Abstract
The advent of novel high-throughput sequencing methods has facilitated identification of non-coding RNAs with fundamental roles in cellular biological and pathological conditions. A group of these consisting of at least 200 nucleotides are called long non-coding RNAs (lncRNAs). Their participation in the pathogenesis of cancer has been highlighted in recent years. Bladder cancer, one of the most prevalent cancers worldwide, exhibits altered expression levels of several lncRNAs. Several in vitro and in vivo studies have assessed the effects of silencing RNAs on cancer cell phenotypes and in vivo tumor growth. For instance, in vitro studies have shown that nuclear paraspeckle assembly transcript 1 (NEAT1), promoter of CDKN1A antisense DNA damage-activated RNA(PANDAR) and metastasis-associated lung adenocarcinoma transcript 1(MALAT1) have oncogenic effects while Maternally expressed 3 (MEG3) and BRAF activated non-coding RNA (BANCR) are tumor suppressors. Analysis of these data will help to identify a panel of lncRNAs that can be potentially used for both early detection and prognosis in bladder cancer patients. Here, we review the roles of several lncRNAs in the oncogenesis, tumor suppression, early detection, and prognosis of bladder cancer.
Keywords: lncRNA, Bladder cancer, Oncogenesis, Tumor suppression, Cancer prognosis
Introduction
The human genome comprises several non-coding RNAs including both microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) that play fundamental regulatory roles in almost every aspect of cellular functions (Dianatpour and Ghafouri-Fard 2017; Nikpayam et al. 2017b; Soudyab et al. 2016). Recently, lncRNAs have gained the attention of researchers in the field of cancer biology because of their differential expression in tumoral tissues compared with the corresponding non-tumoral tissues, and it is clear they play a role in carcinogenesis (Iranpour et al. 2016; Nikpayam et al. 2017a; Sarrafzadeh et al. 2017a, b; Tasharrofi et al. 2016). Several single nucleotide polymorphisms (SNPs) within lncRNAs influence the risk of cancer in different populations (Khorshidi et al. 2017a, b; Taheri et al. 2017a, b). More importantly, lncRNAs are involved in tumor initiation and progression by modulating the self-renewal and differentiation cancer stem cell (CSC) (Huang et al. 2017). Considering the crucial role of CSCs in tumor recurrence and metastasis, as well as the advent of novel CSC-targeted therapies (Tabarestani and Ghafouri-Fard 2012), expression analysis of lncRNAs in cancers is likely to be of practical value.
Bladder cancer is one of the most frequent cancers worldwide (Ghafouri-Fard et al. 2014). Despite the diverse treatment options for patients with bladder cancer (surgery, chemotherapy and radiation therapy), their survival rate has not significantly improved. The high incidence of bladder cancer and its notable recurrence and mortality rates have prompted researchers to find biomarkers that can facilitate its early detection, and improve its prognosis and/or treatment responses in patients (Ghafouri-Fard et al. 2014).
The emergence of high-throughput transcriptome sequencing techniques has assisted in the identification of versatile bladder cancer biomarkers including lncRNAs whose aberrant expression in bladder tissues contributes in tumorigenesis (Zhang et al. 2013).
LncRNAs function in bladder carcinogenesis
LncRNAs can influence oncogenesis and tumor formation in bladder tissue. These functions and their pattern of expression in tumoral tissues (compared with normal tissues) can be divided to two distinct groups summarized in Table 1.
Table 1.
Genomic location, function and expression pattern of lncRNAs in bladder cancer
| LncRNA | Chromosomal location | Expression pattern in bladder cancer | Involvement in other cancers | Function / characteristics |
|---|---|---|---|---|
| Linc00346 | 13q34 | Up-regulated | – | Promotes tumor progression via regulating PI3K/AKT signaling pathways |
| MALAT1 | 11q13.1 | Up-regulated | B-cell neoplasms, bladder, breast, cervical, colon, endometrial, gallbladder, hepatocelluar carcinoma, kidney, liver, lung laryngeal and oral squamous cell carcinoma, nasopharyngeal carcinoma, neuroblastoma, osteosarcoma, pancreas, prostate, uterus, glioblastoma, renal cell carcinoma, multiple myeloma | Promotes cancer cell proliferation, invasion, and metastasis, an important mediator of TGF-β-induced EMT |
| UCA1 | 19p13.12 | Up-regulated | Squamous carcinoma, hepatocellular carcinoma, gastric cancer | Regulates cell cycle distribution via CREB through PI3-K dependent pathway, increases chemoresistance of bladder cancer cells by regulating Wnt signaling |
| PCAT1 | 8q24.21 | Up-regulated | Prostate cancer | Regulates BRCA2 and controls homologous recombination in cancer, contribute to cell proliferation |
| HOTAIR | 12q13.13 | Up-regulated | B-cell neoplasms, breast, cervical, colorectal, ovarian, Esophageal squamous cell cancer, gastrointestinal, hepatocelluar carcinoma, pancreas | Epigenetically silences gene expression via LSD1/CoREST and PRC2 |
