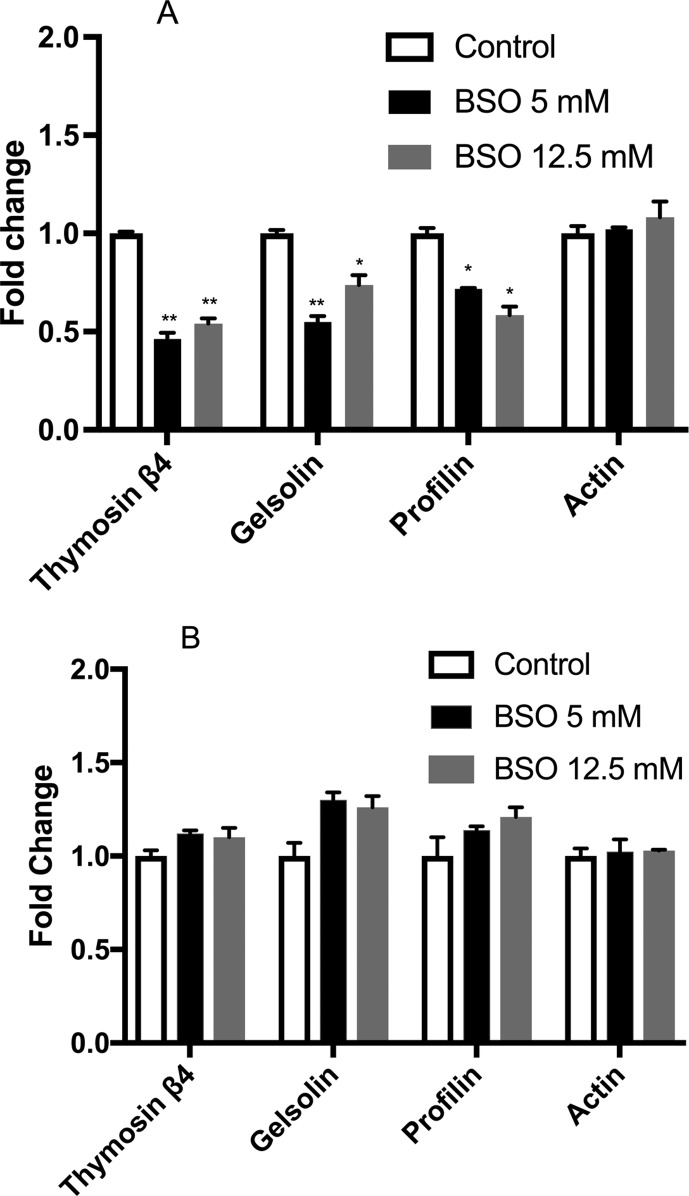
Gene expression of the actin-binding proteins thymosin β4, gelsolin, profilin and actin was evaluated after treatment with 5 and 12.5 mM BSO for 24 and 48 h. The GSH depletion for 24 h induced an important decrease in the expression of thymosin β4, gelsolin and profiling (Figure 4A). However, 48 h of GSH depletion show that this sub-expression is lost (Figure 4B).
Figure 4. Gene expression. (A) Gene expression in MSN cells treated with 5 and 12.5 mM BSO for 24 h (black and grey bars, respectively). (B) Gene expression in MSN cells treated with 5 and 12.5 mM BSO for 48 h (black and grey bars, respectively). The bars represent fold changes versus the control (open bars); all control values are set to 1. All data were previously normalized to the housekeeping gene RPL32. Statistical significance was determined by Student’s t-test. *p<0.05. Experiments were performed three times.
Protein expression levels of actin-binding proteins under GSH depletion
The protein expression of thymosin β4, gelsolin, profilin and actin were evaluated after treatment with 5 and 12.5 mM BSO for 24 and 48 h. Analysis showed that gelsolin and thymosin β4 had a 20% reduction in their expression at 24 h after 5 mM treatment. For the treatment with 12.5 mM, however, we only observed an effect on the profilin protein. For 48 h of treatment with 5 mM, we observed only a slight decrease for thymosin β4 and profilin (Figure 5A,B).
Figure 5. Protein expression. (A) Protein expression in MSN cells treated with 5 and 12.5 mM BSO for 24 h. and 48 h (black and grey bars, respectively). (B) Representative western blots of MSN cells treated with 5 and 12.5 mM BSO for 24 h. and 48 h (black and grey bars, respectively). The bars represent fold changes versus the control (open bars); all control values are set to 1. All data were previously normalized to the β-tubulin. Experiments were performed three times.
