Abstract
Four and a half LIM domain (FHL) protein family members, FHL1 and FHL2, are multifunctional proteins that are enriched in cardiac muscle. Although they both localize within the cardiomyocyte sarcomere (titin N2B), they have been shown to have important yet unique functions within the context of cardiac hypertrophy and disease. Studies in FHL1-deficient mice have primarily uncovered mitogen-activated protein kinase (MAPK) scaffolding functions for FHL1 as part of a novel biomechanical stretch sensor within the cardiomyocyte sarcomere, which acts as a positive regulator of pressure overload-mediated cardiac hypertrophy. New data have highlighted a novel role for the serine/threonine protein phosphatase (PP5) as a deactivator of the FHL1-based biomechanical stretch sensor, which has implications in not only cardiac hypertrophy but also heart failure. In contrast, studies in FHL2-deficient mice have primarily uncovered an opposing role for FHL2 as a negative regulator of adrenergic-mediated signaling and cardiac hypertrophy, further suggesting unique functions targeted by FHL proteins in the “stressed” cardiomyocyte. In this review, we provide current knowledge of the role of FHL1 and FHL2 in cardiac muscle as it relates to their actions in cardiac hypertrophy and cardiomyopathy. A specific focus will be to dissect the pathways and protein-protein interactions that underlie FHLs’ signaling role in cardiac hypertrophy as well as provide a comprehensive list of FHL mutations linked to cardiac disease, using evidence gained from genetic mouse models and human genetic studies.
Keywords: Cardiomyocyte, Four and half LIM domain, Hypertrophy, Cardiomyopathy, Signaling, Sarcomere, Transcription, Stretch sensor
Introduction
The four and a half LIM domain protein (FHL) family, also known as skeletal muscle LIM (SLIM), is part of the larger LIM only subclass of proteins. LIM domain proteins are important mediators of protein-protein interactions, and thus, they are thought to act as docking sites for multi-protein complex assembly based on their highly conserved cysteine-rich zinc finger-like interaction motifs (Sanchez-Garcia and Rabbitts 1994). FHL proteins are structurally composed of four LIM domains and an N-terminal half LIM domain, and to date, four family members exist, which include FHL1, FHL2, FHL3, and FHL4 that display unique developmental and organ-specific expression patterns (Chu et al. 2000). FHL1 is the only FHL family member that is regulated by alternative splicing resulting in three isoforms (FHL1A (full-length), FHL1B (lacks the last LIM domain and contains a nuclear localization/export signal and a RBP-J binding region), and FHL1C (lack the last 2 LIM domains and contain a RBP-J binding region)) (Shathasivam et al. 2010). FHL1, FHL2, and FHL3 are enriched in striated muscle, with FHL2 exhibiting the most restricted expression pattern in cardiac muscle (Chu et al. 2000). Based on their LIM domains, FHLs are also known to interact with many binding proteins, which range from ion channels to structural proteins (Shathasivam et al. 2010); however, in the heart, they have been shown to have a particularly high affinity to the spring element of the sarcomeric protein titin, which regulates compliance of muscle, as well as signal transducers of the mitogen-activated protein kinase (MAPK)/extracellular-regulated kinase (ERK) with a distinct importance in stress and adrenergic-based cardiac hypertrophy (Raskin et al. 2012; Sheikh et al. 2008). This review will highlight the unique roles of FHL1 and FHL2 in cardiac muscle hypertrophy and disease, with a focus on the signaling pathways and functions that intersect their role in cardiac muscle as well as the implications of FHL mutations in human cardiac diseases.
A role for FHL1 in cardiac hypertrophy
Emerging evidence points to a pivotal role for FHL1 in cardiac hypertrophy (Sheikh et al. 2008). Cardiac hypertrophy is described as an adaptive remodeling response to increased cardiac workload whereby cardiac muscle cells physically increase in size (due to increased protein synthesis and sarcomeric re-organization), resulting in an overall increase in cardiac muscle mass (Nakamura and Sadoshima 2018). Although adaptive during early postnatal cardiac growth/development, cardiac hypertrophy can become maladaptive in settings of sustained stress and injury (e.g., pressure overload, myocardial infarction, ischemia) and in the context of certain sarcomeric genetic mutations (e.g., hypertrophic cardiomyopathy-causing mutations) leading to pathological cardiac hypertrophy, disease, and heart failure (Nakamura and Sadoshima 2018). Initial studies linking FHL1 to cardiac hypertrophy were based on expression studies performed in hypertrophic cardiomyopathy models and patients. These studies revealed that amongst the FHL family members, FHL1 was selectively and significantly upregulated in settings of pathological cardiac hypertrophy, signaling, and disease (Chu et al. 2000). FHL1 expression was found to be upregulated in vivo in adult mouse hearts subjected to pressure overload and adult mouse hearts from Gαq transgenic mice exhibiting hypertrophic cardiomyopathy as well as in vitro in neonatal rat ventricular cardiomyocytes treated with hypertrophic agonists (phenylephrine, angiotensin II (Ang II)) and signaling (Gαq adenovirus) pathways (Sheikh et al. 2008). These findings were found to be relevant to humans as FHL1 was significantly upregulated in hearts from patients exhibiting hypertrophic cardiomyopathy (Hwang et al. 1997, 2000; Lim et al. 2001). These results altogether highlighted a role for FHL1 in cardiac hypertrophy that was further reinforced in studies performed in a genetic mouse model deficient in FHL1.
FHL1 genetic mouse model studies
FHL1 drives pressure overload-mediated cardiac hypertrophy and modulates progression of hypertrophic cardiomyopathy
A unique role for FHL1 in pathological cardiac hypertrophy was demonstrated using FHL1-deficient mice. FHL1-deficient mice were viable and displayed a normal life span with normal cardiac size and function; however, they displayed a blunted hypertrophic response and preserved left ventricular function after pressure overload by transverse aortic constriction (TAC) (Sheikh et al. 2008). At both 1 week and 5 weeks post-TAC, FHL1-deficient mice demonstrated significantly reduced left ventricular weight/body weight (LV/BW) ratios, smaller myocyte area, and reduced fetal gene expression (atrial natriuretic peptide (ANP), β-myosin heavy chain (β-MHC), skeletal α-actin (SK α-actin)) compared to wild-type (WT) mice (Sheikh et al. 2008). Further analysis through echocardiography revealed preserved LV systolic function and wall thickness at levels comparable to sham-operated mice (Sheikh et al. 2008). A link to Gαq-mediated hypertrophic signaling was revealed after crossing FHL1-deficient mice with Gαq transgenic mice (a well-established mouse model incorporating both the compensated and decompensated phases of cardiac hypertrophy) (D’Angelo et al. 1997). Double FHL1-deficient/Gαq transgenic mice abrogated the Gαq-mediated hypertrophy, which was revealed through a reduction of LV/BW ratio and fetal gene expression (ANP, β-MHC, SK α-actin) (Sheikh et al. 2008). Cardiac functional analysis through echocardiography showed a restoration of chamber dimension, wall thickness, and systolic function to the same level as WT control mice (Sheikh et al. 2008). This data indicates FHL1 is a critical component of Gαq-mediated hypertrophic signaling and removal is sufficient to prevent the pathological hypertrophic response in this model. Interestingly, FHL2 levels are not impacted in FHL1-deficient mice and may indicate that these proteins operate in distinct pathways (Sheikh et al. 2008), even though they are thought to have opposing roles in the hypertrophic response. FHL1 deletion rescued the pathological hypertrophic response in two distinct models—TAC and Gαq transgenic mice—which provides evidence that it is a mediator of the general hypertrophic response to multiple stimuli.
The pathogenic effect of FHL1 was further confirmed through findings in cardiac myosin-binding protein C heterozygous knockout mice (cMyBP-C hets). The cMyBP-C het model has significantly reduced MyBP-C expression and develops an asymmetric septal hypertrophy with fibrosis by 10 months of age (Carrier et al. 2004). cMyBP-C het mice show greater LV/BW ratios and increased interventricular septal wall thickness compared to WT mice at baseline (Vignier et al. 2014). Gene expression analysis revealed FHL1 levels were significantly elevated in the cMyBP-C het mouse model; however, FHL2 levels remained unchanged (Vignier et al. 2014). The renin-angiotensin system is thought to be a contributor to the progression of cardiac hypertrophy, and thus, irbesartan was exploited (angiotensin AT1 receptor blocker) to inhibit hypertrophy in the cMyBP-C het model (Vignier et al. 2014). Irbesartan treatment successfully normalized the LV/BW ratio and interventricular septal wall thickness to WT levels in cMyBP-C het mice (Vignier et al. 2014). Improved cardiac morphology in the cMyBP-C het mice was accompanied by a reduction of FHL1 gene expression to WT levels as well (Vignier et al. 2014). This study shows a potential pharmacological treatment that could prevent the development of pathological cardiac hypertrophy, and that FHL1 (but not FHL2) functions as a sensitive marker of cardiac morphology in the setting of hypertrophy.