| TUG1 | 22q12.2 | Up-regulated | B-cell neoplasms/non-small cell lung cancer gastric cancer/ osteosarcoma colorectal cancer, esophageal squamous cell carcinoma, gastric cancer, hepatocellular carcinoma, and bladder cancer | Required for retinal development |
| NEAT1 | 11q13.1 | Up-regulated | Oral squamous cell carcinoma, breast hepatocellular carcinoma, lung cancer | A structural component of nuclear paraspeckles, involved in proliferation, cell migration oncogenic roles |
| GAS5 | 1q25.1 | Down-regulated | Breast, kidney, lymphoma, melanoma, prostate, hepatocellular carcinoma, gastric and ovarian | Controls apoptosis, inhibits bladder cancer cell proliferation by regulating CDK6 expression and suppressing the expression of CCL1 |
| Linc00312 | 3p25.3 | Down-regulated | Nasopharyngeal carcinoma, lung carcinomas | Modulation of cell cycle progression and cell apoptosis |
| BANCR | 9q21.11-q21.12 | Down-regulated | Melanoma, non-small cell lung cancer, Colorectal cancer, retinoblastoma, melanoma papillary thyroid carcinoma, gastric cancer, hepatocellular carcinoma | Regulation of cell proliferation, apoptosis, and migration |
| Loc572558 | 9q21.11 | Down-regulated | – | Regulates the p53 signaling pathway |
| HIF1A-AS2 | 14q23.2 | Up-regulated | Kidney cancer, gastriccarcinomas | – |
| Schlap1 | 2q31.3 | Up-regulated | Prostate cancer | Critical for tumor cell metastasis |
| MIR31HG | 9p21.3 | Down-regulated | Breast, colorectal, gastric pancreas | Functions as a cancer-suppressor gene |
| PVT1 | 8q24.21 | Up-regulated | Breast, Burkitt’s lymphomas, ovarian, pancreas, prostate, renal, gastric, hepatocellular carcinoma | Promotes cell proliferation and suppresses cell apoptosis |
| ANRIL | 9p21.3 | Up-regulated | Prostate, hepatocellular carcinoma, lung, ovarian, gastric, breast, esophageal squamous cell carcinoma | Regulates ADIPOR1, VAMP3 and C11ORF10, regulate bladder cancer cell proliferation and apoptosis |
| H19 | 11p15.5 | Up-regulated | Adrenocortical carcinomas, bladder, breast, cervical, gastric, hepatocellular, kidney, liver, lung, ovarian, neuroblastoma | Control of imprinting |
| MDC1-AS | 6p21.33 | Down-regulated | Bladder cancer | Repair of double-strand breaks |
| GHET1 | 7q36.1 | Up-regulated | Gastric cancer | Promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability |
| MEG3 | 14q32.2 | Down-regulated | Acute myeloid leukemia, chronic myeloid leukemia, colon, gastric, hepatocelluar carcinoma, kidney, lung, prostate | Regulation of autophagy activation |
| CCAT2 | 8q24.21 | Up-regulated | Non-small cell lung cancer/ breast colorectal cancer | Control of cell proliferation, migration and apoptosis |
| SNHG16 | 17q25.1 | Up-regulated | Colorectal cancer | Affects genes involved in lipid metabolism |
| DBCCR1 | 9q32–33 | Down-regulated | Lung cancer | Regulates the expression of DBCCR1via DNMT1 |
| TINCR | 19p13.3 | Up-regulated | Squamous cell carcinoma gastric carcinoma | Regulation of cell proliferation and apoptosis |
| PANDAR | 6p21.2 | Up-regulated | Renal cell carcinoma, thyroid, colorectal cancer | Interacts withthe transcription factor NF-YA to limit the expression of pro-apoptotic genes |
| SUMO1P3 | 1q23.2 | Up-regulated | Gastric cancer | – |
| lncRNA-n336928 | – | Up-regulated | – | – |
Data obtained from databases deepbase.sysu.edu.cn/chipbase, diana.imis.athena-innovation.gr, www.lncipedia.org, www.lncrnadb.org, cmbi.bjmu.edu.cn/lncrnadisease, genome.igib.res.in/lncRNome, noncode.org/NONCODERv3, www.valadkhanlab.org
Oncogenic lncRNAs in bladder cancer
Many oncogenic lncRNAs have been shown to exert anti-apoptotic effects. For instance, in vitro studies have shown that nuclear paraspeckle assembly transcript 1 (NEAT1) and promoter of CDKN1A antisense DNA damage-activated RNA (PANDAR) exert pro-proliferative and anti-apoptotic effects in bladder cancer cell lines and enhance cell migration (XianGuo et al. 2016; Zhan et al. 2016a). The trigger for the expression of PANDAR depends on the tumor protein, p53. This lncRNA cooperates with the transcription factor NF-YA to inhibit the expression of pro-apoptotic genes (Guttman and Rinn 2012). The lncRNA hypoxia-inducible factor 1alpha antisense RNA-2 (HIF1A-AS2) is up-regulated in bladder cancer tissues and cells, and its short hairpin RNA-mediated knock-down results in suppression of both cell proliferation and migration while triggering apoptosis. In addition, forced over-expression of HIF1A-AS2 in healthy human uroepithelial cells enhanced cell proliferation and migration while suppressing apoptosis (Chen et al. 2016a). The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been shown to be up-regulated in bladder cancer. Both in vivo and in vitro studies have shown that there is a negative correlation between the expression of MALAT1 and the epithelial cadherin (E-cadherin). MALAT1 silencing prevents transforming growth factor (TGF-β)-induced epithelial-mesenchymal transition (EMT). Furthermore, the effects of MALAT1 knock-down on the inhibition of tumor metastasis have been confirmed in animal models (Fan et al. 2014). Ying et al. (2012) and Fan et al. (2014) have both demonstrated that MALAT1 exerts its role in cancer progression and metastasis by enhancing EMT.