Cell localization of actin-binding proteins: thymosin β4, gelsolin and profilin
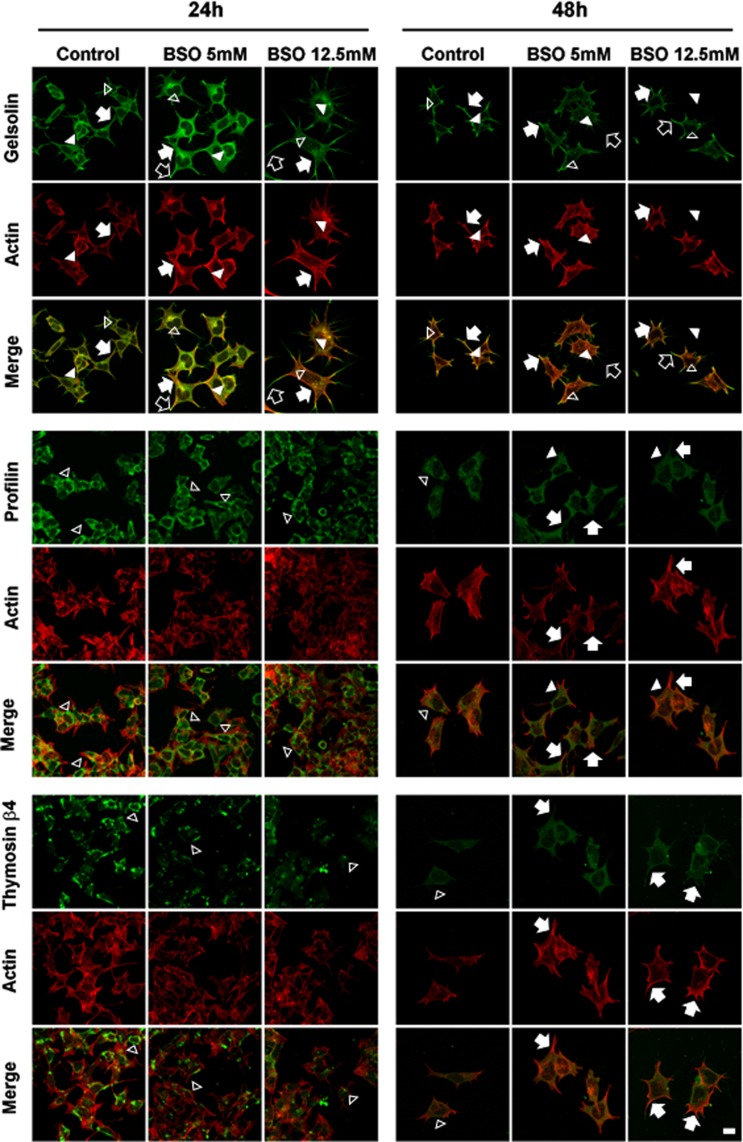
We performed an immunofluorescence analysis of thymosin β4, gelsolin and profilin for a comparison with the actin filament distributions (Figure 6). After 24 h treatment, gelsolin was widely distributed in both the cytoplasm and cytoplasmic projections of the control cells (either lamellipodia or filopodia). In the cytoplasm, we found gelsolin both free (empty arrowhead) and somewhat co-localized with actin filaments (arrowhead), whereas it co-localized with actin filaments when near the membrane and cytoplasmic projections (arrow). After the 5 and 12.5 mM BSO treatments, gelsolin was found free in the cytoplasm (empty arrowhead) and co-localized with actin filaments, similar to the control cells (arrowhead). However, near the cell membrane and in the cytoplasmic projections, gelsolin was strongly co-localized with the actin filaments. This co-localization was more evident in the cytoplasmic projections (arrow). Notably, in the BSO treatments, especially with 12.5 mM, free gelsolin was present at the distal ends of the projections (empty arrow). After 48 h in the control cells, gelsolin was distributed in both the cytoplasm and the cytoplasmic projections; some cytoplasmic gelsolin was free (empty arrowhead), whereas it was found co-localized with actin filaments near the cell membrane (arrowhead) and in the cytoplasmic projections (arrow). After 48 h in the 5 and 12.5 mM BSO treatments, gelsolin was distributed in the cytoplasm (empty arrowhead), in the vicinity of the cell membrane (arrowhead) and in the cytoplasmic projections (arrow). Its co-localization with actin filaments was more evident in the cytoplasmic projections, where it strongly co-localized with the actin filaments (arrow). Similar to the 24 h treatments, free gelsolin was present at the distal end of the cytoplasmic projections or filopodia (empty arrow).
Figure 6. Cell distribution of the actin-binding proteins gelsolin, profilin and thymosin β4. Gelsolin was stained with mouse anti-Gelsolin/FITC-goat anti-mouse (green), Profilin was stained with rabbit anti-Profilin/FITC-goat anti-rabbit (green), Thymosin β4 was stained with goat anti-Thymosin β4/FITC-rabbit anti-goat (green) and actin filaments were stained with Alexa Fluor® phalloidin (red), 40X. Arrows: lamellipodia; arrowheads: filopodia; empty arrow: cone shape; empty arrowheads: cytoplasmic projections and dot-shaped structures.