An additional study using a mouse model with a heterozygous human Arg403Gln missense mutation of α-myosin heavy chain (MHC403/+) focused on the expression of FHL1 during hypertrophy (Christodoulou et al. 2014). The MHC403/+ mutant mice exhibit cardiac dysfunction and manifest hypertrophy, fibrosis, and myocyte disarray with age (Geisterfer-Lowrance et al. 1996). Analysis in the MHC403/+ model using 5′ RNA-seq to detect alternative use of 5′ start sites after hypertrophy showed FHL1 to have the greatest change in 5′ start-site usage during disease (Christodoulou et al. 2014). Further analysis found that FHL1 was also upregulated in the model and there appeared to be a unique higher molecular weight form that was specifically induced (iFHL1) in the MHC403/+ model and other human diseases (HCM, DCM, pressure overload LV hypertrophy, congestive heart failure) (Christodoulou et al. 2014). This iFHL1 was hypothesized to be generated through alternative 5′ start-site usage and plays an important role in the response to cardiac stress; however, the regulatory mechanisms driving this altered transcription remain to be determined (Christodoulou et al. 2014). Analysis of FHL2 in the MHC403/+ mouse model showed that it was also significantly downregulated (Christodoulou et al. 2014). To gain insight into the role that iFHL1 played in this model, the MHC403/+ mouse was crossed with FHL1-deficient mice and assessed for the development of HCM (Christodoulou et al. 2014). Interestingly, MHC403/+/FHL1-deficient mice developed more severe HCM as shown through increased LV hypertrophy, fibrosis, and increase of molecular signatures of pathological hypertrophy (Christodoulou et al. 2014). This would appear opposite to the function of FHL1 deficiency resulting in beneficial outcomes; however, other variables may account for this seemingly conflicting result. FHL2 was shown to decrease in the MHC403/+ mouse model, whereas levels were unchanged in FHL1 null mice (Christodoulou et al. 2014). The loss of FHL2, which is thought to be a negative regulator of cardiac hypertrophy (Kong et al. 2001), may have an overriding effect on the hypertrophic response in the MHC403/+ mouse.
Signaling mechanisms underlying a role for FHL1 in cardiac hypertrophy
FHL1 scaffolds MAPK components to the sarcomeric titin N2B spring element to regulate hypertrophy
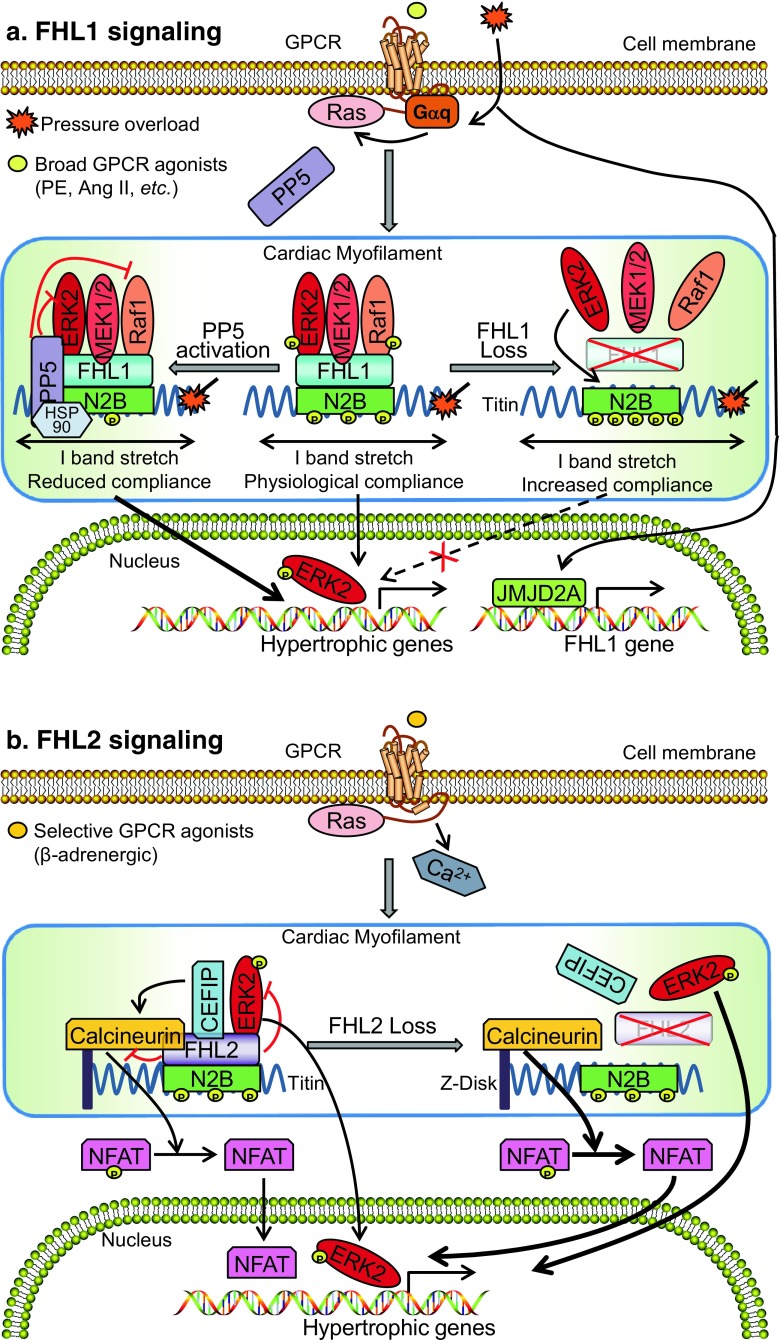
The MAPK/ERK signaling pathway has been shown to play an essential role in stress-induced (pathological) cardiac hypertrophy (Lorenz et al. 2009; Ruppert et al. 2013). Recent studies in FHL1-deficient mice have implicated FHL1 as a component of a novel biomechanical stress sensor complex that scaffolds MAPK signaling molecules to the sarcomeric titin N2B spring element (I band) to regulate stress-induced cardiac hypertrophy (Sheikh et al. 2008) (Fig. 1). Specifically, FHL1 was shown to directly interact with MAPK components (Raf1, MEK2, ERK2) at baseline, and this interaction could be further enhanced when ERK signaling was activated following pressure overload (Purcell et al. 2004; Sheikh et al. 2008). In vitro studies in neonatal rat cardiomyocytes further revealed that overexpression of FHL1 was sufficient to activate MAPK signaling as evidenced by an increase in ERK1/2 phosphorylation, suggesting a role for FHL1 as a positive regulator of MAPK/ERK signaling (Sheikh et al. 2008). In vivo studies in FHL1-deficient mice also revealed that loss of this biomechanical stress sensor complex resulted in a loss of MAPK/ERK signaling associated with a blunted and beneficial (protective) response to stress-induced cardiac hypertrophy (Sheikh et al. 2008). In addition, ex vivo studies performed in cardiac papillary muscles demonstrated that downstream transcriptional targets of MAPK/ERK signaling were also downregulated in FHL1-deficient muscles following acute stretch (Raskin et al. 2012; Sheikh et al. 2008). These studies altogether demonstrate that FHL1 is a positive regulator of MAPK/ERK signaling in response to a hypertrophic stimulus (Sheikh et al. 2008).
Fig. 1.
Schemata of the signaling pathways underlying a role for FHL1 and FHL2 in cardiac hypertrophy. a FHL1 scaffolds MAPK components (Raf-1/MEK2/ERK2) to the sarcomeric titin N2B spring element (biomechanical stress sensor complex), to transmit MAPK signals to regulate muscle compliance and cardiac hypertrophy in a Gq stimulus-specific manner. Loss of FHL1 dissociates this biomechanical stress sensor complex and unmasks phosphorylation sites at titin N2B by ERK2, resulting in increased muscle compliance and a blunted response upon hypertrophic stimulus. PP5 translocates and deactivates this biomechanical stress sensor complex, leading to decreased muscle compliance and heart failure. b FHL2 interacts with EKR2, CEFIP, and calcineurin at the sarcomere to suppress MAPK/ERK and calcineurin/NFAT signaling upon adrenergic-induced cardiac hypertrophy. Loss of FHL2 releases the suppression of the MAPK/ERK and calcineurin/NFAT signaling, leading to increased cardiac hypertrophy. Ang II, angiotensin II; CEFIP, cardiac-enriched FHL2-interacting protein; ERK, extracellular signal-regulated kinase; FHL1, four and a half LIM domain protein-1; FHL2, four and a half LIM domain protein-2; GPCR, G protein-coupled receptor; HSP90, heat shock protein 90; MEK, mitogen-activated protein kinase; NFAT, nuclear factor of activated T cells; PE, phenylephrine; PP5, protein phosphatase 5
FHL1-based biomechanical stress sensor complex is downstream of Gq-mediated hypertrophic signaling
FHL1-mediated MAPK/ERK pathway has been reported as a downstream target of Gαq hypertrophic signaling. Gαq signaling is known to play a critical role in pressure overload-induced cardiac hypertrophy, as cardiac-specific Gαq/Gα11 knockout mice also displayed a blunted response to pressure overload-induced hypertrophy (Dorn and Brown 1999; Wettschureck et al. 2001). Furthermore, studies in cardiac muscle-specific transgenic mice overexpressing Gαq demonstrated that overactivation of cardiac Gαq signaling is sufficient to recapitulate the compensated as well as decompensated phases of cardiac hypertrophy that ensue into heart failure (D'Angelo et al. 1997). Poignant studies in FHL1-deficient mice demonstrated that FHL1 deficiency was sufficient to prevent the cardiomyopathy in Gαq overexpressing transgenic mice (Sheikh et al. 2008). Compared with Gαq transgenic controls, double FHL1-deficient/Gαq transgenic hearts exhibited a significant reduction in LV size and fetal gene expression, which are known to be upregulated with hypertrophy and stress (Sheikh et al. 2008). Although not previously reported in the Gαq transgenic mouse model, elevated ERK1/2 phosphorylation was also observed in Gαq transgenic hearts at baseline (Sheikh et al. 2008). Analysis of double FHL1-deficient/Gαq transgenic hearts further revealed that the blunted response to hypertrophy coincided with a reduction in ERK1/2 phosphorylation (Sheikh et al. 2008). Altogether these findings support the role of FHL1 as a component of a biomechanical stress sensor complex that acts downstream of Gαq to facilitate hypertrophic signaling (Fig. 1).