Another study has demonstrated that with gastric carcinoma high expressed transcript 1 (GHET1, AK123072) silencing leads to inhibition of bladder cancer cells proliferation through the induction of G0/G1 arrest while reversing the EMT in these cells which led to suppression of invasion (Li et al. 2014). The latter effect coincided with decreased vimentin and fibronectin expression and increased E-cadherin expression (Li et al. 2014).
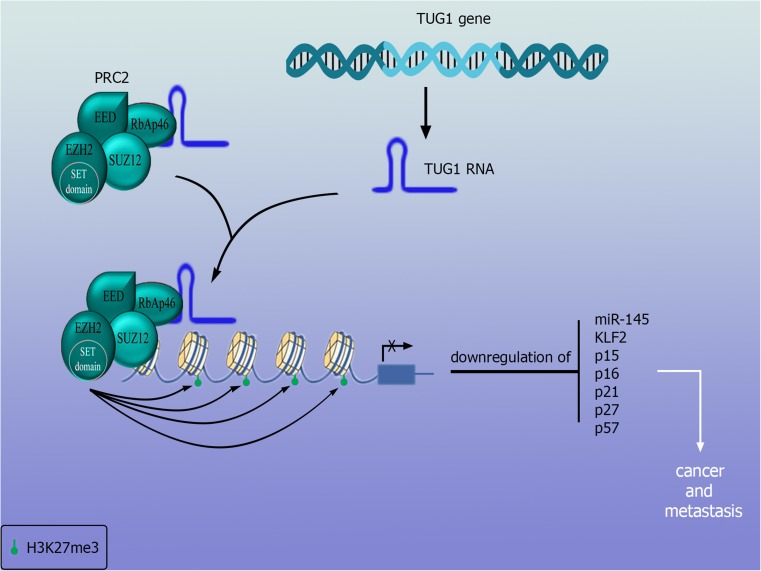
LncRNA taurine up-regulated gene 1 (TUG1) knock- down diminishes cancer cell proliferation and migration capacity without influencing cell cycle distribution and apoptosis (Iliev et al. 2016). An earlier study showed that participation of TUG1 in the invasion of bladder cancer by enhancing EMT is achieved by down-regulation of the microRNA, miR-145, an inhibitor of EMT-inducer ZEB2 (Tan et al. 2015). The participation of TUG1 in cancer development and metastasis is illustrated in Fig. 1. Recently, it was reported that TUG1 silencing suppresses proliferation and triggers apoptosis in bladder cancer cells by inhibiting the Wnt/β-catenin pathway (Liu et al. 2017).
Fig. 1.
TUG1 participation in bladder cancer: TUG1 interacts with PRC2 and EZH2 and promotes methylation of several proteins with inhibitory effects in cell cycle pregreesion
Long intergenic non-coding 00346 (LINC00346) silencing can prevent cell proliferation and migration in bladder cancer and can trigger cell cycle arrest and cell apoptosis. In addition, implantation of these bladder cancer cells into nude mice lowers the rate of tumor growth in mice (Ye et al. 2017).
Silencing of second chromosome locus associated with prostate-1 (SChLAP1) has the same effects as LINC00346 on cell proliferation, invasion and migration. However, there are no reports of its in vivo effects (Zhang et al. 2016).
The lncRNA small nucleolar RNA host gene 16 (SNHG16) is considered to be a tumor suppressor gene in colorectal cancer (Qi et al. 2015). Its overexpression is significantly correlated with aggressive bladder cancer and its knock-down can enhance the effect of chemotherapy in bladder cancer cell lines (Zhu et al. 2011).
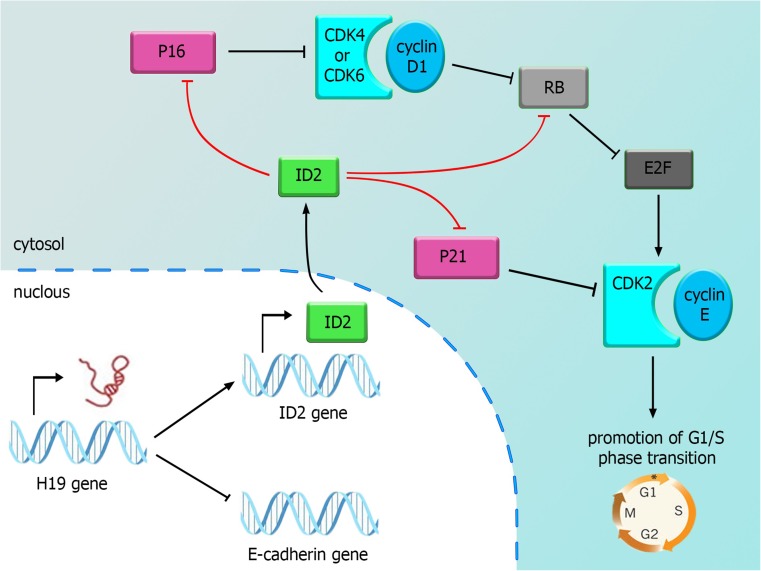
The long non-coding RNA 19 (H19) exerts a crucial role in human tumor growth. Its expression is significantly up-regulated in bladder cancer tissues compared with adjacent normal control tissues. Forced expression of H19 increases bladder cancer cell proliferation coincided with a significant increase in the expression of inhibitor of DNA binding/differentiation 2 (ID2) (Luo et al. 2013a). The consequences of this expression change are depicted in Fig. 2. Another study has shown that elevated levels of H19 increases bladder cancer cell migration in vitro and in vivo. The association of H19 with enhancer of zeste homolog 2 (EZH2) leads to stimulation of the Wnt/β-catenin pathway resulting in down-regulation of E-cadherin. Consequently, H19 promotes bladder cancer metastasis by interacting with EZH2 and suppressing E-cadherin expression (Luo et al. 2013b).