After 24 h of treatments, profilin showed a similar distribution in both the control cells and the cells treated with 5 and 12.5 mM BSO. Profilin was confined in the cytoplasm located away from the cell membrane and did not co-localize with the actin filaments (empty arrowhead). After 48 h of treatment, the control cells exhibited the same behavior, with the protein confined to the cytoplasm without reaching the region of the cytoplasmic membrane (empty arrowhead). In contrast, the distribution of the protein became more widespread in the cells treated with 5 and 12.5 mM BSO (arrowhead), even reaching the cell membrane and co-localizing with actin filaments (arrow).
Thymosin β4 distribution in the control cells was restricted to the cytoplasm, and no co-localization with actin filaments was observed (empty arrowhead), after 24 h of 5 and 12.5 mM BSO treatments, Thymosin β4 also showed the same distribution in the cytoplasm as the control cells (empty arrowhead). After 48 h of treatment, thymosin β4 in the control cells was distributed in the cytoplasm (empty arrowhead). However, after treatment with 5 and 12.5 mM, the distribution of thymosin β4 reached the cytoplasmic projections and was co-localized with the actin filaments (arrow).
Discussion
Considering the essential role of glutathione in physiological cell functions and from the predictions obtained from genomic analysis in GCS-2 cells, unable to synthesize GSH (Rojas et al., 2003; Valverde et al., 2006), it was shown that the remodeling of the actin cytoskeleton and accessory pathways are regulated by the tripeptide. To test this hypothesis we evaluated the role of GSH in regulating the actin cytoskeleton in neuroblastoma MSN cells. We assessed the 5 and 12.5 mM BSO concentrations, which depleted GSH by 70% but did not affect cell viability at 24 or 48 h. These results show that MSN cells are more sensitive to GSH depletion than other tissues (such as those in the kidney or liver) and agree with previous research (Stabel-Burow et al., 1997).
BSO treatment triggers a consistent but not complete GSH depletion. According to the literature, the GSH depletion was partial, suggesting that two pools of GSH are present in MSN cells: one that is easily depleted by BSO and another that is more resistant to depletion (Chen et al., 2005). This finding supported the existence of two sources of GSH.
Having demonstrated the decrease in intracellular GSH levels, we evaluated the redox state in MSN cells after treatment with BSO by measuring the ROS levels and end products of lipid peroxidation. Notably, our aim was to decrease the intracellular GSH level without reaching oxidative stress to determine the role of GSH in regulating the actin cytoskeleton because oxidative stress has already been demonstrated to alter the actin cytoskeleton (Fiaschi et al., 2006; Johansson and Lumberg, 2007; Pierozan et al., 2016).
No changes in the ROS level were found compared to the control condition, suggesting that the depletion of the intracellular GSH level did not induce ROS generation (Table 3). Additionally, we measured lipid peroxidation end products and found that MSN cells treated with 5 and 12.5 mM BSO for 24 h were unable to generate lipid peroxidation end products; in contrast, a slight increase was observed after the 48 h treatment with 12.5 mM BSO. To ensure that the loss of GSH did not induce oxidative stress, we conducted a multiple linear regression analysis to evaluate the influence of ROS and lipid peroxidation due to GSH depletion by BSO treatment on cell viability (Table 4). The analysis showed that neither ROS nor lipid peroxidation appeared to be necessary to predict cell viability.
Thus, the detected ROS and lipid peroxidation levels did not affect cell viability and non-oxidative stress generation could be inferred. This agrees with Franco and Cidioswki (2009), as well as Han et al. (2007), who support the notion of a direct role for GSH independent from oxidative stress. ROS overload may simply be an epiphenomenon associated with the depletion of GSH.
GSC-2 microarray data and MSN gene and protein expression results confirms that the lack of intracellular GSH modulate the gene expression of thymosin β4, gelsolin and profilin. We observed an important decrease in the expression of these genes. However, the microarray data indicated only the down-regulation of thymosin β4 and profilin, while gelsolin was up-regulated. This discrepancy could be due to different cell types used in each study (blastocysts and neuroblasts), or because blastocyst cells were unable to synthesize GSH, with approximately 2% of the normal amount of GSH. Our study never reached the levels of GSH inhibition obtained by previous research (Shi et al., 2000). The same behavior was observed at the protein level, where we observed only a discrete 20% reduction at 24 h that was absent at 48 h.
The confocal microscopy images showed drastic morphological changes in MSN cells when GSH was depleted by BSO treatment. We stained actin filaments and microtubules (another important component of the cytoskeleton) to visualize the distribution of the actin cytoskeleton and, therefore, the cell shape compared to the microtubule distribution. Actin filaments are abundant beneath the plasma membrane, where they form a network and extend throughout the cytoplasm. The control cells showed a characteristic cell shape and were polarized: a lamellipodium and some filopodia were present at one end, while the far end had a cone shape and focal adhesions were present.