FHL1-based biomechanical stress sensor complex modulates titin-based compliance/passive tension after stretch and in cardiac disease
Central to the FHL1 complex is that all of the components within this complex localize to the N2B spring element of titin (Sheikh et al. 2008) (Fig. 1). Cardiac titin is a giant sarcomeric protein that contains molecular springs (PEVK and titin N2B), which act as biomechanical sensors and contribute to passive/diastolic tension after stretch (LeWinter et al. 2007; Linke et al. 1999). Studies in FHL1-deficient cardiac papillary muscles subjected to a maximum 20% stretch displayed a significant reduction in diastolic stress and therefore increased compliance compared to controls (Raskin et al. 2012; Sheikh et al. 2008). Interestingly, FHL1 was found to be upregulated in a PEVK segment (within the cardiac-specific titin N2B isoform) knockout model that developed cardiac hypertrophy and an increase in passive tension (Granzier et al. 2009), further suggesting a role for FHL1 in regulating passive/diastolic tension.
Although the mechanisms of how FHL1 regulates passive/diastolic tension needs to be further dissected, studies in FHL1-deficient mice suggested that saturation of titin N2B phosphorylation may be a key contributor towards this increased compliance (Raskin et al. 2012). Beta-adrenergic pathways (e.g., protein kinase A) are known to activate titin N2B phosphorylation, which in turn increase cardiac muscle compliance (Yamasaki et al. 2002). However, studies in FHL1-deficient cardiac muscles demonstrated that they were unresponsive to isoproterenol-induced increase in muscle compliance (Raskin et al. 2012). Moreover, untreated FHL1-deficient cardiac muscles exhibited a level of muscle compliance similar to isoproterenol-treated control muscles, which exhibit maximal muscle compliance, altogether suggesting that titin N2B phosphorylation may be saturated in FHL1-deficient cardiac muscle upon stretch (Raskin et al. 2012). In vitro kinase studies demonstrated that ERK2 (which is a part of the FHL1 biomechanical stress sensor complex) is a kinase that can directly phosphorylate human titin N2B (Raskin et al. 2012). Furthermore, Ser-3873, Ser-3915, and Ser-3965 residues were critical residues for ERK-mediated human titin N2B phosphorylation as their inactivation was sufficient to block titin N2B phosphorylation both at baseline and in response to adrenergic-mediated signaling (Raskin et al. 2012). A dose-dependent increase in FHL1 levels could significantly reduce ERK2-mediated titin N2B phosphorylation via in vitro phosphorylation assays (Raskin et al. 2012), suggesting that FHL1 may directly interfere with ERK2-mediated titin-N2B phosphorylation. However, recent studies using titin N2B phospho-specific antibodies (including to Ser-3965) in FHL1-deficient mouse hearts revealed a decrease in titin N2B phosphorylation compared to controls (Krysiak et al. 2018), suggesting that titin N2B phosphorylation may be dynamic and hypo-phosphorylated at baseline but hyper-phosphorylated during times of stress/stretch. Future studies assessing the phosphorylation status of titin N2B before and after stretch/stress may provide better insights into how these events correlate to hypertrophic signaling responses. Studies performed in the context of the human hypertrophic cardiomyopathy titin N2B mutation S3799Y that has been previously shown to affect FHL binding also significantly impacted ERK2-mediated titin N2B phosphorylation (Itoh-Satoh et al. 2002; Matsumoto et al. 2005; Raskin et al. 2012), further providing relevance of this mechanism to human cardiomyopathy.
Protein phosphatase 5 can deactivate the FHL1-based biomechanical stress sensor complex to modulate titin-based compliance/passive tension
A recent study has identified protein phosphatase 5 (PP5) as a novel phosphatase regulating the FHL1-based biomechanical stress sensor complex by impacting MAPK signaling and titin-based compliance (Fig. 1). Although PP5 is autoinhibited at baseline, PP5 activation/overexpression causes translocation of PP5 to the FHL1-based biomechanical stress sensor complex as it associates with FHL1 and ERK2 at titin N2B (Krysiak et al. 2018). PP5 activation inhibits MAPK/ERK signaling pathway by dephosphorylating Raf1 and ERK2 in vitro (Krysiak et al. 2018; Mazalouskas et al. 2014; von Kriegsheim et al. 2006). In vivo studies in PP5 transgenic mouse hearts further showed reduced phosphorylation on S338 of Raf compared to WT mouse hearts, which also has further implications in blocking downstream MAPK/ERK signaling (Krysiak et al. 2018). PP5 activation could also significantly reduce titin N2B phosphorylation in neonatal rat cardiomyocytes, while an enzymatically dead catalytic subunit of PP5 (PP5H304A) was unable to reduce titin N2B phosphorylation (Krysiak et al. 2018). In vivo studies further support the role of PP5 in dephosphorylating titin N2B as PP5 transgenic mouse hearts displayed reduced titin N2B phosphorylation at residues S3965, S4043, and S4080 (Krysiak et al. 2018). These molecular mechanisms were demonstrated to be important in driving muscle compliance as PP5 transgenic cardiomyocytes displayed increased passive tension (decrease muscle compliance) (Krysiak et al. 2018), which could be reversed with treatment with exogenous kinases (ERK, protein kinase A, or protein kinase G), which are known to phosphorylate titin N2B (Krysiak et al. 2018). These findings may have important implications in cardiomyopathy and heart failure as PP5 expression is increased in failing human hearts and elderly hypertensive dog hearts with diastolic dysfunction, the latter of which exhibit reduced titin N2B phosphorylation (Krysiak et al. 2018). Further studies are required to better understand the specific role of PP5 in how muscle compliance regulates cardiac disease and whether there are specific implications in stress-induced cardiac hypertrophy settings.
Unique transcriptional actions of FHL1 in cardiac hypertrophy
Given the detrimental role FHL1 plays in the pathological cardiac hypertrophic response, there is a great deal of interest in understanding the mechanisms regulating the expression of FHL1. A study focusing on the histone trimethyllysine demethylase JMJD2A utilized heart-specific deletion (hKO) and overexpression mouse models to understand its role in cardiac hypertrophy. JMJD2A hKO mice had a blunted hypertrophic response to pressure overload through TAC, which mirrored the phenotype observed in FHL1-deficient mice (Sheikh et al. 2008; Zhang et al. 2011). Interestingly, FHL1 mRNA and protein levels were found to be downregulated in the JMJD2A hKO heart after TAC. This raises the possibility that JMJD2A may function as a regulator of FHL1 expression. Additional studies with the Jmjd2a-transgenic (Tg) mice demonstrated a more severe cardiac hypertrophic response than sham controls, which was accompanied by an upregulation of FHL1 mRNA and protein levels (Zhang et al. 2011). To further understand this mechanism, the group performed a chromatin immunoprecipitation assay and revealed JMJD2A binds significantly more to the FHL1 promoter after TAC, with this binding being further increased with the Jmjd2a-Tg mouse heart after TAC. This indicates JMJD2A can drive increased transcription of FHL1 during cardiac stress (TAC). To understand the consequences of increased FHL1 expression, they overexpressed FHL1 in neonatal cardiomyocytes and saw an increase in fetal gene expression (ANP, brain natriuretic peptide (BNP), β-MHC) that can contribute to the hypertrophic response (Zhang et al. 2011). This study proposes a mechanism for regulation of FHL1 that involves an increase in JMJD2A activity after TAC, which increases binding to the FHL1 promoter, drives increased FHL1 expression, and ultimately produces fetal gene expression (Fig. 1). These events contribute to the pathological hypertrophic response during cardiac stress.