Fig. 2.
H19 participation in bladder cancer: H19 overexpression leads to upregulation of ID2 and down-regulation of E-cadherin. The former effect results in suppression of Rbm p16 and p21. The latter leads to metastasis
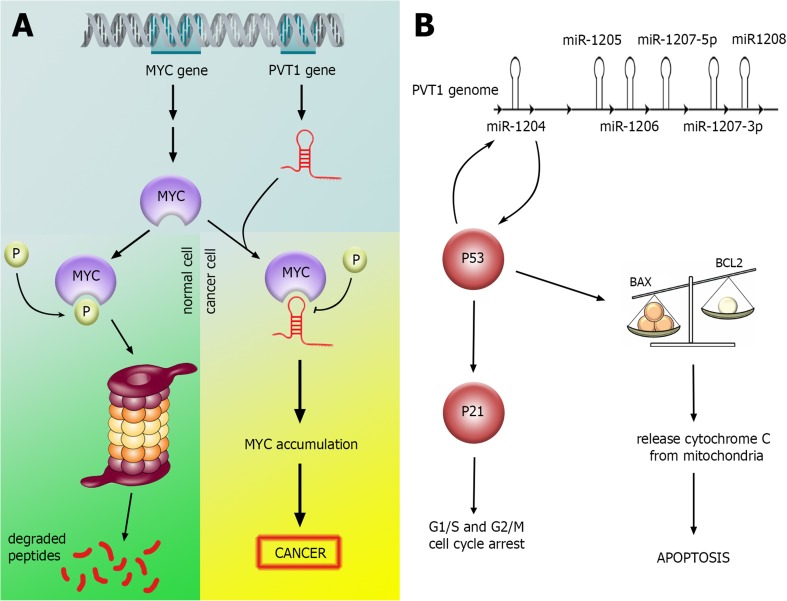
The non-protein coding lncRNAs plasmacytoma variant translocation 1 (PVT1) and colon cancer-associated transcript 2 (CCAT2) are up-regulated in bladder cancer tissues and cells. They both reside in chromosome 8q24 adjacent to MYC proto-oncogene and have pro-proliferative and anti-apoptotic effects in these cells (Li et al. 2016; Zhuang et al. 2015). The interaction of PVT1 with MYC is summarized in Fig. 3.
Fig. 3.
PVT1 participation in bladder cance. a PVT1 overexpression inhibits MYC phosphorylation and its degaradtion in proteasomes. b PVT1 locus acts as a molecular switch for determination of cell fate
Small ubiquitin-like modifier (SUMO) and the SUMO1 pseudogene 3 (SUMO1P3) are other oncogenic lncRNAs whose silencing suppresses proliferation and migration in addition to inducing apoptosis (Zhan et al. 2016b). Also, AB074278 is up-regulated in bladder cancer patients and exerts anti-apoptotic and pro-proliferative effects, possibly through an interaction with EMP1, a tumor suppressor and a negative regulator of cell proliferation (Peter et al. 2014).
While terminal differentiation-induced ncRNA (TINCR) is down-regulated in human squamous cell carcinoma (Kretz et al. 2013), it has been demonstrated to be up-regulated in bladder cancer tissues and cells, and participates in cancer development and progression (Chen et al. 2016b).
HOX transcript antisense RNA (HOTAIR) participates in bladder cancer pathogenesis by suppression of miR-205, a negative-regulator of bladder cell proliferation, migration and invasion (Sun et al. 2015).
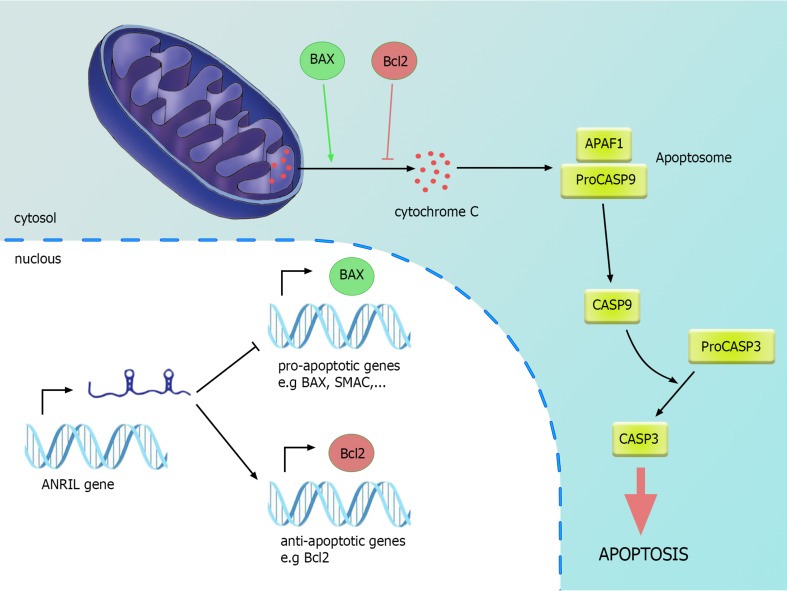
Antisense non-coding RNA in the INK4 locus (ANRIL) overexpression has been demonstrated in bladder cancer compared with the matched adjacent non-tumor tissues. Its silencing resulted in inhibition of cell proliferation and induction of cell apoptosis, together with diminished expression of Bcl-2 and elevated expressions of Bax, cytoplasmic cytochrome c, and Smac and cleaved caspase-9, caspase-3 and PARP. In vivo studies endorsed the effect of ANRIL silencing in the suppression of tumorigenicity of bladder cancer cells in nude mice. The effects of ANRIL in regulating apoptosis through the intrinsic apoptosis pathway are demonstrated in Fig. 4.