The BSO treatments resulted in the loss of the characteristic cell shape and changes in cell polarity. Large cytoplasmic projections were observed along the cell surface, both lamellipodia and filopodia were present and focal adhesions with a re-localization of thymosin β, gelsolin and profilin proteins were detected. Moreover, the cells contained a neurite-like structure. In this case, the structure resembled an axon due to the neuronal origin of the MSN cells. These observations agree with our previous results (Ramos-Espinosa et al., 2012) and with Mollinari et al. (2009), which showed enhanced neurite outgrowth accompanied by increased focal adhesions due to the down-regulation of thymosin β4.
To visualize the changes described above, we used the image processing package Fiji to analyze 25 representative images of the cell shapes in the control and BSO-treated cells. The predominant cell shapes in the control conditions were polarized with a lamellipodium, with filopodia present at one end, whereas the far end had a cone shape. However, the different BSO treatments resulted in the constant and evident loss of the characteristic cell shape, dramatic polarity changes, more evident cytoplasmic projections, the presence of either lamellipodia or filopodia and in some cases filopodia that resembled an axon that gave the cell a neuron-like cell shape. The drastic changes induced by the BSO treatments went beyond the ultrastructure of the actin cytoskeleton. Notably, we found that GSH depletion could alter the regulation of actin cytoskeleton dynamics, thereby causing the formation of neurites in cultured MSN cells. Notably, these changes did not occur in an undifferentiated cell culture, eliminating a possible effect of BSO per se. The neurite formation process is regulated in complex ways. Its growth and guidance depend on well-coordinated cytoskeletal dynamics and occur in conjunction with differentiation and plasticity processes (Wilson et al., 2016). Thus, these results suggest that a decrease in the intracellular GSH content can affect processes involved in the signaling pathway that regulates neurite growth (Bradke and Dotti, 2000a,b; Luo, 2002; Madduri et al., 2009; Wilson et al., 2016).
In conclusion, GSH depletion produced a downregulation of the actin binding proteins profilin, thymosin β4 and gelsolin after 24 h BSO treatments. This down-regulation appears to be sufficient to trigger important changes in their localization and cellular shape in a non-oxidative stress-dependent manner. These results are relevant because exposures to xenobiotics could decrease the levels of GSH and could represent a cofactor that triggers changes in the cytoskeleton to facilitate the acquisition of several disease hallmarks including those related to cancer and neurodegenerative diseases.
Acknowledgments
We thank Dr. Miguel Tapia from the Unidad de Microscopia del Instituto de Investigaciones Biomedicas for his confocal technical assistance and helpful comments. This work was supported by PAPIIT (IN214410). NZF is a recipient of the CONACyT scholarship 231665 and gratefully acknowledges the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México. This publication is part of his doctoral thesis. We thank the technical support of Dr. Maria Alexandra Rodríguez-Sastre.
Footnotes
Associated Editor: Carlos F.M. Menck
References
- Abemayor E, Sidellt N. Human neuroblastoma cell lines as models for the in vitro study of neoplastic and neuronal cell differentiation. Environ Health. 1989;80:3–15. doi: 10.1289/ehp.89803. [DOI] [PMC free article] [PubMed] [Google Scholar]; Abemayor E and Sidellt N (1989) Human neuroblastoma cell lines as models for the in vitro study of neoplastic and neuronal cell differentiation. Environ Health 80:3–15. [DOI] [PMC free article] [PubMed]
- Arias C, Sharma N, Davies P, Shafit-Zagardo B. Okadaic acid induces early changes in microtubule-associated protein 2 and tau phosphorylation prior to neurodegeneration in cultured cortical neurons. J. Neurochem. 1993;61:673–682. doi: 10.1111/j.1471-4159.1993.tb02172.x. [DOI] [PubMed] [Google Scholar]; Arias C, Sharma N, Davies P and Shafit-Zagardo B (1993) Okadaic acid induces early changes in microtubule-associated protein 2 and tau phosphorylation prior to neurodegeneration in cultured cortical neurons. J. Neurochem 61:673-682. [DOI] [PubMed]