A role for FHL2 in cardiac hypertrophy
FHL2 expression is restricted to the heart and is expressed throughout embryonic development into adulthood (Chu et al. 2000). Also, unlike FHL1 that is thought to be a driver of pathological hypertrophy, FHL2 is found to be protective in hypertrophic settings. A study using a 7-day infusion of isoproterenol (β-adrenergic stimulation) to drive pathological cardiac growth found that FHL2 expression was increased through protein analysis immediately after the infusion (Hojayev et al. 2012). This pointed to FHL2 as a component of the hypertrophic response and subsequent experiments utilizing FHL2-deficient mice revealed that FHL2 was protective in the hypertrophic response, as FHL2-deficient mice performed worse than WT controls following isoproterenol challenge (Hojayev et al. 2012). Protein and mRNA analysis of human HCM samples (with MyBPC3 mutations) showed that FHL2 levels are significantly reduced in disease samples compared to controls. Similarly, FHL2 mRNA levels were reduced in two mouse HCM models using MyBPC3 mutation knock-in and knockout strategies (Friedrich et al. 2014). Additional studies using the MHC403/+ mouse, which is an established model of HCM, demonstrated reduced expression of FHL2 in the disease state (Christodoulou et al. 2014). Analysis in this study was performed in a genetically driven HCM model where disease features had developed, and so the reduction of FHL2 expression can be viewed as a mechanism in disease pathogenesis. These results indicate FHL2 plays a protective role in the cardiac hypertrophic response and loss of FHL2 may be a critical marker or driver of disease. Mouse models have helped to better understand the precise role that FHL2 plays in the hypertrophic response to stress.
FHL2 genetic mouse model studies
FHL2 is thought to play a protective role in selective signaling pathways linked to cardiac hypertrophy. Although FHL2-deficient mice (targeting exon 1) display normal cardiac function at baseline and exhibit a similar hypertrophic response to controls following 7 days of pressure overload (Chu et al. 2000), a subsequent study utilizing independent FHL2-deficient mice (targeting exon 2) demonstrated a distinct effect during β-adrenergic-mediated hypertrophy (Kong et al. 2001). Specifically, FHL2-deficient mice challenged with 7 days of continuous infusion of the β-adrenergic agonist, isoproterenol revealed a worsening of cardiac hypertrophy (Kong et al. 2001). Three days after isoproterenol infusion, cardiac analysis revealed that FHL2-deficient mice exhibited a greater increase in heart weight (HW)/BW ratio and increase in ANP (hypertrophic stress marker) when compared to WT mice after isoproterenol infusion (Kong et al. 2001). FHL2-deficient mice similarly demonstrated an exaggerated response to isoproterenol in a more recent study focusing on the functional interaction between FHL2 and calcineurin (Hojayev et al. 2012). Specifically, FHL2-deficient mice displayed a significant increase in HW/BW ratio and BNP (hypertrophic stress marker) following isoproterenol infusion compared to WT (Hojayev et al. 2012). These studies altogether suggest that FHL2 pathways may be uniquely linked to β-adrenergic signaling pathways to play this protective role in the heart.
A mouse model using a cardiac-specific Rho-associated coiled-coil containing kinase knockout mouse model (cROCK2-KO) and two methods of inducing cardiac hypertrophy highlighted the beneficial role FHL2 plays in the hypertrophic response (Okamoto et al. 2013). At baseline, cROCK2-KO mice display normal body weight, cardiac morphology, and cardiac function compared to WT mice (Okamoto et al. 2013). When subjected to either Ang II infusion through an osmotic minipump (functions through activation of RhoA) or TAC, cROCK2-KO mice demonstrated a less severe hypertrophic response than WT mice (Okamoto et al. 2013). This was shown through reduced LV wall thickness, reduced LV mass, and fibrosis in cROCK2-KO mice (Okamoto et al. 2013). Analysis after Ang II infusion revealed that FHL2 was significantly upregulated in cROCK2-KO mice relative to WT mice with Ang II infusion, implicating FHL2 as a mediator of the observed protective phenotype (Okamoto et al. 2013). To better understand if FHL2 was a significant contributor to the protective effects observed in cROCK2-KO mice, haploinsufficient ROCK2 mice were crossed with heterozygous FHL2-deficient mice and infused with Ang II (Okamoto et al. 2013). Double heterozygous mice displayed significantly worse heart weight/tibia length ratios and significantly greater myocyte cross-sectional area than heterozygous ROCK2-deficient mice after Ang II infusion (Okamoto et al. 2013). This demonstrated that reduction of FHL2 was sufficient to remove the resistance to pathological hypertrophy observed in ROCK2-deficient mice.
Signaling mechanisms underlying a role for FHL2 in cardiac hypertrophy
FHL2 suppress hypertrophy in cardiomyocytes through inhibiting MAPK/ERK signaling
Consistent with in vivo studies in FHL2-deficient mice, studies in neonatal rat cardiomyocytes show that FHL2 overexpression leads to a reduction in adrenergic-induced cardiac hypertrophy (Purcell et al. 2004). Mechanistic studies in this system revealed that FHL2, unlike FHL1, may act as a negative regulator of MAPK/ERK signaling and play a protective role in adrenergic-mediated cardiac hypertrophy (Purcell et al. 2004) (Fig. 1). Like FHL1, FHL2 is also bound to the titin N2B region (Lange et al. 2002) and independent studies via yeast two-hybrid studies revealed that FHL2 can also directly interact with ERK2 (Purcell et al. 2004). In vitro studies in neonatal rat cardiomyocytes suggested that FHL2 could potentially inhibit the translocation of phosphorylated ERK2 from the sarcomere to the nucleus, resulting in inhibition of ERK-dependent transcriptional signaling following adrenergic stimulation (Purcell et al. 2004). Consistent with these findings, FHL2 overexpression could reduce MEK1-, GATA4-, and adrenergic-induced hypertrophy also indicative by the diminished upregulation of fetal genes, such as ANP and β-MHC (Purcell et al. 2004). In vivo studies in FHL2 heterozygous-deficient mice also demonstrate that ERK phosphorylation is increased in the setting of Ang II-induced cardiac hypertrophy (Okamoto et al. 2013). The suppressive effect of FHL2 on MAPK/ERK signaling was further supported in studies performed in a cardiac-specific ROCK2 knockout mouse model (Okamoto et al. 2013). Cardiac-specific ROCK2 homozygous-deficient hearts displayed an increase in FHL2 expression coincident with reduced ERK phosphorylation and a blunted response to Ang II and pressure overload-induced cardiac hypertrophy (Okamoto et al. 2013). Studies in ROCK2 heterozygous-deficient mice also revealed an increase in FHL2 expression, reduced ERK phosphorylation, and blunted response to Ang II-induced cardiac hypertrophy (Okamoto et al. 2013). Further studies inAng II-treated FHL2 heterozygous-deficient mice crossed with ROCK2 heterozygous-deficient mice (which reduced FHL2 expression to 50% of WT levels) resulted in an increase in ERK phosphorylation and cardiac hypertrophy to levels equivalent to Ang II-treated WT hearts (Okamoto et al. 2013). Interestingly, these studies suggested that FHL2 co-localized with ROCK2 at the perinuclear region, potentially highlighting new functions of FHL2 and MAPK/ERK signaling outside the sarcomere.
FHL2 is a suppressor of calcineurin/NFAT signaling
Studies in FHL2-deficient mice suggest that FHL2 is a critical regulator of isoproterenol-induced cardiac hypertrophy and calcineurin/ nuclear factor of activated T cell (NFAT) signaling (Wilkins and Molkentin 2004). Calcineurin/NFAT signaling is downstream of β-adrenergic signaling and plays an essential role in isoproterenol-induced myocardial hypertrophy (Wilkins and Molkentin 2004). Calcineurin specifically dephosphorylates NFAT, which then translocates into the nucleus to activate transcription of hypertrophic target genes, such as BNP and regulator of calcineurin 1 isoform 4 (RCAN1.4) (Wilkins and Molkentin 2004). FHL2 and calcineurin intersect at the sarcomere in adult mouse hearts and calcineurin activation can enhance this interaction (Wilkins and Molkentin 2004). In vivo studies in FHL2-deficient mice show that the expression of the NFAT target genes, BNP, and RCAN1.4 are significantly increased in isoproterenol-treated FHL2-deficient hearts, when compared to controls (Wilkins and Molkentin 2004). In vitro studies in neonatal rat cardiomyocytes further revealed that knockdown of FHL2 led to increased transcriptional activation of the calcineurin-mediated NFAT target gene, RCAN1.4 promoter (Wilkins and Molkentin 2004). Overexpression of FHL2 was also sufficient to inhibit the transcriptional activation of the RCAN1.4 promoter and blunt hypertrophic growth in cultured cardiomyocytes induced by constitutively active calcineurin (Wilkins and Molkentin 2004). These studies altogether suggest that the effects of FHL2 in isoproterenol-mediated hypertrophy may be mediated by calcineurin/NFAT signaling (Fig. 1).