Fig. 4.
ANRIL participation in bladder cancer: ANRIL overexpression lead to up-regulation of anti-apoptotic genes while down-regulation of pro-apoptotic genes
Tumor suppressor lncRNAs in bladder tissues
The levels of expression of maternally expressed 3 (MEG3) are decreased in bladder cancer through epigenetic mechanisms (Greife et al. 2014). This change in expression interferes with bladder cancer cell apoptosis while enhancing cell proliferation by induction of autophagy (Ying et al. 2013).
BRAF-activated non-coding RNA (BANCR) expression is also significantly down-regulated in bladder cancer cell lines and patients compared with adjacent non-cancerous tissues. Forced over-expression of BANCR in bladder cancer cells has led to a significant decrease in cell proliferation and migration together with a remarkable increase in apoptosis. However, considering the different expression levels of BANCR in various malignancies, this lncRNA may be involved in different signaling pathways and exert its distinctive role in each cancer (He et al. 2016b).
The lncRNA-growth arrest-specific 5 (GAS5) is regarded as a tumor suppressor in bladder cancer because the knock-down of this gene increases bladder cancer cell proliferation, while its forced overexpression inhibited cell proliferation (Cao et al. 2016). In addition, GAS5 silencing up-regulates CDK6 mRNA and protein levels in bladder cancer cells leading to a significant increase in S phase (Liu et al. 2013). A more recent study has revealed that forced overexpression of GAS5 decreases chemotherapy resistance to doxorubicin, increases doxorubicin-induced apoptosis, and inhibits the expression of the anti-apoptosis protein Bcl-2 (Zhang et al. 2017).
The expression levels of mediator of DNA damage checkpoint protein 1-antisense (MDC1-AS) decrease in bladder cancer. This lncRNA suppresses the development of malignant cell phenotype of bladder cancer cells. It may participate in bladder cancer by enhancing the expression of its antisense tumor-suppressing gene MDC1, which has an important role in the repair of DNA damage (Xue et al. 2015).
LINC00312 expression is also decreased in bladder cancer compared with adjacent unaffected tissues. Forced over-expression of LINC00312 suppresses cell migration and invasion by targeting miR-197-3p. Such an effect was accompanied by low MMP-2 and MMP-9 levels and high TIMP2 level (Wang et al. 2016).
Finally, LOC572558 is another lncRNA whose expression is considerably decreased in bladder cancer and its derived cell lines. Forced expression of LOC572558 suppresses cell proliferation and motility by triggering the arrest of S phase of the cell cycle, and by inducing cell apoptosis in bladder cancer cell lines. Notably, such effects are accompanied by dephosphorylation of AKT, MDM2 and of p53 protein. Consequently, LOC572558 is regarded as a tumor suppressor that exerts its effects through the modulation of the p53 signaling pathway in bladder cancer (Zhu et al. 2016).
The role of lncRNAs in early detection of bladder cancer
Duan et al. recently evaluated the expression of 13 candidate lncRNAs in bladder cancer matched to adjacent healthy tissue. They reported a panel of differentially expressed lncRNAs that was subsequently analyzed using serum samples. Differential expression of three lncRNAs (MEG3, SNHG16 and MALAT1) was observed in the serum of healthy subjects compared to patients with bladder cancer, as well as in patients with benign disease. They suggested this panel might be useful in the diagnosis of bladder cancer (Duan et al. 2016).
The role of lncRNAs in the assessing the prognosis of bladder cancer
Expression of several lncRNAs (NEAT1, PANDAR, PVT1, SUMO1P3, CCAT2 and HIF1A-AS2) in bladder cancer is statistically associated with histological grading and TNM stage of bladder cancer (Chen et al. 2016a; Li et al. 2016; XianGuo et al. 2016; Zhan et al. 2016a, b; Zhuang et al. 2015). In addition, expression of lncRNA-n336928 positively correlates with bladder tumor stage, histological grade, and patient survival (Chen et al. 2015). GHET1 expression is elevated in bladder cancer compared to adjacent unaffected tissues, and its up-regulation correlates with tumor size, using advanced tumor and lymph node status, and poor survival (Li et al. 2014).
Elevated levels of MALAT1 expression are associated with higher grades of histological assessment and tumor stage, as well as lymph node metastasis in these patients (Li et al. 2017). MALAT1 overexpression is indicative of poor survival in these patients (Fan et al. 2014; Li et al. 2017).