- Bains JS, Shaw C. Neurodegenerative disorders in humans: The role of glutathione in oxidative stress-mediated neuronal death. Brain Res Rev. 1997;25:335–358. doi: 10.1016/s0165-0173(97)00045-3. [DOI] [PubMed] [Google Scholar]; Bains JS and Shaw C (1997) Neurodegenerative disorders in humans: The role of glutathione in oxidative stress-mediated neuronal death. Brain Res Rev 25:335–358. [DOI] [PubMed]
- Becker A, Soliman KF. The role of intracellular glutathione in inorganic mercury-induced toxicity in neuroblastoma cells. Neurochem Res. 2009;34:1677–1684. doi: 10.1007/s11064-009-9962-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Becker A and Soliman KF (2009) The role of intracellular glutathione in inorganic mercury-induced toxicity in neuroblastoma cells. Neurochem Res 34:1677-1684. [DOI] [PMC free article] [PubMed]
- Birbach A. Profilin, a multi-modal regulator of neuronal plasticity. BioEssays. 2008;30:994–1002. doi: 10.1002/bies.20822. [DOI] [PubMed] [Google Scholar]; Birbach A (2008) Profilin, a multi-modal regulator of neuronal plasticity. BioEssays 30:994–1002. [DOI] [PubMed]
- Bouaïcha N, Maatouk I. Microcystin-LR and nodularin induce intracellular glutathione alteration, reactive oxygen species production and lipid peroxidation in primary cultured rat hepatocytes. Toxicol Lett. 2004;148:53–63. doi: 10.1016/j.toxlet.2003.12.005. [DOI] [PubMed] [Google Scholar]; Bouaïcha N and Maatouk I (2004) Microcystin-LR and nodularin induce intracellular glutathione alteration, reactive oxygen species production and lipid peroxidation in primary cultured rat hepatocytes. Toxicol Lett148:53–63. [DOI] [PubMed]
- Bradke F, Dotti CG. Differentiated neurons retain the capacity to generate axons from dendrites. Curr Biol. 2000a;10:1467–1470. doi: 10.1016/s0960-9822(00)00807-1. [DOI] [PubMed] [Google Scholar]; Bradke F and Dotti CG (2000a) Differentiated neurons retain the capacity to generate axons from dendrites. Curr Biol 10:1467–1470. [DOI] [PubMed]
- Bradke F, Dotti CG. Establishment of neuronal polarity: Lessons from cultured hippocampal neurons. Curr Opin Neurobiol. 2000b;10:574–581. doi: 10.1016/s0959-4388(00)00124-0. [DOI] [PubMed] [Google Scholar]; Bradke F and Dotti CG (2000b) Establishment of neuronal polarity: Lessons from cultured hippocampal neurons. Curr Opin Neurobiol10:574–581. [DOI] [PubMed]
- Browne RW, Armstrong D. Reduced glutathione and glutathione disulfide. Methods Mol Biol. 1998;108:347–352. doi: 10.1385/0-89603-472-0:347. [DOI] [PubMed] [Google Scholar]; Browne RW and Armstrong D (1998) Reduced glutathione and glutathione disulfide. Methods Mol Biol 108:347–352. [DOI] [PubMed]
- Camera E, Picardo M. Analytical methods to investigate glutathione and related compounds in biological and pathological processes. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;781:181–206. doi: 10.1016/s1570-0232(02)00618-9. [DOI] [PubMed] [Google Scholar]; Camera E and Picardo M (2002) Analytical methods to investigate glutathione and related compounds in biological and pathological processes. J Chromatogr B Analyt Technol Biomed Life Sci 781:181–206. [DOI] [PubMed]
- Chen HHW, Tien Kuo M. Role of glutathione in the regulation of cisplatin resistance in cancer chemotherapy. Metal-based Drugs. 2010:430939–7. doi: 10.1155/2010/430939. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen HHW and Tien Kuo M (2010) Role of glutathione in the regulation of cisplatin resistance in cancer chemotherapy. Metal-based Drugs 430939-7. [DOI] [PMC free article] [PubMed]
- Chen J, Small-Howard A, Yin A, Berry MJ. The responses of Ht22 cells to oxidative stress induced by buthionine sulfoximine (BSO) BMC Neurosci. 2005;6:10. doi: 10.1186/1471-2202-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen J, Small-Howard A, Yin, A and Berry MJ (2005) The responses of Ht22 cells to oxidative stress induced by buthionine sulfoximine (BSO). BMC Neurosci 6:10. [DOI] [PMC free article] [PubMed]
- Cui X, Zhang S, Xu Y, Dang H, Liu C, Wang L, Yang L, Hu J, Liang W, Jiang J, et al. PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma. J Translat Med. 2016;14:137. doi: 10.1186/s12967-016-0884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cui X, Zhang S, Xu Y, Dang H, Liu C, Wang L, Yang L, Hu J, Liang W, Jiang J, et al. (2016) PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma. J Translat Med 14:137. [DOI] [PMC free article] [PubMed]