CEFIP is an agonist of FHL2-mediated calcineurin/NFAT signaling
Recent studies identified the cardiac-enriched FHL2-interacting protein (CEFIP) as a novel FHL2-interacting protein via yeast two-hybrid studies (Dierck et al. 2017). CEFIP is thought to co-localize with FHL2 at the sarcomere in adult mouse heart (Dierck et al. 2017). Although the role of CEFIP in regulating calcineurin/NFAT signaling and hypertrophy appears to be well established, the effect on FHL2 is not linear. CEFIP overexpression (upregulation) alone increased transcription of the NFAT-responsive promoter activity and adrenergic-mediated hypertrophy in treated neonatal rat cardiomyocytes (Dierck et al. 2017). Interestingly, after FHL2 is knocked down (downregulation) in this setting, the NFAT-responsive promoter activity could be further increased to the most maximal levels (Dierck et al. 2017). Conversely, CEFIP knockdown (downregulation) alone resulted in reduced NFAT-responsive promoter activity and adrenergic-mediated cardiac hypertrophy in neonatal rat cardiomyocytes, even in presence of calcineurin overexpression (Dierck et al. 2017). When FHL2 is knocked down in these settings, the NFAT-responsive promoter activity is increased when compared to CEFIP knockdown alone yet decreased when compared to FHL2 knockdown alone (Dierck et al. 2017), suggesting an overriding effect of FHL2 and only partial intersection of CEFIP pathways with FHL2 (Fig. 1).
FHL1 and FHL2 variants are associated with hypertrophic cardiomyopathy
Hypertrophic cardiomyopathy (HCM) mutations are largely found in sarcomeric genes, with the five most common being β-myosin heavy chain (MYH7), cardiac myosin-binding protein C (MYBC3), troponin T (TNNT2), troponin I (TNNI3), and myosin regulatory light chain 2(MYL2) (Friedrich et al. 2012). Increasing evidence points to the contribution that FHL1 mutations play in HCM development and there has been effort to identify these mutations in HCM patient populations without mutations in the classically defined causal genes (Cowling et al. 2011; D’Arcy et al. 2015; Friedrich et al. 2012; Gossios et al. 2013; Hartmannova et al. 2013; Knoblauch et al. 2010; Zhang et al. 2015) (Table 1). A study looking to identify FHL1 mutations in these HCM patient populations reported several FHL1 variants, with two of these predicted to result in a truncated FHL1 protein lacking a large portion of the C-terminus (Friedrich et al. 2012). Analysis of the predicted truncating FHL1 variants through in vitro transduction in neonatal cardiomyocytes resulted in expression of a smaller FHL1 protein at reduced levels (Friedrich et al. 2012). Subsequent analysis of the FHL1 variants in rat-engineered heart tissue resulted in a disruption of contraction force and kinetics (Friedrich et al. 2012). It is intriguing that FHL1 mutations may be linked to HCM; however, in vivo analysis of FHL1 variants via knock-in models is required to more precisely determine their role in human disease.
Table 1.
FHL mutations are associated with various human cardiomyopathies
| FHL gene | Exon location | Genetic change | Amino acid exchange | Type of mutation | Prediction on FHL protein | MAF | Related cardiac disease |
|---|---|---|---|---|---|---|---|
| FHL1 | Exon 3 | c.134delA | p.Ser45fs | Frameshift | FHL1 c-terminal truncated, lacking LIM2–4 domains | Not Known | HCM (Friedrich et al. 2012) |
| Exon 5 | c.441C>T | p.Asp147Asp | Silent mutation | Normal protein product | 0.001636 | HCM (Friedrich et al. 2012) | |
| Exon 5 | c.457T>C | p.Cys153Arg | Missense | FHL1 Zinc finger2 mutation | Not Known | RBM + DCM (Schessl et al. 2008) | |
| Exon 5 | c.459C>A | p.Cys153X | Nonsense | Loss of function; Truncated protein, lacking LIM3–4 domains | Not Known | HCM (Friedrich et al. 2012) | |
| Exon 6 | c.542G>A | p.Trp181X | Nonsense | Truncated FHL1A and FHL1B | 4.652e-5 | HCM (Zhang et al. 2015) | |
| Exon 6 | c.592C>T | p.Gln198X | Nonsense | Truncated FHL1A and FHL1B | Not Known | EDMD + HCM (Gossios et al. 2013) | |
| Exon 6 | c.599_600insT | p.Phe200fs32X | Frameshift | Frameshift of both FHL1A and FHL1B isoforms | Not known | HCM (Hartmannova et al. 2013) | |
| Exon 6 | c.625T>C | p.Cys209Arg | Missense | Truncated FHL1A and FHL1B | Not known | HCM (Knoblauch et al. 2010) | |
| Exon 8 | c.764G>C | p.Cys255Ser | Missense | FHL1A: p.C255 S; FHL1B/C: p.A322P/p.A193, disrupts protein folding and zinc ion binding | Not known | XRDM+HCM (D'Arcy et al. 2015) | |
| Exon 8 | c.764G>C | p.Cys255Ser | Missense | FHL1A: p.C255 S; FHL1B/C: p.A322P/p.A193, disrupts protein folding and zinc ion binding | Not known | EDMD + ARVC (San Roman et al. 2016) | |
| Exon 8 | c.823G>A | p.Asp275Asn | Missense | Mutant FHL1A protein | Not known | HCM (Friedrich et al. 2012) | |
| Exon 8 | c.827G>C | p.Cys276Ser | Missense | Mutate FHL1A protein | Not known | HCM (Friedrich et al. 2012) | |
| FHL2 | Exon 4 | c.142G>A | p.Gly48Ser | Missense | LIM1 domain binding ability change | Not known | DCM (Arimura et al. 2007) |
| Exon 7 | c.512C>T | p.Thr171Met | Missense | LIM3 domain mutation | Not known | HCM (Friedrich et al. 2014) | |
| Exon 7 | c.530G>A | p.Arg177Gln | Missense | LIM3 domain mutation | Not known | HCM (Friedrich et al. 2014) | |
| Exon 7 | c.559G>T | p.Val187Leu | Missense | LIM3 domain mutation | Not known | HCM (Friedrich et al. 2014) | |
| Exon 7 | c.678C>T | p.Asn226Asn | Single nucleotide variant | Normal protein product | Not known | HCM (Friedrich et al. 2014) | |
| Exon 8 | c.804C>T | p.Asp268Asp | Single nucleotide variant | Normal protein product | Not known | HCM (Friedrich et al. 2014) | |
| Exon 8 | c.819C>T | p.Pro273Pro | Single nucleotide variant | Normal protein product | Not known | HCM (Friedrich et al. 2014) |
This table lists the specific human FHL1 and FHL2 mutations identified in various patients with human cardiomyopathies as well as includes the gene location and predicted impact of these mutations on FHL protein isoforms. Minor allele frequencies (MAF) of variants are also indicated based on the population database gnomAD (http://gnomad.broadinstitute.org/). ARVC, arrhythmogenic right ventricular cardiomyopathy; DCM, dilated cardiomyopathy; EDMD, Emery-Dreifuss muscular dystrophy; HCM, hypertrophic cardiomyopathy; RBM, reducing body myopathy; XRDM, X-linked recessive distal myopathy
Due to its location on the X chromosome, FHL1 has been studied as a gender-specific modifier of disease severity in HCM patients (Christodoulou et al. 2014; Ehsan et al. 2017; Friedrich et al. 2012; Hartmannova et al. 2013; San Roman et al. 2016). For example, a frameshift mutation of the FHL1 gene (F200fs32X) was found in three male patients (Hartmannova et al. 2013). This mutation does not affect transcription, splicing, and stability of FHL1 mRNA levels, but rather results in a truncated form of FHL1 protein that is predicted to be missing the LIM3 and LIM4 domains (Hartmannova et al. 2013). These hemizygous male patients display HCM and severe left ventricular diastolic dysfunction in end-stage heart failure. Interestingly, one heterozygous female with the same mutation displayed mild cardiac involvement, which suggests FHL1 plays a role in disease penetration in a gender-specific manner (Hartmannova et al. 2013). It is likely that this truncated form of FHL1 without the capacity for its required protein-protein interactions through LIM domains functions as a toxic protein that prevents proper assembly of complexes in the cell.
FHL2 variants have been identified in HCM patients with no other detectable pathogenic gene mutations (Friedrich et al. 2014). A study identified six mutations (two novel and four known) that were found in distinct HCM populations (Friedrich et al. 2014). Transduction of the FHL2 genetic variants in neonatal rat cardiomyocytes and rat-engineered heart tissue illustrated diverse functions of the variants on hypertrophic gene expression (ANP, BNP, SK α-actin), stability of FHL2 protein, and consequence of expression on tissue function in an engineered system (Friedrich et al. 2014). Analysis was unable to elucidate the mechanisms by which these FHL2 variants may impact HCM development in patients (Friedrich et al. 2014). Further studies are required in model systems such as the mouse where impact of variants on cardiac function and pathogenesis can be more readily defined.
Analysis of a HCM-associated mutation in the titin N2B region (Ser3799Tyr) revealed an increased interaction with FHL2 through a yeast two-hybrid assay (Matsumoto et al. 2005). This is an interesting finding as loss of FHL2 in a knockout mouse model has been shown to increase the severity of the pathological hypertrophic response to β-adrenergic stimulation (Kong et al. 2001). These findings seem to be conflicting; however, the increased interaction between N2B and FHL2 could prevent FHL2 from performing additional roles in the cell or the proper turnover required for sarcomeric homeostasis. FHL1 has been more commonly implicated in HCM, so it would be important to understand the role that FHL1 is playing with this N2B mutation. FHL1 and FHL2 share many binding sites, particularly on N2B, so it is very likely that this mutation is also influencing FHL1 and it is contributing to the HCM.