TUG1 levels elevated in metastatic bladder tumors and are associated with poor survival in muscle-invasive bladder cancer patients (Iliev et al. 2016). However, TINCR expression levels have only been associated with advanced TNM stage (Chen et al. 2016b). On the other hand, low expression of BANCR and MIR31HG positively correlates with TNM stage (He et al. 2016a; He et al. 2016b). Moreover, down-regulation of MEG3 expression is associated with lower recurrence-free survival (Duan et al. 2016). GAS5 down-regulation in bladder cancer is correlated with higher pathological grades and lower disease-free survival (Zhang et al. 2017).
Discussion
In spite of advances in the fields of cancer detection and treatment, bladder cancer mortality rate is persistly high as a result of its high occurrence rate, late diagnosis and resistance to therapies (XianGuo et al. 2016). This necessitates the search for cancer biomarkers with the ability to be applied as therapeutic targets. Aberrant expression of lncRNAs in bladder cancer and their involvement in cancer-related pathways highlights them as suitable targets. A microarray-based expression study showed that 17,112 lncRNAs of 22,074 mRNAs in bladder cancer samples had differentially expressed lncRNAs compared to healthy samples, and that the global pattern of changed expression was more immense than those of protein coding mRNAs (Peter et al. 2014). More recently, expression analysis of lncRNAs in serum or plasma has been suggested as a novel approach for early detection of cancer. Few studies have confirm the feasibility of this method for diagnosis of bladder cancer (Duan et al. 2016). The notable stability of circulating cell-free lncRNAs in different conditions possibly results from their protection by extracellular vesicles, certain folding structures, or by binding to a circulating protein (Duan et al. 2016), and is further evidence for their application as cancer biomarkers. Introduction of a panel of differentially expressed lncRNAs rather than a single lncRNAs is a more promising approach for early detection of cancer.
Several in vitro studies have supported the efficiency of lncRNA knock-down strategies for the induction of apoptosis or inhibition of cell proliferation and migration in bladder cancer cells. Although there is little evidence from animal studies to support this idea, in vivo studies have confirmed the efficiency of this strategy in other cancers (Sun et al. 2016). It is worth emphasizing that several lncRNAs have been demonstrated to exert distinct roles (oncogenic vs. tumor suppressor role) in different cancers, which is perhaps due to the relative significance of certain signaling pathways in each cancer type or the more tissue-specific lncRNAs signature as compared with protein-coding genes. Consequently, the role of each lncRNA should be assessed in distinctive biological contexts. Such expression analyses would pave the way for identifying of the processes leading to bladder cancer initiation and/or progression, such as enhanced cancer cell invasion and proliferation, in addition to factors leading to treatment resistance.
Compliance with ethical standards
Mohammad Taheri declares that he has no conflicts of interest. Mir Davood Omrani declares that he has no conflicts of interest. Soudeh Ghafouri-Fard declares that she has no conflicts of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Cao Q, Wang N, Qi J, Gu Z, Shen H. Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (C-C motif) ligand 1 expression. Mol Med Rep. 2016;13:27–34. doi: 10.3892/mmr.2015.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhuang C, Liu Y, Li J, Dai F, Xia M, Zhan Y, Lin J, Chen Z, He A, Xu W, Zhao G, Guo Y, Cai Z, Huang W. Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Canc Lett. 2016;376:155–164. doi: 10.1016/j.canlet.2016.03.037. [DOI] [PubMed] [Google Scholar]
- Chen T, Xie W, Xie L, Sun Y, Zhang Y, Shen Z, Sha N, Xu H, Wu Z, Hu H, Wu C. Expression of long noncoding RNA lncRNA-n336928 is correlated with tumor stage and grade and overall survival in bladder cancer. Biochem Biophys Res Commun. 2015;468:666–670. doi: 10.1016/j.bbrc.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Chen Z, Liu Y, He A, Li J, Chen M, Zhan Y, Lin J, Zhuang C, Liu L, Zhao G, Huang W, Cai Z. Theophylline controllable RNAi-based genetic switches regulate expression of lncRNA TINCR and malignant phenotypes in bladder cancer cells. Sci Rep. 2016;6:30798. doi: 10.1038/srep30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianatpour A, Ghafouri-Fard S. The role of long non coding RNAs in the repair of DNA double strand breaks. Int J Mol Cell Med. 2017;6:1–12. [PMC free article] [PubMed] [Google Scholar]
- Duan W, Du L, Jiang X, Wang R, Yan S, Xie Y, Yan K, Wang Q, Wang L, Zhang X, Pan H, Yang Y, Wang C. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7:78850–78858. doi: 10.18632/oncotarget.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-β–induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Canc Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- Ghafouri-Fard S, Nekoohesh L, Motevaseli E. Bladder cancer biomarkers: review and update. Asian Pac J Canc Prev. 2014;15:2395–2403. doi: 10.7314/APJCP.2014.15.6.2395. [DOI] [PubMed] [Google Scholar]