- Dedova IV, Nikolaeva OP, Safer D, De La Cruz EM, dos Remedios CG. Thymosin beta4 induces a conformational change in actin monomers. Biophys J. 2006;90:985–992. doi: 10.1529/biophysj.105.063081. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dedova IV, Nikolaeva OP, Safer D, De La Cruz EM and dos Remedios CG (2006) Thymosin beta4 induces a conformational change in actin monomers. Biophys J 90:985–992. [DOI] [PMC free article] [PubMed]
- Dringen R. Metabolism and functions of glutathione in brain. Progr Neurobiol. 2000;62:649–671. doi: 10.1016/s0301-0082(99)00060-x. [DOI] [PubMed] [Google Scholar]; Dringen R (2000) Metabolism and functions of glutathione in brain. Progr Neurobiol 62:649–671. [DOI] [PubMed]
- Fiaschi T, Cozzi G, Raugei G, Formigli L, Ramponi G, Chiarugi P. Redox regulation of beta-actin during integrin-mediated cell adhesion. J Biol Chem. 2006;281:22983–22991. doi: 10.1074/jbc.M603040200. [DOI] [PubMed] [Google Scholar]; Fiaschi T, Cozzi G, Raugei G, Formigli L, Ramponi G and Chiarugi P (2006) Redox regulation of beta-actin during integrin-mediated cell adhesion. J Biol Chem 281:22983–22991. [DOI] [PubMed]
- Franco R, Cidlowski J. Apoptosis and glutathione: Beyond an antioxidant. Cell Death Diff. 2009;16:1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]; Franco R and Cidlowski J (2009) Apoptosis and glutathione: Beyond an antioxidant. Cell Death Diff 16:1303–1314. [DOI] [PubMed]
- Fu X, Cui P, Chen F, Xu J, Gong L, Jiang L, Zhang D, Xiao Y. Thymosin β4 promotes hepatoblastoma metastasis via the induction of epithelial-mesenchymal transition. Mol Med Rep. 2015;12:127–132. doi: 10.3892/mmr.2015.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fu X, Cui P, Chen F, Xu J, Gong L, Jiang L, Zhang D and Xiao Y (2015) Thymosin β4 promotes hepatoblastoma metastasis via the induction of epithelial-mesenchymal transition. Mol Med Rep 12:127-132. [DOI] [PMC free article] [PubMed]
- Han YH, Kim SZ, Kim SH, Park WH. Apoptosis in pyrogallol-treated Calu-6 cells is correlated with the changes of intracellular GSH levels rather than ROS levels. Lung Cancer. 2007;59:301–314. doi: 10.1016/j.lungcan.2007.08.034. [DOI] [PubMed] [Google Scholar]; Han YH, Kim SZ, Kim SH and Park WH (2007) Apoptosis in pyrogallol-treated Calu-6 cells is correlated with the changes of intracellular GSH levels rather than ROS levels. Lung Cancer 59:301–314. [DOI] [PubMed]
- Hartmann A, Speit G. The contribution of cytotoxicity to DNA-effects in the single cell gel test (comet assay) Toxicol Lett. 1997;90:183–188. doi: 10.1016/s0378-4274(96)03847-7. [DOI] [PubMed] [Google Scholar]; Hartmann A and Speit G (1997) The contribution of cytotoxicity to DNA-effects in the single cell gel test (comet assay). Toxicol Lett 90:183–188. [DOI] [PubMed]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Préat T, Knossow M, Guittet E, Carlier MF. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]; Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Préat T, Knossow M, Guittet E and Carlier MF (2004) The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117:611–623. [DOI] [PubMed]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene list using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]; Huang DW, Sherman BT and Lempicki RA (2009) Systematic and integrative analysis of large gene list using DAVID bioinformatics resources. Nat Protoc. 4:44-57. [DOI] [PubMed]
- Johansson M, Lundberg M. Glutathionylation of beta-actin via a cysteinyl sulfenic acid intermediary. BMC Biochem. 2007;8:26. doi: 10.1186/1471-2091-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]; Johansson M and Lundberg M (2007) Glutathionylation of beta-actin via a cysteinyl sulfenic acid intermediary. BMC Biochem 8:26. [DOI] [PMC free article] [PubMed]
- Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mamm Gland Biol Neoplasia. 2007;12:143–152. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]; Kedrin D, van Rheenen J, Hernandez L, Condeelis J and Segall JE (2007) Cell motility and cytoskeletal regulation in invasion and metastasis. J Mamm Gland Biol Neoplasia 12:143–152. [DOI] [PubMed]
- Kleinman WA, Richie JP., Jr Determination of thiols and disulfides using high-performance liquidchromatography with electrochemical detection. J Chromatogr B Biomed Appl. 1995;72:73–80. doi: 10.1016/0378-4347(94)00194-a. [DOI] [PubMed] [Google Scholar]; Kleinman WA and Richie Jr JP (1995) Determination of thiols and disulfides using high-performance liquidchromatography with electrochemical detection. J Chromatogr B Biomed Appl 72:73-80. [DOI] [PubMed]