FHL and other myopathies
FHL1 and FHL2 have been studied largely in the context of the hypertrophic response in muscle and mutations that result in hypertrophic cardiomyopathy (Table 1). Additional studies have implicated FHL1 and FHL2 in other types of cardiac disease such as dilated cardiomyopathy (DCM), arrhythmogenic right ventricular cardiomyopathy (ARVC), and arrhythmias (Table 1). Although often connected to hypertrophic cardiomyopathy, these are distinct cardiac diseases that suggest FHL1 and FHL2 may have a broader role in the cardiomyocyte. This further reveals the complexity of FHL1 and FHL2 signaling and interactions in the heart and provides opportunities to better dissect relevant pathways in cardiac disease.
FHL1 mutations are implicated in myopathies and dystrophies, which has been shown through a FHL1 knockout mouse model and human samples analyzing skeletal muscle (Domenighetti et al. 2014). The link to Emery-Dreifuss muscular dystrophy (EDMD) is particularly interesting as there is a strong cardiac pathology (Cowling et al. 2011). FHL1 mutations contribute to approximately 10% of EDMD cases that have an X-linked inheritance pattern (Pillers and Von Bergen 2016). The cardiac disease is severe in EDMD patients and is a major cause of death (Pillers and Von Bergen 2016). EDMD patients often have arrhythmias and can develop a dilated cardiomyopathy in later stages of disease that progresses to congestive heart failure (Pillers and Von Bergen 2016). Extensive work has been done to characterize FHL1 mutations driving EDMD and has helped localize many mutations to the most distal exons (5–8) of FHL1 and shown that they primarily affect the LIM domain-mediated protein interactions (Cowling et al. 2011). A recent study has linked ARVC with EDMD through a FHL1 mutation. This study identifies a Cys255Ser mutation in a large family with both cardiac dysfunction and neuromuscular disease (San Roman et al. 2016). Family members displayed electrical abnormalities, right ventricular dysfunction, and fibrofatty replacement of myocardium which are common features of ARVC (Ohno 2016; San Roman et al. 2016; Sen-Chowdhry et al. 2010). The involvement of FHL1 in ARVC is an important mechanism for further study. Only a portion of mutations driving ARVC are known, so future work should look in more detail at FHL1 mutations in ARVC patients. Mouse studies and patient-derived-induced pluripotent stem cells (iPSCs) can be useful for dissecting the pathogenesis of this mutation and understand how it contributes to both ARVC and EDMD in this family. Cardiac disease is also seen in reducing body myopathy (RBM), which is an additional severe muscular disease driven by FHL1 mutations (Cowling et al. 2011; Schessl et al. 2008). Patients develop dilated cardiomyopathies; however, cardiac dysfunction is less commonly observed than in EDMD patients because RBM patients often die at an early age from respiratory failure (Cowling et al. 2011). Further characterization of these mutations is required to help dissect the unique FHL1 interactions that are required for normal cardiac function. This can be paired with clinical data to better understand pathways contributing to electrical or structural heart disease.
The diverse cardiac pathologies associated with FHL1 mutations in humans have a strong electrical component, with EDMD and ARVC patients being susceptible to sudden cardiac death. Studies utilizing human samples and animal models have focused on better understanding the role of FHL1 in regulating ion channels and in atrial fibrillation (AF). The voltage-gated potassium channel KCNA5 functions in repolarization of the atria and a study looking for novel KCNA5-interacting proteins identified FHL1 as a positive regulator (Yang et al. 2008). This study provides a possible mechanism for arrhythmias in FHL1 patients outside of its classic signaling pathways, but more work must be done to confirm the relevance of these findings, particularly the electrophysiology data in a more relevant cell type. Patient-derived iPSCs would provide a more relevant system for tracking the expression and function of KCNA5 with mutations in FHL1. An additional study looking for new mechanisms in AF showed upregulation of FHL1 in a porcine atrial pacing-induced AF model (Chen et al. 2007). There is a need to better understand the role of FHL1 in the progression of AF, whether it is an early driver or rather an active signaling protein in the stressed cardiomyocyte. These studies point to FHL1 as a critical factor in the regulation of atrial ion channel function and stress. It will be important to apply these ideas to FHL1 patient mutations to see how these pathways are altered in EDMD or ARVC.
FHL2 has been directly linked to DCM. A study in a Japanese patient population identified a Gly48Ser mutation in FHL2 that associates with familial DCM (Arimura et al. 2007). Assessment of this mutation revealed it disrupts the interaction between FHL2 and the N2B domain of titin through mammalian two-hybrid analysis and causes FHL2 mislocalization in cardiomyocytes and skeletal muscle (Arimura et al. 2007). The importance of FHL2 in DCM is further shown through DCM-causing mutations in titin (Gln4053ter), which revealed decreased FHL2 binding with the N2B region of titin through yeast two-hybrid analysis (Matsumoto et al. 2005). This implicates FHL2 as an important component of DCM development; however, given that FHL1 binds to similar regions of titin N2B, it would be interesting to assess the role that FHL1 plays with this mutation and DCM development.
Previous work has suggested that the titin-FHL2 interaction helps tether metabolic enzymes to the cardiac sarcomere that are essential for the high-energy demand at this cellular location (Lange et al. 2002). Loss of proper localization of these enzymes could be an important contributor to cardiac disease. A study looking at samples from patients with aortic stenosis-driven heart failure observed a decrease of FHL2 levels in the failing heart (Bovill et al. 2009). This coincided with a decrease in metabolic enzyme adenylate kinase and phosphofructokinase 2 (Bovill et al. 2009). These results provide a possible mechanism that may contribute to heart failure; however, further studies are required to better understand the contribution of these altered metabolic enzymes. It is necessary to assess if loss of these enzymes is sufficient to drive heart failure, or if they are just a product of disrupted structure and signaling in the heart.
Conclusions and future directions
From their initial discovery in 1998 (Chan et al. 1998; Lee et al. 1998), FHL1 and FHL2 have emerged as important players and having unique roles in stress and adrenergic-induced cardiac hypertrophy and disease, respectively. The role of FHL1 has been the most well established as a positive regulator of stress-induced cardiac hypertrophy, and it is thought to act as a MAPK scaffolding component of a biomechanical sensor complex at the sarcomeric titin N2B band, which regulates titin N2B phosphorylation/muscle compliance and cardiac hypertrophy during pressure overload and Gαq-mediated stress (Raskin et al. 2012; Sheikh et al. 2008). Although FHL2 is also located at the sarcomere, it may play a more selective and protective role in the heart by acting as a negative regulator of adrenergic-induced cardiac signaling (via MAPK/ERK and calcineurin) and hypertrophy (Hojayev et al. 2012; Purcell et al. 2004). These studies altogether suggest a yin (FHL2)/yang (FHL1) balance between FHL function in the heart during stress, which may be required to keep homeostatic control of cardiac function during stress. Few studies have assessed the impact of both FHL1 and FHL2 in genetic models and hypertrophic settings; however, studies in alpha-myosin heavy chain (MHC403/+) mutant model (which exhibited increased FHL1 and downregulation of FHL2) may provide unique insights and suggest that FHL2 function may dominate and thus explain the worsening cardiomyopathy in this model despite an attempt to reduce FHL1 levels (via cross to FHL1-deficient mice) (Christodoulou et al. 2014). Future studies should explore the role of FHL1 and FHL2 in stress-induced cardiac hypertrophy by the analysis of double FHL1 and FHL2 knockout mice as well as measure expression of both FHL1 and FHL2 in hypertrophic models in order to better gauge the FHL arena to predicate the response to genetic intervention or therapy. It is also intriguing that both FHL1 and FHL2 are bound to the same region of sarcomere (titin N2B) but yet exhibit unique effects on signaling and hypertrophy; thus, future studies focused on better understanding the positive and negative regulators of this complex (e.g., PP5) in the setting of hypertrophic stress may provide new clues as to how to specifically harness the beneficial effects of this complex for therapeutic interventions in human cardiac disease. Human genetic studies have revealed that FHL gene mutations are associated with a number of cardiomyopathies and heart diseases (Table 1); however, the challenge will be to better understand how these mutations impact FHL1 and FHL2 biology in the context of a diseased heart. Future studies employing knock-in mouse models to assess the impact and sufficiency of these mutations to drive human cardiac disease should be a focus.
Funding information
Y.L and J.Z are recipients of the American Heart Association Postdoctoral Fellowship. W.B is a recipient of the National Science Foundation graduate research fellowship. F.S. is supported by grants from NIH/NHLBI (HL09780) and Tobacco-Related Disease Research program (24RT-22).
Compliance with ethical standards
Yan Liang declares that she has no conflict of interest. William H. Bradford declares that he has no conflict of interest. Jing Zhang declares that she has no conflict of interest. Farah Sheikh declares that she has no conflict of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.
None.