- Greife A, Knievel J, Ribarska T, Niegisch G, Schulz WA. Concomitant downregulation of the imprinted genes DLK1 and MEG3 at 14q32.2 by epigenetic mechanisms in urothelial carcinoma. Clin Epigenet. 2014;6:29. doi: 10.1186/1868-7083-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He AB, Chen ZC, Mei HB, Liu YC. Decreased expression of LncRNA MIR31HG in human bladder cancer. Canc Biomark. 2016;17:231–236. doi: 10.3233/CBM-160635. [DOI] [PubMed] [Google Scholar]
- He AB, Liu Y, Chen Z, Li J, Chen M, Liu L, Liao X, Lv Z, Zhan Y, Zhuang C, Lin J, Huang W, Mei H. Over-expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J Exp Clin Canc Res. 2016;35:125. doi: 10.1186/s13046-016-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XX, Xiao R, Pan S, Yang X, Yuan W, Tu Z, Xu M, Zhu Y, Yin Q, Wu Y, Hu W, Shao L, Xiong J, Zhang Q. Uncovering the roles of long non-coding RNAs in cancer stem cells. J Hematol Oncol. 2017;10:62. doi: 10.1186/s13045-017-0428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev R, Kleinova R, Juracek J, Dolezel J, Ozanova Z, Fedorko M, Pacik D, Svoboda M, Stanik M, Slaby O. Overexpression of long non-coding RNA TUG1 predicts poor prognosis and promotes cancer cell proliferation and migration in high-grade muscle-invasive bladder cancer. Tumor Biol. 2016;37:13385–13390. doi: 10.1007/s13277-016-5177-9. [DOI] [PubMed] [Google Scholar]
- Iranpour M, Soudyab M, Geranpayeh L, Mirfakhraie R, Azargashb E, Movafagh A, Ghafouri-Fard S. Expression analysis of four long noncoding RNAs in breast cancer. Tumor Biol. 2016;37:2933–2940. doi: 10.1007/s13277-015-4135-2. [DOI] [PubMed] [Google Scholar]
- Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri-Fard S. ANRIL genetic variants in Iranian breast cancer patients. Cell J. 2017;19:72–78. doi: 10.22074/cellj.2017.4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorshidi HR, Taheri M, Noroozi R, Soudyab M, Sayad A, Ghafouri-Fard S. Investigation of the association of HOTAIR single nucleotide polymorphisms and risk of breast cancer in an Iranian population. Int J Canc Manag. 2017;10:e7498. [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Cui Y, Liu LF, Ren WB, Li QQ, Zhou X, Li YL, Li Y, Bai XY, Zu XB. High expression of long non-coding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin Genitourin Canc. 2017;15:570–576. doi: 10.1016/j.clgc.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Li J, Zhuang C, Liu Y, Chen M, Zhou Q, Chen Z, He A, Zhao G, Guo Y, Wu H, Cai Z, Huang W. shRNA targeting long non-coding RNA CCAT2 controlled by tetracycline-inducible system inhibits progression of bladder cancer cells. Oncotarget. 2016;7:28989–28997. doi: 10.18632/oncotarget.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LJ, Zhu JL, Bao WS, Chen DK, Huang WW, Weng ZL. Long noncoding RNA GHET1 promotes the development of bladder cancer. Int J Clin Exp Pathol. 2014;7:7196–7205. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu H, Cheng H, Li Y, Li X, Zhu C. Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. OncoTarget Ther. 2017;10:2461–2471. doi: 10.2147/OTT.S124595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang W, Jiang J, Bao E, Xu D, Zeng Y, Tao L, Qiu J. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS ONE. 2013;8:e73991. doi: 10.1371/journal.pone.0073991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280:1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- Luo M, Li ZW, Wang W, Zeng YG, Liu ZH, Qiu JX. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Canc Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Nikpayam E, Soudyab M, Tasharrofi B, Sarrafzadeh S, Iranpour M, Geranpayeh L, Mirfakhraie R, Gharesouran J, Ghafouri-Fard S (2017a) Expression analysis of long non-coding ATB and its putative target in breast cancer. Breast Dis 1-10 [DOI] [PubMed]
- Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S. The role of long non-coding RNAs in ovarian cancer. Iran Biomed J. 2017;21:3–15. doi: 10.18869/acadpub.ibj.21.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter S, Borkowska E, Drayton RM, Rakhit CP, Noon AP, Chen W, Catto JW. Identification of differentially expressed long non-coding RNAs in bladder cancer. Clin Canc Res. 2014;7:78850–78858. doi: 10.1158/1078-0432.CCR-14-0706. [DOI] [PubMed] [Google Scholar]
- Qi P, Xu MD, Ni SJ, Shen XH, Wei P, Huang D, Tan C, Sheng WQ, Zhou XY, Du X. Down-regulation of ncRAN, a long non-coding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Mol Carcinogen. 2015;54:742–750. doi: 10.1002/mc.22137. [DOI] [PubMed] [Google Scholar]
- Sarrafzadeh S, Geranpayeh L, Ghafouri-fard S. Expression analysis of long non-coding PCAT-1. Int J Hematol Oncol Stem Cell Res. 2017;11:185–191. [PMC free article] [PubMed] [Google Scholar]
- Sarrafzadeh S, Geranpayeh L, Tasharrofi B, Soudyab M, Nikpayam E, Iranpour M, Mirfakhraie R, Gharesouran J, Ghafouri-Fard S, Ghafouri-Fard S. Expression study and clinical correlations of MYC and CCAT2 in breast cancer patients. Iran Biomed J. 2017;21:303–311. doi: 10.18869/acadpub.ibj.21.5.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudyab M, Iranpour M, Ghafouri-Fard S (2016) The role of long non-coding RNAs in breast cancer. Arch Iran Med 19: 508-517. Doi: 0161907/AIM.0011 [PubMed]