- Ko L, Odawara T, Yen SH. Menadione-induced tau dephosphorylation in cultured human neuroblastoma cells. Brain Res. 1997;760:118–128. doi: 10.1016/s0006-8993(97)00292-8. [DOI] [PubMed] [Google Scholar]; Ko L, Odawara T and Yen SH (1997) Menadione-induced tau dephosphorylation in cultured human neuroblastoma cells. Brain Res 760:118-128. [DOI] [PubMed]
- Lee CW, Vitriol EA, Shim S, Wise AL, Velayutham RP, Zheng JQ. Dynamic localization of G-actin during membrane protrusion in neuronal motility. Curr Biol. 2013;23:1046–1056. doi: 10.1016/j.cub.2013.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee CW, Vitriol EA, Shim S, Wise AL, Velayutham RP and Zheng, JQ (2013) Dynamic localization of G-actin during membrane protrusion in neuronal motility. Curr Biol 23:1046-1056. [DOI] [PMC free article] [PubMed]
- Lee VM, Quinn P, Jennings S, Ng L. NADPH oxidase activity in preeclampsia with immortalized lymphoblasts used as models. Hypertension. 2003;41:925–931. doi: 10.1161/01.HYP.0000062021.68464.9D. [DOI] [PubMed] [Google Scholar]; Lee VM, Quinn P, Jennings S and Ng L (2003) NADPH oxidase activity in preeclampsia with immortalized lymphoblasts used as models. Hypertension 41:925–931. [DOI] [PubMed]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li C and Wong WH (2001) Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci U S A 98: 31-36. [DOI] [PMC free article] [PubMed]
- Lorente G, Syriani E, Morales M. Actin filaments at the leading edge of cancer cells are characterized by a high mobile fraction and turnover regulation by profilin I. PLoS One. 2014;9:e85817. doi: 10.1371/journal.pone.0085817. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lorente G, Syriani E and Morales M (2014) Actin filaments at the leading edge of cancer cells are characterized by a high mobile fraction and turnover regulation by profilin I. PLoS One 9:e85817. [DOI] [PMC free article] [PubMed]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]; Luo L (2002) Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol18:601–635. [DOI] [PubMed]
- Madduri S, Papaloizos M, Gander B. Synergistic effect of GDNF and NGF on axonal branching and elongation in vitro. Neurosci Res. 2009;65:88–97. doi: 10.1016/j.neures.2009.06.003. [DOI] [PubMed] [Google Scholar]; Madduri S, Papaloizos M and Gander B (2009) Synergistic effect of GDNF and NGF on axonal branching and elongation in vitro. Neurosci Res 65:88–97. [DOI] [PubMed]
- McMurray CT. Neurodegeneration: Diseases of the cytoskeleton? Cell Death Diff. 2000;7:861–865. doi: 10.1038/sj.cdd.4400764. [DOI] [PubMed] [Google Scholar]; McMurray CT (2000) Neurodegeneration: Diseases of the cytoskeleton? Cell Death Diff 7:861–865. [DOI] [PubMed]
- Mollinari C, Ricci-Vitiani L, Pieri M, Lucantoni C, Rinaldi AM, Racaniello M, De Maria R, Zona C, Pallini R, Merlo D, Garaci E. Downregulation of themosin β4 in neural progenitor grafts promotes spinal cord regeneration. J Cell Sci. 2009;122:4195–4207. doi: 10.1242/jcs.056895. [DOI] [PubMed] [Google Scholar]; Mollinari C, Ricci-Vitiani L, Pieri M, Lucantoni C, Rinaldi AM, Racaniello M, De Maria R, Zona C, Pallini R, Merlo D and Garaci E (2009) Downregulation of themosin β4 in neural progenitor grafts promotes spinal cord regeneration. J Cell Sci 122:4195- 4207. [DOI] [PubMed]
- Pierozan P, Biasibetti H, Schmitz F, Avila H, Fernandes CG, Pesooa-Pureur R, Wyse AT. Neurotoxicity of methylmercury in isolated astrocytes and neurons: the cytoskeleton as a main target. Mol Neurobiol. 2016;54:5752–5767. doi: 10.1007/s12035-016-0101-2. [DOI] [PubMed] [Google Scholar]; Pierozan P, Biasibetti H, Schmitz F, Avila H, Fernandes CG, Pesooa-Pureur R and Wyse AT (2016) Neurotoxicity of methylmercury in isolated astrocytes and neurons: the cytoskeleton as a main target. Mol Neurobiol 54:5752-5767. [DOI] [PubMed]
- Ramaekers F, Bosman F. The cytoskeleton and disease. J Pathol. 2004;204:351–354. doi: 10.1002/path.1665. [DOI] [PubMed] [Google Scholar]; Ramaekers F and Bosman F (2004) The cytoskeleton and disease. J Pathol 204:351–354. [DOI] [PubMed]
- Ramos-Espinosa P, Rojas E, Valverde M. Differential DNA damage response to UV and hydrogen peroxide depending of differentiation stage in a neuroblastoma model. Neurotoxicology. 2012;33:1086–1095. doi: 10.1016/j.neuro.2012.05.017. [DOI] [PubMed] [Google Scholar]; Ramos-Espinosa P, Rojas E and Valverde M (2012) Differential DNA damage response to UV and hydrogen peroxide depending of differentiation stage in a neuroblastoma model. Neurotoxicology 33:1086–1095. [DOI] [PubMed]