Footnotes
This article is part of a Special Issue on ‘Heart Failure Due to Non-Myofibrillar Defects’ edited by Elisabeth Ehler and Katja Gehmlich.
Yan Liang, William H. Bradford and Jing Zhang contributed equally to this work and should be regarded as joint first authors.
References
- Arimura T, Hayashi T, Matsumoto Y, Shibata H, Hiroi S, Nakamura T, Inagaki N, Hinohara K, Takahashi M, Manatsu SI, Sasaoka T, Izumi T, Bonne G, Schwartz K, Kimura A. Structural analysis of four and half LIM protein-2 in dilated cardiomyopathy. Biochem Biophys Res Commun. 2007;357(1):162–167. doi: 10.1016/j.bbrc.2007.03.128. [DOI] [PubMed] [Google Scholar]
- Bovill E, Westaby S, Crisp A, Jacobs S, Shaw T. Reduction of four-and-a-half LIM-protein 2 expression occurs in human left ventricular failure and leads to altered localization and reduced activity of metabolic enzymes. J Thorac Cardiovasc Surg. 2009;137(4):853–861. doi: 10.1016/j.jtcvs.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Carrier L, Knoll R, Vignier N, Keller DI, Bausero P, Prudhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J, Jr, Schwartz K, Chien KR. Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc Res. 2004;63(2):293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Chan KK, Tsui SK, Lee SM, Luk SC, Liew CC, Fung KP, Waye MM, Lee CY. Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene. 1998;210(2):345–350. doi: 10.1016/S0378-1119(97)00644-6. [DOI] [PubMed] [Google Scholar]
- Chen CL, Lin JL, Lai LP, Pan CH, Huang SK, Lin CS. Altered expression of FHL1, CARP, TSC-22 and P311 provide insights into complex transcriptional regulation in pacing-induced atrial fibrillation. Biochim Biophys Acta. 2007;1772(3):317–329. doi: 10.1016/j.bbadis.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Christodoulou DC, Wakimoto H, Onoue K, Eminaga S, Gorham JM, DePalma SR, Herman DS, Teekakirikul P, Conner DA, McKean DM, Domenighetti AA, Aboukhalil A, Chang S, Srivastava G, McDonough B, De Jager PL, Chen J, Bulyk ML, Muehlschlegel JD, Seidman CE, Seidman JG. 5'RNA-Seq identifies Fhl1 as a genetic modifier in cardiomyopathy. J Clin Invest. 2014;124(3):1364–1370. doi: 10.1172/JCI70108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PH, Ruiz-Lozano P, Zhou Q, Cai C, Chen J. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech Dev. 2000;95(1–2):259–265. doi: 10.1016/S0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- Cowling BS, Cottle DL, Wilding BR, D'Arcy CE, Mitchell CA, McGrath MJ. Four and a half LIM protein 1 gene mutations cause four distinct human myopathies: a comprehensive review of the clinical, histological and pathological features. Neuromuscul Disord. 2011;21(4):237–251. doi: 10.1016/j.nmd.2011.01.001. [DOI] [PubMed] [Google Scholar]
- D'Angelo DD, Sakata Y, Lorenz JN, Boivin GP, Walsh RA, Liggett SB, Dorn GW., 2nd Transgenic Galphaq overexpression induces cardiac contractile failure in mice. Proc Natl Acad Sci U S A. 1997;94(15):8121–8126. doi: 10.1073/pnas.94.15.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy C, Kanellakis V, Forbes R, Wilding B, McGrath M, Howell K, Ryan M, McLean C. X-linked recessive distal myopathy with hypertrophic cardiomyopathy caused by a novel mutation in the FHL1 gene. J Child Neurol. 2015;30(9):1211–1217. doi: 10.1177/0883073814549807. [DOI] [PubMed] [Google Scholar]
- Dierck F, Kuhn C, Rohr C, Hille S, Braune J, Sossalla S, Molt S, van der Ven PFM, Furst DO, Frey N. The novel cardiac z-disc protein CEFIP regulates cardiomyocyte hypertrophy by modulating calcineurin signaling. J Biol Chem. 2017;292(37):15180–15191. doi: 10.1074/jbc.M117.786764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenighetti AA, Chu PH, Wu T, Sheikh F, Gokhin DS, Guo LT, Cui Z, Peter AK, Christodoulou DC, Parfenov MG, Gorham JM, Li DY, Banerjee I, Lai X, Witzmann FA, Seidman CE, Seidman JG, Gomes AV, Shelton GD, Lieber RL, Chen J. Loss of FHL1 induces an age-dependent skeletal muscle myopathy associated with myofibrillar and intermyofibrillar disorganization in mice. Hum Mol Genet. 2014;23(1):209–225. doi: 10.1093/hmg/ddt412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW, 2nd, Brown JH. Gq signaling in cardiac adaptation and maladaptation. Trends Cardiovasc Med. 1999;9(1–2):26–34. doi: 10.1016/S1050-1738(99)00004-3. [DOI] [PubMed] [Google Scholar]
- Ehsan M, Jiang H, Thomson KL, Gehmlich K. When signalling goes wrong: pathogenic variants in structural and signalling proteins causing cardiomyopathies. J Muscle Res Cell Motil. 2017;38(3–4):303–316. doi: 10.1007/s10974-017-9487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich FW, Wilding BR, Reischmann S, Crocini C, Lang P, Charron P, Muller OJ, McGrath MJ, Vollert I, Hansen A, Linke WA, Hengstenberg C, Bonne G, Morner S, Wichter T, Madeira H, Arbustini E, Eschenhagen T, Mitchell CA, Isnard R, Carrier L. Evidence for FHL1 as a novel disease gene for isolated hypertrophic cardiomyopathy. Hum Mol Genet. 2012;21(14):3237–3254. doi: 10.1093/hmg/dds157. [DOI] [PubMed] [Google Scholar]
- Friedrich FW, Reischmann S, Schwalm A, Unger A, Ramanujam D, Munch J, Muller OJ, Hengstenberg C, Galve E, Charron P, Linke WA, Engelhardt S, Patten M, Richard P, van der Velden J, Eschenhagen T, Isnard R, Carrier L. FHL2 expression and variants in hypertrophic cardiomyopathy. Basic Res Cardiol. 2014;109(6):451. doi: 10.1007/s00395-014-0451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen FJ, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272(5262):731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- Gossios TD, Lopes LR, Elliott PM. Left ventricular hypertrophy caused by a novel nonsense mutation in FHL1. Eur J Med Genet. 2013;56(5):251–255. doi: 10.1016/j.ejmg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Granzier HL, Radke MH, Peng J, Westermann D, Nelson OL, Rost K, King NM, Yu Q, Tschope C, McNabb M, Larson DF, Labeit S, Gotthardt M. Truncation of titin's elastic PEVK region leads to cardiomyopathy with diastolic dysfunction. Circ Res. 2009;105(6):557–564. doi: 10.1161/CIRCRESAHA.109.200964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmannova H, Kubanek M, Sramko M, Piherova L, Noskova L, Hodanova K, Stranecky V, Pristoupilova A, Sovova J, Marek T, Maluskova J, Ridzon P, Kautzner J, Hulkova H, Kmoch S. Isolated X-linked hypertrophic cardiomyopathy caused by a novel mutation of the four-and-a-half LIM domain 1 gene. Circ Cardiovasc Genet. 2013;6(6):543–551. doi: 10.1161/CIRCGENETICS.113.000245. [DOI] [PubMed] [Google Scholar]
- Hojayev B, Rothermel BA, Gillette TG, Hill JA. FHL2 binds calcineurin and represses pathological cardiac growth. Mol Cell Biol. 2012;32(19):4025–4034. doi: 10.1128/MCB.05948-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DM, Dempsey AA, Wang RX, Rezvani M, Barrans JD, Dai KS, Wang HY, Ma H, Cukerman E, Liu YQ, Gu JR, Zhang JH, Tsui SK, Waye MM, Fung KP, Lee CY, Liew CC. A genome-based resource for molecular cardiovascular medicine: toward a compendium of cardiovascular genes. Circulation. 1997;96(12):4146–4203. doi: 10.1161/01.CIR.96.12.4146. [DOI] [PubMed] [Google Scholar]
- Hwang DM, Dempsey AA, Lee CY, Liew CC. Identification of differentially expressed genes in cardiac hypertrophy by analysis of expressed sequence tags. Genomics. 2000;66(1):1–14. doi: 10.1006/geno.2000.6171. [DOI] [PubMed] [Google Scholar]
- Itoh-Satoh M, Hayashi T, Nishi H, Koga Y, Arimura T, Koyanagi T, Takahashi M, Hohda S, Ueda K, Nouchi T, Hiroe M, Marumo F, Imaizumi T, Yasunami M, Kimura A. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem Biophys Res Commun. 2002;291(2):385–393. doi: 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- Knoblauch H, Geier C, Adams S, Budde B, Rudolph A, Zacharias U, Schulz-Menger J, Spuler A, Yaou RB, Nurnberg P, Voit T, Bonne G, Spuler S. Contractures and hypertrophic cardiomyopathy in a novel FHL1 mutation. Ann Neurol. 2010;67(1):136–140. doi: 10.1002/ana.21839. [DOI] [PubMed] [Google Scholar]