- Sun L, Xue H, Jiang C, Zhou H, Gu L, Liu Y, Xu C, Xu Q (2016) LncRNA DQ786243 contributes to proliferation and metastasis of colorectal cancer both in vitro and in vivo. Biosci Rep 36. 10.1042/BSR20160048 [DOI] [PMC free article] [PubMed] [Retracted]
- Sun X, Du P, Yuan W, Du Z, Yu M, Yu X, Hu T. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. doi: 10.1038/cddis.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabarestani S, Ghafouri-Fard S. Cancer stem cells and response to therapy. Asian Pac J Canc Prev. 2012;13:5947–5954. doi: 10.7314/APJCP.2012.13.12.5947. [DOI] [PubMed] [Google Scholar]
- Taheri M, Habibi M, Noroozi R, Rakhshan A, Sarrafzadeh S, Sayad A, Omrani MD, Ghafouri-Fard S. HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene. 2017;613:20–24. doi: 10.1016/j.gene.2017.02.031. [DOI] [PubMed] [Google Scholar]
- Taheri M, Pouresmaeili F, Omrani MD, Habibi M, Sarrafzadeh S, Noroozi R, Rakhshan A, Sayad A, Ghafouri-Fard S. Association of ANRIL gene polymorphisms with prostate cancer and benign prostatic hyperplasia in an Iranian population. Biomark Med. 2017;11:413–422. doi: 10.2217/bmm-2016-0378. [DOI] [PubMed] [Google Scholar]
- Tan J, Qiu K, Li M, Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589:3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Tasharrofi B, Soudyab M, Nikpayam E, Iranpour M, Mirfakhraie R, Sarrafzadeh S, Geranpayeh L, Azargashb E, Sayad A, Ghafouri-Fard S. Comparative expression analysis of hypoxia-inducible factor-alpha and its natural occurring antisense in breast cancer tissues and adjacent noncancerous tissues. Cell Biochem Func. 2016;34:572–578. doi: 10.1002/cbf.3230. [DOI] [PubMed] [Google Scholar]
- Wang Y-Y, Wu Z-Y, Wang G-C, Liu K, Niu X-B, Gu S, Meng J-S. LINC00312 inhibits the migration and invasion of bladder cancer cells by targeting miR-197-3p. Tumor Biol. 2016;37:14553–14563. doi: 10.1007/s13277-016-5303-8. [DOI] [PubMed] [Google Scholar]
- XianGuo C, ZongYao H, Jun Z, Song F, GuangYue L, LiGang Z, KaiPing Z, YangYang Z, ChaoZhao L (2016) Promoting progression and clinicopathological significance of NEAT1 over-expression in bladder cancer. Oncotarget. 10.18632/oncotarget.10084
- Xue Y, Ma G, Zhang Z, Hua Q, Chu H, Tong N, Yuan L, Qin C, Yin C, Zhang Z, Wang M. A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer. Oncotarget. 2015;6:484–493. doi: 10.18632/oncotarget.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Ding W, Wang N, Huang H, Pan Y, Wei A. Long noncoding RNA linc00346 promotes the malignant phenotypes of bladder cancer. Biochem Biophys Res Commun. 2017;491:79–84. doi: 10.1016/j.bbrc.2017.07.045. [DOI] [PubMed] [Google Scholar]
- Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol BioSyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol BioSyst. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- Zhan YH, Lin J, Liu Y, Chen M, Chen X, Zhuang C, Liu L, Xu W, Chen Z, He A, Zhang Q, Sun X, Zhao G, Huang W. Up-regulation of long non-coding RNA PANDAR is associated with poor prognosis and promotes tumorigenesis in bladder cancer. J Exp Clin Canc Res. 2016;35:83. doi: 10.1186/s13046-016-0354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan YH, Liu Y, Wang C, Lin J, Chen M, Chen X, Zhuang C, Liu L, Xu W, Zhou Q, Sun X, Zhang Q, Zhao G, Huang W. Increased expression of SUMO1P3 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Oncotarget. 2016;7:16038–16048. doi: 10.18632/oncotarget.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo Y, Song Y, Shang C. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Canc Chemother Pharmacol. 2017;79:49–55. doi: 10.1007/s00280-016-3194-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi Z, Nan Y, Li M. Inhibiting malignant phenotypes of the bladder cancer cells by silencing long noncoding RNA SChLAP1. Int Urol Nephrol. 2016;48:711–716. doi: 10.1007/s11255-016-1230-2. [DOI] [PubMed] [Google Scholar]
- Zhang QA, Su M, Lu GM, Wang JD. The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Canc. 2013;12:101. doi: 10.1186/1476-4598-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dai B, Zhang H, Shi G, Shen Y, Ye D. Long non-coding RNA LOC572558 inhibits bladder cancer cell proliferation and tumor growth by regulating the AKT–MDM2–p53 signaling axis. Canc Lett. 2016;380:369–374. doi: 10.1016/j.canlet.2016.04.030. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yu M, Li Z, Kong C, Bi J, Li J, Gao Z, Li Z. ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology. 2011;77:510. doi: 10.1016/j.urology.2010.09.022. [DOI] [PubMed] [Google Scholar]
- Zhuang C, Li J, Liu Y, Chen M, Yuan J, Fu X, Zhan Y, Liu L, Lin J, Zhou Q, Xu W, Zhao G, Cai Z, Huang W. Tetracycline-inducible shRNA targeting long non-coding RNA PVT1 inhibits cell growth and induces apoptosis in bladder cancer cells. Oncotarget. 2015;6:41194–41203. doi: 10.18632/oncotarget.5880. [DOI] [PMC free article] [PubMed] [Google Scholar]