- Reynolds CP, Biedler JL, Spengler BA, Reynolds DA, Ross RA, Frenkel EP, Smith RG. Characterization of human neuroblastoma cell lines established before and after therapy. J Natl Cancer Inst. 1986;76:375–387. [PubMed] [Google Scholar]; Reynolds CP, Biedler JL, Spengler BA, Reynolds DA, Ross RA, Frenkel EP and Smith RG (1986) Characterization of human neuroblastoma cell lines established before and after therapy. J Natl Cancer Inst 76:375–387. [PubMed]
- Rojas E, Shi ZZ, Valverde M, Paules RS, Habib GM, Lieberman MW. Survival and changes in gene expression in cells unable to synthesize glutathione. BioFactors. 2003;17:13–19. doi: 10.1002/biof.5520170102. [DOI] [PubMed] [Google Scholar]; Rojas E, Shi ZZ, Valverde M, Paules RS, Habib GM and Lieberman MW (2003) Survival and changes in gene expression in cells unable to synthesize glutathione. BioFactors 17:13–19. [DOI] [PubMed]
- Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]; Schägger H (2006). Tricine-SDS-PAGE. Nat Protoc 1:16–22. [DOI] [PubMed]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfed A, Schmid B, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfed A, Schmid B et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9:676–682. [DOI] [PMC free article] [PubMed]
- Shi ZZ, Osei-Frimpong J, Kala G, Kala SV, Barrios RJ, Habib GM, Lukin DJ, Danney CM, Matzuk MM, Lieberman MW. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc Natl Acad Sci U S A. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi ZZ, Osei-Frimpong J, Kala G, Kala SV, Barrios RJ, Habib GM, Lukin DJ, Danney CM, Matzuk MM and Lieberman MW (2000) Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc Natl Acad Sci U S A 97:5101–5106. [DOI] [PMC free article] [PubMed]
- Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistence. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]; Siddik ZH (2003). Cisplatin: Mode of cytotoxic action and molecular basis of resistence. Oncogene 22:7265-7279. [DOI] [PubMed]
- Stabel-Burow J, Kleu A, Schuchmann S, Heinemann U. Glutathione levels and nerve cell loss in hippocampal cultures from trisomy 16 mouse- a model of Down syndrome. Brain Res. 1997;765:313–318. doi: 10.1016/s0006-8993(97)00480-0. [DOI] [PubMed] [Google Scholar]; Stabel-Burow J, Kleu A, Schuchmann S and Heinemann U (1997) Glutathione levels and nerve cell loss in hippocampal cultures from trisomy 16 mouse- a model of Down syndrome. Brain Res 765:313-318. [DOI] [PubMed]
- Sun HQ, Yamamoto M, Mejillano M, Yin HL. Regulatory protein. J Biol Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]; Sun HQ, Yamamoto M, Mejillano M and Yin HL (1999) Regulatory protein. J Biol Chem 274:33179–33182. [DOI] [PubMed]
- Valverde M, Rojas E, Kala SV, Kala G, Lieberman MW. Survival and cell death in cells constitutively unable to synthesize glutathione. Mutat Res. 2006;594:172–180. doi: 10.1016/j.mrfmmm.2005.08.004. [DOI] [PubMed] [Google Scholar]; Valverde M, Rojas E, Kala SV, Kala G and Lieberman MW (2006) Survival and cell death in cells constitutively unable to synthesize glutathione. Mutat Res 594:172–180. [DOI] [PubMed]
- Wang D, Lippard SJ. Cellular processing of platinium anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]; Wang D and Lippard SJ (2005) Cellular processing of platinium anticancer drugs. Nat Rev Drug Discov 4:307-320 [DOI] [PubMed]
- Wilson C, Terman JR, Gonzalez-Billault C, Ahmed G. Actin filaments - a target for redox regulation. Cytoskeleton. 2016;73:577–595. doi: 10.1002/cm.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wilson C, Terman JR, Gonzalez-Billault C and Ahmed G (2016) Actin filaments - a target for redox regulation. Cytoskeleton 73:577–595. [DOI] [PMC free article] [PubMed]
- Winder SJ, Ayscough KR. Actin-binding proteins. J Cell Sci. 2005;118:651–654. doi: 10.1242/jcs.01670. [DOI] [PubMed] [Google Scholar]; Winder SJ and Ayscough KR (2005) Actin-binding proteins. J Cell Sci 118:651–654. [DOI] [PubMed]
- Ying GX, Liu X, Wang WY, Wang Y, Dong JH, Jin HF, Huang C, Zhou CF. Regulated transcripts in the hippocampus following transections of the entorhinal afferents. Biochem Biophys Res Commun. 2004;322:210–216. doi: 10.1016/j.bbrc.2004.07.099. [DOI] [PubMed] [Google Scholar]; Ying GX, Liu X, Wang WY, Wang Y, Dong JH, Jin HF, Huang C and Zhou CF (2004) Regulated transcripts in the hippocampus following transections of the entorhinal afferents. Biochem Biophys Res Commun 322:210–216. [DOI] [PubMed]