- Kong Y, Shelton JM, Rothermel B, Li X, Richardson JA, Bassel-Duby R, Williams RS. Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to beta-adrenergic stimulation. Circulation. 2001;103(22):2731–2738. doi: 10.1161/01.CIR.103.22.2731. [DOI] [PubMed] [Google Scholar]
- Krysiak J, Unger A, Beckendorf L, Hamdani N, von Frieling-Salewsky M, Redfield MM, Dos Remedios CG, Sheikh F, Gergs U, Boknik P, Linke WA. Protein phosphatase 5 regulates titin phosphorylation and function at a sarcomere-associated mechanosensor complex in cardiomyocytes. Nat Commun. 2018;9(1):262. doi: 10.1038/s41467-017-02483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer BW, Perriard JC, Ehler E. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci. 2002;115(Pt 24):4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- Lee SM, Tsui SK, Chan KK, Garcia-Barcelo M, Waye MM, Fung KP, Liew CC, Lee CY. Chromosomal mapping, tissue distribution and cDNA sequence of four-and-a-half LIM domain protein 1 (FHL1) Gene. 1998;216(1):163–170. doi: 10.1016/S0378-1119(98)00302-3. [DOI] [PubMed] [Google Scholar]
- LeWinter MM, Wu Y, Labeit S, Granzier H. Cardiac titin: structure, functions and role in disease. Clin Chim Acta. 2007;375(1–2):1–9. doi: 10.1016/j.cca.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lim DS, Roberts R, Marian AJ. Expression profiling of cardiac genes in human hypertrophic cardiomyopathy: insight into the pathogenesis of phenotypes. J Am Coll Cardiol. 2001;38(4):1175–1180. doi: 10.1016/S0735-1097(01)01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA, Rudy DE, Centner T, Gautel M, Witt C, Labeit S, Gregorio CC. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. J Cell Biol. 1999;146(3):631–644. doi: 10.1083/jcb.146.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz K, Schmitt JP, Schmitteckert EM, Lohse MJ. A new type of ERK1/2 autophosphorylation causes cardiac hypertrophy. Nat Med. 2009;15(1):75–83. doi: 10.1038/nm.1893. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi T, Inagaki N, Takahashi M, Hiroi S, Nakamura T, Arimura T, Nakamura K, Ashizawa N, Yasunami M, Ohe T, Yano K, Kimura A. Functional analysis of titin/connectin N2-B mutations found in cardiomyopathy. J Muscle Res Cell Motil. 2005;26(6–8):367–374. doi: 10.1007/s10974-005-9018-5. [DOI] [PubMed] [Google Scholar]
- Mazalouskas MD, Godoy-Ruiz R, Weber DJ, Zimmer DB, Honkanen RE, Wadzinski BE. Small G proteins Rac1 and Ras regulate serine/threonine protein phosphatase 5 (PP5)∙extracellular signal-regulated kinase (ERK) complexes involved in the feedback regulation of Raf1. J Biol Chem. 2014;289(7):4219–4232. doi: 10.1074/jbc.M113.518514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Sadoshima J (2018) Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol. 10.1038/s41569-018-0007-y [DOI] [PubMed]
- Ohno S. The genetic background of arrhythmogenic right ventricular cardiomyopathy. J Arrhythm. 2016;32(5):398–403. doi: 10.1016/j.joa.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto R, Li Y, Noma K, Hiroi Y, Liu PY, Taniguchi M, Ito M, Liao JK. FHL2 prevents cardiac hypertrophy in mice with cardiac-specific deletion of ROCK2. FASEB J. 2013;27(4):1439–1449. doi: 10.1096/fj.12-217018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillers DA, Von Bergen NH. Emery-Dreifuss muscular dystrophy: a test case for precision medicine. Appl Clin Genet. 2016;9:27–32. doi: 10.2147/TACG.S75028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell NH, Darwis D, Bueno OF, Muller JM, Schule R, Molkentin JD. Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. Mol Cell Biol. 2004;24(3):1081–1095. doi: 10.1128/MCB.24.3.1081-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin A, Lange S, Banares K, Lyon RC, Zieseniss A, Lee LK, Yamazaki KG, Granzier HL, Gregorio CC, McCulloch AD, Omens JH, Sheikh F. A novel mechanism involving four-and-a-half LIM domain protein-1 and extracellular signal-regulated kinase-2 regulates titin phosphorylation and mechanics. J Biol Chem. 2012;287(35):29273–29284. doi: 10.1074/jbc.M112.372839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert C, Deiss K, Herrmann S, Vidal M, Oezkur M, Gorski A, Weidemann F, Lohse MJ, Lorenz K. Interference with ERK(Thr188) phosphorylation impairs pathological but not physiological cardiac hypertrophy. Proc Natl Acad Sci U S A. 2013;110(18):7440–7445. doi: 10.1073/pnas.1221999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Roman I, Navarro M, Martinez F, Albert L, Polo L, Guardiola J, Garcia-Molina E, Munoz-Esparza C, Lopez-Ayala JM, Sabater-Molina M, Gimeno JR. Unclassifiable arrhythmic cardiomyopathy associated with Emery-Dreifuss caused by a mutation in FHL1. Clin Genet. 2016;90(2):171–176. doi: 10.1111/cge.12760. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10(9):315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Schessl J, Zou Y, McGrath MJ, Cowling BS, Maiti B, Chin SS, Sewry C, Battini R, Hu Y, Cottle DL, Rosenblatt M, Spruce L, Ganguly A, Kirschner J, Judkins AR, Golden JA, Goebel HH, Muntoni F, Flanigan KM, Mitchell CA, Bonnemann CG. Proteomic identification of FHL1 as the protein mutated in human reducing body myopathy. J Clin Invest. 2008;118(3):904–912. doi: 10.1172/JCI34450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen-Chowdhry S, Morgan RD, Chambers JC, McKenna WJ. Arrhythmogenic cardiomyopathy: etiology, diagnosis, and treatment. Annu Rev Med. 2010;61:233–253. doi: 10.1146/annurev.med.052208.130419. [DOI] [PubMed] [Google Scholar]
- Shathasivam T, Kislinger T, Gramolini AO. Genes, proteins and complexes: the multifaceted nature of FHL family proteins in diverse tissues. J Cell Mol Med. 2010;14(12):2702–2720. doi: 10.1111/j.1582-4934.2010.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh F, Raskin A, Chu PH, Lange S, Domenighetti AA, Zheng M, Liang X, Zhang T, Yajima T, Gu Y, Dalton ND, Mahata SK, Dorn GW, 2nd, Brown JH, Peterson KL, Omens JH, McCulloch AD, Chen J. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest. 2008;118(12):3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignier N, Le Corvoisier P, Blard C, Sambin L, Azibani F, Schlossarek S, Delcayre C, Carrier L, Hittinger L, Su JB. AT1 blockade abolishes left ventricular hypertrophy in heterozygous cMyBP-C null mice: role of FHL1. Fundam Clin Pharmacol. 2014;28(3):249–256. doi: 10.1111/fcp.12031. [DOI] [PubMed] [Google Scholar]
- von Kriegsheim A, Pitt A, Grindlay GJ, Kolch W, Dhillon AS (2006) Regulation of the Raf-MEK-ERK pathway by protein phosphatase 5. Nat Cell Biol 8(9):1011–1016. 10.1038/ncb1465 [DOI] [PubMed]
- Wettschureck N, Rutten H, Zywietz A, Gehring D, Wilkie TM, Chen J, Chien KR, Offermanns S. Absence of pressure overload induced myocardial hypertrophy after conditional inactivation of Galphaq/Galpha11 in cardiomyocytes. Nat Med. 2001;7(11):1236–1240. doi: 10.1038/nm1101-1236. [DOI] [PubMed] [Google Scholar]
- Wilkins BJ, Molkentin JD. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem Biophys Res Commun. 2004;322(4):1178–1191. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
- Yamasaki R, Wu Y, McNabb M, Greaser M, Labeit S, Granzier H. Protein kinase A phosphorylates titin’s cardiac-specific N2B domain and reduces passive tension in rat cardiac myocytes. Circ Res. 2002;90(11):1181–1188. doi: 10.1161/01.RES.0000021115.24712.99. [DOI] [PubMed] [Google Scholar]
- Yang Z, Browning CF, Hallaq H, Yermalitskaya L, Esker J, Hall MR, Link AJ, Ham AJ, McGrath MJ, Mitchell CA, Murray KT. Four and a half LIM protein 1: a partner for KCNA5 in human atrium. Cardiovasc Res. 2008;78(3):449–457. doi: 10.1093/cvr/cvn038. [DOI] [PubMed] [Google Scholar]
- Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J Clin Invest. 2011;121(6):2447–2456. doi: 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BQ, Si N, Liu DF. Identification of a novel four and a half LIM domain 1 mutation in a Chinese male presented with hypertrophic cardiomyopathy and mild skeletal muscle hypertrophy. Chin Med J. 2015;128(16):2269–2270. doi: 10.4103/0366-6999.162493. [DOI] [PMC free article] [PubMed] [Google Scholar]